Abstract
Filamentous actinobacteria are widely used as microbial cell factories to produce valuable secondary metabolites, including the vast majority of clinically relevant antimicrobial compounds. Secondary metabolites are typically encoded by large biosynthetic gene clusters, which allow for a modular approach to generating diverse compounds through recombination. Protoplast fusion is a popular method for whole genome recombination that uses fusion of cells that are transiently wall-deficient. This process has been applied for both inter- and intraspecies recombination. An important limiting step in obtaining diverse recombinants from fused protoplasts is regeneration of the cell wall, because this forces the chromosomes from different parental lines to segregate, thereby preventing further recombination. Recently, several labs have gained insight into wall-deficient bacteria that have the ability to proliferate without their cell wall, known as L-forms. Unlike protoplasts, L-forms can stably maintain multiple chromosomes over many division cycles. Fusion of such L-forms would potentially allow cells to express genes from both parental genomes while also extending the time for recombination, both of which can contribute to an increased chemical diversity. Here, we present a perspective on how L-form fusion has the potential to become a platform for novel compound discovery and may thus help to overcome the antibiotic discovery void.
Keywords: secondary metabolites, actinomycetes, protoplast fusion, novel compound discovery, cell wall-deficiency, heteroploidy
1. Introduction
Microorganisms have been used as a chassis to produce beneficial compounds like antibiotics, growth promoters, enzymes, inflammatory drugs, and protease inhibitors. Of these, antibiotics have made a tremendous impact on our lives due to their application in clinical and veterinary settings. Considering the global challenge of increased antimicrobial resistance, there is an urgent need for the discovery of new compounds. Antibiotics are produced via complex pathways encoded by large biosynthetic gene clusters (BGCs). These clusters not only include the necessary genes for biosynthesis of the antibiotic, but also those required for regulation of gene expression, export, and resistance to the antimicrobial compound. Streptomycetes are well-known for their ability to produce a large variety of antibiotics and may carry up to 70 BGCs per genome [1]. Notably, many of these gene clusters are silent or only poorly expressed, which so far has limited the successful exploitation of some of these metabolites as new antimicrobial agents. Genetic engineering of these BGCs for optimal production can be challenging, for instance, due to the presence of cryptic functions and the non-tractability of natural strains that harbor these BGCs. This has been one of the reasons behind the void in novel compound discovery with very few structurally new classes of antibiotics being introduced in the market in the past decades [2]. In contrast, genomic data analysis has unearthed 33,351 putative BGCs in 1154 microbial genomes [3]. The biosynthetic capabilities are thus plentiful, but our capacity to harness them is limited.
Protoplast fusion is a commonly used method to introduce a BGC or entire chromosome into a recipient cell for further genetic manipulation or directed evolution approaches. Its application for genome shuffling has resulted in strains with higher productivity, prominent examples being that of clavulanic acid [4], cephalosporin [5], diverse enzymatic activity [6], and novel compounds (indolizomycin [7]). This method has also led to the activation of silent BGCs [8]. However, protoplast fusion has potential disadvantages, such as the requirement for multiple fusion and regeneration phases, a short time frame during which recombination can occur due to the necessity for protoplasts to regenerate their cell wall, and lastly the instability of recombinants. From this perspective, we take a deeper look into the benefits and hurdles of protoplast fusion and then propose the use of permanent cell-wall-deficient forms as an alternative system for novel compound discovery.
2. History of Bacterial Protoplast Fusion
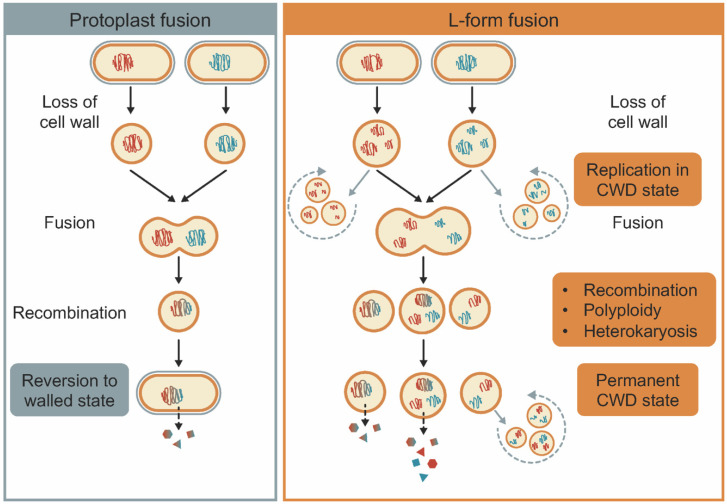
The need for protoplast fusion mainly arose from the difficulty of introducing DNA into cells, especially Gram-positive bacteria. The Gram-positive cell wall typically consists of many interconnected layers of peptidoglycan and teichoic acids [9]. Methods like electroporation and conjugation help alleviate the hurdles of passing through the multilayered cell wall but are restricted in terms of size of DNA that can be transformed. Hence, an alternative was found by stripping away the cell wall altogether. The basic set-up of protoplast fusion involves four consecutive steps: (i) protoplast formation, (ii) protoplast fusion, (iii) recombination between chromosomes within the fused cells, and (iv) reversion of protoplasts to walled cells (Figure 1) [10,11,12]. Protoplasts are obtained by treating walled cells with lysozyme, which degrades the major component of the cell wall, peptidoglycan (PG). The subsequent exposure of protoplasts to crowding agents, such as polyethylene glycol (PEG), causes protoplasts to aggregate. This in turn forces their membranes in close proximity with one another, causing them to fuse. Electrofusion and laser-induced fusion are also commonly used methods to obtain fused cells [12]. Fusion is followed by a brief period of recombination, during which the fusants are maintained in an osmotically protected environment, allowing them to regenerate their cell wall without lysis. In order to select for the desired recombinants, the cells are finally grown in a selective medium.
Figure 1.
The general process of protoplast fusion compared with L-form fusion. Both methods share some steps like the loss of cell wall and cell–cell fusion but differ in other key steps. Fusion in L-forms can lead to multiple possibilities due to polyploidy in daughter cells, whereas reversion to a walled state is essential in protoplast fusion. Differences in chromosome size are for depiction purposes only and not to scale.
Because protoplast fusion is non-specific, it has been used for recombination within and between different species. Industrial strains like Lactobacillus species have been a common target for genome shuffling via protoplast fusion, resulting in increased yield of lactic acid and improved acid tolerance [13]. Within species fusion may also be combined with random mutagenesis to increase genetic variation, followed by screening for the desired phenotype [14]. For instance, improved degradation of the toxic pesticide pentachlorophenol was achieved in Sphingobium chrolophenolicum via protoplast fusion generating cells that had become more resistant [15]. Fusion between phylogenetically distant bacteria has also been performed with Streptomyces griseus and Micromonospora sp. where recombinants displayed characteristics of both parents. More specifically, recombinants had a colony morphology like Micromonospora but with the carbohydrate and amino acid utilization abilities of S. griseus [16]. Notably, protoplast fusion is not limited to microbial cells, but has also been successfully used with plants to improve particular traits [10,17].
Taken together, protoplast fusion as a means for genome shuffling has provided several new biological activities and strains with improved traits or growth dynamics [18].
3. Disadvantages of Protoplast Fusion
The process of protoplast fusion introduces entire genomes into a single cellular compartment, allowing them to recombine. Protoplasts carrying multiples chromosomes are typically unable to be sustained for long periods of time and are thus unstable. This instability was observed when the mycaminose-producer Streptomyces fradiae was fused with a picronolide-producing Streptomyces strain [19]. The fusant transiently produced a novel macrolide antibiotic that was not formed by either parent that was hypothesized to have been due to a diploid state of the fusant. Unfortunately, the ability to produce this new compound was rapidly lost after subsequent culturing. Another disadvantage of fusing protoplasts is the limited period during which the chromosomes can fuse with one another to generate recombinants. This is due to the regeneration of a cell wall, which starts immediately after cells have fused. In some fusions, the efficiency of obtaining a recombinant with activity different compared to the parental lines is not quantitatively the same. Protoplasts of the raffinose utilizer Micromonaspora carbonaceae and Streptomyces griseus were fused by treatment with PEG [16]. Interestingly, only 2 out of 24 fusants revealed different antimicrobial activity compared to their parents. Moreover, protoplast regeneration can be unsuccessful due to inadequate media supplements, rendering the fusants non-viable. Fusants often die without the addition of sucrose, proline, and microelements in specific proportions to the selection media [20]. Lastly, rebuilding the cell wall in fusants producing cell wall-targeting agents may be complicated. This may lead to lower recombinant efficiencies following protoplast fusion [21]. An alternative method would thus be valuable, especially for generating increased phenotypic diversity and stabilizing fusant strains.
4. Cell-Wall-Deficient L-Forms as an Attractive Alternative for Protoplasts
While protoplasts are transiently wall-deficient, many bacteria can be forced into a permanent wall-deficient state as so-called L-forms [22,23]. They are variants of bacteria that have an altered cell wall organization, which when made irreversible on account of mutations in the genome result in stable cell wall deficiency. They may be referred to by other names in literature such as L-phase, L-variants, and L-organisms, with the “L” signifying the place of discovery, i.e., the Lister Institute [24]. Since their discovery, L-form research has contributed to the fields of bacterial cell biology [25,26], biotechnology [27], host-associated symbiotic and pathogenic interactions [28,29], and the origin of life [23,30]. Unlike protoplasts, cell-wall-deficient L-forms are able to propagate without their cell wall [31]. Proliferation of L-forms is based on biophysical principles, whereby an imbalance in the cell surface-to-volume ratio of cells causes membrane blebbing, tubulation, and vesicle formation. The imbalance required for this mode of proliferation can be obtained by increasing synthesis of fatty acids [32], which act as building blocks for the membrane, and by increasing the fluidity of the membrane [33]. Many (if not all) bacteria can be forced into an L-form state, including antibiotic-producing actinobacteria [34,35,36,37,38,39,40]. In most cases, loss of the cell wall is triggered by exposing walled cells to a combination of lytic enzymes and antibiotics in an osmotically protective environment. However, formation of wall-less cells has also been observed as a mechanism to escape stress [41], such as osmotic stress in filamentous actinomycetes [38], heat stress in E. coli [42], and nutrient stress in Mycobacterium bovis [43]. One of the important consequences of wall deficiency is that the resulting cells are polyploid. Because DNA segregation is likely unregulated during this phase, division of L-forms leads to the uneven distribution of chromosomes among progeny (Figure 1). While some cells inherit a single chromosome, others will inherit multiple chromosomes, with different ratios of the parental chromosomal types.
The structural similarity of L-forms to protoplasts makes then amenable to fusion using PEG or electrofusion. Increased membrane fluidity of L-form cells may also facilitate fusion in the absence of such inducing conditions. Indeed, we have observed spontaneous fusion between L-forms (our unpublished results). In contrast to protoplasts, L-form fusion is not followed by regeneration of the cell wall and the subsequent generation of haploid cells (Figure 1). Three potential outcomes are possible after fusion: (i) recombination between chromosomes, (ii) polyploidy where multiple copies of the recombinant genome are present, and (iii) heterokaryosis where both parental genomes are maintained. L-forms could thus overcome the problem of the short recombination time and fusants that only transiently produce a new metabolite due a diploid state prior to cell wall regeneration [19]. The use of L-forms instead allows for maintenance of heterokaryotic genomes in the same cytoplasmic compartment thereby prolonging opportunities for genetic variation to arise through recombination, resulting in novel production phenotypes. The presence of multiple chromosomes can have beneficial effects in terms of productivity (antibiotics, enzymes), fermentation, and biocontrol. This can happen via various routes such as (i) masking of deleterious alleles by a dominant or complimentary copy, (ii) interactions between genes that were previously not in the same environment, and (iii) gene dosage effects (multiple copies of a biosynthetic gene). It has been shown for Streptomyces specifically that interactions between different species can induce antibiotic production and activity [44,45]. By introducing genomes of different species in the same compartment using L-forms, one could potentially achieve a similar effect. Metabolite-inducing interactions that are governed by transcriptional regulators or epigenetic modifiers are good candidates to test [46,47]. The introduction of multiple copies of a gene cluster or induction of gene duplication is already in practice for increased antibiotic production [48,49]. For instance, actinorhodin production by S. coelicolor was increased 20-fold after tandem amplification of the act gene cluster, resulting in nine copies in the genome [48]. In the context of antibiotic production, L-forms are ideal for production of compounds targeting the cell wall since they are naturally resistant. This reduces loss of biomass during production. L-forms also do not show the formation of inclusion bodies, which can be a useful trait for overproducing proteins that are toxic to the cell [27]. Lastly, L-forms can be easily stored at −80 degrees for extended periods of time. It is yet unclear though if subculturing after revival has an effect on the genomes of the hybrid L-forms.
Collectively, the use of CWD bacteria alleviates most of the disadvantages of protoplast fusion. However, L-form fusion itself does have certain lose ends namely, the time and mutations required to achieve wall-deficient propagation, heterogeneity of fusants, and metabolite production levels. Obtaining a stable L-form strain from a wild-type requires multiple rounds of exposure to cell wall-targeting agents. During this exposure, mutations can arise that help alleviate the stress of surviving and growing without a cell wall such [41,50]. Recent studies are shedding light on the metabolic requirements for maintaining a wall-deficient state in a range of bacteria, thus accelerating the process of generating L-forms. The second issue of fusant heterogeneity when using L-forms can make phenotype screening and quantification difficult. Interestingly, the availability of single cell metabolomics, microfluidics, and encapsulation techniques can help solve this by isolating individual fusants immediately after fusion [51,52]. The fusants can then be tracked over time for certain biochemical activity to identify stable phenotypes. Lastly, studies attempting to use L-forms for overproduction of metabolites have resulted in mixed outcomes depending on the species of L-form used. The production of tetracycline by S. viridifaciens L-forms was lower than the walled parent [40], while on the other hand P. mirabilis L-forms engineered to express a variety of enzymes, antibodies, and polypeptides were able to produce a higher yield than their walled parent [27,53]. A better understanding of L-form growth and its physiology, division, and chromosome dynamics will help to develop this model as an attractive system for identifying novel compounds.
Taken together by providing the extended opportunities for recombination and maintenance of heterokaryons, the wall-deficient lifestyle provides an opportunity for novel compound discovery. The more we understand about growth, division, and DNA segregation in these systems, the better we can apply them for large scale bioprocessing as well.
Acknowledgments
We thank the members of the Claessen lab for fruitful discussions.
Author Contributions
Conceptualization, S.S., D.E.R., A.K. and D.C.; literature review, S.S. and G.Ö.B.; writing-original draft, S.S.; writing-review and editing S.S., G.Ö.B., D.E.R., A.K. and D.C. All authors have read and agreed to the published version of the manuscript.
Funding
S.S. is supported by an NWA Startimpuls from the Origins Centre, the Netherlands. D.C. is supported by an NWO Vici grant.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Belknap K.C., Park C.J., Barth B.M., Andam C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020;10:2003. doi: 10.1038/s41598-020-58904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silver L.L. Challenges of antibacterial discovery. Clin. Microbiol. Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J., et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi K.P., Kim K.H., Kim J.W. Strain improvement of clavulanic acid producing Streptomyces clavuligerus; Proceedings of the 10th International Symposium Biology Actinomycetes (ISBA); Beijing, China. 27–30 May 1997; [Google Scholar]
- 5.Hamlyn P.F., Ball C. Genetics of Industrial Microorganisms. American Society for Microbiology; Washington, DC, USA: 1979. [Google Scholar]
- 6.Okanishi M., Suzuki N., Furuta T. Variety of hybrid characters among recombinants obtained by interspecific protoplast fusion in Streptomycetes. Biosci. Biotechnol. Biochem. 1996;60:1233–1238. doi: 10.1271/bbb.60.1233. [DOI] [Google Scholar]
- 7.Gomi S., Ikeda D., Nakamura H., Naganawa H., Yamashita F., Hotta K., Kondo S., Okami Y., Umezawa H., Iitaka Y. Isolation and structure of a new antibiotic, indolizomycin, produced by a strain SK2-52 obtained by interspecies fusion treatment. J. Antibiot. (Tokyo) 1984;37:1491–1494. doi: 10.7164/antibiotics.37.1491. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen C.T., Dhakal D., Pham V.T., Nguyen H.T., Sohng J.-K. Recent advances in strategies for activation and discovery/characterization of cryptic biosynthetic gene clusters in Streptomyces. Microorganisms. 2020;8:616. doi: 10.3390/microorganisms8040616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopal M., Walker S. Protein and Sugar Export and Assembly in Gram-positive Bacteria. Springer; Heidelberg, Germany: 2015. Envelope structures of Gram-positive bacteria; pp. 1–44. [Google Scholar]
- 10.Peberdy J.F. Protoplast fusion—A tool for genetic manipulation and breeding in industrial microorganisms. Enzyme Microb. Technol. 1980;2:23–29. doi: 10.1016/0141-0229(80)90004-6. [DOI] [Google Scholar]
- 11.Gokhale D.V., Puntambekar U.S., Deobagkar D.N. Protoplast fusion: A tool for intergeneric gene transfer in bacteria. Biotechnol. Adv. 1993;11:199–217. doi: 10.1016/0734-9750(93)90041-K. [DOI] [PubMed] [Google Scholar]
- 12.Gong J., Zheng H., Wu Z., Chen T., Zhao X. Genome shuffling: Progress and applications for phenotype improvement. Biotechnol. Sustain. Hum. Soc. 2009;27:996–1005. doi: 10.1016/j.biotechadv.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Patnaik R., Louie S., Gavrilovic V., Perry K., Stemmer W.P.C., Ryan C.M., del Cardayré S. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 2002;20:707–712. doi: 10.1038/nbt0702-707. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood D.A., Wright H.M. Factors affecting recombinant frequency in protoplast fusions of Streptomyces coelicolor. Microbiology. 1979;111:137–143. doi: 10.1099/00221287-111-1-137. [DOI] [PubMed] [Google Scholar]
- 15.Dai M., Copley S.D. Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl. Environ. Microbiol. 2004;70:2391–2397. doi: 10.1128/AEM.70.4.2391-2397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imada C., Okanishi M., Okami Y. Intergenus protoplast fusion between Streptomyces and Micromonospora with reference to the distribution of parental characteristics in the fusants. J. Biosci. Bioeng. 1999;88:143–147. doi: 10.1016/S1389-1723(99)80192-3. [DOI] [PubMed] [Google Scholar]
- 17.Lazar G.B. Protoplasts 1983. Springer; Berlin, Germany: 1983. Recent developments in plant protoplast fusion and selection technology; pp. 61–67. [Google Scholar]
- 18.Adrio J.-L., Demain A.L. Recombinant organisms for production of industrial products. Bioeng. Bugs. 2010;1:116–131. doi: 10.4161/bbug.1.2.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda H., Inoue M., Tanaka H., Omura S. Interspecific protoplast fusion among macrolide-producing streptomycetes. J. Antibiot. (Tokyo) 1984;37:1224–1230. doi: 10.7164/antibiotics.37.1224. [DOI] [PubMed] [Google Scholar]
- 20.Marcone G.L., Carrano L., Marinelli F., Beltrametti F. Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. (Tokyo) 2010;63:83–88. doi: 10.1038/ja.2009.127. [DOI] [PubMed] [Google Scholar]
- 21.Baltz R.H., Matsushima P. Protoplast fusion in Streptomyces: Conditions for efficient genetic recombination and cell regeneration. Microbiology. 1981;127:137–146. doi: 10.1099/00221287-127-1-137. [DOI] [PubMed] [Google Scholar]
- 22.Allan E.J., Hoischen C., Gumpert J. Advances in Applied Microbiology. Volume 68. Academic Press; Cambridge, MA, USA: 2009. Chapter 1 Bacterial L-Forms; pp. 1–39. [DOI] [PubMed] [Google Scholar]
- 23.Errington J., Mickiewicz K., Kawai Y., Wu L.J. L-form bacteria, chronic diseases and the origins of life. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20150494. doi: 10.1098/rstb.2015.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klieneberger E. The natural occurrence of pleuropneumonia-like organism in apparent symbiosis with Streptobacillus moniliformis and other bacteria. J. Pathol. Bacteriol. 1935;40:93–105. doi: 10.1002/path.1700400108. [DOI] [Google Scholar]
- 25.Hoischen C., Gura K., Luge C., Gumpert J. Lipid and fatty acid composition of cytoplasmic membranes from Streptomyces hygroscopicus and its stable protoplast-type L form. J. Bacteriol. 1997;179:3430–3436. doi: 10.1128/JB.179.11.3430-3436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui R.A., Hoischen C., Holst O., Heinze I., Schlott B., Gumpert J., Diekmann S., Grosse F., Platzer M. The analysis of cell division and cell wall synthesis genes reveals mutationally inactivated ftsQ and mraY in a protoplast-type L-form of Escherichia coli. FEMS Microbiol. Lett. 2006;258:305–311. doi: 10.1111/j.1574-6968.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- 27.Gumpert J., Hoischen C. Use of cell wall-less bacteria (L-forms) for efficient expression and secretion of heterologous gene products. Curr. Opin. Biotechnol. 1998;9:506–509. doi: 10.1016/S0958-1669(98)80037-2. [DOI] [PubMed] [Google Scholar]
- 28.Somerson N.L., Ehrman L., Kocka J.P., Gottlieb F.J. Streptococcal L-forms isolated from Drosophila paulistorum semispecies cause sterility in male progeny. Proc. Natl. Acad. Sci. USA. 1984;81:282–285. doi: 10.1073/pnas.81.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markova N. L-form bacteria cohabitants in human blood: Significance for health and diseases. Discov. Med. 2017;23:305–313. [PubMed] [Google Scholar]
- 30.Sladek T.L. A hypothesis for the mechanism of mycoplasma evolution. J. Theor. Biol. 1986;120:457–465. doi: 10.1016/S0022-5193(86)80039-X. [DOI] [PubMed] [Google Scholar]
- 31.Mercier R., Kawai Y., Errington J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife. 2014;3:e04629. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercier R., Kawai Y., Errington J. Excess membrane synthesis drives a primitive mode of cell proliferation. Cell. 2013;152:997–1007. doi: 10.1016/j.cell.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 33.Mercier R., Domínguez-Cuevas P., Errington J. Crucial role for membrane fluidity in proliferation of primitive cells. Cell Rep. 2012;1:417–423. doi: 10.1016/j.celrep.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Dienes L. L Organisms of Klieneberger and Streptobacillus moniliformis. J. Infect. Dis. 1939;65:24–42. doi: 10.1093/infdis/65.1.24. [DOI] [Google Scholar]
- 35.Jass J., Phillips L.E., Allan E.J., Costerton J.W., Lappin-Scott H.M. Growth and adhesion of Enterococcus faecium L-forms. FEMS Microbiol. Lett. 1994;115:157–162. doi: 10.1111/j.1574-6968.1994.tb06631.x. [DOI] [PubMed] [Google Scholar]
- 36.Leaver M., Domínguez-Cuevas P., Coxhead J.M., Daniel R.A., Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 37.Glover W.A., Yang Y., Zhang Y. Insights into the molecular basis of L-Form formation and survival in Escherichia coli. PLoS ONE. 2009;4:e7316. doi: 10.1371/journal.pone.0007316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramijan K., Ultee E., Willemse J., Zhang Z., Wondergem J.A.J., van der Meij A., Heinrich D., Briegel A., van Wezel G.P., Claessen D. Stress-induced formation of cell-wall-deficient cells in filamentous actinomycetes. Nat. Commun. 2018;9:5164. doi: 10.1038/s41467-018-07560-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Studer P., Staubli T., Wieser N., Wolf P., Schuppler M., Loessner M.J. Proliferation of Listeria monocytogenes L-form cells by formation of internal and external vesicles. Nat. Commun. 2016;7:13631. doi: 10.1038/ncomms13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Innes C.M.J., Allan E.J. Induction, growth and antibiotic production of Streptomyces viridifaciens L-form bacteria. J. Appl. Microbiol. 2001;90:301–308. doi: 10.1046/j.1365-2672.2001.01243.x. [DOI] [PubMed] [Google Scholar]
- 41.Claessen D., Errington J. Cell wall deficiency as a coping strategy for stress. Trends Microbiol. 2019;27:1025–1033. doi: 10.1016/j.tim.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 42.Markova N., Slavchev G., Michailova L., Jourdanova M. Survival of Escherichia coli under lethal heat stress by L-form conversion. Int. J. Biol. Sci. 2010;6:303–315. doi: 10.7150/ijbs.6.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slavchev G., Michailova L., Markova N. Stress-induced L-forms of Mycobacterium bovis: A challenge to survivability. New Microbiol. 2013;36:157–166. [PubMed] [Google Scholar]
- 44.Abrudan M.I., Smakman F., Grimbergen A.J., Westhoff S., Miller E.L., van Wezel G.P., Rozen D.E. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc. Natl. Acad. Sci. USA. 2015;112:11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrand S., Bohni N., Schnee S., Schumpp O., Gindro K., Wolfender J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Plants Pharm. Shelf Recent Trends Leads Find. Bioprod. 2014;32:1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Williams R.B., Henrikson J.C., Hoover A.R., Lee A.E., Cichewicz R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 47.Yang X.-L., Awakawa T., Wakimoto T., Abe I. Induced biosyntheses of a novel butyrophenone and two aromatic polyketides in the plant pathogen Stagonospora nodorum. Nat. Prod. Bioprospect. 2013;3:141–144. doi: 10.1007/s13659-013-0055-2. [DOI] [Google Scholar]
- 48.Murakami T., Burian J., Yanai K., Bibb M.J., Thompson C.J. A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA. 2011;108:16020–16025. doi: 10.1073/pnas.1108124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haginaka K., Asamizu S., Ozaki T., Igarashi Y., Furumai T., Onaka H. Genetic approaches to generate hyper-producing strains of goadsporin: The relationships between productivity and gene duplication in secondary metabolite biosynthesis. Biosci. Biotechnol. Biochem. 2014;78:394–399. doi: 10.1080/09168451.2014.885824. [DOI] [PubMed] [Google Scholar]
- 50.Kawai Y., Mercier R., Mickiewicz K., Serafini A., Sório de Carvalho L.P., Errington J. Crucial role for central carbon metabolism in the bacterial L-form switch and killing by β-lactam antibiotics. Nat. Microbiol. 2019;4:1716–1726. doi: 10.1038/s41564-019-0497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blainey P.C. The future is now: Single-cell genomics of bacteria and archaea. FEMS Microbiol. Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zenobi R. Single-Cell metabolomics: Analytical and biological perspectives. Science. 2013;342:1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 53.Klessen C., Schmidt K.H., Gumpert J., Grosse H.H., Malke H. Complete secretion of activable bovine prochymosin by genetically engineered L forms of Proteus mirabilis. Appl. Environ. Microbiol. 1989;55:1009–1015. doi: 10.1128/AEM.55.4.1009-1015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]