Abstract
Worldwide there has been a significant increase in the incidence of oropharyngeal squamous cell carcinoma (OPSCC) etiologically attributed to oncogenic human papillomavirus (HPV). Reliable and accurate identification and detection tools are important as the incidence of HPV-related cancer is on the rise. Several HPV detection methods for OPSCC have been developed and each has its own advantages and disadvantages in regard to sensitivity, specificity, and technical difficulty. This review summarizes our current knowledge of molecular methods for detecting HPV in OPSCC, including HPV DNA/RNA polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), p16 immunohistochemistry (IHC), and DNA/RNA in situ hybridization (ISH) assays. This summary may facilitate the selection of a suitable method for detecting HPV infection, and therefore may help in the early diagnosis of HPV-related carcinoma to reduce its mortality, incidence, and morbidity.
Keywords: Human papillomavirus (HPV), Molecular detection, Oropharyngeal squamous cell carcinoma (OPSCC)
1. Introduction
Oropharyngeal squamous cell carcinoma (OPSCC) originates from the non-keratinizing stratified mucosal epithelium in the upper aerodigestive tract (Suciu et al., 2014). OPSCC is caused primarily by exposure to tobacco and alcohol, yet there is a significant increase in the incidence of OPSCC worldwide, etiologically attributed to oncogenic human papillomavirus (HPV). The relationship between HPV and head and neck cancer has been discussed by many researchers reporting the presence of HPV DNA in head and neck cancer patients and in healthy oral mucosa. The proportion of HPV-related OPSCC worldwide ranges from 32% to 73%. This variation is due to several factors such as sexual habits, geographic differences, and variation in the molecular techniques used to define HPV in OPSCC (Lin et al., 2011; Oji and Chukwuneke, 2012; Mehanna et al., 2013; Gupta and Johnson, 2014).
Based on its carcinogenic potential, HPV can be classified into high-risk and low-risk types. Low-risk types such as HPV6, 11, 42, 43, and 44 usually result in benign lesions, while high-risk types such as HPV16, 18, 31, 33, 35, 45, 51, 52, 56, 58, 59, 68, 73, and 82 can cause malignant tumors. The diameter of the virus is about 52–55 nm. It is characterized as non-enveloped double-stranded DNA and consists of 8000 bp-long circular DNA molecules (Muñoz et al., 2003; Gupta and Gupta, 2015). The genome consists of six “early” regions (E1, E2, E4, E5, E6, and E7), which specifically encode viral DNA replication proteins and transformation, and two “late” regions (L1 and L2) that encode structural proteins of the virus (Zheng and Baker, 2006). Four messenger RNA (mRNA) isoforms of the E6 protein have been detected in epithelial cells infected by HPV16. These isoforms, E6*I, FLE6, E6*II, and E6*X, are evidence that other viral proteins could have additional important functions. Two other mRNA isoforms have been observed in cells infected by HPV18 (Graham, 2010). Among the listed genes, E6 and E7 genes are responsible for severe infection leading to cancer (de Villiers et al., 2004). E6 targets tumor protein 53 (p53), and E7 targets retinoblastoma protein (pRb) tumor suppressors. Changes in apoptosis, DNA repair mechanisms, and cell cycle control due to earlier alteration eventually lead to overexpression of tumor suppressor gene (p16) (Babiker et al., 2013).
Several HPV detection methods for OPSCC samples have been developed at the DNA, mRNA, and protein levels. Each technique has its own strengths and limitations. Based on a recent review article, p16 immunohistochemistry (IHC), polymerase chain reaction (PCR), DNA in situ hybridization (ISH), and the loop-mediated isothermal amplification (LAMP) assay are the techniques most commonly used for HPV testing in OPSCC (Qureishi et al., 2017). It is important to have a reliable and accurate identification and detection method for HPV in OPSCC as it is relevant to prognosis and treatment strategies for patients (Bhargava et al., 2010). Information on the HPV status of patients will aid clinicians in patient management. The objective of this review was to summarize current knowledge on the molecular detection methods of HPV in OPSCC. The mechanism, strengths, limitations, sensitivity, and specificity of each method are summarized in Table 1.
Table 1.
Summary of HPV detection methods for OPSCC
| Method | Mechanism | Strength | Weakness | Sensitivity | Specificity |
| (%) | (%) | ||||
| HPV DNA PCR | Use Taq DNA polymerase and specific primers in a standard PCR reaction to amplify the target DNA fragment in the sample | Highly sensitive and specific | Unable to distinguish “clinically significant” HPV infections (i.e., HPV transcriptional active infection in tumor cells with expression of E6 and E7) | 94–100 | 74–92 |
| qPCR | The amount of the nucleic acid present in the sample is quantified using: (1) fluorescent dyes that non-specifically intercalate with double-stranded DNA, and (2) sequence-specific DNA probes consisting of fluorescently labelled reports (Abreu et al., 2012) | Able to detect transcriptionally active HPV; providing an estimation of HPV viral load; highly sensitive and specific; no post-PCR processing | Laborious; instrument is too costly; multiplexing is still limited | >95 | 100 |
| Droplet digital PCR | Based on water-oil emulsion droplet technology for measuring the absolute copy number of nucleic acid targets without external standards | Highly specific and sensitive; requiring minimal amounts of nucleic acids for detection; providing absolute target quantification without reference to the standard/calibration curve; able to detect rare mutations and quantify gene mutations; technically feasible; lower sensitivity to PCR inhibitors; more precise and reproducible data compared to qPCR; cost-effective | Need further research | 100 | 100 |
| LAMP | Rely on auto-cycling strand displacement DNA synthesis Bst DNA polymerase enzyme with high strand displacement activity and a set of two specially designed inner and outer primers (Notomi et al., 2000) | Highly specific and sensitive; cost-effective; rapid; robust | False-positive results in subsequent reactions | 99 | >90 |
| p16 IHC | Involve alteration of p16 mRNA expression resulting from p16 gene methylation of the promoter region. Methylation results in cascade events of pRb and E2F followed by p16 upregulation, which is highly associated and is unique for HPV-positive oropharyngeal carcinoma (Lewis et al., 2012) | Highly sensitive; cost-effective; widely available | Lack of specificity (false positive results); time-consuming; result interpretation requires a pathologist for confirmation; different p16ʏINK4aһ cut-offs to detect HPV-transformed OPSCC | 91–97 | 78–88 |
| DNA ISH | Labeled probes hybridize to target HPV DNA sequences in the tumor cell nucleus, and the probes may be either HPV-specific or cocktail-specific to detect different high-risk HPV types simultaneously | Highly specific; cost-effective; allowing a reliable identification of viral physical state | Limited sensitivity (false negative results are particularly likely with low HPV copy numbers); signal interpretation can be subjective due to lack of clean, clear staining signals; time-consuming; requiring a large amount of purified DNA; uncertain performance in detection of less common high-risk HPV subtypes | 76–92 | 78–96 |
| HPV E6/E7 mRNA ISH | Involve reverse transcription of mRNA into cDNA and subsequent amplification of target DNA sequences of interest using PCR | Highly specific and sensitive; allowing detection of transcriptionally active HPV | Require dedicated equipment and workflow; cost-prohibitive for some clinical laboratories; require technical expertise that is not routinely available in pathology laboratories; interpretation is somewhat subjective based on amplification curve analysis | >93 | >90 |
cDNA: complementary DNA; E2F: E2 factor; HPV: human papillomavirus; IHC: immunohistochemistry; ISH: in situ hybridization; LAMP: loop-mediated isothermal amplification; OPSCC: oropharyngeal squamous cell carcinoma; PCR: polymerase chain reaction; qPCR: quantitative PCR; pRb: retinoblastoma protein
2. Molecular detection assays
2.1. PCR
During recent decades, there have been huge advances in nucleic acid diagnostic tests. Molecular tests are preferred because of their rapidity, high sensitivity and specificity. They are able to pick up low copy numbers or low concentrations of sequences, and specificity is provided through a probe that is designed specifically to complement the target gene (Sigma-Aldrich, 2018). These assays have been used extensively to detect the presence of HPV in a broad range of clinical samples, such as oral rinse, plasma, fine needle aspirates, fresh tissue, and formalin-fixed paraffin-embedded (FFPE) tissue (Zarei et al., 2007). PCR is a selective target amplification assay able to increase the HPV sequences present in biological specimens exponentially and reproducibly. Theoretically, after 30 amplification cycles, the amplification process can produce one billion copies of a single double-stranded DNA molecule. The sensitivity and specificity of PCR-based methods can vary, depending on the DNA extraction procedures, site and type of clinical sample, sample transportation and storage, primer set, and performance of the DNA polymerase used in the reaction.
To improve sensitivity and specificity, the one-step PCR has been modified. Nested PCR is a combined PCR assay that uses a combination of two consensus primers. For example, MY09/11, GP5+/GP6+, PGMY09/11, and SPF10 LiPA primers target consensus sequences of variable lengths (different primer sets target sequences of different lengths) within the HPV L1 gene (Mirghani et al., 2014). In nested PCR, two primer sets are used in two rounds of PCR amplification. The first PCR amplification yields a larger amplicon size (464 bp), while in the second PCR amplification, the amplicon size is reduced to 155 bp. This short target increases the specificity of the assay and can reduce false-negative results caused by short DNA fragments originating from fixation-induced DNA degradation in FFPE samples, for example. FFPE samples are easily degraded due to extensive cross-linking of proteins to DNA. Thus, to overcome this problem, the use of a primer that yields a small amplicon will increase the chances of it being detected (Candotto et al., 2017). In a parallel study by Jalouli et al. (2015), nested PCR showed an increase in HPV detection in FFPE samples in terms of positivity rate, efficiency rate, and sensitivity compared to a single PCR assay. Multiplex PCR allows simultaneous detection in one amplification tube. Simultaneous detection could minimize the use of reagents and reduce errors compared to a single-plex assay. Wasserman et al. (2017) showed that multiplex PCR was able to detect a high prevalence (79%) of high-risk HPV in HPV-positive saliva samples.
Overall, PCR offers high sensitivity (>95%) and specificity (>85%) in HPV detection in OPSCC (Schache et al., 2011; Tawe et al., 2018). PCR is one of the simplest and most direct methods to use. The PCR amplicons also can be used for downstream techniques like sequencing and cloning. However, the assay has a few drawbacks. It can easily be affected by contamination of the sample by even trace amounts of DNA, which can produce misleading results. In addition, PCR amplification requires post-amplification analysis either by gel electrophoresis, restriction-fragment length polymorphism (RFLP), direct sequencing or hybridization with type-specific oligonucleotide probes using various hybridization formats such as dot blot, Southern blot, microtiter enzyme-linked immunosorbent assay (ELISA) plate, reverse line blot strip assays, and microchip format assays (Coser et al., 2011). PCR machines are quite expensive and PCR is incapable of distinguishing transcriptionally active or inactive HPV in HPV-positive samples (Schache et al., 2013).
2.2. Quantitative PCR
Quantitative PCR (qPCR) provides an assessment of HPV detection and quantification of viral load (Coser et al., 2011). A tumor with a high viral load could be a favorable prognostic factor. If the viral load is higher than the median value, it may indicate that there is virus replication in the tumor cells (Mellin et al., 2002). This assay uses the accumulation of a fluorescent reporter molecule to monitor the PCR progression and DNA concentration. It offers a shorter time and fewer manual steps than conventional PCR due to the elimination of post-amplification detection procedures. qPCR is more reproducible, rapid, and applicable to clinical samples than other PCR-based assays (Wang et al., 2014). In addition, qPCR offers a sensitive and specific detection and quantification of HPV subtypes in a wide range of samples including fresh tissue, frozen tissue, FFPE tissue, and cellular samples. Several researchers have demonstrated the presence of HPV DNA in p16INK4a-positive salivary oral rinse samples using qPCR with high sensitivity (>90%) and specificity (100%) (Ritari et al., 2012; Chai et al., 2015; Zil-e-Rubab et al., 2018). The improved sensitivity and specificity reported in their studies could be due to the use of strain-specific primers instead of primers that target the conserved L1 open reading frame (ORF) which contains degenerate primer sequences that may lower sensitivity (Fontaine et al., 2007; Chai et al., 2015). Ahn et al. (2014) reported a sensitivity of 52.8% for saliva, 67.3% for plasma, and 76.1% when using combined saliva and plasma samples, for detection of HPV16 E6/E7 DNA using qPCR in pre-treatment patients. Dang et al. (2015) demonstrated that 33 of 100 cancer patients were positive for any type of HPV in oral rinse samples using qPCR.
qPCR can determine how much of a specific DNA sequence or gene is present in the sample. It allows for both detection and quantification of the viral load in real time, while it is being synthesized (Arney and Bennett, 2010). The two common methods used to detect and quantify the product include fluorescent dyes that non-specifically intercalate with double-stranded DNA and sequence-specific DNA probes consisting of fluorescently-labelled reports (Abreu et al., 2012). The drawbacks of this method are that the detection probe is expensive, and although post-amplification analysis is not required, the real-time machine is very expensive (Duncan et al., 2013).
2.3. Quantitative reverse-transcription PCR
HPV E6/E7 mRNA detection by quantitative reverse-transcription PCR (RT-qPCR) is highly correlated with improved patient survival in OPSCC and is considered to be the “gold standard” test for classifying an OPSCC as being positive for transcriptionally active high-risk HPV (Mirghani et al., 2014; Bishop et al., 2015). These transcripts are associated with cellular genotoxic damage and gene expression changes that cause cancer (Robinson et al., 2012). Several studies have reported the detection of HPV E6/E7 mRNA in OPSCC using this technique. Deng et al. (2013) demonstrated that HPV E6/E7 transcripts were detected in 27.8% (15/54) of HPV-positive tumor samples, while Holzinger et al. (2012) detected HPV E6/E7 transcripts in 50% of OPSCC tumor samples. HPV E6/E7 mRNA PCR is less favorable for routine screening as it is technically demanding, needing fresh-frozen tissue which is fragile and is of limited availability (Yu et al., 2012). The RNA isolation process for RT-qPCR is laborious as it requires additional sample preparation steps and more tumor cells than HPV ISH and p16 IHC (Mirghani et al., 2014).
2.4. Droplet digital PCR
Droplet digital PCR (ddPCR) is the latest technique for the detection of oncogenic HPV. It is currently the most accurate, rapid, and sensitive method to quantify HPV viral load in OPSCC (Biron et al., 2016; Albano et al., 2017; Antonsson et al., 2018; Stevenson et al., 2020). ddPCR involves partitioning a single nucleic acid sample into up to 20 000 discrete water-in-oil droplets and performing PCR analysis on each droplet independently, using custom-designed HPV16 L1 or E6-specific primers and probe sets, with the results reported digitally and quantitatively. This technique offers better precision, accuracy, and reproducibility than RT-qPCR. It also allows quantification of the absolute amount of target present in samples, while dealing with competing targets and DNA degradation. It can be used to detect rare mutations, to quantify gene expression even with a low copy number, and can be multiplexed for higher efficiency. ddPCR quantification indicated that a higher viral load correlates with improved survival in HPV-positive oropharyngeal tumors (Stevenson et al., 2020; Veyer et al., 2020).
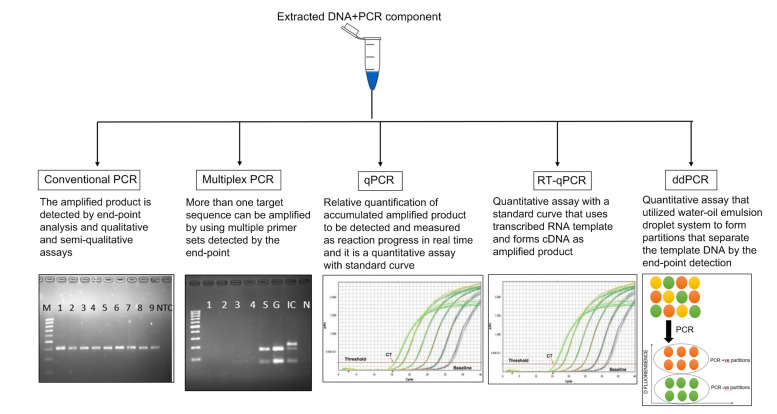
Several studies have made use of ddPCR for detection of oncogenic HPV E6/E7 mRNA in OPSCC (Isaac et al., 2017; Stevenson et al., 2020; Veyer et al., 2020). HPV E6/E7 mRNA has been detected in fresh tissues of OPSCC and oropharyngeal swab specimens, and was found to have 100% and 98% sensitivity, respectively, compared to p16 IHC using target RNA 20%–50% lower concentration than reported for RT-qPCR. Because of its high sensitivity and specificity, this technique is suitable for assessing the HPV16 viral load in more sample types, including oral/oropharyngeal swabs as opposed to fresh tissue, compared to assessments based on the commonly used IHC p16 marker (Carcopino et al., 2012). Researchers found that ddPCR analysis of oropharyngeal swabs is a quantitative, rapid, and cost-effective tool for minimally invasive oncogenic detection of HPV (Isaac et al., 2017). In the available literature, it is the most sensitive and reliable tool for detection of HPV in OPSCC without a tissue biopsy, and has many potential applications for both diagnosis and disease control (Ahn et al., 2014). A schematic diagram of PCR-based assays including conventional PCR, qPCR, RT-qPCR, and ddPCR for detection of HPV in OPSCC is shown in Fig. 1.
Fig. 1.
A schematic diagram of PCR-based amplification assays for detection of HPV
HPV: human papillomavirus; PCR: polymerase chain reaction; qPCR: quantitative PCR; RT-qPCR: quantitative reverse-transcription PCR; ddPCR: droplet digital PCR
2.5. Loop-mediated isothermal amplification assay
LAMP is a technique that applies isothermal conditions for amplification of the target DNA. LAMP was introduced 20 years ago (Notomi et al., 2000) and since then has been used extensively in nucleic acid research and in clinical applications as a screening tool. It is a single tube technique that amplifies a few copies of DNA into a billion copies within an hour. Since its establishment, the LAMP assay has proved to be a simple, time-efficient, and cost-effective method for the detection of high-risk HPV. LAMP applies four to six primers specially designed to recognize six to eight distinct regions of a target gene, hence its high efficiency and precision. The use of four compulsory primers which are a forward primer (F3), backward primer (B3), forward inner primer (FIP), and a backward inner primer (BIP) increases specificity. An additional loop primer can increase the amplification speed and efficiency, thereby reducing the reaction time of the original LAMP reaction (Mori et al., 2013). The LAMP amplification technique is very simple and straightforward to perform. The amplification can be completed within an hour using simple and inexpensive equipment, such as a water bath and heating block, which is available in most laboratories (Dhama et al., 2014). A semiskilled person provided with comprehensive protocols can effectively perform the assay (Abdullahi et al., 2015). In addition, LAMP is more sensitive than PCR, with a 10-to 100-fold higher sensitivity and a detection limit of 0.01–10.00 plaque forming units (PFUs) of virus (Parida et al., 2008). One of the unique features of LAMP is that this technique has an ability to amplify genes in a poorly processed or non-processed sample, eliminating the DNA extraction step. Bst polymerase enzyme has high resistance to some PCR inhibitors that are often present in saliva and blood. This enzyme also helps to eliminate the need for extensive sample purification (Hamzan et al., 2018).
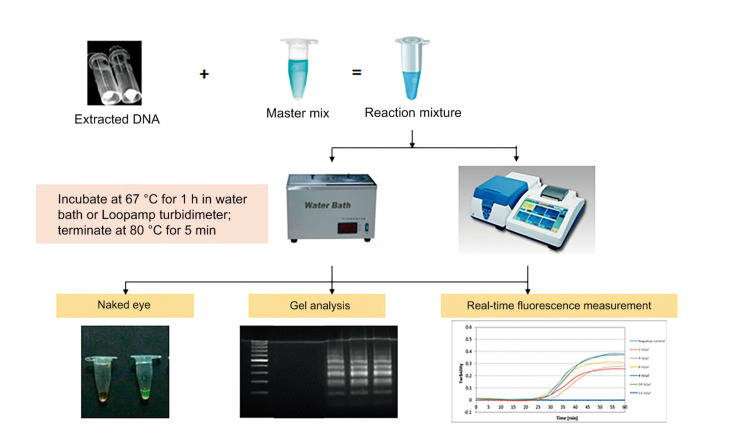
The amplified target can be visualized using the naked eye because of the large amount of magnesium pyrophosphate by-product produced during the reaction, and can be followed by agarose gel electrophoresis to confirm the result (Mori et al., 2001). The presence or absence of the target DNA can be clearly observed by color changes, and later determined by its optical density using a spectrophotometer at 650 nm. Naked eye detection is extremely simple, inexpensive, and reliable, and can prevent cross-contamination between samples. In addition, real-time monitoring of LAMP can be accomplished through spectrophotometric analysis using a real-time turbidity meter. A real-time turbidity meter was first developed and commercialized by Mori et al. (2004). This machine is capable of maintaining the LAMP reaction solutions in commercially available 0.2-mL PCR tubes at the optimum temperature and continuously measuring the turbidity of multiple samples (Notomi et al., 2015). The highly efficient and sensitive LAMP assay has some drawbacks. For example, it can cause false-positive results in subsequent reactions due to a small amount of aerosolized amplification product. Fig. 2 shows the LAMP process for detection of HPV in OPSCC.
Fig. 2.
A schematic diagram of the LAMP process for detection of HPV in OPSCC
LAMP: loop-mediated isothermal amplification; HPV: human papillomavirus; OPSCC: oropharyngeal squamous cell carcinoma
Several LAMP assays have been reported for the detection of HPV in OPSCC (Chen et al., 2015; Livingstone et al., 2016; Rohatensky et al., 2018). Livingstone et al. (2016) reported LAMP sensitivity of 99.4% and specificity of 93.2%, which were highly comparable to those of PCR for the detection and subtyping of clinical OPSCC samples. A recent study showed that the LAMP reaction was able to detect viral DNA down to a copy number of 105 for HPV16, 103 for HPV18, 104 for HPV31, and 105 for HPV35, with 100% specificity and without DNA purification (Rohatensky et al., 2018).
3. Detection using hybridization techniques
3.1. DNA in situ hybridization
ISH is another widely used technique to detect the presence of HPV, particularly in fixed tumor tissue samples. This technique uses an antisense probe to bind to a complementary HPV DNA target on tumor cells. The presence of HPV DNA in ISH during the hybridization process is evaluated using a microscope to search for punctate dot-like hybridization signals within the nuclei of epithelial cells (Snijders et al., 2010). Determination of the integration status of HPV DNA by ISH has been a routine practice because of its cost effectiveness and feasibility. DNA ISH allows a reliable identification of the physical state of the virus. The appearance of HPV DNA either in integrated or episomal form can be differentiated through the appearance of punctuate or diffuse signals (Anneroth et al., 1987). Generally, sample tissues are treated for hybridization using histochemistry to fix the target transcripts in place and to increase probe access. The probe hybridizes to the target sequence at an elevated temperature and then the excess probe is washed away (in the case of an unhybridized excess RNA probe, following previous hydrolysis using RNase). The probe, which is labeled with either radio-, fluorescent-, or antigen-labeled bases (e.g., digoxigenin), is then identified and quantified in the tissue using either autoradiography, fluorescence microscopy, or IHC, respectively. ISH can also use two or more radioactively labeled probes or other non-radioactive labels to detect two or more transcripts simultaneously.
The effectiveness of this assay is limited by its low sensitivity at low viral loads. The differentiation of HPV DNA by its appearance is often difficult to interpret and can be time-consuming. This can happen because of unusual staining characteristics or when the samples are small, such as small core biopsies and cell blocks which need extra interpretation time. Subsequently, these problems could present a false-positive result, especially if the observation is done under low magnification. It was suggested to examine the DNA ISH slides at high magnification to avoid missing weak or focal staining. Automated DNA ISH could be used to reduce the time consumption of manual DNA ISH. Another drawback of this procedure is that it is relatively insensitive. The sensitivity was once reported to be 63.6% for DNA ISH, compared to 81.8% in p16 IHC (Khor et al., 2013). Nevertheless, the specificity is high. DNA ISH also requires relatively large amounts of purified DNA. A low DNA copy number could cause weak staining in a DNA ISH assay (Bray et al., 2018).
3.2. RNA in situ hybridization
RNA ISH uses probes that are complementary to E6/E7 mRNA as the gold standard for HPV detection in tumor tissue (Schache et al., 2011). At present, HPV mRNA ISH has a great advantage compared with DNA ISH because of its ability to detect the presence of transcriptionally active HPV, which indicates the existence of HPV-related oncogenesis. RNA ISH was demonstrated to have higher sensitivity than DNA ISH (Bishop et al., 2012; Schache et al., 2013; Mirghani et al., 2015). It correlated more strongly with p16 immunostaining, produced results with strong signals, and was easily interpreted due to a brighter and more diffuse signal than that generally seen with the DNA ISH assay. Another study reported its sensitivity as 97% and specificity as 93%, and proved that this assay could untangle the HPV status in 88% of negative DNA ISH cases. RNA ISH allows direct visualization of viral transcripts from a processed tissue section. The improvement of viral detection due to natural target amplification by viral mRNA transcription has also been identified. Standardization can be enhanced by the use of an automated staining platform that can reduce turnaround time and enhance reproducibility (Saetiew et al., 2011).
RNA ISH could serve as a reliable standalone test to clarify the presence of HPV due to a strong association with the HPV-associated OPSCC biomarker, the p16 protein (Bishop et al., 2012). ISH for high-risk HPV E6/E7 mRNA is a highly specific, highly sensitive tool for HPV detection in OPSCC. Because of HPV-independent mechanisms, the p16 protein may be overexpressed. Therefore, all p16 IHC-positive OPSCCs should be considered for retesting using mRNA ISH to check transcriptionally active HPVs. This is particularly relevant when considering de-escalated treatment strategies for patients with HPV-positive tumors and providing favorable results for this subgroup of patients (Randén-Brady et al., 2019). The disadvantages of RNA ISH are that the probe is expensive and is not readily available because it requires extensive space, infrastructure, and equipment, and the process requires more technical expertise (Garibyan and Avashia, 2013). PCR requires nucleic acid extraction from FFPE samples and more specific techniques to proceed for fragmented or ruptured RNA after FFPE sample processing. The use of frozen material is technically demanding and labour-intensive for a routine process (Mirghani et al., 2013).
4. p16 immunohistochemistry
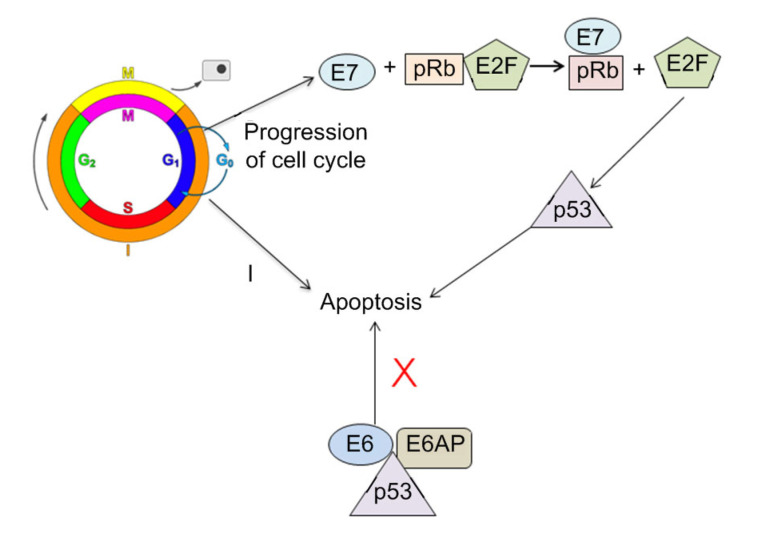
Overexpression of p16 is accepted as a surrogate diagnostic biomarker of transcriptionally active oncogenic HPV infection in OPSCC (Ang et al., 2010). HPV oncoproteins (E6 and E7) are responsible for causing genetic alterations in HPV+OPSCC (Akagi et al., 2014). E6 contains zinc-binding motifs, which form complexes with the host cell p53 tumor suppressor protein that causes p53 degradation (Hewitt et al., 2014). Degradation of p53 leads to a decrease of p21, a downstream target in the pathway. Since p53 functions in regulating cell growth and tumor suppression, the loss of p53 results in deregulation of the cell cycle and promotes mutation, chromosomal instability, and carcinogenesis of the host genome. On the other hand, E7 oncoprotein forms complexes with proteins in the pRb gene family, which are negative regulators of cell growth. pRb1 regulates cell cycle progression by binding and disrupting the function of transcription factors, such as E2 factor (E2F) (Suresh, 2016). Hypo-phosphorylated forms of pRb are thought to block cell cycle progression. HPV E7 binds to hypo-phosphorylated forms of pRb and releases free E2F transcription factors and others, such as p16, into the cell, which consequently leads to transcriptional activation of several genes involved in cell proliferation. The release of these transcription factors causes a cascade of events in the cell as part of its progression through the cell cycle (Singhi and Westra, 2010). Downstream upregulation of p16 is seen when pRb is made ineffective by E7. Once HPV is integrated, E6 and E7 work in concert to effectively transform epithelial cells and this action enables the progression to cancer. The degradation of pRb automatically enhances expression of p16INK4a, which can be a hallmark marker for HPV-related oncogenic activity and malignant transformation in OPSCC (Weiss et al., 2012). Fig. 3 shows the mechanisms of E6 and E7 proteins involved in the development of HPV-associated cancers.
Fig. 3.
Mechanisms of HPV E6 and E7 proteins in the development of HPV-associated cancers
E7 and E6 react with the tumor suppressor gene products pRb and p53 in host cell proteins, respectively, resulting in carcinogenesis. E2F: E2 factor; E6AP: E6-associated protein; HPV: human papillomavirus
Detection of the p16INK4a surrogate marker by p16 IHC is commonly used as a standalone test for the diagnosis of OPSCC. IHC typically detects antigens in tumor tissue sections by immunological and chemical reactions. Several studies have reported a strong correlation between p16 overexpression and HPV-associated OPSCC and prognosis (Schache et al., 2011; Lewis et al., 2012). Another study reported that the presence of HPV DNA in tumors correlated well with the expression of p16INK4a (κ=0.80; 95% confidence interval (CI), 0.73–0.87) (Ang et al., 2010). A pooled analysis of p16 data reported a sensitivity of 94%, specificity of 83%, and positive predictive value of 93% for high-risk HPV E6/E7 expression (Goot-Heah et al., 2012).
The low cost and feasibility of p16 testing on FFPE tissue have made p16 IHC the most favored test compared to ISH and PCR-based assays (Lewis et al., 2012). This method demonstrated very good agreement with the gold standard HPV E6/E7 mRNA expression (Kim et al., 2018). However, it is moderately specific and could lead to a false-positive result, since p16 overexpression that represents the loss of pRb could also occur through mechanisms other than oncogenic HPV E7 expression (Fischer et al., 2010). Few studies have reported that HPV DNA is not present in 8% to 20% of p16-positive OPSCCs (Rietbergen et al., 2014; Mirghani et al., 2015). Thus, they have recommended that HPV status in all p16-positive cases should be confirmed by another method (Mirghani et al., 2016; Volpi et al., 2018; Randén-Brady et al., 2019). Thus, using p16 IHC as a standalone test for HPV detection can mislead diagnostic and treatment approaches, particularly when considering de-escalation (Rietbergen et al., 2014). Furthermore, p16 requires additional cost and could lead to diagnostic lags in health care systems with limited pathologists, because of the need for additional human resources (Hewitt et al., 2014).
5. Diagnostic algorithms: why is detection of HPV in OPSCC important?
OPSCC containing transcriptionally active high-risk HPV in its tumor cells (HPV-positive OPSCCs) is classified as a distinct clinical entity according to the recent World Health Organization (WHO) classification of head and neck tumors (El-Naggar et al., 2017). Its risk factors, demographic, morphological, molecular, and clinical profiles differ substantially from other types of head and neck squamous cell carcinomas (HNSCCs) (Lewis et al., 2018). Apart from the prevailing evidence of HPV16 as a causative factor in more than 90% of cases, this subset of patients is less likely to contain smokers and alcohol users, and more likely to be associated with oral sex as a major risk factor. They are typically younger (50–56 years old) white males of higher economic standing (Gillison et al., 2008). Histopathological results of these cases commonly show non-keratinizing squamous cell carcinoma as opposed to HPV-negative tumors, with a site predilection for tongue and palatine tonsils.
Prognosis wise, a higher three-year overall survival rate in HPV-positive OPSCCs (82.4% vs. 52.7% in HPV-negative lesions) and rates of progression-free survival (73.7% vs. 43.4%) were reported. There is 58% reduction in risk of death in the HPV-positive OPSCC group, regardless of whether these patients were treated with concurrent systemic therapy (cisplatin) combined with standard-fractionation radiotherapy or accelerated-fractionation radiotherapy. Furthermore, the cumulative incidence of second primary tumors among patients with HPV-positive tumors was significantly lower. They concluded that the HPV status of the tumor was the major determinant of overall survival rate and its higher rate reflected increased intrinsic sensitivity to radiation or better radiosensitization (Ang et al., 2010). Current guidelines by the National Comprehensive Cancer Network® (NCCN, 2020) on diagnostic workup for cancer of the oropharynx involving the base of the tongue, tonsil, posterior pharyngeal wall, and soft palate include testing newly diagnosed OPSCC patients for high-risk HPV, either from the primary tumor or from cervical nodal metastases, using p16 IHC with a 70% nuclear and cytoplasmic staining cutoff (Lewis et al., 2018; NCCN, 2020).
The incorporation of more than one testing method may further increase the sensitivity of detection results (NCCN, 2020). A combination of two diagnostic algorithms commonly used was p16 IHC as the firstline assay, followed by HPV DNA/RNA PCR or HPV DNA ISH. In an algorithm described by Chai et al. (2016), PCR was able to confirm the presence of 92.9% HPV16 DNA and 60.0% HPV16 RNA using oral fluid samples in p16INK4a-positive tumors. This study indicated that p16INK4a positivity in a tumor is strongly associated with HPV16 infection. Schache et al. (2011) applied the combination of p16 IHC with DNA and RNA qPCR, which demonstrated high sensitivity (97%) and specificity (94%) of DNA qPCR when compared to the RNA qPCR. In predicting tumor p16 positivity using saliva, HPV RNA PCR has a lower sensitivity compared to HPV DNA PCR. There are two possible reasons for these results: these may be due to the unstable nature of RNA or no E6/E7 mRNA expression in patients with HPV DNA tumors. Another study compared the detection of HPV DNA in p16-positive samples in which qPCR successfully detected 54% HPV DNA, while ISH failed to detect any HPV DNA in p16-positive samples. This study suggested that p16 immunoreactivity and HPV genotyping by qPCR may be useful markers of HPV infection in OPSCC (Kouketsu et al., 2016).
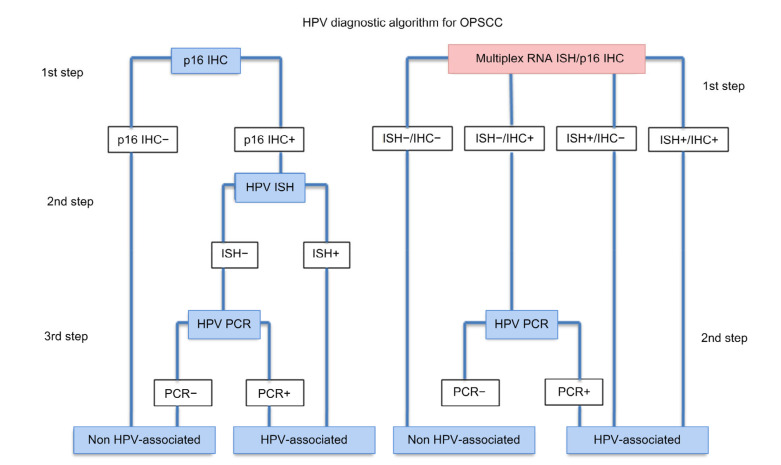
Rooper et al. (2016) described a two-step technique comprising p16 IHC, followed by HPV DNA ISH and HPV RNA ISH, using tumor tissue. RNA ISH has been reported to be a highly sensitive and specific platform capable of clarifying the status of OPSCC tumor tissue HPV that is p16-positive for IHC, but HPV-negative for DNA ISH. Mirghani et al. (2011) demonstrated a false-negative result through a combination of p16 and DNA ISH where the overexpression of p16 was related to a non-viral mechanism. Later, they suggested using RNA ISH as a potential algorithm in line with p16 because of its high sensitivity (Mirghani et al., 2015). Lewis et al. (2012) successfully identified 94% of additional HPV-positive cases in samples that were negatively amplified using ISH assays, but p16-positive in IHC assays. Rietbergen et al. (2014) reported that detection of HPV by PCR in addition to p16 is crucial for a better identification of HPV-related OPSCC. Combined testing of p16INK4a IHC and HPV DNA PCR significantly improves specificity, while retaining high sensitivity. According to their studies, the pooled sensitivities of p16INK4a IHC, HPV DNA PCR, HPV DNA ISH, and p16INK4a IHC/HPV DNA PCR combined testing were 94% (95% CI, 91%–97%), 98% (95% CI, 94%–100%), 85% (95% CI, 76%–92%), and 93% (95% CI, 87%–97%), respectively. The pooled specificities were 83% (95% CI, 78%–88%), 84% (95% CI, 74%–92%), 88% (95% CI, 78%–96%), and 96% (95% CI, 89%–100%), respectively. The combined testing of p16INK4a IHC/HPV DNA PCR showed a sensitivity similar to either p16INK4a IHC or HPV DNA PCR alone, but was significantly more specific than either separate test (Prigge et al., 2017). Marino et al. (2020) recently established a novel multiplex HPV RNA ISH/p16 IHC assay to detect both HPV E6/E7 transcripts and p16INK4a overexpression simultaneously. There are advantages from combining various detection assays to achieve an accurate and reliable HPV status; however, it is technically inconvenient, may produce discordant results, and requires more cost and time. Fig. 4 shows an algorithm for the detection of HPV in FFPE tissue from head and neck biopsies.
Fig. 4.
HPV diagnostic algorithm for OPSCC
Reprinted from Marino et al. (2020), with permission from Springer Nature via Copyright Clearance Center’s RightsLink service. HPV: human papillomavirus; IHC: immunohistochemistry; ISH: in situ hybridization; OPSCC: oropharyngeal squamous cell carcinoma; PCR: polymerase chain reaction
According to the NCCN (2020), p16 IHC staining and PCR-based assays are highly reliable in terms of sensitivity, while ISH has the highest specificity. Thus, they recommended integration of several testing methods including both PCR (higher sensitivity, lower specificity) and ISH (less sensitivity, higher specificity) for an equivocal p16 or uncertain clinical scenario (Singhi and Westra, 2010; Snow and Laudadio, 2010; Lewis et al., 2018; NCCN, 2020). Despite significant heterogeneity in patient populations, sample size, HPV detection methods, tumor stage and treatment, comorbidity, and the inclusion of various other prognostic factors in the analysis, the survival benefit of HPV-positive OPSCC has been maintained across almost all studies. For HPV-positive tumors, significant decreases in the risk of progression and disease-related death were confirmed in large prospective studies where OPSCC patients were uniformly staged and treated (Lewis et al., 2018).
6. Conclusions
The HPV status of a primary or metastatic OPSCC may have consequences for treatment, staging, and even for therapy. The recommendation for regular HPV testing currently reflects its role as an important prognostic predictor for OPSCC patients. The choice of a suitable method for HPV detection has become increasingly complex. Detection can be accomplished either by PCR, qPCR, LAMP, IHC, ISH, or a combination of these methods. Despite the availability of various techniques, molecular tests have always been the gold standard and are the most commonly used. Detecting the presence of HPV oncogene E6/E7 mRNA transcripts is regarded as the gold standard, and p16 as a surrogate biomarker in clinical settings. However, each of the listed methods has its pros and cons. To choose the best techniques for detection, it is important to consider the type of specimen that will be used and the availability of equipment and skilled personnel. Strong financial resources are needed as some of the reagents and probes are very expensive. The guidelines for the identification of HPV in cervical carcinoma are widely available, but no specific consensus on the gold standard for HPV testing in OPSCC has yet been reached. None of the developed assays seems to have both very high specificity and sensitivity. Therefore, multimodal testing that integrates two HPV detection methods could help to reliably identify patients with transcriptionally active high-risk HPV-positive OPSCC and to avoid the possibility of false-positive and false-negative cases (Marino et al., 2020). The use of highly sensitive, specific, and accurate methods is critical, especially when considering de-escalation treatment approaches for HPV-positive OPSCC patients.
Acknowledgments
We thank Ms. Syaidatul Akmal SYAIFUDDIN from School of Dental Sciences, Universiti Sains Malaysia for her efforts in editing the figures.
Footnotes
Project supported by the Universiti Sains Malaysia Research University Grant (1001/PPSG/8012345)
Contributors: Fatin Hazwani FAUZI and Nurul Izzati HAMZAN analyzed the literature and prepared the first draft of the manuscript. Nurhayu Ab RAHMAN and Siti SURAIYA edited and checked the final version. Suharni MOHAMAD designed, revised, edited, and checked the final version. All authors have read and approved the final manuscript and, therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines: Fatin Hazwani FAUZI, Nurul Izzati HAMZAN, Nurhayu Ab RAHMAN, Siti SURAIYA, and Suharni MOHAMAD declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Abdullahi UF, Naim R, Wan Taib WR, et al. Loop-mediated isothermal amplification (LAMP), an innovation in gene amplification: bridging the gap in molecular diagnostics; a review. Indian J Sci Technol. 2015;8(17):1–12. [Google Scholar]
- 2.Abreu ALP, Souza RP, Gimenes F, et al. A review of methods for detect human Papillomavirus infection. Virol J, 9:262. 2012 doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn SM, Chan JYK, Zhang Z. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus–related head and neck cancer. JAMA Otolaryngol Head Neck Surg. 2014;140(9):846–854. doi: 10.1001/jamaoto.2014.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akagi K, Li JF, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24(2):185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albano PM, Holzinger D, Salvador C, et al. Low prevalence of human papillomavirus in head and neck squamous cell carcinoma in the northwest region of the Philippines. PLoS ONE. 2017;12(2):e0172240. doi: 10.1371/journal.pone.0172240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anneroth G, Batsakis J, Luna M. Review of the literature and a recommended system of malignancy grading in oral squamous cell carcinomas. Scand J Dent Res. 1987;95(3):229–249. doi: 10.1111/j.1600-0722.1987.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 8.Antonsson A, Knight L, Panizza BJ, et al. HPV-16 viral load in oropharyngeal squamous cell carcinoma using digital PCR. Acta Oto-Laryngol. 2018;138(9):843–847. doi: 10.1080/00016489.2018.1461239. [DOI] [PubMed] [Google Scholar]
- 9.Arney A, Bennett KM. Molecular diagnostics of human papillomavirus. Lab Med. 2010;41(9):523–530. doi: 10.1309/lm75wgjsvmi7vvef. [DOI] [Google Scholar]
- 10.Babiker AY, Eltom FM, Abdalaziz MS, et al. Screening for high risk human papilloma virus (HR-HPV) subtypes, among Sudanese patients with oral lesions. Int J Clin Exp Med. 2013;6(4):275–281. [PMC free article] [PubMed] [Google Scholar]
- 11.Bhargava A, Saigal S, Chalishazar M. Histopathological grading systems in oral squamous cell carcinoma: a review. J Int Oral Health. 2010;2(4):1–10. [Google Scholar]
- 12.Biron VL, Kostiuk M, Isaac A, et al. Detection of human papillomavirus type 16 in oropharyngeal squamous cell carcinoma using droplet digital polymerase chain reaction. Cancer. 2016;122(10):1544–1551. doi: 10.1002/cncr.29976. [DOI] [PubMed] [Google Scholar]
- 13.Bishop JA, Ma XJ, Wang HW, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36(12):1874–1882. doi: 10.1097/PAS.0b013e318265fb2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishop JA, Lewis JS Jr, Rocco JW, et al. HPV-related squamous cell carcinoma of the head and neck: an update on testing in routine pathology practice. Semin Diagn Pathol. 2015;32(5):344–351. doi: 10.1053/j.semdp.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 16.Candotto V, Lauritano D, Nardone M, et al. HPV infection in the oral cavity: epidemiology, clinical manifestations and relationship with oral cancer. Oral Implantol. 2017;10(3):209–220. doi: 10.11138/orl/2017.10.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carcopino X, Henry M, Mancini J, et al. Significance of HPV 16 and 18 viral load quantitation in women referred for colposcopy. J Med Virol. 2012;84(2):306–313. doi: 10.1002/jmv.23190. [DOI] [PubMed] [Google Scholar]
- 18.Chai RC, Lambie D, Verma M, et al. Current trends in the etiology and diagnosis of HPV-related head and neck cancers. Cancer Med. 2015;4(4):596–607. doi: 10.1002/cam4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai RC, Lim BH, Frazer I, et al. A pilot study to compare the detection of HPV-16 biomarkers in salivary oral rinses with tumour p16INK4a expression in head and neck squamous cell carcinoma patients. BMC Cancer, 16:178. 2016 doi: 10.1186/s12885-016-2217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen HW, Weissenberger G, Atkins E, et al. Highly sensitive loop-mediated isothermal amplification for the detection of Leptospira . Int J Bacteriol, 2015:147173. 2015 doi: 10.1155/2015/147173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coser J, da Rocha Boeira T, Fonseca ASK, et al. Human papillomavirus detection and typing using a nested-PCR-RFLP assay. Braz J Infect Dis. 2011;15(5):467–472. doi: 10.1016/S1413-8670(11)70229-X. [DOI] [PubMed] [Google Scholar]
- 22.Dang J, Feng QH, Eaton KD, et al. Detection of HPV in oral rinse samples from OPSCC and non-OPSCC patients. BMC Oral Health, 15:126. 2015 doi: 10.1186/s12903-015-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng ZY, Hasegawa M, Kiyuna A, et al. Viral load, physical status, and E6/E7 mRNA expression of human papillomavirus in head and neck squamous cell carcinoma. Head Neck. 2013;35(6):800–808. doi: 10.1002/hed.23034. [DOI] [PubMed] [Google Scholar]
- 24.de Villiers EM, Fauquet C, Broker TR, et al. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Dhama K, Karthik K, Chakraborty S, et al. Loop-mediated isothermal amplification of DNA (LAMP): a new diagnostic tool lights the world of diagnosis of animal and human pathogens: a review. Pak J Biol Sci. 2014;17(2):151–166. doi: 10.3923/pjbs.2014.151.166. [DOI] [PubMed] [Google Scholar]
- 26.Duncan LD, Winkler M, Carlson ER, et al. p16 immunohistochemistry can be used to detect human papillomavirus in oral cavity squamous cell carcinoma. J Oral Maxillofac Surg. 2013;71(8):1367–1375. doi: 10.1016/j.joms.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 27.El-Naggar AK, Chan JKC, Grandis JR, et al. WHO Classification of Head and Neck Tumours. World Health Organization Classification of Tumours. IARC Press, Lyon, France; 2017. pp. 136–138. [Google Scholar]
- 28.Fischer CA, Kampmann M, Zlobec I, et al. p16 expression in oropharyngeal cancer: its impact on staging and prognosis compared with the conventional clinical staging parameters. Ann Oncol. 2010;21(10):1961–1966. doi: 10.1093/annonc/mdq210. [DOI] [PubMed] [Google Scholar]
- 29.Fontaine V, Mascaux C, Weyn C, et al. Evaluation of combined general primer-mediated PCR sequencing and type-specific PCR strategies for determination of human papillomavirus genotypes in cervical cell specimens. J Clin Microbiol. 2007;45(3):928–934. doi: 10.1128/JCM.02098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garibyan L, Avashia N. Research techniques made simple: polymerase chain reaction (PCR) J Invest Dermatol. 2013;133(3):e6. doi: 10.1038/jid.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillison ML, D'Souza G, Westra W, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100(6):407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 32.Goot-Heah K, Kwai-Lin T, Froemming GRA, et al. Human papilloma virus 18 detection in oral squamous cell carcinoma and potentially malignant lesions using saliva samples. Asian Pac J Cancer Prev. 2012;13(12):6109–6113. doi: 10.7314/apjcp.2012.13.12.6109. [DOI] [PubMed] [Google Scholar]
- 33.Graham SV. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5(10):1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta B, Johnson NW. Systematic review and meta-analysis of association of smokeless tobacco and of betel quid without tobacco with incidence of oral cancer in South Asia and the Pacific. PLoS ONE. 2014;9(11):e113385. doi: 10.1371/journal.pone.0113385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta S, Gupta S. Role of human papillomavirus in oral squamous cell carcinoma and oral potentially malignant disorders: a review of the literature. Indian J Dent. 2015;6(2):91–98. doi: 10.4103/0975-962X.155877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamzan NI, Fauzi FH, Taib H, et al. Simple and rapid detection of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans by loop-mediated isothermal amplification assay. Bangladesh J Med Sci. 2018;17(3):402–410. doi: 10.3329/bjms.v17i3.36995. [DOI] [Google Scholar]
- 37.Hewitt SM, Baskin DG, Frevert CW, et al. Controls for immunohistochemistry: the Histochemical Society’s standards of practice for validation of immunohistochemical assays. J Histochem Cytochem. 2014;62(10):693–697. doi: 10.1369/0022155414545224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holzinger D, Schmitt A, Dyckhoff G, et al. Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res. 2012;72(19):4993–5003. doi: 10.1158/0008-5472.can-11-3934. [DOI] [PubMed] [Google Scholar]
- 39.Isaac A, Kostiuk M, Zhang H, et al. Ultrasensitive detection of oncogenic human papillomavirus in oropharyngeal tissue swabs. J Otolaryngol Head Neck Surg, 46:5. 2017 doi: 10.1186/s40463-016-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jalouli M, Jalouli J, Ibrahim SO, et al. Comparison between single PCR and nested PCR in detection of human papilloma viruses in paraffin-embedded OSCC and fresh oral mucosa. In Vivo. 2015;29(1):65–70. [PubMed] [Google Scholar]
- 41.Khor GH, Froemming GRA, Zain RB, et al. DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma. Int J Med Sci. 2013;10(12):1727–1739. doi: 10.7150/ijms.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KY, Lewis JS, Jr, Chen Z. Current status of clinical testing for human papillomavirus in oropharyngeal squamous cell carcinoma. J Pathol Clin Res. 2018;4(4):213–226. doi: 10.1002/cjp2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouketsu A, Sato I, Abe S, et al. Detection of human papillomavirus infection in oral squamous cell carcinoma: a cohort study of Japanese patients. J Oral Pathol Med. 2016;45(8):565–572. doi: 10.1111/jop.12416. [DOI] [PubMed] [Google Scholar]
- 44.Lewis JS, Jr, Chernock RD, Ma XJ, et al. Partial p16 staining in oropharyngeal squamous cell carcinoma: extent and pattern correlate with human papillomavirus RNA status. Mod Pathol. 2012;25(9):1212–1220. doi: 10.1038/modpathol.2012.79. [DOI] [PubMed] [Google Scholar]
- 45.Lewis JS, Jr, Beadle B, Bishop JA, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med. 2018;142(5):559–597. doi: 10.5858/arpa.2017-0286-CP. [DOI] [PubMed] [Google Scholar]
- 46.Lin WJ, Jiang RS, Wu SH, et al. Smoking, alcohol, and betel quid and oral cancer: a prospective cohort study. J Oncol, 2011:525976. 2011 doi: 10.1155/2011/525976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livingstone DM, Rohatensky M, Mintchev P, et al. Loop mediated isothermal amplification (LAMP) for the detection and subtyping of human papillomaviruses (HPV) in oropharyngeal squamous cell carcinoma (OPSCC) J Clin Virol. 2016;75:37–41. doi: 10.1016/j.jcv.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 48.Marino FZ, Ronchi A, Stilo M, et al. Multiplex HPV RNA in situ hybridization/p16 immunohistochemistry: a novel approach to detect papillomavirus in HPV-related cancers. A novel multiplex ISH/IHC assay to detect HPV. Infect Agents Cancer, 15:46. 2020 doi: 10.1186/s13027-020-00310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehanna H, Beech T, Nicholson T, et al. Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer–systematic review and meta-analysis of trends by time and region. Head Neck. 2013;35(5):747–755. doi: 10.1002/hed.22015. [DOI] [PubMed] [Google Scholar]
- 50.Mellin H, Dahlgren L, Munck-Wikland E, et al. Human papillomavirus type 16 is episomal and a high viral load may be correlated to better prognosis in tonsillar cancer. Int J Cancer. 2002;102(2):152–158. doi: 10.1002/ijc.10669. [DOI] [PubMed] [Google Scholar]
- 51.Mirghani H, Moreau F, Lefèvre M, et al. Human papillomavirus type 16 oropharyngeal cancers in lymph nodes as a marker of metastases. Arch Otolaryngol Head Neck Surg. 2011;137(9):910–914. doi: 10.1001/archoto.2011.141. [DOI] [PubMed] [Google Scholar]
- 52.Mirghani H, Ferchiou F, Moreau F, et al. Oropharyngeal cancers: significance of HPV16 detection in neck lymph nodes. J Clin Virol. 2013;57(2):120–124. doi: 10.1016/j.jcv.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 53.Mirghani H, Amen F, Moreau F, et al. Human papilloma virus testing in oropharyngeal squamous cell carcinoma: what the clinician should know. Oral Oncol. 2014;50(1):1–9. doi: 10.1016/j.oraloncology.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 54.Mirghani H, Casiraghi O, Amen F, et al. Diagnosis of HPV-driven head and neck cancer with a single test in routine clinical practice. Mod Pathol. 2015;28(12):1518–1527. doi: 10.1038/modpathol.2015.113. [DOI] [PubMed] [Google Scholar]
- 55.Mirghani H, Casiraghi O, Guerlain J, et al. Diagnosis of HPV driven oropharyngeal cancers: comparing p16 based algorithms with the RNAscope HPV-test. Oral Oncol. 2016;62:101–108. doi: 10.1016/j.oraloncology.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Mori Y, Nagamine K, Tomita N, et al. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 57.Mori Y, Kitao M, Tomita N, et al. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Meth. 2004;59(2):145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother. 2013;19(3):404–411. doi: 10.1007/s10156-013-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muñoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 60.NCCN National Comprehensive Cancer Network. Cancer of the oropharynx. Version 1.2021, Nov. 16. https://www. nccn.org/professionals/physician_gls/pdf/head-and-neck_blocks.pdf.2020. [Google Scholar]
- 61.Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Notomi T, Mori Y, Tomita N, et al. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015;53(1):1–5. doi: 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 63.Oji C, Chukwuneke F. Poor oral hygiene may be the sole cause of oral cancer. J Maxillofac Oral Surg. 2012;11(4):379–383. doi: 10.1007/s12663-012-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parida M, Sannarangaiah S, Dash PK, et al. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18(6):407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prigge ES, Arbyn M, von Knebel Doeberitz M, et al. Diagnostic accuracy of p16INK4a immunohistochemistry in oropharyngeal squamous cell carcinomas: a systematic review and meta-analysis. Int J Cancer. 2017;140(5):1186–1198. doi: 10.1002/ijc.30516. [DOI] [PubMed] [Google Scholar]
- 66.Qureishi A, Mawby T, Fraser L, et al. Current and future techniques for human papilloma virus (HPV) testing in oropharyngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2017;274(7):2675–2683. doi: 10.1007/s00405-017-4503-1. [DOI] [PubMed] [Google Scholar]
- 67.Randén-Brady R, Carpén T, Jouhi L, et al. In situ hybridization for high-risk HPV E6/E7 mRNA is a superior method for detecting transcriptionally active HPV in oropharyngeal cancer. Hum Pathol. 2019;90:97–105. doi: 10.1016/j.humpath.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 68.Rietbergen MM, Snijders PJF, Beekzada D, et al. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int J Cancer. 2014;134(10):2366–2372. doi: 10.1002/ijc.28580. [DOI] [PubMed] [Google Scholar]
- 69.Ritari J, Hultman J, Fingerroos R, et al. Detection of human papillomaviruses by polymerase chain reaction and ligation reaction on universal microarray. PLoS ONE. 2012;7(3):e34211. doi: 10.1371/journal.pone.0034211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson M, Schache A, Sloan P, et al. HPV specific testing: a requirement for oropharyngeal squamous cell carcinoma patients. Head Neck Pathol. 2012;6(S1):83–90. doi: 10.1007/s12105-012-0370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohatensky MG, Livingstone DM, Mintchev P, et al. Assessing the performance of a loop mediated isothermal amplification (LAMP) assay for the detection and subtyping of high-risk suptypes of human papilloma virus (HPV) for oropharyngeal squamous cell carcinoma (OPSCC) without DNA purification. BMC Cancer, 18:166. 2018 doi: 10.1186/s12885-018-4087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rooper LM, Gandhi M, Bishop JA, et al. RNA in-situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry but HPV negative by DNA in-situ hybridization. Oral Oncol. 2016;55:11–16. doi: 10.1016/j.oraloncology.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 73.Saetiew C, Limpaiboon T, Jearanaikoon P, et al. Rapid detection of the most common high-risk human papillomaviruses by loop-mediated isothermal amplification. J Virol Met. 2011;178(1-2):22–30. doi: 10.1016/j.jviromet.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 74.Schache AG, Liloglou T, Risk JM, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res. 2011;17(19):6262–6271. doi: 10.1158/1078-0432.CCR-11-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schache AG, Liloglou T, Risk JM, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108(6):1332–1339. doi: 10.1038/bjc.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sigma-Aldrich qPCR Technical Guide. https://www. gene-quantification.de/SIAL-qPCR-Technical-Guide.pdf.2018. [Google Scholar]
- 77.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116(9):2166–2173. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 78.Snijders PJ, Heideman DAM, Meijer CJLM. Methods for HPV detection in exfoliated cell and tissue specimens. APMIS. 2010;118(6-7):520–528. doi: 10.1111/j.1600-0463.2010.02621.x. [DOI] [PubMed] [Google Scholar]
- 79.Snow AN, Laudadio J. Human papillomavirus detection in head and neck squamous cell carcinomas. Adv Anat Pathol. 2010;17(6):394–403. doi: 10.1097/PAP.0b013e3181f895c1. [DOI] [PubMed] [Google Scholar]
- 80.Stevenson A, Wakeham K, Pan J, et al. Droplet digital PCR quantification suggests that higher viral load correlates with improved survival in HPV-positive oropharyngeal tumours. J Clin Virol, 129:104505. 2020 doi: 10.1016/j.jcv.2020.104505. [DOI] [PubMed] [Google Scholar]
- 81.Suciu M, Morariu SH, Ormenisan A, et al. Oral squamous cell carcinoma of the maxilla, a second malignancy after a right ethmoido-maxillary chondrosarcoma. Rom J Morphol Embryol. 2014;55(S3):1247–1251. [PubMed] [Google Scholar]
- 82.Suresh MJ. Detection of p16INK4a in Oropharyngeal and Upper Respiratory Tract Squamous Cell Carcinoma. Master T. Chengalpattu Medical College, India. https:// 1library.net/document/oz1g1dvz-detection-oropharyngeal-upper-respiratory-tract-squamous-cell-carcinoma.html; 2016. [Google Scholar]
- 83.Tawe L, Grover S, Narasimhamurthy M, et al. Molecular detection of human papillomavirus (HPV) in highly fragmented DNA from cervical cancer biopsies using double-nested PCR. MethodsX. 2018;5:569–578. doi: 10.1016/j.mex.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Veyer D, Wack M, Mandavit M, et al. HPV circulating tumoral DNA quantification by droplet-based digital PCR: a promising predictive and prognostic biomarker for HPV-associated oropharyngeal cancers. Int J Cancer. 2020;147(4):1222–1227. doi: 10.1002/ijc.32804. [DOI] [PubMed] [Google Scholar]
- 85.Volpi CC, Ciniselli CM, Gualeni AV, et al. In situ hybridization detection methods for HPV16 E6/E7 mRNA in identifying transcriptionally active HPV infection of oropharyngeal carcinoma: an updating. Hum Pathol. 2018;74:32–42. doi: 10.1016/j.humpath.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 86.Wang XY, Seo DJ, Lee MH, et al. Comparison of conventional PCR, multiplex PCR, and loop-mediated isothermal amplification assays for rapid detection of Arcobacter species. J Clin Microbiol. 2014;52(2):557–563. doi: 10.1128/JCM.02883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wasserman JK, Rourke R, Purgina B, et al. HPV DNA in saliva from patients with SCC of the head and neck is specific for p16-positive oropharyngeal tumours. J Otolaryngol Head Neck Surg, 46:3. 2017 doi: 10.1186/s40463-016-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weiss D, Koopmann M, Basel T, et al. Cyclin A1 shows age-related expression in benign tonsils, HPV16-dependent overexpression in HNSCC and predicts lower recurrence rate in HNSCC independently of HPV16. BMC Cancer, 12:259. 2012 doi: 10.1186/1471-2407-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu DJ, Chen Y, Wu SH, et al. Simultaneous detection and differentiation of human papillomavirus genotypes 6, 11, 16 and 18 by AllGlo quadruplex quantitative PCR. PLoS ONE. 2012;7(11):e48972. doi: 10.1371/journal.pone.0048972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zarei M, Moradie A, Hamkar R, et al. Detection of human papillomavirus DNA sequences in oral lesions using polymerase chain reaction. Acta Med Iran. 2007;45(3):177–182. [Google Scholar]
- 91.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zil-e-Rubab , Baig S, Zaman U, et al. Human papilloma virus 16/18: fabricator of trouble in oral squamous cell carcinoma. Int J Infect Dis. 2018;69:115–119. doi: 10.1016/j.ijid.2018.02.003. [DOI] [PubMed] [Google Scholar]