Abstract
This paper presents a review of deep learning (DL)-based medical image registration methods. We summarized the latest developments and applications of DL-based registration methods in the medical field. These methods were classified into seven categories according to their methods, functions and popularity. A detailed review of each category was presented, highlighting important contributions and identifying specific challenges. A short assessment was presented following the detailed review of each category to summarize its achievements and future potential. We provided a comprehensive comparison among DL-based methods for lung and brain registration using benchmark datasets. Lastly, we analyzed the statistics of all the cited works from various aspects, revealing the popularity and future trend of DL-based medical image registration.
Keywords: deep learning, medical image registration, review
1. Introduction
Image registration, also known as image fusion or image matching, is the process of aligning two or more images based on image appearances. Medical image registration seeks to find an optimal spatial transformation that best aligns the underlying anatomical structures. Medical image registration is used in many clinical applications such as image guidance (Taylor and Stoianovici 2003, Sarrut 2006, Yang et al 2011b, De Silva et al 2016), motion tracking (Fu et al 2011, Yang et al 2012), segmentation (Han et al 2008, Yang et al 2011c, 2014, 2016, Yang and Fei 2012, Fu et al 2017), dose accumulation (Velec et al 2011, Bender et al 2012, Chao et al 2012, Andersen et al 2013, Samavati et al 2016, Chetty and Rosu-Bubulac 2019), image reconstruction (Qiao et al 2006, Li et al 2010, Dang et al 2014, McClelland et al 2017) and so on. As table 1 shows, medical image registration is a broad topic which can be grouped from various perspectives. From an input image point of view, registration methods can be divided into unimodal, multimodal, interpatient, intra-patient (e.g. same- or different-day) registration. From a deformation model point of view, registration methods can be divided in to rigid, affine and deformable methods. From a region of interest (ROI) perspective, registration methods can be grouped according to anatomical sites such as brain, lung registration and so on. From an image pair dimension perspective, registration methods can be divided into 3D to 3D, 3D to 2D and 2D to 2D/3D.
Table 1.
Registration categories of different aspects.
| Aspects | Registration Categories |
|---|---|
| Input image types | Unimodal, Multimodal, Interpatient, Intra-patient (same/different day) |
| Deformation model types | Rigid, Affine, Deformable |
| ROI | Brain, Thorax, Lung, Abdomen, Pelvic, etc |
| Image pair dimensions | 3D/3D, 3D/2D, 2D/2D, 2D/3D |
Different applications and registration methods face different challenges. For multi-modal image registration, it is difficult to design accurate image similarity measures due to the inherent appearance differences between different imaging modalities. Inter-patient registration can be tricky since the underlying anatomical structures are different across patients. Different-day intra-patient registration is challenging due to image appearance changes caused by metabolic processes, bowel movement, patient gaining/losing weight and so on. It is crucial for the registration to be computationally efficient in order to provide real-time image guidance. Examples of such application include 3D-MR to 2D/3D-US prostate registration to guide brachytherapy catheter placement and 3D-CT to 2D x-ray registration in intraoperative surgeries. For segmentation and dose accumulation, it is important to ensure the registration has high spatial accuracy. Motion tracking can be used for motion management in radiotherapy such as patient-setup and treatment planning. Motion tracking could also be used to assess respiratory function through 4D-CT lung registration and to access cardiac function through myocardial tissue tracking. In addition, motion tracking could be used to compensate for irregular motion in image reconstruction. In terms of deformation model, rigid transformation is often too simple to represent the actual tissue deformation while free-form transformation is ill-conditioned and hard to regularize. One limitation of 2D-2D registration is it ignores the out-of-plane deformation. Nevertheless, 3D-3D registration is usually computationally demanding, resulting in slow registration.
Many methods have been proposed to deal with the above-mentioned challenges. Popular registration methods include optical flow (Yang et al 2008, 2011a), demons (Vercauteren et al 2009), ANTs (Avants et al 2011), HAMMER (Shen 2007), ELASTIX (Klein et al 2010) and so on. Scale invariant feature transform (SIFT) and mutual information (MI) have been proposed for multi-modal image similarity calculation (Gong et al 2014). For 3D image registration, GPU has been adopted to accelerate the computational speed (Shams et al 2010). Multiple transformation regularization methods including spatial smoothing (Yang et al 2010), diffeomorphic (Vercauteren et al 2009), spline-based (Szeliski and Coughlan 1997), FE-based (Brock et al 2005) and other deformable models have been proposed. Though medical image registration has been extensively studied, it remains a hot research topic. The field of medical image registration has been evolving rapidly with hundreds of papers published each year. Recently, DL-based methods have changed the landscape of medical image processing research and achieved the-state-of-art performances in many applications (Fu et al 2018, Harms et al 2019, Dong et al 2019a, 2019c, Wang et al 2019a, 2019b, 2019c, 2019d, 2019f, Lei et al 2019b, 2019d, 2019e, 2019f, 2019g, Liu et al 2019c, 2019d). However, deep learning (DL) in medical image registration has not been extensively studied until the past three to four years. Though several review papers on DL in medical image analysis have been published (Litjens et al 2017, Shen et al 2017, Ker et al 2018, Liu et al 2018, Meyer et al 2018, Maier et al 2019, Sahiner et al 2019, Zhang and Sejdic 2019), there are very few review papers that are specific to DL in medical image registration (Haskins et al 2019b). The goal of this paper is to summarize the latest developments, challenges and trends in DL-based medical image registration methods. With this survey, we aim to
Summarize the latest developments in DL-based medical image registration.
Highlight contributions, identify challenges and outline future trends.
Provide detailed statistics on recent publications from different perspectives.
2. Deep learning
2.1. Convolutional neural network
Convolutional neural network (CNN) is a class of deep neural networks with regularized multilayer perceptron. CNN uses convolution operation in place of general matrix multiplication in simple neural networks. The convolutional filters and operations in CNN make it suitable for visual imagery signal processing. Because of its excellent feature extraction ability, CNN is one of the most successful models for image analysis. Since the breakthrough of AlexNet (Krizhevsky et al 2012), many variants of CNN have been proposed and have achieved the-state-of-art performances in various image processing tasks. A typical CNN usually consists of multiple convolutional layers, max pooling layers, batch normalization layers, dropout layers, a sigmoid or softmax layer. In each convolutional layer, multiple channels of feature maps were extracted by sliding trainable convolutional kernels across the input feature maps. Hierarchical features with high-level abstraction are extracted using multiple convolutional layers. These feature maps usually go through multiple fully connected layer before reaching the final decision layer. Max pooling layers are often used to reduce the image sizes and to promote spatial invariance of the network. Batch normalization is used to reduce internal covariate shift among the training samples. Weight regularization and dropout layers are used to alleviate data overfitting. The loss function is defined as the difference between the predicted and the target output. CNN is usually trained by minimizing the loss via gradient back propagation using optimization methods.
Many different types of network architectures have been proposed to improve the performance of CNN (Litjens et al 2017). U-Net proposed by Ronneberger et al is among one of the most used network architectures (Ronneberger et al 2015). U-Net was originally used to perform neuronal structures segmentation. U-Net adopts symmetrical contractive and expansive paths with skip connections between them. U-Net allows effective feature learning from a small number of training datasets. Later, He et al proposed a residual network (ResNet) to ease the difficulty of training deep neural networks (He et al 2016). The difficulty in training deep networks is caused by gradient degradation and vanishing. They reformulated the layers as learning residual functions instead of directly fitting a desired underlying mapping. Inspired by residual network, Huang et al later proposed a densely connected convolutional network (DenseNet) by connecting each layer to every other layer (Huang et al 2017). Inception module was first used in GoogLeNet to alleviate the problem of gradient vanishing and allow for more efficient computation of deeper networks (Szegedy et al 2015). Instead of performing convolution using a kernel with fixed size, an inception module uses multiple kernels of different sizes. The resulting feature maps were concatenated and processed by the next layer. Recently, attention gate was used in CNN to improve performance in image classification and segmentation (Schlemper et al 2018). Attention gate could learn to suppress irrelevant features and highlight salient features useful for a specific task.
2.2. Autoencoder
An autoencoder (AE) is a type of neural network that learns to copy its input to its output without supervision (Pierre 2012). An AE usually consists of an encoder which encodes the input into a low-dimensional latent state space and a decoder which restores the original input from the low-dimensional latent space. To prevent an AE from learning an identity function, regularized AEs were invented. Examples of regularized AEs include sparse AE, denoising AE and contractive AE (Tschannen et al 2018). Recently, convolutional AE (CAE) was proposed to combine CNN with traditional AEs (Chen et al 2017). CAE replaces the fully connected layer in traditional AE with convolutional layers and transpose-convolutional layers. CAE has been used in multiple medical image processing tasks such as lesion detection, segmentation, image restoration (Litjens et al 2017). Different from above-mentioned AEs, variational AE (VAE) is a generative model that learns latent representation using a variational approach (Hjelm et al 2016). VAE has been used for anomaly detection (Zimmerer et al 2018) and image generation (Dosovitskiy and Brox 2016).
2.3. Recurrent neural network
A recurrent neural network (RNN) is a type of neural network that was used to model dynamic temporal behavior (Giles et al 1994). RNN is widely used for natural language processing (Chung et al 2014). Unlike feedforward networks such as CNN, RNN is suitable for processing temporal signal. The internal state of RNN was used to model and ‘memorize’ previously processed information. Therefore, the output of RNN was dependent on not only its immediate input but also its input history. Long short-term memory (LSTM) is one type of RNN which has been used in image processing tasks (Bakker 2002). Recently, Cho et al proposed a simplified version of LSTM, called gated recurrent unit (Cho et al 2014).
2.4. Reinforcement learning
Reinforcement learning (RL) is a type of machine learning that focused on predicting the best actions to take given its current state in an environment (Thrun 1992). RL is usually modeled as a Markov decision process using a set of environment states and actions. An artificial agent is trained to maximize its cumulative expected rewards. The training process often involves an exploration-exploitation tradeoff. Exploration means to explore the whole space to gather more information while exploitation means to explore the promising areas given current information. Q-learning is a model-free RL algorithm, which aims to learn a Q function that models the action-reward relationship. Bellman equation is often used in Q-learning for reward calculation. The Bellman equation calculates the maximum future reward as the immediate reward the agent gets for entering the current state plus a weighted maximum future reward for the next state. For image processing, the Q function is often modeled as CNN, which could encode input images as states and learn the Q function via supervised training (Ghesu et al 2016, Liao et al 2016, Krebs et al 2017, Miao et al 2017).
2.5. Generative adversarial network
A typical generative adversarial network (GAN) consists of two competing networks, a generator and a discriminator (Goodfellow et al 2014). The generator is trained to generate artificial data that approximate a target data distribution from a low-dimensional latent space. The discriminator is trained to distinguish the artificial data from actual data. The discriminator encourages the generator to predict realistic data by penalizing unrealistic predictions via learning. Therefore, the discriminative loss could be considered as a dynamic network-based loss term. The generator and discriminator both are getting better during training to reach Nash equilibrium. Multiple variants of GAN include conditional GAN (cGan) (Mirza and Osindero 2014), InfoGan (Chen et al 2016), CycleGAN (Zhu et al 2017), StarGan (Choi et al 2018) and so on. In medical imaging, GAN has been used to perform image synthesis for inter- or intra-modality, such as MR to synthetic CT (Lei et al 2019d, 2019e), CT to synthetic MR (Dong et al 2019a, Lei et al 2019a), CBCT to synthetic CT (Harms et al 2019), non-attenuation correction (non-AC) PET to CT (Dong et al 2019d), low-dose PET to synthetic full-dose PET (Lei et al 2019b), non-AC PET to AC PET (Dong et al 2019b), low-dose CT to full-dose CT (Wang et al 2019e) and so on. In medical image registration, GAN is usually used to either provide additional regularization or translate multi-modal registration to unimodal registration. Out of medical imaging, GAN has been widely used in many other fields including science, art, games and so on.
3. Deep learning in medical image registration
DL-based registration methods can be classified according to DL properties, such as network architectures (CNN, RL, GAN etc), training process (supervised, unsupervised etc), inference types (iterative, one-shot prediction), input image sizes (patch-based, whole image-based), output types (dense transformation, sparse transformation on control points, parametric regression of transformation model etc) and so on. In this paper, we classified DL-based medical image registration methods according to its methods, functions and popularity in to seven categories, including (1) RL-based methods, (2) Deep similarity-based methods, (3) Supervised transformation predication, (4) Unsupervised transformation prediction, (5) GAN in medical image registration, (6) Registration validation using DL, and (7) Other learning-based methods. In each category, we provided a comprehensive table, listing all the surveyed works belonging to this category and summarizing their important features.
Before we delve into the details of each category, we provided a detailed overview of DL-based medical image registration methods with their corresponding components and features in figure 1. The purpose of figure 1 is to give the readers an overall understanding of each category by putting its important features side by side with each other. CNN was initially designed to process highly structured datasets such as images, which are usually expressed by regular grid-sampling data points. Therefore, almost all cited methods have utilized convolutional kernels in their DL design. This explains why the CNN module is in the middle of figure 1.
Figure 1.
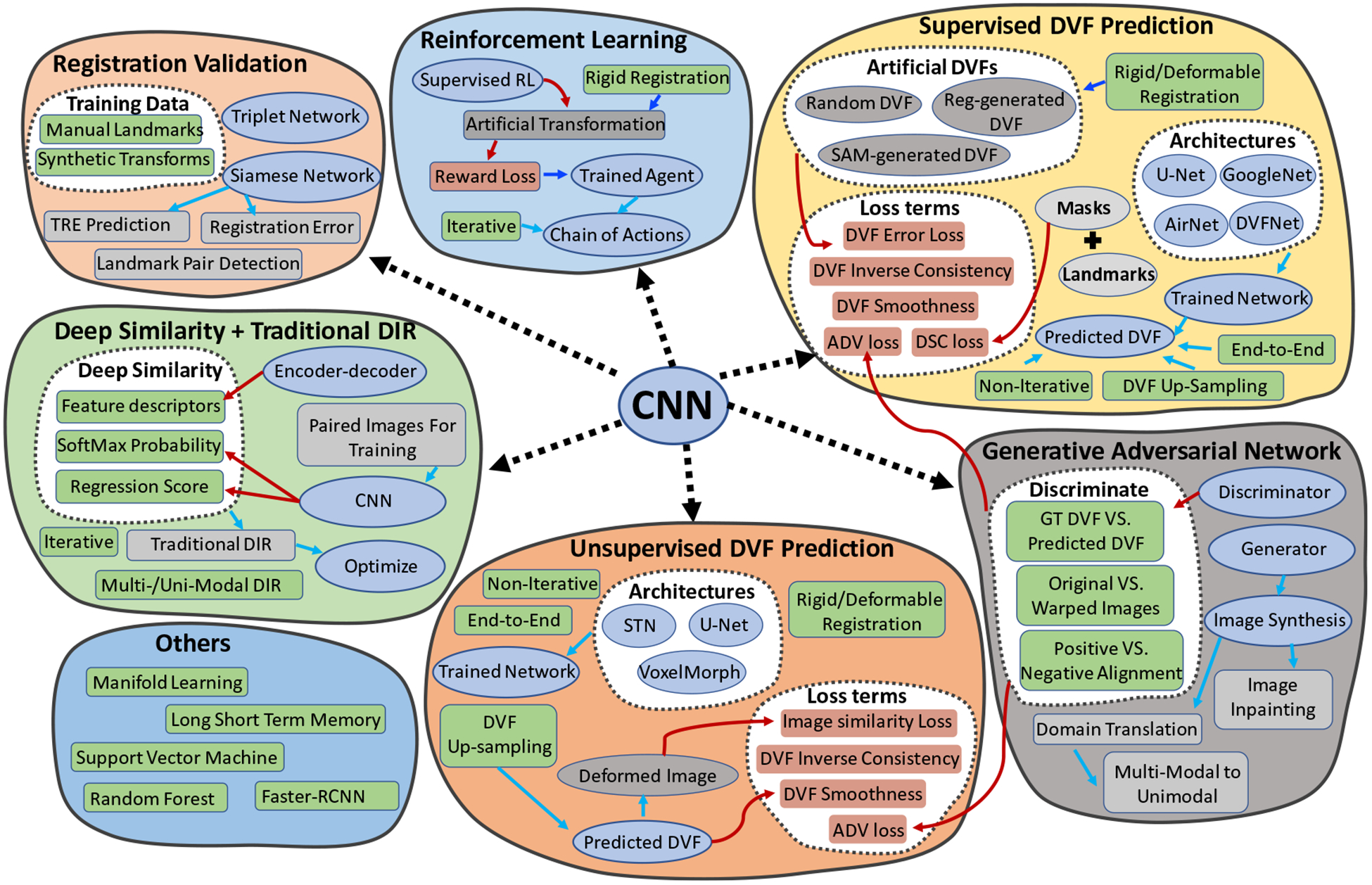
Overview of seven categories of DL-based methods in medical image registration.
Works cited in this review were collected from various databases, including Google Scholar, PubMed, Web of Science, Semantic Scholar and so on. To collect as many works as possible, we used a variety of keywords including but not limited to machine learning, DL, learning-based, CNN, image registration, image fusion, image alignment, registration validation, registration error prediction, motion tracking, motion management and so on. We totally collected over 150 papers that are closely related to DL-based medical image registration. Most of these works were published between the year of 2016 and 2019. The number of publications is plotted against year by stacked bar charts in figure 2. Number of papers were counted by categories. The total number of publications has grown dramatically over the last few years. Figure 2 shows a clear trend of increasing interest in supervised transformation prediction (SupCNN) and unsupervised transform prediction (UnsupCNN). Meanwhile, GAN are gradually gaining popularity. On the other hand, the number of papers of RL-based medical image registration has decreased in 2019, which may indicate decreasing interest in RL for medical image registration. The ‘DeepSimilarity’ in figure 2 represents the category of deep similarity-based registration methods. The number of papers in this category has also increased, however, only slightly as compared to ‘SupCNN’ and ‘UnsupCNN’ categories. In addition, more and more studies were published on using DL for medical image registration validations.
Figure 2.
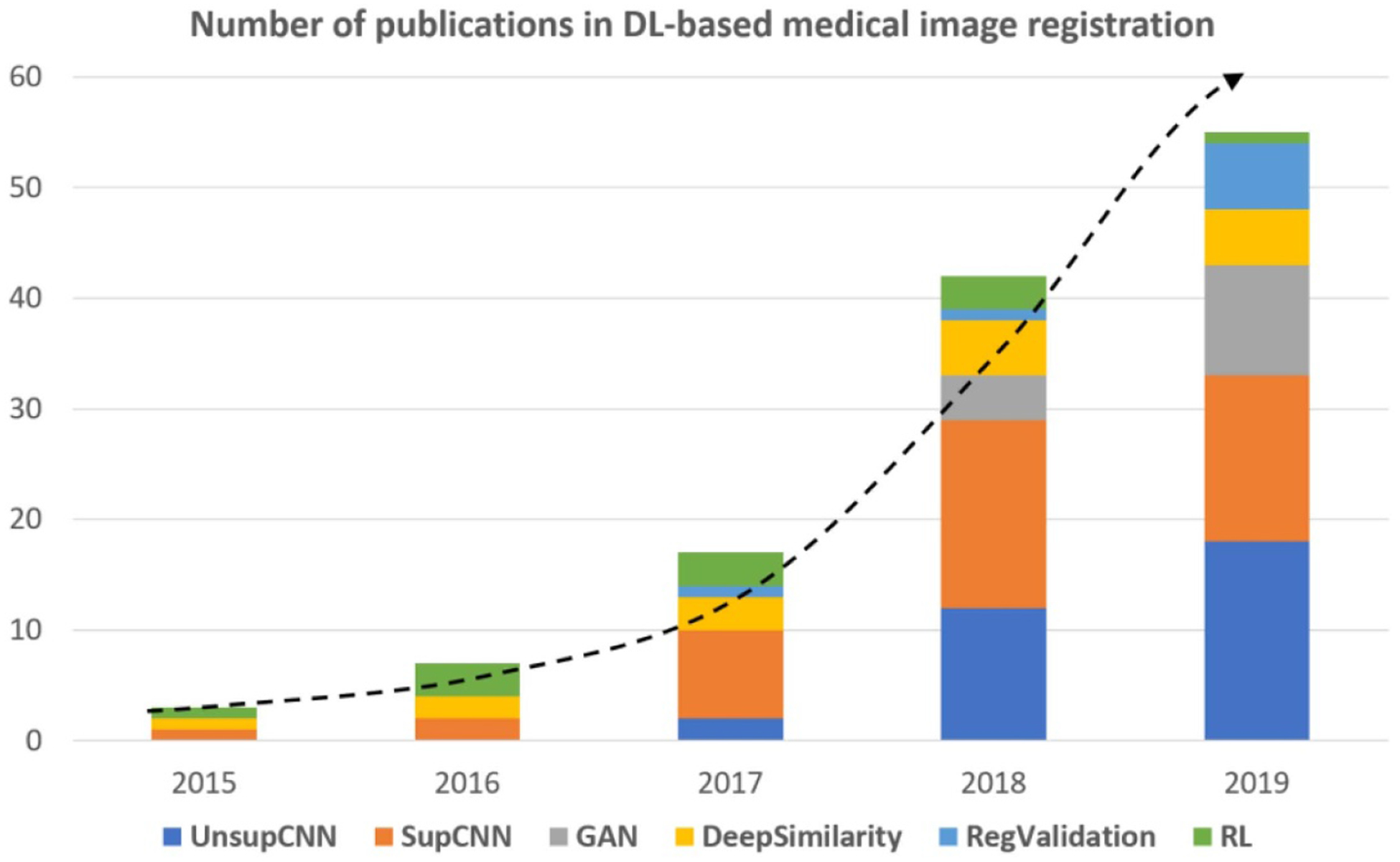
Overview of number of publications in DL-based medical image registration. The dotted line indicates increased interest in DL-based registration methods over the years. ‘DeepSimilarity’ is the category of using DL-based similarity measures in traditional registration frameworks. ‘RegValidation’ represents the category of using DL for registration validation.
3.1. Deep similarity-based methods
Conventional intensity-based similarity metrics include sum-of-square distance (SSD), mean square distance (MSD), (normalized) cross correlation (CC), and (normalized) mutual information (MI). Generally, conventional similarity measures work quite well for unimodal image registration where the image pair shares the same intensity distribution such as CT-CT, MR-MR image registration. However, noise and artifacts in images such as US and CBCT often cause conventional similarity measures to perform poorly even in unimodal image registration. Metrics such as SSD and MSD does not work for multi-modal image registration. To develop a similarity measure for multi-modality image registration, handcrafted descriptors such as MI were proposed. To improve its performance, a variety of MI variants such as correlation ratio-based MI (Gong et al 2017), contextual conditioned MI (Rivaz et al 2014) and modality independent neighborhood descriptor (MIND) (Heinrich et al 2012) have been proposed. Recently, CNN has achieved huge success in tasks such as image classification and segmentation problems. However, CNN has not been widely used in image registration tasks until the last three to four years. To take the advantage of CNN, several groups tried to replace the traditional image similarity measures such as SSD, MAE and MI with DL-based similarity measures, achieving promising registration results. In the following section, we described several important works that attempted to use DL-based similarity measures in medical image registration.
3.1.1. Overview of works.
Table 2 shows a list of selected references that belong to this category. Cheng et al proposed a deep similarity learning network to train a binary classifier (Cheng et al 2018). The network was trained to learn the correspondence of two image patches from CT-MR image pair. The continuous probabilistic value was used as the similarity score. Similarly, Simonovsky et al proposed a 3D similarity network using a few aligned image pairs (Simonovsky et al 2016). The network was trained to classify whether an image pair is aligned or not. They observed that hinge loss performed better than cross entropy. The learnt deep similarity metric was then used to replace MI in traditional deformable image registration (DIR) for brain T1-T2 registration. It is important to ensure the smoothness of first order derivative in order to fit the deep similarity metrics into traditional DIR frameworks. The gradient of the deep similarity metric with respect to transformation was calculated using chain rule. They found out that high overlap of neighboring patches led to smoother and more stable derivatives. They have trained the network using IXI brain datasets and tested it using a completely independent datasets called ALBERTs in order to show the good generality of the learnt metric. They showed that the learnt deep similarity metric outperformed MI by a significant margin.
Table 2.
Overview of deep similarity-based methods.
| References | ROI | Dimension | Modality | Transformation | Supervision |
|---|---|---|---|---|---|
| (Simonovsky et al 2016) | Brain | 3D-3D | T1-T2 | Deformable | Supervised |
| (Wu et al 2016) | Brain | 3D-3D | MR | Deformable | Unsupervised |
| (So and Chung 2017) | Brain | 3D-3D | MR | Rigid, Deformable | Supervised |
| (Cheng et al 2018) | Brain | 2D-2D | MR-CT | Rigid | Supervised |
| (Haskins et al 2019a) | Prostate | 3D-3D | MR-US | Rigid | Supervised |
| (Sedghi et al 2018) | Brain | 2D-2D | MR | Rigid | Weakly Supervised |
| (Ferrante et al 2019) | Brain, HN, Abdomen | 3D-3D | MR, CT | Deformable | Weakly Supervised |
Compared to CT-MR and T1-T2 image registration, MR-US image registration is more challenging due to the fundamental imaging acquisition differences between MR and US. A DL-based similarity measure is desired for MR-US image registration. Haskins et al proposed to use CNN to predict the target registration error (TRE) between 3D MR and transrectal US (TRUS) images (Haskins et al 2019a). The predicted TRE was used as image similarity metric for MR-US rigid registration. TREs obtained from expert-aligned images were used as ground truth. The CNN was trained to regress to the TRE as similarity prediction. The learnt metric was non-smooth and non-convex, which hinders gradient-based optimization. To address this issue, they performed multiple TRE predictions throughout the optimization. The average TRE estimate was used as the similarity metric to mitigate the non-convex problem and to expand the capture range. They claimed that the learnt similarity metric outperformed MI and its variant MIND (Heinrich et al 2012).
In previous works, accurate image alignment is needed for deep similarity metrics learning. However, it is very difficult to obtain well aligned multi-modal image pairs for network training. The quality of image alignment could affect the accuracy of the learnt deep similarity metrics. To mitigate this problem, Sedghi et al used special data augmentation techniques called dithering and symmetrizing to discharge the need for well-aligned images for deep metric learning (Sedghi et al 2018). The learnt deep metric outperformed MI on 2D brain image registration. Though they managed to relax the absolute accuracy of image alignment in network training, roughly-aligned image pairs were still necessary. To eliminate the need for aligned image pairs, Wu et al proposed to use stacked AEs (SAE) to learn intrinsic feature representations by unsupervised learning (Wu et al 2016). The convolutional SAE could encode an image to obtain low-dimensional feature representations for image similarity calculation. The learnt feature representations were used in Demons and HAMMER to perform brain image DIR. They showed that the image registration performance has improved consistently using the learnt feature representations in terms of the Dice similarity coefficient (DSC). To test the generality of the learnt feature representation, they reused network trained using LONI dataset on ADNI datasets. The results were comparable to the case of learning feature representation from the same datasets.
It was shown that combining multi-metric measures could produce more robust registration results compared to using the metrics individually. Ferrante et al used support vector machine (SVM) to learn weights of an aggregation of similarity measures including anatomical segmentation maps and displacement vector labels (Ferrante et al 2019). They have showed that the multi-metric outperformed conventional single-metric approaches. To deal with the non-convex of the aggregated similarity metric, they optimized a regularized upper bound of the loss using CCCP algorithm (Yuille and Rangarajan 2003). One limitation of this method was that segmentation masks of the source images were needed at testing stage.
3.1.2. Assessments.
Deep similarity metric has shown its potential to outperform traditional similarity metrics in medical image registration. However, it is difficult to ensure that its derivative is smooth for optimization. The above-mentioned measures of using a large overlap (Simonovsky et al 2016) or performing multiple TRE predictions (Haskins et al 2019a) are computationally demanding and only mitigate the problem of non-convex derivatives. Well-aligned image pairs are difficult to obtain for deep similarity network training. Though Wu et al (2016) has demonstrated that deep similarity network could be trained in an unsupervised manner, they only tested on unimodal image registration. Extra experiments on multi-modal images need to be performed to show its effectiveness. The biggest limitation of this category maybe that the registration process still inherits the iterative nature of traditional DIR frameworks, which slows the registration process. As more and more papers on direct transformation prediction emerge, it is expected that this category will be less attractive in the future.
3.2. RL in medical image registration
One disadvantage of the previous category is that the registration process is iterative and time-consuming. It is desired to develop a method to predict transformation in one shot. However, one shot transformation prediction is very difficult due to the high dimensionality of the output parameter space. RL has recently gained a lot of attention since the publications from Mnih et al (2015) and Silver et al (2016). They combined RL with DNN to achieve human-level performances on Atari and Go. Inspired by the success of RL, and to circumvent the challenge of high dimensionality in one shot transformation prediction, several groups proposed to combine CNN with RL to decompose the registration task into a sequence of classification problems. The strategy is to find a series of actions, such as rotation and translation along certain axis by a certain value, to iteratively improve image alignment.
3.2.1. Overview of works.
Table 3 shows a list of selected references that used RL in medical image registration. Liao et al was one of the first to explore RL in medical image registration (Liao et al 2016). The task was to perform 3D-3D, rigid, cone beam CT (CBCT)-CT image registration. Specific challenges of the registration include large differences in field of views (FOVs) between the CT and CBCT in spine registration and the severe streaking artifacts in CBCT. An artificial agent was trained using a greedy supervised approach to perform rigid image registration. The artificial agent was modelled using CNN, which took raw images as input and output the next optimal action. The action space consists of 12 candidate transformations, which are ±1 mm of translation and ±1 degree of rotation along the x, y, and z axis, respectively. Ground truth alignment were obtained using iterative closest point registration of expert-defined spine landmarks and epicardium segmentation, followed by visual inspection and manual editing. Data augmentation was used to artificially de-align the image pair with known transformations. Different from Mnih et al who trained their network with repeated trial and error, Liao et al trained the network with greedy supervision, where the reward can be calculated explicitly via a recursive function. They showed that the network training process with supervision was a magnitude more efficient than the training process of Mnih et al’s network. They also claimed their network could reliably overcome local maxima, which was challenging for generic optimization algorithms when the underlying problem was non-convex.
Table 3.
Overview of RL in medical image registration.
| References | ROI | Dimension | Modality | Transformation |
|---|---|---|---|---|
| (Ghesu et al 2016; Ghesu et al 2019) | Cardiac, HN | 2D, 3D | MR, CT, US | NA |
| (Krebs et al 2017) | Prostate | 3D-3D | MR | Deformable |
| (Liao et al 2016) | Spine, Cardiac | 3D-3D | CT-CBCT | Rigid |
| (Ma et al 2017) | Chest, Abdomen | 2D-2D | CT-Depth Image | Rigid |
| (Miao et al 2017) | Spine | 3D-2D | 3DCT-Xray | Rigid |
| (Zheng et al 2018) | Spine | 3D-2D | 3DCT-Xray | Rigid |
| (Sun et al 2019) | Nasopharyngeal | 2D-2D | MR-CT | Rigid with scaling |
Motivated by Liao et al (2016), Miao et al proposed a multi-agent system with an auto attention mechanism to rigidly register 3D-CT with 2D x-ray spine image (Miao et al 2017). Reliable 2D-3D image registration could map the pre-operative 3D data to real-time 2D x-ray images by image fusion. To deal with various image artifacts, they proposed to use an auto-attention mechanism to detect regions with reliable visual cues to drive the registration. In addition, they used a dilated FCN-based training mechanism to reduce the degree of freedom of training data to improve the training efficiency. They have outperformed single agent-based and optimization-based methods in terms of TRE.
Sun et al proposed to use an asynchronous RL algorithm with customized reward function for 2D MR-CT image registration (Sun et al 2019). They used datasets from 99 patients diagnosed as nasopharyngeal carcinoma. Ground truth image alignments were obtained using toolbox Elastix (Klein et al 2010). Different from previous works, Sun et al incorporated scaling factor into the action space. The action space consists of eight candidate transformations including ±1 pixel for translation, ±1 degree for rotation and ±0.05 for scaling. CNN was used to encode image states and LSTM was used to encode hidden states between neighboring frames. Their results were better than that obtained using Elastix in terms of TRE when the initial image alignment was poor. The use of actor-critic scheme (Grondman et al 2012) allowed the agent to explore transformation parameter spaces freely and avoided local minima when the initial alignment was poor. On the contrary, when the initial image alignment was good, Elastix was slightly better than their method. In the inference phase, a Monte Carlo rollout strategy was proposed to terminate the searching path to reach a better action.
All of the above-mentioned methods focused on rigid registration since rigid transformation could be represented by a low-dimensional parametric space, such as rotation, translation and scaling. However, non-rigid, free-form transformation model has high dimensionality and non-linearity which would result in a huge action space. To deal with this problem, Krebs et al proposed to build a statistical deformation model (SDM) with a low-dimensional parametric space (Krebs et al 2017). Principal component analysis (PCA) was used to construct SDM on B-spline deformation vector field (DVF). Modes of the PCA of the displacement were used as the unknow vectors for the agents to optimize. They evaluated the method on inter-subject MR prostate image registration in both 2D and 3D. The method achieved DSC scores of 0.87 and 0.80 for 2D and 3D, respectively.
Ghesu et al proposed to use RL to detect 3D-landmarks in medical images (Ghesu et al 2016). This method was mentioned since it belongs to the category of RL and the detected landmarks could be used for landmark-based image registration. They reformulated the landmark detection task as a behavioral problem for the network to learn. To deal with local minima problem, a multi-scale approach was used. Experiments on 3D-CT scans were conducted to compare with another five methods. The results showed that the detection accuracy was improved by 20%–30% while being 2–3 orders of magnitude faster.
3.2.2. Assessment.
The biggest limitation of RL-based image registration is that the transformation model is highly constrained to low-dimensionality. As a result, most of the RL-based registration methods used rigid transformation models. Though Krebs et al has applied RL to non-rigid image registration by predicting a low-dimensional parametric space of statistical deformation model, the accuracy and flexibility of the deformation model is highly constrained and may not be adequate to represent the actual deformation. RL-based image registration methods have shown its usefulness in enhancing the robustness of many algorithms in multi-modal image registration tasks. Despite the usefulness of RL, statistics indicates loss of popularity of this category, evidenced by the decreasing number of papers in 2019. As the techniques advance, more and more direct transformation predication methods are proposed. The accuracy of the direct transformation prediction methods is constantly improving, achieving comparable accuracy to top traditional DIR methods. Therefore, the advantage of casting registration as a sequence of classification problems in RL-based registration methods is gradually vanishing.
3.3. Supervised transformation predication
Both deep similarity-based and RL-based registration methods are iterative methods in order to avoid the challenges of one-shot transformation prediction. Despite the difficulties, several groups have attempted to train networks to directly infer the final transformation in a single forward prediction. The challenges include (1) high dimensionality of the output parametric space, (2) lack of training datasets with ground truth transformations and (3) regularization of the predicted transformation. Methods including ground truth transformation generation, image re-sampling and transformation regularization methods have been proposed to overcome these challenges. Table 4 shows a list of selected references that used supervised transformation prediction for medical image registration.
Table 4.
Overview of supervised transformation prediction methods.
| References | ROI | Dimension | Patch-based | Modality | Transformation |
|---|---|---|---|---|---|
| (Miao et al 2016) | Implant, TEE | 2D-3D | Yes | Xray | Rigid |
| (Pei et al 2017) | Cranial | 2D-3D | No | CBCT-Xray | Deformable |
| (Rohé et al 2017) | Cardiac | 3D-3D | No | MR | Deformable |
| (Uzunova et al 2017) | Cardiac, Brain | 2D-2D | No | MR | Deformable |
| (Yang et al 2017) | Brain | 3D-3D | Yes | MR | Deformable |
| (Cao et al 2018b) | Brain | 3D-3D | Yes | MR | Deformable |
| (Cao et al 2018a) | Pelvic | 3D-3D | Yes | MR-CT | Deformable |
| (Hering et al 2019) | Cardiac | 2D-2D | No | MR | Deformable |
| (Hu et al 2018a; Hu et al 2018b) | Prostate | 3D-3D | No | MR-US | Deformable |
| (Lv et al 2018) | Abdomen | 2D-2D | Yes | MR | Deformable |
| (Salehi et al 2018) | Brain | 3D-3D/2D | No | MR | Rigid |
| (Sentker et al 2018) | Lung | 3D-3D | Yes | CT | Deformable |
| (Sloan et al 2018) | Brain | 2D-2D | No | T1-T2 | Rigid |
| (Sun et al 2018) | Liver | 2D-2D | Yes | CT-US | Affine |
| (Yan et al 2018) | Prostate | 3D-3D | No | MR-US | Rigid + Affine |
| (Eppenhof et al 2018; Eppenhof and Pluim 2019) | Lung | 3D-3D | No | CT | Deformable |
| (Fan et al 2019b) | Brain | 3D-3D | Yes | MR | Deformable |
| (Foote et al 2019) | Lung | 3D-2D | No | CT | Deformable |
| (Kori and Krishnamurthi 2019) | Brain | 3D-3D | No | T1, T2, Flair | Affine |
| (Liu et al 2019a) | Skull, Upper Body | 2D-2D | No | DRR-Xray | Deformable |
| (Sokooti et al 2017; Sokooti et al 2019b; Onieva Onieva et al 2018) | Lung | 3D-3D | Yes | CT | Deformable |
3.3.1. Overview of works.
3.3.1.1. Ground truth transformation generation.
For supervised transformation prediction, it is important to generate many image pairs with known transformations for network training. Numerous data augmentation techniques were proposed for artificial transformations generation. Generally, these artificial transformation generation methods can be classified into three groups: (1) random transformation, (2) traditional registration-generated transformation and (3) model-based transformations.
3.3.1.1.1. Random transformation generation.
Salehi et al aimed to speed up and improve the capture range of 3D-3D and 2D-3D rigid image registration of fetal brain MR scans (Salehi et al 2018). CNN was used to predict both rotation and translation parameters. The network was trained using datasets generated by randomly rotating and translating the original 3D images. Both MSE and geodesic distance were used for loss function calculation. Geodesic distance is the distance between two points on a unit sphere. They have showed significant improvement after combining the geodesic distance loss with the MSE loss. Sun et al used expert aligned CT-US image pairs as ground truth (Sun et al 2018). Known artificial affine transformations were used to synthesize training datasets. The network was trained to predict the affine parameters. They have trained network which worked for simulated CT-US registration. However, it does not work on real CT-US pairs due to the vast appearance differences between the simulated and the real US. They have tried multiple methods to counter-act overfitting, such as deleting dropout layers, less complex network, parameter regularization and weight decay. Unfortunately, none of them worked.
Eppenhof et al proposed to train a CNN using synthetic random transformations to perform 3D-CT lung DIR (Eppenhof et al 2018). The output of the network was DVF on a thin plate spline transform grid. MSE between the predicted DVF and the ground truth DVF was used as loss function. They achieved 4.02 ± 3.08 mm TRE on DIRLAB (Castillo et al 2009), which was much worse than 1.36 ± 0.99 mm (Berendsen et al 2014) of the traditional DIR method. They later improved their method to use a U-Net architecture (Eppenhof and Pluim 2019). The network was trained on whole image. Images were down-sampled to fit into GPU memory. Again, synthetic random transformation was used to train the network. Affine pre-registration was required prior to CNN transformation prediction. They managed to reduce the TRE from 4.02 ± 3.08 mm to 2.17 ± 1.89 mm on DIRLAB datasets. Despite the slightly worse TRE than traditional DIR methods, they have demonstrated the possibility of direct transformation prediction using CNN.
3.3.1.1.2. Traditional registration-generated transformations.
Later, several groups tried to use traditional registration methods to register an image pair to generate ‘ground truth’ transformations for the network to learn. The rationale is that random transformation generation might be too different from the true transformation, which might deteriorate the performance of network.
Sentker et al used DVF generated from traditional DIRs including PlastiMatch (Modat et al 2010), NiftyReg (Shackleford et al 2010) and VarReg (Werner et al 2014) as ground truth (Sentker et al 2018). MSE between the predicted and the ground truth DVF was used as loss function to train a network for 3D-CT lung registration. On DIRLAB (Castillo et al 2009) datasets, they achieved better TRE using DVFs generated by VarReg as compared to PlastiMatch and NiftyReg. Results showed that their CNN-based registration method was comparable to the original traditional DIR in terms of TRE. The best TRE values they have achieved on DIRLAB is 2.50 ± 1.16 mm. Fan et al proposed a BIRNet to perform brain image registration using dual supervision (Fan et al 2019b). Ground truth transformations were obtained using existing registration methods. MSE between the ground truth and the predicted transformations were used as loss function. They used not only the original image but also its difference and gradient images as input to the network.
3.3.1.1.3. Model-based transformation generation.
Uzunova et al aimed to generate a large and diverse set of training image pairs with known transformations from a few sample images (Uzunova et al 2017). They proposed to learn highly expressive statistical appearance models (SAM) from a few training samples. Assuming Gaussian distribution for the appearance parameters, they synthesized huge amounts of realistic ground truth training datasets. FlowNet (Dosovitskiy et al 2015) architecture was used to register 2D MR cardiac images. For comparison, they have generated ground truth transformations using three different methods, which are affine registration-generated, randomly-generated and the proposed SAM-generated transformations. They showed that CNN learnt from the SAM-generated transformation outperformed CNN learnt from randomly generated and affine registration-generated transformation.
Sokooti et al generated artificial DVFs using model-based respiratory motion to simulate ground truth DVF for 3D-CT lung image registration (Sokooti et al 2019b). For comparison, random transformations were also generated using single frequency and mixed frequencies. They tested different combinations of various network structures including U-Net whole image, multi-view based and U-Net advanced. The multi-view and U-Net advanced all used patch-based training. TRE and Jacobian determinant were used as evaluation metrics. After comparison, they claimed that the realistic model-based transformation performed better compared to random transformations in terms of TRE. On average, they achieved TRE of 2.32 mm and 1.86 mm for SPREAD and DIRLAB datasets, respectively.
3.3.1.2. Supervision methods.
As neural network develops, many new supervision terms such as ‘supervised’, ‘unsupervised’, ‘deeply supervised’, ‘weakly supervised’, ‘dual supervised’, ‘self-supervised’ have emerged. Generally, neural network learns to perform a certain task by minimizing a predefined loss function via optimization. These terms refer to how the training datasets are prepared and how the networks are trained using the datasets. In the following paragraph, we briefly describe the definition of each supervision strategy in the context of DL-based image registration.
The learning process of a neural network is supervised if the desired output is already known in the training datasets. Supervised network means the network is trained with the ground truth transformation, which is a dense DVF for free deformation and a parametric vector of 6 for rigid transformation. On the other hand, unsupervised learning has no target output available in the training datasets, which means the desired DVFs or target transformation parameters are absent in the training datasets. Unsupervised network was also referred to self-supervised network since the warped image is generated from one of the input image pair and compared to another input image for supervision. Deep supervision usually means that the differences between outputs from multiple layers and the desired outputs are penalized during training whereas normal supervision only penalizes the difference between the final output and the desired output. In this manner, supervision was extended to deep layers of the network. Weak supervision represents scenario where ground truth other than the exact desired output is available in the training datasets and used to calculate the loss function. For example, a network is called weakly supervised if corresponding anatomical structural masks or landmark pairs, not the desired dense DVF, are used to train the network for direct dense DVF prediction. Dual supervision means that the network is trained using both supervised and unsupervised loss functions.
3.3.1.2.1. Weak supervision.
Methods that use ground truth transformation generation were mainly supervised method for direct transformation prediction. Weakly supervised transformation prediction has also been explored. Instead of using artificially-generated transformations, Hu et al proposed to use higher-level correspondence information such as labels of anatomical organs for network training (Hu et al 2018b). They argued that such anatomical labels were more reliable and practical to obtain. They trained a CNN to perform deformable MR-US prostate image registration. The network was trained using weakly supervised method, meaning that only corresponding anatomical labels, not dense voxel-level spatial correspondence, were used for loss calculation. The anatomical labels were required only in the training stage for loss calculation. Labels were not required in inference stage to facilitate fast registration. Similarly, Hering et al combined the complementary information from segmentation labels and image similarity to train a network (Hering et al 2019). They showed significant higher DSC scores than using only image similarity loss or segmentation label loss in 2D MR cardiac DIR.
3.3.1.2.2. Dual supervision.
Technically, dual supervision is not strictly defined. It usually means the network was trained using two types of important loss functions. Cao et al used dual supervision which includes a MR-MR loss and a CT-CT loss (Cao et al 2018a). Prior to network training, they transformed the multi-modality to unimodality registration by using pre-aligned counterpart images, for MR-CT registration. The MR has a pre-aligned CT and CT has a pre-aligned MR. The loss function has a dual similarity loss including MR-MR and CT-CT loss. They showed that the dual-modality similarity performed better than SyN (Avants et al 2008) and single modality similarity in terms of DSC and average surface distance (ASD) in pelvic image registration. Liu et al used representation learning to learn feature-based descriptors with probability maps of confidence level (Liu et al 2019a). Then, the learnt descriptor pairs across the image were used to build a geometric constraint using Hough voting or RANSAC. The network was trained using both supervised synthetic transformations and an unsupervised descriptor image similarity loss. Similarly, Fan et al combined both supervised and unsupervised loss terms for dual supervision in MRI brains image registration (Fan et al 2019b).
3.3.2. Assessment.
In recent two to three years, we have seen a huge interest in supervised CNN direct transformation prediction, evidenced by an increasing number of publications. Though direct transformation prediction has yet to outperform the state-of-the-art traditional DIR methods, the registration accuracy has improved greatly. Some methods have achieved comparable registration accuracy to the traditional DIR methods. Ground truth transformation generation will continue to play an important role in network training. Limitations of using artificially generated image pair with known ground truth transformations include (1) the generated transformation might not reflect the true physiological motion, (2) the generated transformation might not capture the large range of variations of actual image registration scenarios and (3) the artificially generated image pairs in the training stage are different from the actual image pair in the inference stage. To deal with the first limitations, we can use various transformation generation models. Adequate data augmentation could be performed to mitigate the second limitation. Domain adaption (Ferrante et al 2018, Zheng et al 2018) could be used to account for the domain difference between the artificially-generated and the true images. Image registration is an ill-posed problem, the ground truth transformation could help to constrain the final transformation prediction. Combinations of different loss functions and DVF regularization methods have also been examined to improve the accuracy of registration. We expect DL-based registration of this category to keep growing in the future.
3.4. Unsupervised transformation prediction
It is desired to develop unsupervised image registration methods to overcome the lack of training datasets with known transformations. However, it is difficult to define proper loss function of the network without ground truth transformations. In 2015, Jaderberg et al proposed a spatial transformer network (STN) which explicitly allows spatial manipulation of data within the network (Jaderberg et al 2015). Importantly, the spatial transformer network was a differentiable module that can be inserted in to existing CNN architectures. The publication of STN has inspired many unsupervised image registration methods since STN enables image similarity loss calculation during the training process. A typical unsupervised transformation prediction network for DIR takes an image pair as input and directly output dense DVF, which was used by STN to warp the moving image to generate warped images. The warped images were then compared to fixed images to calculate image similarity loss. DVF smoothness constraint was normally used to regularize the predicted DVF.
3.4.1. Overview of works.
Table 5 shows a list of selected references that performed unsupervised transformation prediction. Yoo et al proposed to use a CAE to encode image to a vector to calculate similarity, called feature-based similarity which is different from handcrafted feature similarity such as SIFT (Yoo et al 2017). They showed this feature-based similarity measure was better than intensity-based similarity measure for DIR. They have combined the deep similarity metrics and STN for unsupervised transformation estimation in 2D electron microscopy (EM) neural tissue image registration. Balakrishnan et al proposed an unsupervised CNN-based DIR method for MR brain atlas-based registration (Balakrishnan et al 2018, 2019). They used a U-Net like architecture and named it ‘VoxelMorph’. In the training, the network penalized the differences in image appearances with the help of STN. Smoothness constraint was used to penalize local spatial variations in the predicted transformation. They have achieved comparable performance to ANT (Avants et al 2011) registration method in terms of DSC score of multiple anatomical structures. Later, they extended their method to leverage auxiliary segmentations available in the training data. A DSC loss function was added to the original loss functions in the training stage. Segmentation labels were not required during testing. They investigated unsupervised brain registration, with and without segmentation label DSC loss. Their results showed that the segmentation loss could help yield improved DSC scores. The performance is comparable to ANT and NiftyReg, while being x150 faster than ANTs and x40 faster than NiftyReg.
Table 5.
Overview of unsupervised transformation prediction methods.
| References | ROI | Dimension | Patch-based | Modality | Transformation |
|---|---|---|---|---|---|
| (Ghosal and Rayl 2017) | Brain | 3D-3D | No | MR | Deformable |
| (Shan et al 2017) | Brain, Liver | 2D-2D | No | MR, CT | Deformable |
| (Vos et al 2017) | Cardiac | 2D-2D | No | MR | Deformable |
| (Yoo et al 2017) | Neural tissue | 2D-2D | No | EM | Deformable |
| (Chee and Wu 2018) | Brain | 3D-3D | No | MR | Affine |
| (Fan et al 2018; Fan et al 2019a) | Brain | 3D-3D | Yes | MR | Deformable |
| (Ferrante et al 2018) | Lung, Cardiac | 2D-2D | No | MR, Xray | Deformable |
| (Kearney et al 2018) | HN | 3D-3D | Yes | CT | Deformable |
| (Krebs et al 2018) | Cardiac | 3D-3D | No | MR | Deformable |
| (Li and Fan 2018) | Brain | 3D-3D | No | MR | Deformable |
| (Qin et al 2018; Mahapatra et al 2018a) | Cardiac | 2D-2D | No | MR | Deformable |
| (Sheikhjafari et al 2018) | Cardiac | 2D-2D | No | MR | Deformable |
| (Shu et al 2018) | Neuron tissue | 2D-2D | Yes | EM | Affine |
| (Stergios et al 2018) | Lung | 3D-3D | No | MR | Deformable |
| (Sun and Zhang 2018) | Brain | 3D-3D | No | MR-US | Deformable |
| (de Vos et al 2019) | Cardiac, Lung | 3D-3D | Yes | MR, CT | Affine and Deformable |
| (Zhang 2018) | Brain | 3D-3D | No | MR | Deformable |
| (Balakrishnan et al 2018; Dalca et al 2018; Balakrishnan et al 2019) | Brain | 3D-3D | No | MR | Deformable |
| (Elmahdy et al 2019a; Elmahdy et al 2019b) | Prostate | 3D-3D | Yes | CT | Deformable |
| (Fan et al 2019a) | Brain, Pelvic | 3D-3D | Yes | MR, CT | Deformable |
| (Jiang et al 2019) | Lung | 2D-2D | Yes | CT | Deformable |
| (Kim et al 2019) | Liver | 3D-3D | No | CT | Deformable |
| (Kuang 2019; Kuang and Schmah 2019) | Brain | 3D-3D | No | MR | Deformable |
| (Zhao etal 2019) | Liver | 3D-3D | No | CT | Deformable |
| (Lei et al 2019c) | Abdomen | 3D-3D | Yes | CT | Deformable |
| (Mahapatra et al 2018c) | Retina | 2D-2D | No | FA | Deformable |
| (Nguyen-Duc et al 2019) | Neural tissue | 2D-2D | No | EM | Deformable |
| (Yu et al 2019b) | Abdominopelvic | 3D-3D | Yes | CT-PET | Deformable |
| (Fechter and Baltas 2019) | Lung, Cardiac | 3D-3D | Yes | CT, MR | Deformable |
| (Lei et al 2020) | Abdomen | 3D-3D | Yes | CT | Deformable |
Like (Balakrishnan et al 2019), Qin et al also used segmentation as complementary information for cardiac MR image registration (Qin et al 2018). They found out that the feature learnt by registration CNN could be used in segmentation as well. The predicted DVF was used to deform the masks of moving image to generate masks of the fixed image. They trained a joint segmentation and registration model for cardiac cine image registration and proved that the joint mode could generate better results than the separate models alone in both segmentation and registration tasks. Similar idea has been explored in (Mahapatra et al 2018b) as well. They claimed registration and segmentation are complementary functions and combining them can improve each other’s performance.
Later, Zhang et al proposed a network with trans-convolutional layers for end-to-end DVF prediction in MR brain DIR (Zhang 2018). They focused on the diffeomorphic mapping of the transformation. To encourage smoothness and avoid folding of the predicted transformation, they proposed an inverse-consistent regularization term to penalize the difference between two transformations from the respective inverse mappings. The loss function consists of an image similarity loss, a transformation smoothness loss, an inverse consistent loss and an anti-folding loss. Their method has outperformed Demons and Syn, in terms of DSC score, sensitivity, positive predictive value, average surface distance and Hausdorff distance. A similar idea was proposed by Kim et al who used cycle consistent loss to enforce DVF regularization (Kim et al 2019). They also used identity loss where the output DVF should be zero if the moving and fixed image are the same image.
For 3D-CT image registration, Lei et al used an unsupervised CNN to perform abdominal image registration (Lei et al 2019c). They used a dilated inception module to extract multi-scale motion features for robust DVF prediction. Apart from the image similarity loss and DVF smoothness loss, they integrated a discriminator to provide additional adversarial loss for DVF regularization. Vos et al proposed an unsupervised affine and DIR framework by stacking multiple CNN into a larger network (de Vos et al 2019). The network was tested on cardiac cine MRI and 3D CT lung image registration. They showed their method was comparable to conventional DIR method while being several orders of magnitude faster. Like (de Vos et al 2019), Zhao et al cascaded affine and deformable networks for CT liver DIR (Zhao et al 2019). Recently, Jiang et al proposed a multi-scale framework with unsupervised CNN for 3D CT lung DIR (Jiang et al 2019). They cascaded three CNN models with each model focusing on its own scale level. The network was trained using image patches to optimize an image similarity loss and a DVF smoothness loss. They showed that network trained on SPARE datasets could generalize to a different DIRLAB datasets. In addition, the same trained network also performed well on CT-CBCT and CBCT-CBCT registration without retraining or fine-tuning. They achieved an average TRE of 1.66 ± 1.44 mm on DIRLAB datasets. Fu et al proposed an unsupervised method for 3D-CT lung DIR (Fu et al 2020). They first performed whole-image registration on down-sampled image using a CoarseNet to warp the moving image globally. Then, image patches of the globally warped moving image were registered to the image patches of the fixed image using a patch-based FineNet. They also incorporated a discriminator to provide adversarial loss by penalizing unrealistic warped images. Vessel enhancement was performed prior to DIR to improve the registration accuracy. They have achieved an average TRE of 1.59 ± 1.58 mm, which outperformed some traditional DIR methods. Interestingly, both Jiang et al and Fu et al have achieved better TRE values using unsupervised methods than the supervised methods in (Eppenhof and Pluim 2019) and (Sentker et al 2018).
3.4.2. Assessment.
Compared to supervised transformation prediction, unsupervised methods effectively alleviate the problem of lack of training datasets. Various regularization terms have been proposed to encourage plausible transformation prediction. Several groups have achieved comparable or even better results in terms of TRE on DIRLAB 3D-CT lung DIR. However, most of the methods in this category focused on unimodality registration. There has been a lack of investigation in multi-modality image registration using unsupervised methods. To provide additional supervision, several groups have combined supervised with unsupervised methods for transformation prediction (Fan et al 2019b). The combination seems beneficial; however, more investigation was needed to justify its effectiveness. Given the promising results of the unsupervised methods, we expect a continuous growth of interest in this category.
3.5. GAN in medical image registration
The use of GAN in medical image registration can be generally categorized in two groups: (1) to provide additional regularization of the predicted transformation; (2) to perform cross-domain image mapping. Table 6 shows a list of selected references that utilized GAN to aid the registration.
Table 6.
Overview of registration methods using GAN.
| References | ROI | Dimension | Patch-based | Modality | Transformation |
|---|---|---|---|---|---|
| (Fan et al 2018) | Brain | 3D-3D | Yes | MR | Deformable |
| (Hu et al 2018a) | Prostate | 3D-3D | No | MR-US | Deformable |
| (Yan et al 2018) | Prostate | 3D-3D | No | MR-US | Deformable |
| (Salehi et al 2018) | Brain | 3D-3D | No | MR | Rigid |
| (Elmahdy et al 2019b) | Prostate | 3D-3D | Yes | CT | Deformable |
| (Fan et al 2019a) | Brain, Pelvic | 3D-3D | Both | MR, CT | Deformable |
| (Lei et al 2019c) | Abdomen | 3D-3D | Yes | CT | Deformable |
| (Fu et al 2020) | Lung | 3D-3D | Yes | CT | Deformable |
| (Mahapatra et al 2018a; Mahapatra et al 2018b; Mahapatra et al 2018c) | Retina, Cardiac | 2D-2D | No | FA, Xray | Deformable |
| (Qin et al 2019) | Lung, Brain | 2D-2D | No | T1-T2, CT-MR | Deformable |
3.5.1. Overview of works.
3.5.1.1. GAN-based regularization.
Since image registration is an ill-posed problem, it is crucial to have adequate regularization to encourage plausible transformations and to prevent unrealistic transformations such as tissue folding. Commonly used regularization terms include DVF smoothness constraint, anti-folding constraint and inverse consistency constraint. However, it remains ambiguous whether these constraints are adequate for proper regularization. Recently, GAN-based regularization terms have been introduced to the realm of image registration. The idea is to train an adversarial network to introduce a network-based loss for transformation regularization. In the literature, discriminators were trained to distinguish three types of inputs, including (1) whether a transformation is predicted or ground truth, (2) whether an image is realistic or warped by predicted transformation, (3) whether an image pair alignment is positive or negative.
Yan et al trained an adversarial network to tell whether an image was deformed using ground truth transformation or predicted transformation (Yan et al 2018). Randomly generated transformations from manually aligned image pairs were used as ground truth to train a network to perform MR-US prostate image registration. The trained discriminator could provide not only an adversarial loss for regularization but also a discriminator score for alignment evaluation. Fan et al used a discriminator to distinguish whether an image pair were well aligned (Fan et al 2019a). In unimodal image registration, they have defined a positive image alignment case as weighted linear combination of the fixed and the moving images. In multi-modal image registration case, positive image alignments were pre-defined using paired MR and CT images. They performed on MR brain images for unimodal registration and on pelvic CT-MR for multi-modal registration. They have showed that the performance increased with the adversarial loss. Lei et al used a discriminator to judge whether the warped image is realistic enough to the original images (Lei et al 2019c). Fu et al used a similar idea and showed that the inclusion of adversarial loss could improve registration accuracy in 3D-CT lung DIR (Fu et al 2020).
The above GAN-based methods have tried to introduce regularization from the image or transformation appearance perspective. Differently, Hu et al tried to introduce biomechanical constraints to 3D MR-US prostate image registration by discriminating whether a transformation is predicted or generated by finite element analysis (Hu et al 2018a). Instead of adding the adversarial loss to existing smoothness loss, they replaced the smoothness loss with the adversarial loss. They showed that their method could predict physically plausible deformation without any other smoothness penalty.
3.5.1.2. GAN-based cross-domain image mapping.
For multi-modal image registration, progresses have been made by using deep similarity metrics in traditional DIR frameworks. Using iterative methods, several works have outperformed the-state-of-art MI similarity measures. However, in terms of direct transformation prediction, multi-modal image registration has not benefited from DL as much as unimodal image registration has. This is mainly due to the vast appearance differences between different modalities. To overcome this challenge, GAN has been used to translate multi-modal to unimodal image registration by mapping images from one modality to another.
Salehi et al trained a CNN using T2-weighted images to perform fetal brain MR registration. They tested the network on T1-weighted images by first mapping the T1 to T2 image domain using a conditional GAN (Salehi et al 2018). They showed the trained network generalized well on the synthesized T2 images. Qin et al used an unsupervised image-to-image translation framework to cast multi-modal to unimodal image registration (Qin et al 2019). The image to image translation method assumes the images could be decomposed into content code and style code. They have showed comparable results to MIND and Elastix on BraTs datasets in terms of RMSE of DVF error. On COPDGene datasets, they outperformed MIND and Elastix in terms of DICE, mean contour distance (MCD) and Hausdorff distance. Mahapatra et al combined cGan (Mirza and Osindero 2014) and registration network together to directly predict both DVF and warped image (Mahapatra et al 2018c). They implicitly transformed image in one modality to another modality. They outperformed Elastix on 2D retinal image registration in terms of Hausdorff distance, MAD and MSE. Elmahdy et al claimed that inpainting gas pockets in the rectum could enhance rectum and seminal vesicle registration (Elmahdy et al 2019b). They used GAN to detect and inpaint rectum gas pocket prior to image registration.
3.5.2. Assessment.
GAN has been shown to be promising in medical image registration via either novel adversarial loss or image domain translation. For adversarial losses, GAN could provide learnt network-based regularizations that are complementary to traditional handcrafted regularization terms. For image domain translation, GAN effectively cast the more challenging multi-modal registration to unimodal image registration, which allows many existing unimodal registration algorithms to be applied to multi-modal image registration. However, the absolute intensity mapping accuracy of GAN is yet to be investigated. GAN has also been applied to deep similarity metric learning in registration and alignment validation. As evidenced by the trend in figure 2, we expect to see more papers using GAN in image registration tasks in the future.
3.6. Registration validation using DL
The performance of image registration could be evaluated using image similarity metrics such as SSD, NCC and MI. However, the image similarity metrics only evaluate the overall alignment on the whole image. To have a deeper insight into local registration accuracy, we usually rely on manual landmark pair selection. Nevertheless, manual landmark pair selection is time-consuming, subjective and error-prone especially when many landmarks were to be selected. Fu et al used a Siamese network for large quantity landmark pair detection on 3D-CT lung images (Fu et al 2019). The network was trained using the manual landmark pairs from DIRLAB datasets. They performed experiments comparisons, showing that the network could outperform human in landmark pair detection. Neylon et al proposed to use a deep neural network to predict TRE for given image similarity metrics (Neylon et al 2017). The network was trained using patient-specific biomechanical models of head-neck anatomy. They demonstrated that the network could rapidly and accurately quantify registration performance.
3.6.1. Overview of works.
Table 7 shows a list of selected references that used deep learning to aid registration validation. Eppenhof et al proposed a TRE alternative to assess DIR registration accuracy. They used synthetic transformations as ground truth to avoid the need for manual annotations (Eppenhof and Pluim 2018). The ground truth error map was the L2 difference between ground truth transformations and the predicted transformations. They trained a network to robustly estimate registration errors with sub-voxel accuracy. Galib et al predicted an overall registration error index, which is the ratio between good alignment sub-volumes and poor alignment sub-volumes (Galib et al 2019). They justified the choice of threshold TRE of 3.5 mm as a cutoff value of good and bad alignment. Their network was trained using manually labeled landmarks from DIRLAB. Sokooti et al proposed a random forest regression method for quantitative error prediction of DIR (Sokooti et al 2019a). They used both intensity-based features such as MIND and registration-based features such as transformation Jacobian determinant. Dubost et al used ventricle DSC score to evaluate brain registration (Dubost et al 2019). The ventricle was segmented using DL-based method.
Table 7.
Overview of registration validation methods using DL.
| References | ROI | Dimension | Modality | End point |
|---|---|---|---|---|
| (Neylon et al 2017) | HN | 3D | CT | TRE prediction |
| (Eppenhof and Pluim 2018) | Lung | 3D | CT | Registration error |
| (Dubost et al 2019) | Brain | 3D | MRI | DSC score |
| (Fu et al 2019) | Lung | 3D | CT | Landmark Pairs |
| (Galib et al 2019) | Lung | 3D | CT | Registration error |
| (Sokooti et al 2019a) | Lung | 3D | CT | Registration error |
3.6.2. Assessment.
The number of papers using DL for registration evaluation has increased significantly in 2019. Most works treated registration error prediction as a supervised regression problem. Network was trained using manually annotated datasets. It is important to make sure the ground truth datasets are of high quality. Most of existing methods focused on lung because benchmark datasets with manual landmark pairs exists for 3D CT lung such as DIRLAB. It would be interesting to see the method be applied on many other treatment sites. Unsupervised registration error prediction is another interesting research topic to eliminate the need for manual annotated datasets.
3.7. Other learning-based methods in medical image registration
Table 8 shows a list of other methods that utilized deep learning to aid the registration such as LSTM, transfer learning, FasterRCNN and so on. Jiang et al proposed to use CNN to lean and infer expressive sparse multi-grid configurations prior to B-spline coefficient optimization (Jiang and Shackleford 2018). Liu et al used a ten-layer FCN for image synthesis without GAN to transform multimodal to unimodal registration among T1-weighted, T2-weighed, and proton density images (Liu et al 2019b). Then, they used Elastix software with SSD similarity metric for the registration of brain phantom and IXI datasets. They outperformed MI similarity index. Wright et al proposed to use LSTM network to predict a rigid transformation and an isotropic scaling factor for MR-US fetal brain registration (Wright et al 2018). Bashiri et al used Laplacian eigenmap as a manifold learning method to implement a multi-modal to unimodal image translation in 2D brain image registration (Bashiri et al 2019).
Table 8.
Overview of other DL-based image registration methods.
| References | ROI | Dimension | Modality | Transformation | Methods |
|---|---|---|---|---|---|
| (Jiang and Shackleford 2018) | Lung | 3D-3D | CT | Deformable | Multi-grid Inference |
| (Wright et al 2018) | Brain | 3D-3D | MR-US | Rigid | LSTM |
| (Bashiri et al 2019) | Brain | 2D-2D | CT, T1, T2, PD | Rigid | Manifold Learning |
| (Xia et al 2019) | Brain, Abdomen | 2D-3D | CT-PET, CT-MRI | Deformable | CAE, DSCNN |
| (Yu et al 2019a) | Spine | 3D-2D | 3DCT-Xray | Rigid | FasterRCNN |
| (Liu et al 2019b) | Brain | 2D-2D | T1-T2, T1-PD | Deformable | FCN |
| (Zheng et al 2018) | Spine | 3D-2D | 3DCT-Xray | Rigid | Domain adaptation |
| (Mahapatra and Ge 2019) | Chest, Brain | 2D-2D | Xray, MRI | Deformable | Transfer Learning |
Mahapatra and Ge proposed to use transfer learning to reuse part of the network weights that were learned from chest x-ray registration on MRI brain image registration. In the transfer learning, the weights of the last few layers were updated iteratively based on the output of a discriminator while the rest of the network weights were kept constant (Mahapatra and Ge 2019). Yu et al proposed to use FasterRCNN (Ren et al 2017) for vertebrae bounding box detection (Yu et al 2019a). The detected bounding box was then matched to doctor-annotated bounding box on the x-ray image. Zheng et al proposed a domain adaptation module to cope with the domain variance between synthetic data and real data (Zheng et al 2018). The adaptation module can be trained using a few paired real and synthetic data. The trained module could be plugged into the network to transfer the real features to approach the synthetic features. Since network was trained on synthetic data, the network should perform well on synthetic data. Hence, it is reasonable to transfer the real data features to synthetic features.
4. Benchmark
Benchmarking is important for readers to understand through comparison the advantages and disadvantages of each method. For image registration, both registration accuracy and computational time could be benchmarked. However, researchers have been reporting registration accuracies more than the computational speed. Computational speed is largely dependent on the hardware, which is often different from group to group. According to the statistics of the cited works, the top two ROIs of registration are brain and lung. Therefore, we summarized the registration datasets for brain, registration accuracies for lung.
4.1. Lung
DIRLAB is one of the most cited public datasets for 4D-CT chest image registration studies (Castillo et al 2009). DIRLAB provides 300 manually selected landmark pairs for end-exhalation and end-inhalation phases. This dataset was frequently used for 4D-CT lung registration benchmarking. To provide the readers a better understanding of the latest DL-based registration, we have listed the TREs of three top performing traditional methods and seven DL-based lung registration methods. Table 9 shows that DL-based lung registration methods have not outperformed the top traditional DIR methods yet in terms of TRE. However, DL-based DIR methods have been making substantial improvement over the years, with Fu et al and Jiang et al almost achieving comparable TRE to top traditional methods. TREs of traditional DIR on case 8 were consistently better than that of the DL-based DIR. Case 8 is one of the most challenging cases in the DIRLAB datasets with impaired image quality and significant lung motion. This phenomenon suggests that the robustness and competency of DL-based DIR need to be further improved. Table 10 lists the workstation configurations and computational time of registration for several DL-based methods.
Table 9.
Comparison of target registration error (TRE) values among different methods on DIRLAB datasets, TRE unit: (mm).
| Top traditional methods | DL methods | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Set | Before registration | (Heinrich et al 2013) | (Berendsen et al 2014) | (Staring et al 2010) | (Eppenhof and Pluim 2019) | (de Vos et al 2019) | (Sentker et al 2018) | (Fu et al 2020) | (Sokooti et al 2019b) | (Jiang et al 2019) | (Fechter and Baltas 2019) |
| 1 | 3.89 ±2.78 | 0.97 ± 0.5 | 1.00 ± 0.52 | 0.99 ± 0.57 | 1.45 ± 1.06 | 1.27 ± 1.16 | 1.20 ± 0.60 | 0.98 ± 0.54 | 1.13 ± 0.51 | 1.20 ± 0.63 | 1.21 ± 0.88 |
| 2 | 4.34 ± 3.90 | 0.96 ± 0.5 | 1.02 ±0.57 | 0.94 ± 0.53 | 1.46 ± 0.76 | 1.20 ± 1.12 | 1.19 ±0.63 | 0.98 ± 0.52 | 1.08 ± 0.55 | 1.13 ±0.56 | 1.13 ± 0.65 |
| 3 | 6.94 ± 4.05 | 1.21 ± 0.7 | 1.14 ±0.89 | 1.13 ± 0.64 | 1.57 ± 1.10 | 1.48 ± 1.26 | 1.67 ± 0.90 | 1.14 ±0.64 | 1.33 ± 0.73 | 1.30 ±0.70 | 1.32 ± 0.82 |
| 4 | 9.83 ±4.86 | 1.39 ± 1.0 | 1.46 ± 0.96 | 1.49 ± 1.01 | 1.95 ± 1.32 | 2.09 ± 1.93 | 2.53 ±2.01 | 1.39 ± 0.99 | 1.57 ± 0.99 | 1.55 ±0.96 | 1.84 ± 1.76 |
| 5 | 7.48 ± 5.51 | 1.72 ± 1.6 | 1.61 ± 1.48 | 1.77 ± 1.53 | 2.07 ± 1.59 | 1.95 ± 2.10 | 2.06 ± 1.56 | 1.43 ± 1.31 | 1.62 ± 1.30 | 1.72 ± 1.28 | 1.80 ± 1.60 |
| 6 | 10.89 ± 6.9 | 1.49 ± 1.0 | 1.42 ± 0.89 | 1.29 ±0.85 | 3.04 ± 2.73 | 5.16 ± 7.09 | 2.90 ± 1.70 | 2.26 ± 2.93 | 2.75 ± 2.91 | 2.02 ± 1.70 | 2.30 ± 3.78 |
| 7 | 11.03 ± 7.4 | 1.58 ± 1.2 | 1.49 ± 1.06 | 1.26 ± 1.09 | 3.41 ± 2.75 | 3.05 ± 3.01 | 3.60 ± 2.99 | 1.42 ± 1.16 | 2.34 ± 2.32 | 1.70 ± 1.03 | 1.91 ± 1.65 |
| 8 | 15.0 ±9.01 | 2.11 ± 2.4 | 1.62 ± 1.71 | 1.87 ±2.57 | 2.80 ± 2.46 | 6.48 ± 5.37 | 5.29 ± 5.52 | 3.13 ± 3.77 | 3.29 ± 4.32 | 2.64 ± 2.78 | 3.47 ± 5.00 |
| 9 | 7.92 ± 3.98 | 1.36 ± 0.7 | 1.30 ±0.76 | 1.33 ± 0.98 | 2.18 ± 1.24 | 2.10 ± 1.66 | 2.38 ± 1.46 | 1.27 ± 0.94 | 1.86 ± 1.47 | 1.51 ±0.94 | 1.47 ± 0.85 |
| 10 | 7.3 ±6.35 | 1.43 ± 1.6 | 1.50 ± 1.31 | 1.14 ±0.89 | 1.83 ± 1.36 | 2.09 ± 2.24 | 2.13 ± 1.88 | 1.93 ± 3.06 | 1.63 ± 1.29 | 1.79 ± 1.61 | 1.79 ± 2.24 |
| Mean | 8.46 ± 5.48 | 1.43 ± 1.3 | 1.36 ± 0.99 | 1.32 ± 1.24 | 2.17 ± 1.89 | 2.64 ± 4.32 | 2.50 ± 1.16 | 1.59 ± 1.58 | 1.86 ±2.12 | 1.66 ± 1.44 | 1.83 ± 2.35 |
Table 10.
Workstation configurations and computational time.
| References | Configurations | Input image size (pixel) | Computation time |
|---|---|---|---|
| (Eppenhof and Pluim 2019) | Intel Xeon CPU E5-2640 v4 with 512 GB memory, Nvidia Titan XP graphics card with 12 GB memory | 128 × 128 × 128 | ~3.3 min |
| (de Vos et al 2019) | Intel Xeon E5-1620 3.60 GHz CPU with 4 cores (8 threads), and 32 GB of memory, NVIDIA Titan-X GPU | Not available | <1 s |
| (Sentker et al 2018) | Intel Xeon CPU E5-1620 and Nvidia Titan Xp GPU. | Not available | A few seconds |
| (Fu et al 2020) | NVIDIA Tesla V100 GPU with 32 GB of memory | ~250 × 200 × 100 | <1 min |
| (Sokooti et al 2019b) | Nvidia Titan XP GPU with 12 GB of memory | ~273 × 273 × 273 | <3 s |
| (Jiang et al 2019) | CPU of Intel Xeon with 64 GB memory and NVIDIA Quadro P4000 GPU | 256 × 256 × 96 | <2 s |
| (Fechter and Baltas 2019) | Intel Xeon CPU with 8 cores, 60 GB memory and Nvidia Titan XP GPU | Not available | ~4 min |
4.2. Brain
Brain image registration has much wider options in databases than lung image registration. As a result, authors were not consistent on which database to use for training and testing and what metrics to use for validations. To facilitate benchmarking, we have listed a number of works on brain image registration in table 11, which presents the datasets, the registration transformation model and the evaluation metrics. DSC of multiple ROI is the most commonly used evaluation metric. MI and surface distance measures are the next frequently used evaluation metrics.
Table 11.
Benchmark datasets and evaluation metrics used in brain registration.
| References | Datasets | Transformation | Evaluation metrics |
|---|---|---|---|
| (Liu et al 2019b) | IXI | Deformable | TRE, MI |
| (Kuang and Schmah 2019) | MindBoggle-101 | Deformable | DSC |
| (Kori and Krishnamurthi 2019) | BRATS, ALBERT | Affine | DSC, MI, SSIM, MSE |
| (Ferrante et al 2019) | IBSR | Deformable | DSC, MI, NCC, SAD, DWT |
| (Fan et al 2019b) | LONI, LPBA40, IBSR, CUMC, MGH, IXI | Deformable | DSC, ASD |
| (Balakrishnan et al 2019) | OASIS, ABIDE, ADHD, MCIC, PPMI, HABS, Harvard GSP | Deformable | DSC |
| (Zhang 2018) | ADNI | Deformable | DSC, SEN, PPV, ASD, HD |
| (Sloan et al 2018) | OASIS, IXI, ISLES | Rigid | MSE |
| (Sedghi et al 2018) | IXI | Rigid | MAE of degree and translation |
| (Li and Fan 2018) | ADNI | Deformable | DSC |
| (Cao et al 2018b) | LONI, ADNI, IXI | Deformable | DSC, ASD |
| (Yang et al 2017) | OASIS, IBIS, LPBA, IBSR, MGH, CUMC | Deformable | Target overlap |
| (Shan et al 2017) | LPBA | Deformable | TRE, JACC |
| (Ghosal and Rayl 2017) | IXI | Deformable | SSD, PSNR, SSIM |
| (Wu et al 2016) | LONI, ADNI | Deformable | DSC |
| (Simonovsky et al 2016) | IXI, ALBERT | Deformable | DSC, JACC |
5. Statistics
After careful study of each category, it is important to step back and look at the whole picture. Out of the 150 + papers cited, more than half of the papers were aimed at direct transformation prediction using either supervised or unsupervised transformation prediction. The category of deep similarity-based methods accounts for 14% of all methods while the category of GAN account for 10% of all methods. Publications from the same group (Conference papers which were extended into journal papers) were counted only once if there were no substantial differences in content. One paper may belong to multiple categories. For example, unsupervised CNN method could use GAN generated loss for additional transformation regularization. Details percentages are shown in figure 3.
Figure 3.
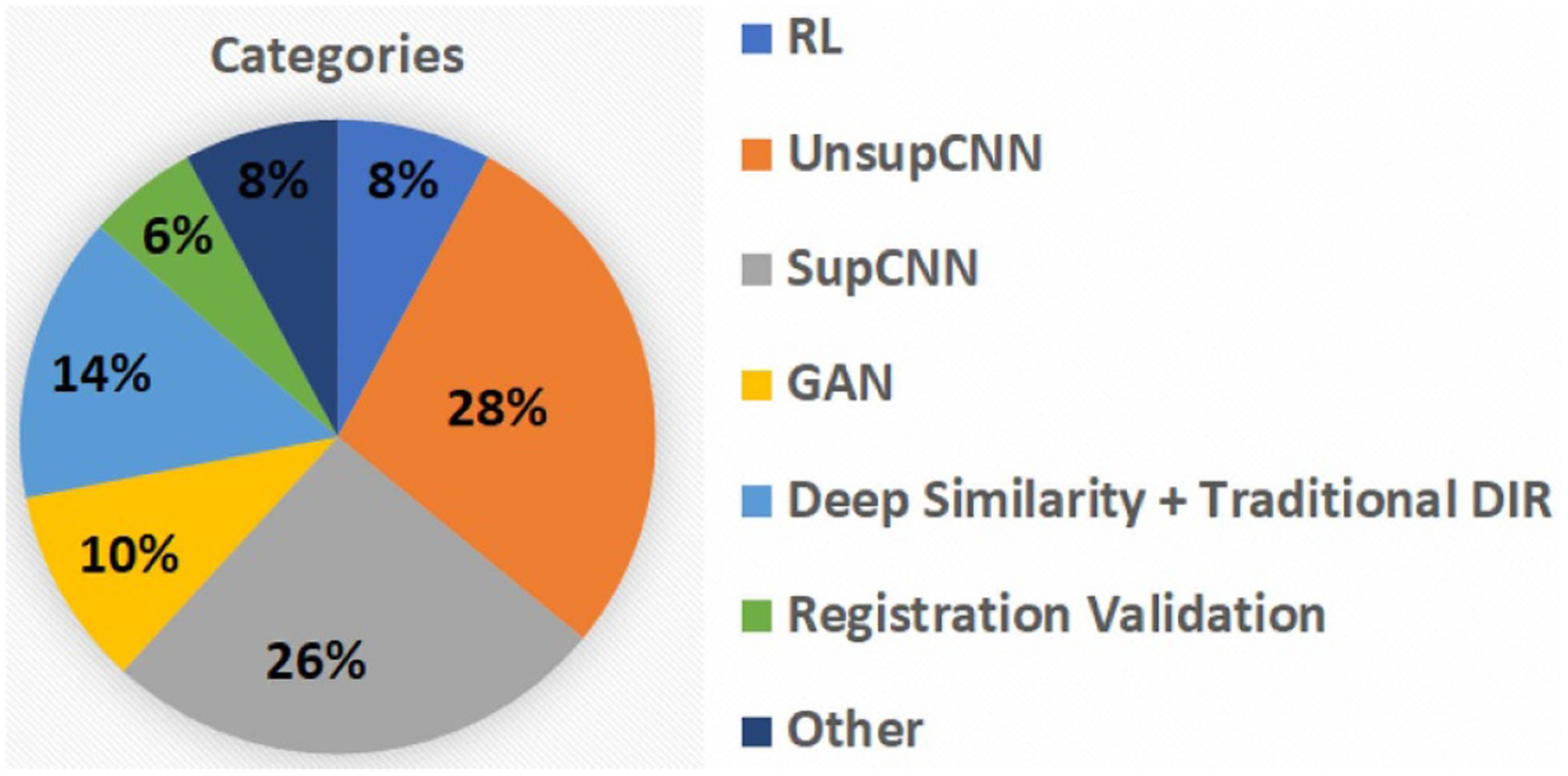
Percentage pie chart of different categories.
Besides the number of papers, we have also analyzed the percentage distributions of many other attributes including input image pair dimension, transformation model, image domain, patch-based training, DL frameworks and ROI of the cited works. The percentage distributions were shown in figure 4. 60% of the works were solving 3D-3D registration problems. The 2D-3D image registration works are mostly to register 3D-CT to 2D x-ray images for intraoperation image guidance. The percentages of the number of deformable, rigid and affine registration papers are 72%, 19% and 9%, respectively. Most of the rigid registration papers are for intra-patient brain and spine alignment. There are more publications on unimodal than multi-modal image registration. Due to the superior performance of DL-based similarity measures to traditional similarity measures, the number of DL-based multi-modality image registration papers is increasing and accounts for 41% of all the papers. Patch-based training was often adopted to save GPU memory. Figure 4 shows that 70% of all works used whole image-based training. The 70% includes not only 3D-3D but also 2D-3D and 2D-2D image registrations. Almost all 2D-2D registration used whole image-based training since 2D images are much less memory demanding than 3D images. Therefore, for 3D-3D image registration, there are roughly the same number of works that used whole image-based training and patch-based training. In terms of DL frameworks, Tensorflow is the leading framework which accounts for more than half of all papers. Pytorch is the second most popular DL framework which accounts for a quarter of all papers. Early works used Caffe and Theano, which was used less and less over the years as compared to Tensorflow and Pytorch. Theano has officially ceased development after version 1.0. The DL toolbox of Matlab is the least used framework perhaps due to licensing. In terms of the ROI, MR brain and CT lung are the most studied sites. Brain is the top registration target in all works. The reason for the wide adoption of brain include its clinical importance, its availability of public datasets and its relative simplicity of registration.
Figure 4.
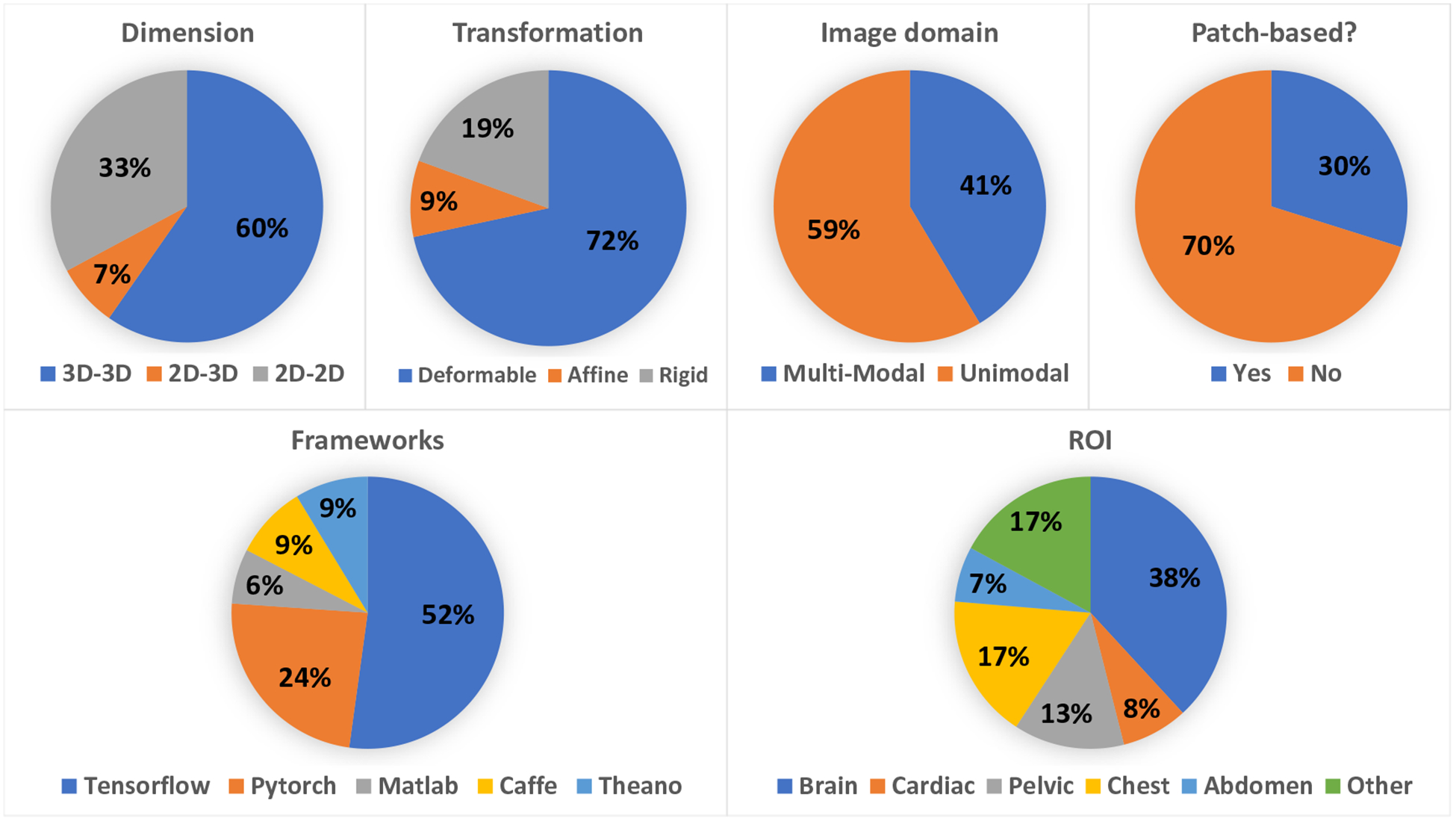
Percentage pie chart of various attributes of DL-based image registration methods.
6. Discussion
Though image registration has been extensively studied, DL-based medical image registration is a relatively new research area. We have collected over 150 papers, most of which were published in the last three to four years. We generally classify these methods into seven non-exclusive categories. Many methods could be classified into multiple categories. For example, GAN was mostly used in combination with supervised or unsupervised transformation prediction methods as an auxiliary regularization or image pre-processing step. Supervised and unsupervised methods were combined for dual supervision in some works. DL-based registration validation methods were included in this review because methods in this category often involve learning a deep similarity metric, therefore, could be used for image registration. RL and deep similarity-based methods are iterative whereas supervised and unsupervised based methods are non-iterative. For iterative methods, multiple works have reported that deep similarity metrics have superior performance to handcrafted intensity-based image similarity metric. For non-iterative methods, DL-based methods have yet to outperform traditional DIR methods. Take lung registration for example, the best performing DL-based methods are only comparable to the-state-of-art traditional DIR methods in terms of TRE. However, DL-based direct transformation methods are generally order of magnitude faster than traditional DIR methods. This is mainly due to the non-iterative nature and the powerful GPU utilized. A common feature that is used in both traditional DIR and DL-based methods is multi-scale strategy. Multi-scale registration could help the optimization avoid local maxima and allow large deformation registration. Regarding network generality, Fu et al and Jiang et al both showed that network trained using one set of datasets could be readily applied to an independent set of datasets given that the two image domains are close to each other.
6.1. Whole image-based vs. patch-based transformation prediction
Whole image-based training and patch-based training have their own advantages and disadvantages. Due to limited GPU memory, the original images were often down-sampled to avoid memory overflow in whole image-based training. The down-sampling process could cause information loss and limit the registration accuracy. On the other hand, whole image training allows large inception field which enables registration of large deformations and mitigate the problem of local maxima in registration. Unless data augmentation is used, whole image-based training usually suffers from shortage of training datasets. On the contrary, patch-based training were not affected by the shortage of training datasets as much since many image patches could be sampled from the original images. In addition, patch-based training usually has better performance locally than whole-image based training. Recently, several groups combined whole-image training with patch-based training as a multi-scale approach for image registration (Lei et al 2019c, Fu et al 2020). They have achieved promising results in terms of registration accuracy. One challenge with patch-based image registration is the patch fusion process, which stack many image patches to generate the final whole-image transformation. The patch fusion process could generate grid-like artifacts along the edges of the patches. One way to mitigate the problem is to use large patch overlap prior to patch fusion. However, it would make the inference process computationally inefficient. Another method is to use a non-parametric registration model for transformation prediction. One such example is LDDMM model used in QuickSilver (Yang et al 2017). Instead of directly predicting final spatial transformation, QuickSilver predict the momentum of the LDDMM model. The LDDMM model can generate diffeomorphic spatial transformation without the need of smooth momentum predictions.
6.2. Loss functions
Despite large variations in details, loss function definitions of the cited works share many common features. Almost all loss function definitions consist of one or more combinations of the following six types of losses, which are (1) intensity-based image appearance loss, (2) deep similarity-based image appearance loss, (3) transformation smoothness constraint, (4) transformation physical fidelity loss, (5) transformation error loss with respect to ground truth transformation and (6) adversarial loss. Intensity-based image appearance loss includes SSD, MSE, MAE, MI, MIND, SSIM, CC and its variants. Deep similarity-based image appearance loss usually calculates the correlation between the learnt feature-based image descriptors. Transformation smoothness constraints usually involve the calculation of the first and second orders of spatial derivatives of predicted transformation. Transformation physical fidelity loss includes inverse consistency loss, negative Jacobian determinant loss, identity loss, anti-folding loss and so on. Transformation error loss was the error between predicted and ground truth transformations, which was only valid for supervised transformation prediction. Adversarial loss was the trainable network-based loss. Some auxiliary loss terms include the DSC loss of the anatomical labels or TRE loss of pre-selected landmark pairs.
6.3. Challenges and opportunities
One of the most common challenges for supervised DL-based methods is the lack of training datasets with known transformations. This problem could be alleviated by various data augmentation methods. However, the data augmentation methods could introduce additional errors such as the bias of unrealistic artificial transformations and image domain shifts between training and testing stages. Several groups have demonstrated good generality of the trained network by applying them to datasets different from the training datasets. This inspired us to think that transfer learning may be used to alleviate the problem of lack of training data. Surprisingly, transfer learning has not been used in medical image registration. For unsupervised methods, efforts were made to combine different kinds of regularization terms to constrain the predicted transformation. However, it is difficult to investigate the relative importance of each regularization term. Researchers are still trying to find an optimal set of transformation regularization terms that could help generate not only physically plausible but also physiologically realistic deformation field for a certain registration task. This is partially due to the lack of registration validation methods. Due to the unavailability of ground truth transformation between an image pair, it is hard to compare the performances of different registration methods. Therefore, registration validation methods are equally important as registration methods. We have observed an increased number of papers focusing on registration validation in 2019. More research on registration validation methods is desired in order to reliably evaluate the performances of different registration methods under different parametric configurations.
6.4. Trends
Judging from the statistics of the cited works, there is a clear trend of direct transformation prediction for fast image registration. So far, supervised and unsupervised transformation prediction methods are almost equally studied with a close number of publications in either category. Either supervised or unsupervised methods have their own advantages and disadvantages. We speculate that more research will be focused on combining supervised and unsupervised methods in the future. GAN-based methods have been gradually gaining popularity since GAN could be used to not only introduce additional regularizations but also perform image domain translation to cast multi-modal to unimodal image registration. We should see a steady growth of GAN-based medical image registration. New transformation regularization techniques have always been a hot topic due to the ill-posedness of the registration problem.
Acknowledgments
This research is supported in part by the National Cancer Institute of the National Institutes of Health under Award Number R01CA215718, and Dunwoody Golf Club Prostate Cancer Research Award, a philanthropic award provided by the Winship Cancer Institute of Emory University.
Footnotes
Disclosures
The authors declare no conflicts of interest.
References
- Andersen ES, Noe KØ, Sørensen TS, Nielsen SK, Fokdal L, Paludan M, Lindegaard JC and Tanderup K 2013. Simple DVH parameter addition as compared to deformable registration for bladder dose accumulation in cervix cancer brachytherapy Radiother. Oncol 107 52–57 [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M and Gee JC 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain Med. Image Anal 12 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A and Gee JC 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration Neuroimage 54 2033–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B 2002. Reinforcement learning with long short-term memory Advances in Neural Information Processing Systems 14 (NIPS 2001) ed Dietterich TG, Becker S and Ghahramani Z (Cambridge, MA: MIT Press; ) pp 1475–82 [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J and Dalca AV 2018. An unsupervised learning model for deformable medical image registration 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (Salt Lake City, UT, 18–23 June 2018) (Piscataway, NJ: IEEE; ) pp 9252–60 [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J and Dalca AV 2019. VoxelMorph: a learning framework for deformable medical image registration IEEE Trans. Med. Imaging 38 1788–800 [DOI] [PubMed] [Google Scholar]
- Bashiri FS, Baghaie A, Rostami R, Yu ZY and D’Souza RM 2019. Multi-modal medical image registration with full or partial data: a manifold learning approach J. Imaging 5 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender ET, Hardcastle N and Tome WA 2012. On the dosimetric effect and reduction of inverse consistency and transitivity errors in deformable image registration for dose accumulation Med. Phys 39 272–80 [DOI] [PubMed] [Google Scholar]
- Berendsen F, Kotte ANT, Viergever M and Pluim JP 2014. Registration of organs with sliding interfaces and changing topologies Proc.SPIE 9034 90340E [Google Scholar]
- Brock KK, Sharpe MB, Dawson LA, Kim SM and Jaffray DA 2005. Accuracy of finite element model-based multi-organ deformable image registration Med. Phys 32 1647–59 [DOI] [PubMed] [Google Scholar]
- Cao X, Yang J, Wang L, Xue Z, Wang Q and Shen D 2018a. Deep learning based inter-modality image registration supervised by intra-modality similarity Machine Learning in Medical Imaging. MLMI 2018. Lecture Notes in Computer Science, ed Shi Y, Suk HI and Liu M (Berlin: Springer; ) vol 11046 pp 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XH, Yang JH, Zhang J, Wang Q, Yap PT and Shen DG 2018b. Deformable image registration using a cue-aware deep regression network IEEE Trans. Biomed. Eng 65 1900–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo R, Castillo E, Guerra R, Johnson VE, McPhail T, Garg AK and Guerrero T 2009. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets Phys. Med. Biol 54 1849–70 [DOI] [PubMed] [Google Scholar]
- Chao M, Penagaricano J, Yan Y, Moros EG, Corry P and Ratanatharathorn V 2012. Voxel-based dose reconstruction for total body irradiation with helical tomotherapy Int. J. Radiat. Oncol. Biol. Phys 82 1575–83 [DOI] [PubMed] [Google Scholar]
- Chee E and Wu Z 2018. AIRNet: self-supervised affine registration for 3D medical images using neural networks (arXiv:1810.02583) [Google Scholar]
- Chen M, Shi X, Zhang Y, Wu D and Guizani M 2017. Deep features learning for medical image analysis with convolutional AE neural network IEEE Trans. Big Data ( 10.1109/TBDATA.2017.2717439) [DOI] [Google Scholar]
- Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I and Abbeel P 2016. InfoGAN: Interpretable representation learning by information maximizing generative adversarial nets (arXiv:1606.03657v1) [Google Scholar]
- Cheng X, Zhang L and Zheng Y 2018. Deep similarity learning for multimodal medical images Comput. Methods Biomech. Biomed. Eng.: Imaging Visualization 6 248–52 [Google Scholar]
- Chetty IJ and Rosu-Bubulac M 2019. Deformable registration for dose accumulation Semin. Radiat. Oncol 29 198–208 [DOI] [PubMed] [Google Scholar]
- Cho K, Merrienboer B, Gülçehre Ç, Bahdanau D, Bougares F, Schwenk H and Bengio Y 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation (arXiv:1406.1078) [Google Scholar]
- Choi Y, Choi M, Kim M, Ha J, Kim S and Choo J 2018. StarGAN: unified generative adversarial networks for multi-domain image-to-image translation 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition (Salt Lake City, UT, 18–23 June 2018) (Piscataway, NJ: IEEE; ) pp 8789–97 [Google Scholar]
- Chung J, Gülçehre Ç, Cho K and Bengio Y 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling (arXiv:1412.3555) [Google Scholar]
- Dalca AV, Balakrishnan G, Guttag JV and Sabuncu MR 2018. Unsupervised learning for fast probabilistic diffeomorphic registration Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science vol 11070, ed Frangi A, Schnabel J, Davatzikos C, Alberola-López C and Fichtinger G (Berlin: Springer; ) pp 729–38 [Google Scholar]
- Dang H, Wang AS, Sussman MS, Siewerdsen JH and Stayman JW 2014. dPIRPLE: a joint estimation framework for deformable registration and penalized-likelihood CT image reconstruction using prior images Phys. Med. Biol 59 4799–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva T, Uneri A, Ketcha MD, Reaungamornrat S, Kleinszig G, Vogt S, Aygun N, Lo SF, Wolinsky JP and Siewerdsen JH 2016. 3D-2D image registration for target localization in spine surgery: investigation of similarity metrics providing robustness to content mismatch Phys. Med. Biol 61 3009–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M and Isgum I 2019. A DL framework for unsupervised affine and deformable image registration Med. Image Anal 52 128–43 [DOI] [PubMed] [Google Scholar]
- Dong X, Lei Y, Tian S, Wang T, Patel P, Curran WJ, Jani AB, Liu T and Yang X 2019a. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network Radiother. Oncol 141 192–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Lei Y, Wang T, Higgins K, Liu T, Curran WJ, Mao H, Nye JA and Yang X 2019b. Deep learning-based attenuation correction in the absence of structural information for whole-body PET imaging Phys. Med. Biol 65 055011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, Liu T and Yang X 2019c. Automatic multiorgan segmentation in thorax CT images using U-net-GAN Med. Phys 46 2157–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, Mao H, Nye JA and Yang X 2019d. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging Phys. Med. Biol 64 215016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosovitskiy A and Brox T 2016. Generating images with perceptual similarity metrics based on deep networks (arXiv:1602.02644v2) [Google Scholar]
- Dosovitskiy A, Fischer P, Ilg E, Häusser P, Hazirbas C, Golkov V, Smagt P, Cremers D and Brox T 2015. FlowNet: learning optical flow with convolutional networks 2015 IEEE Int. Conf. on Computer Vision (ICCV) (Santiago, Chile, 7–13 December 2015) (Piscataway, NJ: IEEE; ) pp 2758–66 [Google Scholar]
- Dubost F et al. 2019. Automated image registration quality assessment utilizing deep-learning based ventricle extraction in clinical data (arXiv:1907.00695) [Google Scholar]
- Elmahdy MS et al. 2019a. Robust contour propagation using DL and image registration for online adaptive proton therapy of prostate cancer Med. Phys 46 3329–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmahdy MS, Wolterink JM, Sokooti H, Išgum I and Staring M 2019b. Adversarial optimization for joint registration and segmentation in prostate CT radiotherapy Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Lecture Notes in Computer Science vol 11769, ed Shen D et al. (Berlin: Springer; ) pp 366–74 [Google Scholar]
- Eppenhof KAJ, Lafarge MW, Moeskops P, Veta M and Pluim JPW 2018. Deformable image registration using convolutional neural networks Proc. SPIE 10574 105740S [Google Scholar]
- Eppenhof KAJ and Pluim JPW 2018. Error estimation of deformable image registration of pulmonary CT scans using convolutional neural networks J. Med. Imaging 5 024003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppenhof KAJ and Pluim JPW 2019. Pulmonary CT registration through supervised learning with convolutional neural networks IEEE Trans. Med. Imaging 38 1097–105 [DOI] [PubMed] [Google Scholar]
- Fan J, Cao X, Xue Z, Yap PT and Shen D 2018. Adversarial similarity network for evaluating image alignment in DL based registration Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science vol 11070, ed Frangi A, Schnabel J, Davatzikos C, Alberola-López C and Fichtinger G (Berlin: Springer; ) pp 739–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JF, Cao XH, Wang Q, Yap PT and Shen DG 2019a. Adversarial learning for mono- or multi-modal registration Med. Image Anal 58 101545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JF, Cao XH, Yap EA and Shen DG 2019b. BIRNet: brain image registration using dual-supervised fully convolutional networks Med. Image Anal 54 193–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechter T and Baltas D 2019. One shot learning for deformable medical image registration and periodic motion tracking (arXiv:1907.04641) [DOI] [PubMed] [Google Scholar]
- Ferrante E, Dokania PK, Silva RM and Paragios N 2019. Weakly supervised learning of metric aggregations for deformable image registration IEEE J. Biomed. Health 23 1374–84 [DOI] [PubMed] [Google Scholar]
- Ferrante E, Oktay O, Glocker B and Milone DH 2018. On the adaptability of unsupervised CNN-based deformable image registration to unseen image domains Machine Learning in Medical Imaging. MLMI 2018. Lecture Notes in Computer Science vol 11046, ed Shi Y, Suk HI and Liu M (Berlin: Springer; ) pp 294–302 [Google Scholar]
- Foote MD, Zimmerman BE, Sawant A and Joshi SC 2019. Real-Time 2D-3D deformable registration with deep learning and application to lung radiotherapy targeting Information Processing in Medical Imaging. IPMI 2019. Lecture Notes in Computer Science vol 11492, ed Chung A, Gee J, Yushkevich P and Bao S (Berlin: Springer; ) pp 265–76 [Google Scholar]
- Fu Y, Lei Y, Wang T, Higgins K, Bradley JD, Curran WJ, Liu T and Yang X 2020. LungRegNet: an unsupervised deformable image registration method for 4D-CT lung Med. Phys 47 1763–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Liu S, Li H and Yang D 2017. Automatic and hierarchical segmentation of the human skeleton in CT images Phys. Med. Biol 62 2812–33 [DOI] [PubMed] [Google Scholar]
- Fu Y et al. 2018. A novel MRI segmentation method using CNN-based correction network for MRI-guided adaptive radiotherapy Med. Phys 45 5129–37 [DOI] [PubMed] [Google Scholar]
- Fu YB, Chui CK, Teo CL and Kobayashi E 2011. Motion tracking and strain map computation for quasi-static magnetic resonance elastography Medical Image Computing and Computer-Assisted Intervention – MICCAI 2011. Lecture Notes in Computer Science vol 6891, ed Fichtinger G, Martel A and Peters T (Berlin: Springer; ) pp 428–35 [DOI] [PubMed] [Google Scholar]
- Fu YB, Wu X, Thomas AM, Li HH and Yang DS 2019. Automatic large quantity landmark pairs detection in 4DCT lung images Med. Phys 46 4490–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galib SM, Lee HK, Guy CL, Riblett MJ and Hugo GD 2019. A fast and scalable method for quality assurance of deformable image registration on lung CT scans using convolutional neural networks Med. Phys 47 99–109 [DOI] [PubMed] [Google Scholar]
- Ghesu F, Georgescu B, Zheng Y, Grbic S, Maier A, Hornegger J and Comaniciu D 2019. Multi-scale deep reinforcement learning for real-time 3D-landmark detection in CT scans IEEE Trans. Pattern Anal. Mach. Intell 41 176–89 [DOI] [PubMed] [Google Scholar]
- Ghesu FC, Georgescu B, Mansi T, Neumann D, Hornegger J and Comaniciu D 2016. An artificial agent for anatomical landmark detection in medical images Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Lecture Notes in Computer Science vol 9902, ed Ourselin S et al. (Cham: Springer; ) pp 229–37 [Google Scholar]
- Ghosal S and Rayl N 2017. Deep deformable registration: enhancing accuracy by fully convolutional neural net Pattern Recogn. Lett 94 81–86 [Google Scholar]
- Giles CL, Kuhn GM and Williams RJ 1994. Dynamic recurrent neural networks: theory and applications IEEE Trans. Neural Networks 5 153–6 [Google Scholar]
- Gong L, Wang H, Peng C, Dai Y, Ding M, Sun Y, Yang X and Zheng J 2017. Non-rigid MR-TRUS image registration for image-guided prostate biopsy using correlation ratio-based mutual information Biomed. Eng. Online 16 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong M, Zhao S, Jiao L, Tian D and Wang S 2014. A novel coarse-to-fine scheme for automatic image registration based on SIFT and mutual information IEEE Trans. Geosci. Remote Sens 52 4328–38 [Google Scholar]
- Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville AC and Bengio Y 2014. Generative adversarial nets (arXiv:1406.2661v1) [Google Scholar]
- Grondman I, Busoniu L, Lopes GAD and Babuska R 2012. A survey of actor-critic reinforcement learning: standard and natural policy gradients IEEE Trans. Syst. Man Cybern. Part C (Applications and Reviews) 42 1291–307 [DOI] [PubMed] [Google Scholar]
- Han X, Hoogeman MS, Levendag PC, Hibbard LS, Teguh DN, Voet P, Cowen AC and Wolf TK 2008. Atlas-based auto-segmentation of head and neck CT images Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008. Lecture Notes in Computer Science vol 5242, ed Metaxas D, Axel L, Fichtinger G and Székely G (Berlin: Springer; ) pp 434–41 [DOI] [PubMed] [Google Scholar]
- Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, Curran WJ, Liu T and Yang X 2019. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography Med. Phys 46 3998–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins G, Kruecker J, Kruger U, Xu S, Pinto PA, Wood BJ and Yan PK 2019a. Learning deep similarity metric for 3D MR-TRUS image registration Int. J. Comput. Ass. Rad 14 417–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins G, Kruger U and Yan P 2019b. Deep learning in medical image registration: a survey (arXiv:1903.02026) [Google Scholar]
- He K, Zhang X, Ren S and Sun J 2016. Deep residual learning for image recognition 2016 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) (Las Vegas, NV, 27–30 June 2016) (Piscataway, NJ: IEEE; ) pp 770–8 [Google Scholar]
- Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady SM and Schnabel JA 2012. MIND: modality independent neighbourhood descriptor for multi-modal deformable registration Med. Image Anal 16 1423–35 [DOI] [PubMed] [Google Scholar]
- Heinrich MP, Jenkinson M, Brady M and Schnabel JA 2013. MRF-based deformable registration and ventilation estimation of lung CT IEEE Trans. Med. Imaging 32 1239–48 [DOI] [PubMed] [Google Scholar]
- Hering A, Kuckertz S, Heldmann S and Heinrich MP 2019. Enhancing label-driven deep deformable image registration with local distance metrics for state-of-the-art cardiac motion tracking Bildverarbeitung für die Medizin 2019. Informatik aktuell ed Handels H et al (Berlin: Springer; ) pp 309–14 [Google Scholar]
- Hjelm RD, Plis SM and Calhoun VC 2016. Variational AEs for feature detection of magnetic resonance imaging data (arXiv:1603.06624) [Google Scholar]
- Hu Y, Gibson E, Ghavami N, Bonmati E, Moore CM, Emberton M, Vercauteren T, Noble JA and Barratt DC 2018a. Adversarial deformation regularization for training image registration neural networks Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science vol 11070, ed Frangi A, Schnabel J, Davatzikos C, Alberola-López C and Fichtinger G (Berlin: Springer; ) pp 774–82 [Google Scholar]
- Hu YP et al. 2018b. Weakly-supervised convolutional neural networks for multimodal image registration Med. Image Anal 49 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu Z, Maaten L and Weinberger KQ 2017. Densely connected convolutional networks 2017 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) (Honolulu, HI, 21–26 July 2017) (Piscataway, NJ: IEEE; ) pp 2261–9 [Google Scholar]
- Jaderberg M, Simonyan K, Zisserman A and Kavukcuoglu K 2015. Spatial transformer networks (arXiv:1506.02025) [Google Scholar]
- Jiang P and Shackleford JA 2018. CNN driven sparse multi-level B-spline image registration 2018 IEEE/CVF Conf. on Computer Vision and Pattern Recognition (Salt Lake City, UT, 18–23 June 2018) (Salt Lake City, UT, 18–23 June 2018) (Piscataway, NJ: IEEE; ) pp 9281–9 [Google Scholar]
- Jiang Z, Yin FF, Ge Y and Ren L 2019. A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration Phys. Med. Biol 65 015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney V, Haaf S, Sudhyadhom A, Valdes G and Solberg TD 2018. An unsupervised convolutional neural network-based algorithm for deformable image registration Phys. Med. Biol 63 185017. [DOI] [PubMed] [Google Scholar]
- Ker J, Wang LP, Rao J and Lim T 2018. Deep learning applications in medical image analysis IEEE Access 6 9375–89 [Google Scholar]
- Kim B, Kim J, Lee J-G, Kim DH, Park SH and Ye JC 2019. Unsupervised deformable image registration using cycle-consistent CNN Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. Lecture Notes in Computer Science vol 11769, ed Shen D (Berlin: Springer; ) pp 166–74 [Google Scholar]
- Klein S, Staring M, Murphy K, Viergever MA and Pluim JP 2010. elastix: a toolbox for intensity-based medical image registration IEEE Trans. Med. Imaging 29 196–205 [DOI] [PubMed] [Google Scholar]
- Kori A and Krishnamurthi G 2019. Zero shot learning for multi-modal real time image registration (arXiv:1908.06213) [Google Scholar]
- Krebs J, Mansi T, Delingette H, Zhang L, Ghesu FC, Miao S, Maier AK, Ayache N, Liao R and Kamen A 2017. Robust non-rigid registration through agent-based action learning Medical Image Computing and Computer Assisted Intervention – MICCAI 2017. Lecture Notes in Computer Science vol 10433, ed Descoteaux M et al. (Berlin: Springer; ) pp 344–52 [Google Scholar]
- Krebs J, Mansi T, Mailhe B, Ayache N and Delingette H 2018. Unsupervised probabilistic deformation modeling for robust diffeomorphic registration Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, DLMIA 2018, ML-CDS 2018. Lecture Notes in Computer Science vol 11045, ed Stoyanov D (Berlin: Springer; ) pp 101–9 [Google Scholar]
- Krizhevsky A, Sutskever I and Hinton GE 2012. ImageNet classification with deep convolutional neural networks Commun. ACM 60 84–90 [Google Scholar]
- Kuang D 2019. On reducing negative Jacobian determinant of the deformation predicted by deep registration networks (arXiv:1907.00068) [Google Scholar]
- Kuang D and Schmah T 2019. FAIM – a ConvNet method for unsupervised 3D medical image registration Machine Learning in Medical Imaging. MLMI 2019. Lecture Notes in Computer Science vol 11861, ed Suk HI, Liu M, Yan PP and Lian C (Berlin: Springer; ) pp 646–54 [Google Scholar]
- Lei Y et al. 2019a. CT prostate segmentation based on synthetic MRI-aided deep attention fully convolution network Med. Phys. Inform 47 530–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, Mao H, Nye JA and Yang X 2019b. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks Phys. Med. Biol 64 215017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Fu Y, Harms J, Wang T, Curran WJ, Liu T, Higgins K and Yang X 2019c. 4D-CT deformable image registration using an unsupervised deep convolutional neural network Artificial Intelligence in Radiation Therapy. AIRT 2019. Lecture Notes in Computer Science vol 11850, ed Nguyen D, Xing L and Jiang S (Berlin: Springer; ) pp 26–33 [Google Scholar]
- Lei Y, Fu Y, Wang T, Liu Y, Patel P, Curran WJ, Liu T and Yang X 2020. 4D-CT deformable image registration using multiscale unsupervised DL Phys. Med. Biol 65 085003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Harms J, Wang T, Liu Y, Shu HK, Jani AB, Curran WJ, Mao H, Liu T and Yang X 2019d. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks Med. Phys 46 3565–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y. et al. MRI-based synthetic CT generation using semantic random forest with iterative refinement. Phys. Med. Biol. 2019e;64:085001. doi: 10.1088/1361-6560/ab0b66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y et al. 2019f. Learning-based CBCT correction using alternating random forest based on auto-context model Med. Phys 46 601–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y et al. 2019g. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net Med. Phys 46 3194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H and Fan Y 2018. Non-rigid image registration using self-supervised fully convolutional networks without training data 2018 IEEE 15th Int. Symp. on Biomedical Imaging (ISBI 2018) (Washington, DC, 4–7 April 2018) (Piscataway, NJ: IEEE; ) pp 1075–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Jia X, Lewis JH, Gu X, Folkerts M, Men C and Jiang SB 2010. Real-time volumetric image reconstruction and 3D tumor localization based on a single x-ray projection image for lung cancer radiotherapy Med. Phys 37 2822–6 [DOI] [PubMed] [Google Scholar]
- Liao R, Miao S, Tournemire P, Grbic S, Kamen A, Mansi T and Comaniciu D 2016. An artificial agent for robust image registration (arXiv:1611.10336) [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B and Sanchez CI 2017. A survey on DL in medical image analysis Med. Image Anal 42 60–88 [DOI] [PubMed] [Google Scholar]
- Liu C, Ma LH, Lu ZM, Jin XC and Xu JY 2019a. Multimodal medical image registration via common representations learning and differentiable geometric constraints Electron. Lett 55 316–18 [Google Scholar]
- Liu XL, Jiang DS, Wang MN and Song ZJ 2019b. Image synthesis-based multi-modal image registration framework by using deep fully convolutional networks Med. Biol. Eng. Comput 57 1037–48 [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen X, Wang ZF, Wang ZJ, Ward RK and Wang XS 2018. Deep learning for pixel-level image fusion: recent advances and future prospects Inform. Fusion 42 158–73 [Google Scholar]
- Liu Y, Lei Y, Wang T, Kayode O, Tian S, Liu T, Patel P, Curran WJ, Ren L and Yang X 2019c. MRI-based treatment planning for liver stereotactic body radiotherapy: validation of a DL-based synthetic CT generation method Br. J. Radiol 92 20190067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a DL-based liver synthetic CT generation method. Phys. Med. Biol. 2019d;64:145015. doi: 10.1088/1361-6560/ab25bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Yang M, Zhang J and Wang XY 2018. Respiratory motion correction for free-breathing 3D abdominal MRI using CNN-based image registration: a feasibility study Br. J. Radiol 91 20170788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Wang J, Singh V, Tamersoy B, Chang Y-J, Wimmer A and Chen T 2017. Multimodal image registration with deep context reinforcement learning Medical Image Computing and Computer Assisted Intervention – MICCAI 2017. Lecture Notes in Computer Science vol 10433, ed Descoteaux M et al. (Berlin: Springer; ) pp 240–8 [Google Scholar]
- Mahapatra D, Antony B, Sedai S and Garnavi R 2018a. Deformable medical image registration using generative adversarial networks 2018 IEEE 15th Int. Symp. on Biomedical Imaging (ISBI 2018) Washington, DC, 4–7 April 2018 (Piscataway, NJ: IEEE; ) pp 1449–53 [Google Scholar]
- Mahapatra D and Ge Z 2019. Combining transfer learning and segmentation information with GANs for training data independent image registration (arXiv:1903.10139) [Google Scholar]
- Mahapatra D, Ge ZY, Sedai S and Chakravorty R 2018b. Joint registration and segmentation of x-ray images using generative adversarial networks Machine Learning in Medical Imaging. MLMI 2018. Lecture Notes in Computer Science vol 11046, ed Shi Y, Suk HI and Liu M (Berlin: Springer; ) pp 73–80 [Google Scholar]
- Mahapatra D, Sedai S and Garnavi R 2018c. Elastic registration of medical images with GANs (arXiv:1805.02369) [Google Scholar]
- Maier A, Syben C, Lasser T and Riess C 2019. A gentle introduction to DL in medical image processing Z Med. Phys 29 86–101 [DOI] [PubMed] [Google Scholar]
- McClelland JR et al. 2017. A generalized framework unifying image registration and respiratory motion models and incorporating image reconstruction, for partial image data or full images Phys. Med. Biol 62 4273–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Noblet V, Mazzara C and Lallement A 2018. Survey on DL for radiotherapy Comput. Biol. Med 98 126–46 [DOI] [PubMed] [Google Scholar]
- Miao S, Piat S, Fischer PW, Tuysuzoglu A, Mewes PW, Mansi T and Liao R 2017. Dilated FCN for multi-agent 2D/3D medical image registration (arXiv:1712.01651) [Google Scholar]
- Miao S, Wang ZJ and Liao R 2016. A CNN regression approach for real-time 2D/3D registration IEEE Trans. Med. Imaging 35 1352–63 [DOI] [PubMed] [Google Scholar]
- Mirza M and Osindero S 2014. Conditional generative adversarial nets (arXiv:1411.1784) [Google Scholar]
- Mnih V et al. 2015. Human-level control through deep reinforcement learning Nature 518 529–33 [DOI] [PubMed] [Google Scholar]
- Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC and Ourselin S 2010. Fast free-form deformation using graphics processing units Comput. Methods Programs Biomed 98 278–84 [DOI] [PubMed] [Google Scholar]
- Neylon J, Min YG, Low DA and Santhanam A 2017. A neural network approach for fast, automated quantification of DIR performance Med. Phys 44 4126–38 [DOI] [PubMed] [Google Scholar]
- Nguyen-Duc T, Yoo I, Thomas L, Kuan A, Lee WC and Jeong WK 2019. Weakly supervised learning in deformable EM image registration using slice interpolation 2019 IEEE 16th Int. Symp. on Biomedical Imaging (ISBI 2019) (Venice, Italy, 8–11 April 2019) (Piscataway, NJ: IEEE; ) pp 670–3 [Google Scholar]
- Onieva Onieva J, Marti-Fuster B, Pedrero de la Puente M and San José Estépar R 2018. Diffeomorphic lung registration using deep CNNs and reinforced learning Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO 2018, BIA 2018, TIA 2018. Lecture Notes in Computer Science vol 11040, ed Stoyanov D (Berlin: Springer; ) pp 284–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YR, Zhang YG, Qin HF, Ma GY, Guo YK, Xu TM and Zha HB 2017. Non-rigid craniofacial 2D-3D registration using CNN-based regression Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2017, ML-CDS 2017. Lecture Notes in Computer Science vol 10553, ed Cardoso M (Berlin: Springer; ) pp 117–25 [Google Scholar]
- Pierre B 2012. AEs, unsupervised learning, and deep architectures Proc. of ICML Workshop on Unsupervised and Transfer Learning, PMLR 27 pp 37–49 [Google Scholar]
- Qiao F, Pan T, Clark JW Jr. and Mawlawi OR 2006. A motion-incorporated reconstruction method for gated PET studies Phys. Med. Biol 51 3769–83 [DOI] [PubMed] [Google Scholar]
- Qin C, Bai W, Schlemper J, Petersen SE, Piechnik SK, Neubauer S and Rueckert D 2018. Joint learning of motion estimation and segmentation for cardiac MR image sequences (arXiv:1806.04066) [Google Scholar]
- Qin C, Shi BB, Liao R, Mansi T, Rueckert D and Kamen A 2019. Unsupervised deformable registration for multi-modal images via disentangled representations Information Processing in Medical Imaging. IPMI 2019. Lecture Notes in Computer Science vol 11492, ed Chung A, Gee J, Yushkevich P and Bao S (Berlin: Springer; ) pp 249–61 [Google Scholar]
- Ren S, He K, Girshick R and Sun J 2017. Faster R-CNN: towards real-time object detection with region proposal networks IEEE Trans. Pattern Anal. Mach. Intell 39 1137–49 [DOI] [PubMed] [Google Scholar]
- Rivaz H, Karimaghaloo Z, Fonov VS and Collins DL 2014. Nonrigid registration of ultrasound and MRI using contextual conditioned mutual information IEEE Trans. Med. Imaging 33 708–25 [DOI] [PubMed] [Google Scholar]
- Rohé -M-M, Datar M, Heimann T, Sermesant M and Pennec X 2017. SVF-Net: learning deformable image registration using shape matching Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. Lecture Notes in Computer Science vol 10433, ed Descoteaux M (Berlin: Springer; ) pp 266–74 [Google Scholar]
- Ronneberger O, Fischer P and Brox T 2015. U-Net: convolutional networks for biomedical image segmentation (arXiv:1505.04597) [Google Scholar]
- Sahiner B, Pezeshk A, Hadjiiski LM, Wang XS, Drukker K, Cha KH, Summers RM and Giger ML 2019. Deep learning in medical imaging and radiation therapy Med. Phys 46 e1–e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi SSM, Khan S, Erdogmus D and Gholipour A 2018. Real-time deep registration with geodesic loss for image-to-template rigid registration (arXiv:1803.05982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavati N, Velec M and Brock KK 2016. Effect of deformable registration uncertainty on lung SBRT dose accumulation Med. Phys 43 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrut D 2006. Deformable registration for image-guided radiation therapy Z Med. Phys 16 285–97 [DOI] [PubMed] [Google Scholar]
- Schlemper J, Oktay O, Schaap M, Heinrich MP, Kainz B, Glocker B and Rueckert D 2018. Attention gated networks: learning to leverage salient regions in medical images Med. Image Anal 53 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedghi A, Luo J, Mehrtash A, Pieper SD, Tempany CM, Kapur T, Mousavi P and Wells WM 2018. Semi-supervised deep metrics for image registration (arXiv:1804.01565) [Google Scholar]
- Sentker T, Madesta F and Werner R 2018. GDL-FIRE4D: deep learning-based fast 4D CT image registration Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Lecture Notes in Computer Science vol 11070, ed Frangi A et al. (Berlin: Springer; ) pp 765–73 [Google Scholar]
- Shackleford JA, Kandasamy N and Sharp GC 2010. On developing B-spline registration algorithms for multi-core processors Phys. Med. Biol 55 6329–51 [DOI] [PubMed] [Google Scholar]
- Shams R, Sadeghi P, Kennedy RA and Hartley RI 2010. A survey of medical image registration on multicore and the GPU IEEE Signal Process Mag. 27 50–60 [Google Scholar]
- Shan S, Guo X, Yan W, Chang EIC, Fan Y and Xu Y 2017. Unsupervised end-to-end learning for deformable medical image registration (arXiv:1711.08608) [Google Scholar]
- Sheikhjafari A, Noga M, Punithakumar K and Ray N 2018. Unsupervised deformable image registration with fully connected generative neural network Submission to 1st Conf. on Medical Imaging with Deep Learning (MIDL 2018) (https://openreview.net/pdf?id=HkmkmW2jM) [Google Scholar]
- Shen D 2007. Image registration by local histogram matching Pattern Recogn. 40 1161–72 [Google Scholar]
- Shen D, Wu G and Suk H-I 2017. Deep learning in medical image analysis Annu. Rev. Biomed. Eng 19 221–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Chen X, Xie Q and Han H 2018. An unsupervised network for fast microscopic image registration Proc. SPIE 10581 105811D [Google Scholar]
- Silver D et al. 2016. Mastering the game of Go with deep neural networks and tree search Nature 529 484–9 [DOI] [PubMed] [Google Scholar]
- Simonovsky M, Gutiérrez-Becker B, Mateus D, Navab N and Komodakis N 2016. A deep metric for multimodal registration Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016. Lecture Notes in Computer Science vol 9902, ed Ourselin S et al. (Berlin: Springer; ) pp 10–18 [Google Scholar]
- Sloan JM, Goatman KA and Siebert JP 2018. Learning rigid image registration - utilizing convolutional neural networks for medical image registration BIOIMAGING 2 89–99 [Google Scholar]
- So RWK and Chung ACS 2017. A novel learning-based dissimilarity metric for rigid and non-rigid medical image registration by using Bhattacharyya Distances Pattern Recogn. 62 161–74 [Google Scholar]
- Sokooti H, Bdd V, Berendsen FF, Ghafoorian M, Yousefi S, Lelieveldt BPF, Išgum I and Staring M 2019b. 3D convolutional neural networks image registration based on efficient supervised learning from artificial deformations (arXiv:1908.10235) [Google Scholar]
- Sokooti H, de Vos B, Berendsen F, Lelieveldt BPF, Išgum I and Staring M 2017. Nonrigid image registration using multi-scale 3D convolutional neural networks Medical Image Computing and Computer Assisted Intervention—MICCAI 2017. Lecture Notes in Computer Science vol 10433, ed Descoteaux M et al. (Berlin: Springer; ) pp 232–9 [Google Scholar]
- Sokooti H, Saygili G, Glocker B, Lelieveldt BPF and Staring M 2019a. Quantitative error prediction of medical image registration using regression forests Med. Image Anal 56 110–21 [DOI] [PubMed] [Google Scholar]
- Staring M, Klein S, Reiber JHC, Niessen WJ and Stoel BC 2010. Pulmonary image registration with elastix using a standard intensity-based algorithm Medical Image Analysis for the Clinic: A Grand Challenge 2010 (Scotts Valley, CA: CreateSpace Independent Publishing Platform; ) pp 73–80 [Google Scholar]
- Stergios C, Mihir S, Maria V, Guillaume C, Marie-Pierre R, Stavroula M and Nikos P 2018. Linear and deformable image registration with 3D convolutional neural networks Image Analysis for Moving Organ, Breast, and Thoracic Images. RAMBO 2018, BIA 2018, TIA 2018. Lecture Notes in Computer Science vol 11040, ed Stoyanov D (Berlin: Springer; ) pp 13–22 [Google Scholar]
- Sun L and Zhang S 2018. Deformable MRI-ultrasound registration Using 3D convolutional neural network Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation. POCUS 2018, BIVPCS 2018, CuRIOUS 2018, CPM 2018. Lecture Notes in Computer Science vol 11042, ed Stoyanov D (Berlin: Springer; ) pp 152–8 [Google Scholar]
- Sun S, Hu J, Yao M, Hu J, Yang X, Song Q and Wu X 2019. Robust multimodal image registration using deep recurrent reinforcement learning Computer Vision—ACCV 2018. Lecture Notes in Computer Science vol 11362, ed Jawahar C, Li H, Mori G and Schindler K (Berlin: Springer; ) pp 511–26 [Google Scholar]
- Sun Y, Moelker A, Niessen WJ and van Walsum T 2018. Towards robust CT-ultrasound registration using deep learning methods Understanding and Interpreting Machine Learning in Medical Image Computing Applications. MLCN 2018, DLF 2018, IMIMIC 2018. Lecture Notes in Computer Science vol 11038, ed Stoyanov D (Berlin: Springer; ) pp 43–51 [Google Scholar]
- Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V and Rabinovich A 2015. Going deeper with convolutions 2015 IEEE Conf. on Computer Vision and Pattern Recognition (CVPR) (Boston, MA, 7–12 June 2015) (Piscataway, NJ: IEEE; ) pp 1–9 [Google Scholar]
- Szeliski R and Coughlan J 1997. Spline-based image registration Int. J. Comput. Vis 22 199–218 [Google Scholar]
- Taylor RH and Stoianovici D 2003. Medical robotics in computer-integrated surgery IEEE Trans. Rob. Autom 19 765–81 [Google Scholar]
- Thrun S 1992. Efficient exploration in reinforcement learning Technical Report (Pittsburgh, PA: Carnegie Mellon University; ) [Google Scholar]
- Tschannen M, Bachem O and Lucic M 2018. Recent advances in AE-based representation learning (arXiv:1812.05069) [Google Scholar]
- Uzunova H, Wilms M, Handels H and Ehrhardt J 2017. Training CNNs for image registration from few samples with model-based data augmentation Medical Image Computing and Computer Assisted Intervention—MICCAI 2017. Lecture Notes in Computer Science vol 1043, ed Descoteaux M et al. (Berlin: Springer; ) pp 223–31 [Google Scholar]
- Velec M, Moseley JL, Eccles CL, Craig T, Sharpe MB, Dawson LA and Brock KK 2011. Effect of breathing motion on radiotherapy dose accumulation in the abdomen using deformable registration Int. J. Radiat. Oncol. Biol. Phys 80 265–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren T, Pennec X, Perchant A and Ayache N 2009. Diffeomorphic demons: efficient non-parametric image registration NeuroImage 45 S61–S72 [DOI] [PubMed] [Google Scholar]
- Vos B, Berendsen FF, Viergever MA, Staring M and Išgum I 2017. End-to-end unsupervised deformable image registration with a convolutional neural network Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2017, ML-CDS 2017. Lecture Notes in Computer Science, ed Cardoso M (Berlin: Springer; ) vol 10553 pp 204–12 [Google Scholar]
- Wang B et al. 2019a. Deeply supervised 3D fully convolutional networks with group dilated convolution for automatic MRI prostate segmentation Med. Phys 46 1707–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ghavidel BB, Beitler JJ, Tang X, Lei Y, Curran WJ, Liu T and Yang X 2019b. Optimal virtual monoenergetic image in ‘TwinBeam’ dual-energy CT for organs-at-risk delineation based on contrast-noise-ratio in head-and-neck radiotherapy J. Appl. Clin. Med. Phys 20 121–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T et al. 2019c. Dosimetric study on learning-based cone-beam CT correction in adaptive radiation therapy Med. Dosim 44 e71–e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T et al. 2019d. A learning-based automatic segmentation and quantification method on left ventricle in gated myocardial perfusion SPECT imaging: a feasibility study J. Nucl. Cardiol 27 976–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Lei Y, Tian Z, Dong X, Liu Y, Jiang X, Curran WJ, Liu T, Shu HK and Yang X 2019e. Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy J. Med. Imaging 6 043504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Manohar N, Lei Y, Dhabaan A, Shu HK, Liu T, Curran WJ and Yang X 2019f. MRI-based treatment planning for brain stereotactic radiosurgery: dosimetric validation of a learning-based pseudo-CT generation method Med. Dosim 44 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner R, Schmidt-Richberg A, Handels H and Ehrhardt J 2014. Estimation of lung motion fields in 4D CT data by variational non-linear intensity-based registration: A comparison and evaluation study Phys. Med. Biol 59 4247–60 [DOI] [PubMed] [Google Scholar]
- Wright R, Khanal B, Gomez A, Skelton E, Matthew J, Hajnal JV, Rueckert D and Schnabel JA 2018. LSTM spatial co-transformer networks for registration of 3D fetal US and MR brain images Data Driven Treatment Response Assessment and Preterm, Perinatal, and Paediatric Image Analysis. PIPPI 2018, DATRA 2018. Lecture Notes in Computer Science vol 11076, ed Melbourne A (Berlin: Springer; ) pp 149–59 [Google Scholar]
- Wu GR, Kim M, Wang Q, Munsell BC and Shen DG 2016. Scalable high-performance image registration framework by unsupervised deep feature representations learning IEEE Trans. Biomed 63 1505–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia KJ, Yin HS and Wang JQ 2019. A novel improved deep convolutional neural network model for medical image fusion Cluster Comput. 22 1515–27 [Google Scholar]
- Yan P, Xu S, Rastinehad AR and Wood BJ 2018. Adversarial image registration with application for MR and TRUS image fusion Machine Learning in Medical Imaging. MLMI 2018. Lecture Notes in Computer Science, ed Shi Y, Suk HI and Liu M (Berlin: Springer; ) vol 11046 pp 197–204 [Google Scholar]
- Yang D, Brame S, El Naqa I, Aditya A, Wu Y, Goddu SM, Mutic S, Deasy JO and Low DA 2011a. Technical note: DIRART–A software suite for deformable image registration and adaptive radiotherapy research Med. Phys 38 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Goddu SM, Lu W, Pechenaya OL, Wu Y, Deasy JO, El Naqa I and Low DA 2010. Technical note: deformable image registration on partially matched images for radiotherapy applications Med. Phys 37 141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Li H, Low DA, Deasy JO and El Naqa I 2008. A fast inverse consistent deformable image registration method based on symmetric optical flow computation Phys. Med. Biol 53 6143–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Akbari H, Halig L and Fei B 2011b. 3D non-rigid registration using surface and local salient features for transrectal ultrasound image-guided prostate biopsy Proc. SPIE 7964 79642V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X and Fei B 2012. 3D prostate segmentation of ultrasound images combining longitudinal image registration and machine learning Proc. SPIE 8316 83162O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ghafourian P, Sharma P, Salman K, Martin D and Fei B 2012. Nonrigid registration and classification of the kidneys in 3D dynamic contrast enhanced (DCE) MR images Proc. SPIE 8314 83140B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kwitt R, Styner M and Niethammer M 2017. Quicksilver: fast predictive image registration—a DL approach NeuroImage 158 378–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Rossi PJ, Jani AB, Mao H, Curran WJ and Liu T 2016. 3D transrectal ultrasound (TRUS) prostate segmentation based on optimal feature learning framework Proc. SPIE 9784 97842F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schuster D, Master V, Nieh P, Fenster A and Fei B 2011c. Automatic 3D segmentation of ultrasound images using atlas registration and statistical texture prior Proc. SPIE 7964 796432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu N, Cheng G, Zhou Z, Yu DS, Beitler JJ, Curran WJ and Liu T 2014. Automated segmentation of the parotid gland based on atlas registration and machine learning: a longitudinal MRI study in head-and-neck radiation therapy Int. J. Radiat. Oncol. Biol. Phys 90 1225–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo I, Hildebrand DGC, Tobin WF, Lee WCA and Jeong W-K 2017. ssEMnet: serial-section electron microscopy image registration using a spatial transformer network with earned features Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2017, ML-CDS 2017. Lecture Notes in Computer Science vol 10553, ed Cardoso M (Berlin: Springer; ) pp 249–57 [Google Scholar]
- Yu HC et al. 2019a. A novel framework for 3D-2D vertebra matching 2019 IEEE Conference on Multimedia Information Processing and Retrieval (MIPR) (San Jose, CA, 28–30 March 2019) (Piscataway, NJ: IEEE; ) pp 121–6 [Google Scholar]
- Yu HJ, Zhou XR, Jiang HY, Kang HJ, Wang ZG, Hara T and Fujita H 2019b. Learning 3D non-rigid deformation based on an unsupervised DL for PET/CT image registration Proc. SPIE 10953 109531X [Google Scholar]
- Yuille AL and Rangarajan A 2003. The concave-convex procedure Neural Comput. 15 915–36 [DOI] [PubMed] [Google Scholar]
- Zhang J 2018. Inverse-consistent deep networks for unsupervised deformable image registration (arXiv:1809.03443) [Google Scholar]
- Zhang ZW and Sejdic E 2019. Radiological images and machine learning: trends, perspectives, and prospects Comput. Biol. Med 108 354–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lau T, Luo J, Chang EI-C and Xu Y 2019. Unsupervised 3D end-to-end medical image registration with volume tweening network IEEE J. Biomed. Health Inform 24 1394–404 [DOI] [PubMed] [Google Scholar]
- Zheng JN, Miao S, Wang ZJ and Liao R 2018. Pairwise domain adaptation module for CNN-based 2-D/3-D registration J. Med. Imaging 5 021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Park T, Isola P and Efros AA 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks 2017 IEEE Int. Conf. on Computer Vision (ICCV) (Venice, Italy, 22–29 October 2017) (Piscataway, NJ: IEEE; ) pp 2242–51 [Google Scholar]
- Zimmerer D, Kohl SAA, Petersen J, Isensee F and Maier-Hein KH 2018. Context-encoding variational AE for unsupervised anomaly detection (arXiv:1812.05941) [Google Scholar]


