Abstract
The complete chloroplast genome of Stephania tetrandra was sequenced and assembled for the first time. The chloroplast genome is 159,974 bp in length, containing a large single-copy (LSC) region of 90,539 bp and a small single-copy region (SSC) of 20,735 bp, separated by a pair of inverted repeats (IRs) of 24,350 bp. The genome contains 113 unique genes, including 79 protein-coding genes (PCGs), 30 tRNA genes, and four rRNA genes. Among them, 15 genes have one intron each and three genes contain two introns. The overall GC content is 37.8%, while the corresponding values of LSC, SSC, and IR regions are 35.8, 32.4, and 43.7%, respectively. Phylogenetic analysis showed that S. tetrandra is more closely related to the clade of two species within Stephania, providing new insight into the evolution of Menispermaceae.
Keywords: Complete chloroplast genome, Stephania tetrandra, Menispermaceae
The genus Stephania belongs to Menispermaceae, which has about 60 species in the world. This genus mainly occurs in the tropical and subtropical regions of Asia and Africa, with a few species are found in Oceania (Luo et al. 2008). Thirty-seven species of Stephania have been recorded from China (Luo et al. 2008). The plants of this genus are rich in alkaloids and are often used as pharmaceutical ingredients (Semwal et al. 2010). In this study, we sequenced and assembled the chloroplast genome of S. tetrandra for the first time. It will facilitate a deeper understanding and exploitation of this group.
Fresh leaves of S. tetrandra were collected from Anfu, Jiangxi, China (GPS: 27°17′57.328″, 114°18′43.902″). Herbarium voucher (Voucher No. JZ2020050201) is deposited in Medicinal Herbarium, Jiangxi University of Traditional Chinese Medicine, Nanchang, China. Total genomic DNA was extracted using the modified CTAB method (Doyle and Doyle 1987) and sequenced on an Illumina NovaSeq platform with paired-end reads of 150 bp. The GetOrganelle pipeline (Jin et al. 2020) was performed for the de novo assembly of complete chloroplast genome. Genes were annotated using PGA (Qu et al. 2019) and visually checked in Geneious version 8.0.2 (Kearse et al. 2012) using chloroplast genome of S. kwangsiensis (GenBank accession NC_048524) as a reference. The predicted transfer RNAs (tRNAs) were confirmed by tRNAscan-SE 2.0 (Lowe and Chan 2016). Finally, the complete chloroplast genome with annotations was submitted to GenBank (accession MT859132). Raw reads were deposited in the GenBank Sequence Read Archive (SRA SRR12717620).
The size of complete chloroplast genome of S. tetrandra is 159,974 bp with high coverage (mean 1484 ×). It has a typical quadripartite structure, including a large single-copy (LSC) region of 90,539 bp, a small single-copy region (SSC) of 20,735 bp, and a pair of inverted repeats (IRs) of 24,350 bp. There are 79 protein-coding genes (PCGs), 30 tRNA genes, and four rRNA genes. Among these genes, 15 of them (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) are single-intron genes, and three genes (clpP, rps12, and ycf3) contain two introns. The overall GC content is 37.8%, while the GC content of LSC, SSC, and IR regions are 35.8, 32.4, and 43.7%, respectively.
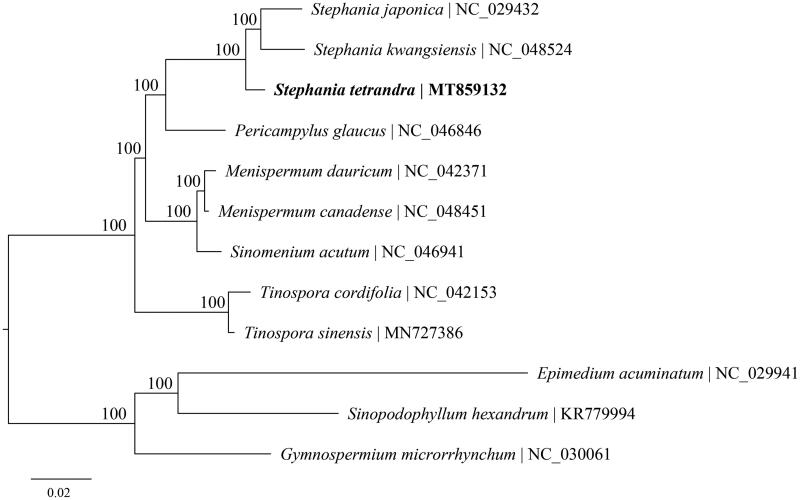
To identify the phylogenetic relationship of S. tetrandra in Menispermaceae, the phylogenetic tree including S. tetrandra, two other species of Stephania, four other genera of Menispermaceae, and three outgroups (Gymnospermium microrrhynchum, Sinopodophyllum hexandrum, and Epimedium acuminatum) of Berberidaceae were reconstructed using complete chloroplast genomes. The sequences were aligned using MAFFT version 7.017 plugin (Katoh et al. 2002) and visually checked in Geneious. Phylogenetic analysis was performed by RAxML version 8.2 (Stamatakis 2014) using 1000 replicates of a rapid bootstrap analysis with GTRGAMMA substitution model. The phylogenetic relationships among all sampled species were fully resolved with maximum support, showing S. tetrandra is more closely related to the clade of two species within Stephania, and Stephania is grouped with Pericampylus (Figure 1). The chloroplast genome obtained in this study could provide essential data to determine the evolutionary relationship of Menispermaceae.
Figure 1.
Maximum-likelihood phylogenetic tree based on complete cp genomes. Numbers close to each node are bootstrap support values.
Funding Statement
This work was supported by the Project of Chinese Medicine from Health Commission of Jiangxi Province [2016A046]; The Scientific Research Foundation for the First-class Discipline of Chinese Medicine Program of Jiangxi University of Traditional Chinese Medicine [JXSYLXK-ZHYAO028]; and Traditional Chinese Medicine Public Health Special Project [201207002].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession numbers [KR779994, NC_029941, NC_030061, MN727386, NC_029432, NC_042153, NC_042371, NC_046846, NC_046941, NC_048451, and NC_048524]. The complete chloroplast genome generated for this study has been deposited in GenBank with accession number MT859132. All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: SRR12717620.
References
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y ,Yi TS ,Li DZ.. 2020. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. Genome biology 21(1): 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XR, Chen T, Gilbert MG.. 2008. Menispermaceae. In: Wu ZY, Revan P, Hong DY, editors. Flora of China 7. St. Louis, Beijing: Science Press Beijing, Missourii Botanical Garden Press; p. 1–31. [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semwal DK, Badoni R, Semwal R, Kothiyal SK, Singh GJP, Rawat U.. 2010. The genus Stephania (Menispermaceae): chemical and pharmacological perspectives. J Ethnopharmacol. 132(2):369–383. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession numbers [KR779994, NC_029941, NC_030061, MN727386, NC_029432, NC_042153, NC_042371, NC_046846, NC_046941, NC_048451, and NC_048524]. The complete chloroplast genome generated for this study has been deposited in GenBank with accession number MT859132. All high-throughput sequencing data files are available from the GenBank Sequence Read Archive (SRA) accession number: SRR12717620.