Abstract
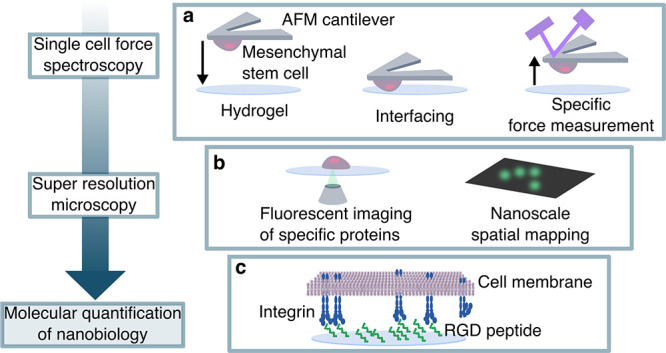
A common approach to tailoring synthetic hydrogels for regenerative medicine applications involves incorporating RGD cell adhesion peptides, yet assessing the cellular response to engineered microenvironments at the nanoscale remains challenging. To date, no study has demonstrated how RGD concentration in hydrogels affects the presentation of individual cell surface receptors. Here we studied the interaction between human mesenchymal stem cells (hMSCs) and RGD-functionalized poly(ethylene glycol) hydrogels, by correlating macro- and nanoscale single-cell interfacial quantification techniques. We quantified RGD unbinding forces on a synthetic hydrogel using single cell atomic force spectroscopy, revealing that short-term binding of hMSCs was sensitive to RGD concentration. We also performed direct stochastic optical reconstruction microscopy (dSTORM) to quantify the molecular interactions between integrin α5β1 and a biomaterial, unexpectedly revealing that increased integrin clustering at the hydrogel-cell interface correlated with fewer available RGD binding sites. Our complementary, quantitative approach uncovered mechanistic insights into specific stem cell-hydrogel interactions, where dSTORM provides nanoscale sensitivity to RGD-dependent differences in cell surface localization of integrin α5β1. Our findings reveal that it is possible to precisely determine how peptide-functionalized hydrogels interact with cells at the molecular scale, thus providing a basis to fine-tune the spatial presentation of bioactive ligands.
Keywords: AFM, single cell force spectroscopy, dSTORM, RGD, PEG hydrogel, integrin α5β1
Synthetic hydrogels are a highly tailorable and advantageous class of biomaterials for many regenerative medicine applications. Because of their high level of hydration and ability for the researcher to specify the desired structure and properties, they have a range of diverse applications, such as targeted delivery of therapeutics, larger scaffolds to support replacement tissue, or model systems to study disease progression or drug tolerance. Because of the relative ease of fabrication and functionalization, synthetic hydrogels can be engineered to promote specific cellular interactions at multiple length scales.1,2 As unfunctionalized synthetic hydrogels are often bioinert, a common approach is to incorporate cell adhesion peptides to mimic the composition of the native extracellular matrix (ECM), facilitating cell adhesion and migration.2,3 However, determining the optimal peptide concentration and presentation of these molecules for a specific application is not trivial,4−6 yet peptide availability has been shown to have significant implications on downstream cell behavior following surface receptor reorganization.7 Although great strides have been made in engineering a broad range of highly sophisticated bioactive hydrogels, our understanding of the nanoscale cellular response of individual receptor interactions to these materials is very limited. In recent years, a variety of exciting techniques have emerged,8,9 enabling the precise investigation of single cell-material interactions at molecular length scales. We hypothesize that leveraging a selection of these techniques in parallel with conventional approaches for studying cellular interaction with biomaterials will elucidate previously unquantified nanoscale observations, thus informing the rational design of synthetic bioactive hydrogels for a variety of applications. To demonstrate the value of such an approach, we investigated how presenting a controlled amount of cell-adhesive peptide within a versatile hydrogel platform affected interfaced human mesenchymal stem cells (hMSCs) from the macro- to nanoscale. This was accomplished by observing the migration speed of individual cells on a 2D hydrogel surface, measuring overall cell-hydrogel adhesions and individual rupture forces of cell-peptide interactions using single cell force spectroscopy (SCFS), and visualizing individual presentation and induced clustering of integrins on the cell membrane interfaced with the hydrogel using direct stochastic optical reconstruction microscopy (dSTORM).
For this study, we used hMSCs, which are highly researched for use in regenerative medicine applications. This includes the direct use of hMSCs against chronic inflammation,10,11 as delivery agents for gene therapies,12 or encapsulated in engineered tissue systems for regeneration of many tissues in the body.13−15 They are broadly investigated due to their low immunogenicity, ability to migrate to sites of injury, and immunomodulatory actions.16 However, hMSC therapies have a low translation to clinic due to several limitations, including low cell retention and cell survival rates. A major hurdle to improving cell engraftment is the lack of understanding of specific molecular interactions between cells and the material substrate with which they are delivered. Understanding the underlying nanomechanical and molecular spatial presentation of cell adhesion receptors is hypothesized to aid design of engineered biomaterials. Integrins are cell-adhesion receptors that cluster at the membrane at points of force generation, such as focal adhesions hence they are critical to cell adhesion and motility. In particular, nanoscale spacing of integrin ligands was shown to affect integrin activation and subsequently focal adhesion dynamics.7,17 Moreover, integrin α5β1 has also been implicated in hMSC adhesion and migration through fibronectin binding18 and is thus considered an important target for improved hMSC engraftment in regenerative medicine applications.
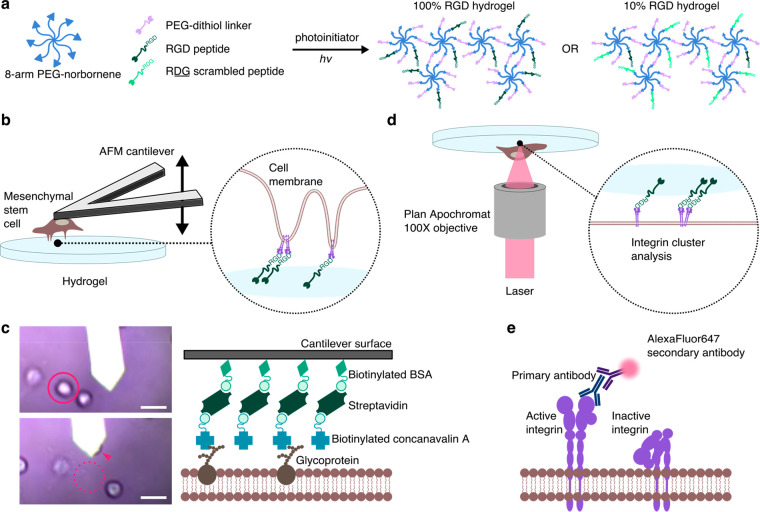
To demonstrate our technological approach, we used RGD peptide-functionalized hydrogels interfaced with hMSCs as a model system. Poly(ethylene glycol) (PEG) hydrogels based on 8-arm, 20 000 Da PEG-norbornene were photo-cross-linked with varying concentrations of a covalently tethered linear cell-adhesive RGD peptide (CGGRGDSP), where the RGD sequence is present in fibronectin and a few other extracellular matrix proteins that bind several cell-surface receptors, notably integrin α5β1.19,20 The integrin-binding RGDSP motif was previously used to facilitate cellular attachment to PEG-based hydrogels,21 and tethered RGD peptides in PEG hydrogels were found to be necessary for encapsulated hMSC survival.6 We chose to use the linear RGDSP peptide due to its higher affinity for binding integrin α5β1, compared to most forms of cyclic RGD.22−24 PEG-based thiol-norbornene “photoclick” hydrogels present a highly versatile and well-controlled platform to probe cell-material interactions.25 The 8-arm PEG macromers were cross-linked into a network with nondegradable PEG-1000 dithiol linker and tethered RGD peptide at a molar ratio of 2:1 linker:peptide in the presence of photoinitiator and UV light (Figure 1a). A concentration of 6.8 mM RGD was used to generate a hydrogel with a high level of RGD peptide availability, referred to here as the 100% RGD, “high” binding hydrogel (Table 1). A 10% RGD hydrogel was also generated by replacing 90% of the RGD peptide with a scrambled version of the peptide, RDG (CGGRDGSP), providing a “low” binding hydrogel. Similarly, a 0% RGD hydrogel was generated using 100% RDG peptide and used as a control where indicated.
Figure 1.
Schematic of stem cell-hydrogel interfacing. (a) 8-arm PEG-norbornene (20 kDa) is cross-linked with nondegradable PEG-dithiol (1000 Da), with tethered 6.8 mM cell-adhesive RGD peptide (CGGRGDSP) or nonadhesive RDG scrambled peptide (CGGRDGSP), in the presence of photoinitiator and 365 nm light to generate a 3D photo-cross-linked hydrogel network. (b) hMSCs bound to an AFM tip are brought into contact with hydrogels of varying RGD concentration, permitting analysis of specific unbinding events by single cell force spectroscopy (SCFS). (c) hMSCs attach to functionalized cantilevers through concanavalin-A—cell membrane glycoprotein interactions. Scale bar = 30 μm. (d) hMSCs adhered to hydrogels are fixed and immunolabeled, and surface localization of the RGD binding integrin α5β1 is imaged using the super-resolution imaging technique direct stochastic optical reconstruction microscopy (dSTORM). (e) Integrins in their active, extended conformation are immunolabeled with a primary antibody and visualized by detecting blinking of secondary antibody-bound AlexaFluor647.
Table 1. RGD Peptide-Functionalized Hydrogel Specificationsa.
| 100% RGD | 10% RGD | 0% RGD | |
|---|---|---|---|
| % RGD peptide | 100 | 10 | 0 |
| % scrambled peptide (RDG) | 0 | 90 | 100 |
| RGD concentration (mM) | 6.8 | 0.68 | 0 |
RGD peptide was added at a concentration of 6.8 mM corresponding to the 100% RGD gel. This was further mixed with RDG (scrambled) peptide to dilute RGD concentration to 10% and 0% RGD, to keep total peptide concentration consistent between hydrogel conditions.
To investigate single cell behavior on hydrogels at the macro scale, live cell migration was monitored using live tracking microscopy, which delivers indispensable insights into the study of cell-material interactions. The migration paths of individual cells were tracked in order to verify the macroscopic behavior of single cells on PEG-RGD hydrogels, and to correlate these observations with more detailed micro- and nanoscale measurements. To approach the nanoscale mechanical and spatial quantification of cell surface receptors in cell-material interfacing, we combined two intrinsically quantitative molecular scale techniques to characterize the cell surface interactions of hMSCs with the RGD peptide-functionalized hydrogels; SCFS and dSTORM.
SCFS is an atomic force microscopy (AFM) technique which provides molecular insight into single cell interactions with materials.26,27 SCFS was previously applied in just a handful of studies to measure unbinding forces between single cells and materials to identify molecular interactions and elucidate initial adhesion mechanisms,9 but to the best of our knowledge has not yet been used on hydrogels with tailored cell-adhesive interactions. It is a valuable tool for characterizing cell-surface interactions with a highly sensitive but broad range of measurable forces (10 pN to 100 nN), and precise spatial and temporal control.28,29 In our experimental setup, single hMSCs were adhered to functionalized AFM cantilevers and brought into contact with the hydrogels (Figure 1b). Upon retraction of the hMSC-bound cantilever, individual unbinding events could be precisely measured. The attachment of the hMSCs to the cantilever was mediated by concanavalin-A-cell membrane glycoprotein interactions (Figure 1c).
dSTORM is a single molecule fluorescent imaging technique, which offers the ability to quantify the presence of specific protein targets at nanoscale spatial resolutions, typically around 20–50 nm. Since its introduction in 2008,30 dSTORM has enabled precise visualization and quantification of many cellular components of varying morphologies, cell surface receptors, receptor clustering, and even receptor conformation.31−34 Typically, dSTORM is carried out on very thin samples, however here we describe an adapted setup enabling the dSTORM imaging on cells interacting with a hydrogel. The availability of the RGD-binding cell surface receptor, integrin α5β1,19,20 was analyzed on the surface of hMSCs in contact with the RGD hydrogels by imaging the inverted hydrogels and focusing the laser at the interfacing membrane (Figure 1d). Antibody receptor labeling was used to detect cell surface integrin α5β1 in their active, extended conformation (Figure 1e). These combined techniques provide mechanistic insights into how hMSCs interact with materials at the nanoscale via reorganization of the cell surface receptors that regulate their attachment and subsequent migration.
Results/Discussion
RGD Availability Dictates hMSC Migration on Peptide-Functionalized Hydrogels
Photo-cross-linked PEG hydrogels provide a very versatile tailorable platform, therefore the specific formulation used for these studies was carefully considered. We chose to employ a relatively stiff hydrogel with a Young’s modulus around 20 kPa (Figure S1a), similar to the stiffness of muscle tissue.35 Hydrogels with modulus in this range previously showed enhanced hMSC attachment to RGD peptide hydrogels, compared to softer gels.5 We additionally confirmed that total peptide content in each hydrogel condition was of similar magnitude (Figure S1b). This previous study5 also found that a minimum of 0.5 mM RGD peptide was necessary for cell attachment, which is similar to our “low” 10% RGD hydrogel with 0.68 mM peptide, whereas much higher levels of RGD presentation, such as 5 mM (similar to our “high” 100% RGD hydrogel with 6.8 mM peptide), promoted a spread morphology and enhanced proliferation for hMSCs.36 We measured similarly high numbers of cells attached to both the 100% and 10% RGD hydrogels (Figure S2), which facilitated further investigation and comparison of the hydrogels via SCFS and dSTORM. No cells remained bound to 0% RGD hydrogels after 24 h and were thus not included in the migration experiments, and this further confirmed the nonbinding nature of the RDG scrambled peptide.
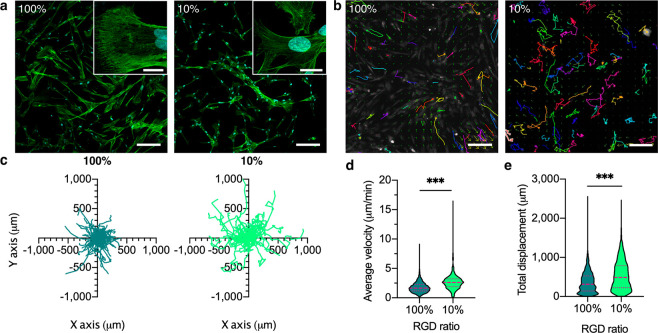
A critical role of hMSCs in vivo is their ability to migrate to sites of injury.37 This migration is affected by hMSC adhesion to its substrate. Observing the hMSCs bound to high and low RGD hydrogels revealed that cellular morphology was markedly affected by the amount of RGD, exhibiting a more spread morphology on 100% RGD, and spindle-shape on 10% RGD (Figure 2a). We next characterized the migration of individual hMSCs to determine if the concentration of the RGD cell-adhesion peptide had an effect on the macro scale behavior of the cells. We carried out live fluorescent imaging over a period of 6 h (Figure 2b) and found that hMSCs on high and low binding hydrogels displayed no directional preference (Figure 2c). Interestingly, the hMSCs on the 10% RGD hydrogels traveled across the hydrogel surface much faster, with a median velocity of 2.6 μm/min, compared to 1.7 μm/min on 100% RGD hydrogels (Figure 2d). These velocities were both higher than hMSCs on tissue culture plastic (TCP) (Figure S3). Furthermore, the hMSCs on 10% RGD hydrogels traveled farther, with a 488 μm median displacement compared to 314 μm on 100% RGD hydrogels (Figure 2e). These experiments demonstrated and confirmed that availability of the RGD peptide influences morphology and migration behavior.
Figure 2.
Migration analysis of hMSCs on RGD hydrogels. (a) Representative confocal images of hMSCs bound to hydrogels, labeled for actin (green) and nuclei (blue). Scale bar = 200 μm main image, 20 μm inset. (b) Representative images of CellTracker Orange-labeled hMSC trajectories on the hydrogels, tracked over 6 h. Scale bar = 200 μm. (c) Migration directionality plots of hMSCs. N = 71 tracks per condition. (d) Average velocity of hMSCs on hydrogels with different RGD concentration. N = 748–1203 tracks from n = 3 hydrogel replicates per condition. (e) Total displacement of hMSCs on hydrogels. N = 748–1353 cells from n = 3 hydrogel replicates per condition. Nonparametric two-tailed t test with Mann–Whitney post hoc test. Violin plots represent median ± IQR. ***p < 0.001.
Mapping hMSC-RGD Unbinding Forces Using Single Cell Force Spectroscopy
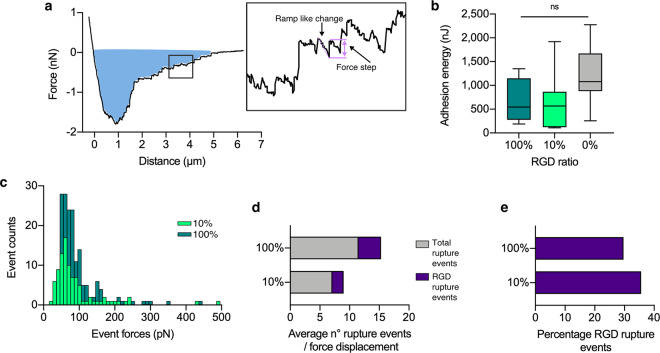
SCFS was utilized to investigate the influence of RGD concentration on adhesion forces between the hMSCs and the hydrogels, thus providing molecular level insight to explain differences in migration velocity. During a SCFS measurement, changes in cantilever deflection (force) can be measured as the cell is pulled away and completely detached from the hydrogel (Figure 3a). Here, we have quantified the nano- and picoscale adhesion between the hMSC and the hydrogel relative to RGD concentration. First, we quantified the adhesion energy required to fully detach the hMSC from the hydrogel, from integrating the area under the retraction curve (annotated blue area in Figure 3a), on the hydrogels with varying RGD concentration (Figure 3b). The mean total adhesion energy of the hMSC on the 0% RGD hydrogel was not significantly different from the 100% and 10% RGD hydrogels (Figure 3b), which would appear contrary to the fact that no cells adhered to these hydrogels in culture. However, SCFS involves physically pushing the cell into the hydrogel surface which results in substantial nonspecific adhesion to the hydrogel polymer network (Figure S4), which we attribute to electrostatic interactions between the cell and hydrogel.38 The mean adhesion energies for 100% and 10% RGD hydrogels are not significantly different, indicating that the availability of RGD does not have a measurable impact on overall adhesion of a single cell. However, looking within each force–distance curve it is possible to deconvolute important adhesion events occurring at the molecular level. Within the force–distance curve for hMSC interaction with the hydrogel, typical rupture events are observed (Figure 3A, inset box), notably a force step (or “jump”) preceded by a ramp-like change in force.28 These total rupture events were quantified (Figure 3c) and a Gaussian fit was applied to the distribution of forces up to 200 pN. There was a mean event force of 67 ± 4 pN (mean ± SD) on the 100% RGD hydrogel, and 60 ± 3 pN on the 10% RGD hydrogel. For the 0% RGD hydrogel there was a greatly reduced number of overall events with a random distribution of the associated forces, therefore no fit could be applied (Figure S5a). Furthermore, when the hMSCs were incubated with soluble RGD peptide, effectively blocking the available surface integrins, the number of rupture events were significantly reduced, indicating that these events were indeed due to cellular interaction with hydrogel-bound RGD (Figure S5b). To further characterize the cause of this specific force rupture event, we performed SCFS on RGD peptide chemisorbed via the thiols of the terminal cysteines on a gold substrate (Au-RGD) compared to a clean gold (Au) substrate to quantify rupture events in the presence of only RGD without the underlying hydrogel (Figure S5c). On the Au-RGD surface the mean force was quantified at 61 ± 2 pN, while on the Au substrate there were a negligible number of events and thus the distribution was unable to be fitted (Figure S5c). Hence, the mean rupture force of 61 ± 2 pN can be attributed to an unbinding event between the hMSC and RGD. To determine if the mean rupture force of 67 ± 4 pN (between the hMSC and the 100% RGD hydrogel) is representative of an individual ligand–receptor unbinding event, the relationship between the rupture force and retraction speed was explored. A shorter contact time of 50 ms was employed to increase the sensitivity of the force measurement to single-molecule interactions,39 with a range of retraction velocities (0.1–20 μm/s). Only rupture events which were a single, discrete force step to the zero-force level were analyzed to ensure the measurement was a single-molecule unbinding event (Figure S6a), and as less than 30% of the force–distance curves yielded this discrete force event, there was an approximately 85% probability that this rupture event was the unbinding of a single-bond rupture.40 Using this single-molecule binding approach, the mean rupture force was quantified as 63 ± 8 pN at the same retraction velocity (1 μm/s) as the longer adhesion measurements (67 ± 4 pN) on the 100% RGD hydrogel. For the Au-RGD surfaces, even with the very short contact time the number of rupture events were too frequent to extract single unbinding events; conversely, for the 10% RGD hydrogel the number of rupture events were too few for meaningful statistical analysis. Furthermore, the mean rupture forces recorded on 100% RGD hydrogels under these single-molecule binding conditions revealed a linear relationship with the logarithmic increasing retraction speed (Figure S6b). According to the Bell/Evans theory of kinetic bond rupture, there is a linear relationship between this rupture force and the effective loading rate, which is the product of the effective spring constant and the retraction velocity.41 Hence, observing this linear relationship indicates that the observed rupture forces are the result of a single bond rupture event. Therefore, it was proposed that a single unbinding event occurred at 63 ± 8 pN specifically between the hMSC and the RGD peptide cross-linked to the PEG hydrogel. This is within the range reported for integrin-ECM unbinding forces which vary between 40 to 140 pN dependent on loading rate,39,42 and comparable to the rupture force for α5β1/FN7-10 complex, reported as 69 pN for a similar loading rate.43
Figure 3.
Single cell force spectroscopy of hMSCs detaching from the hydrogel surface. (a) Representative force–distance retraction curve when the hMSC-functionalized AFM tip is retracted from the surface of 100% RGD hydrogel. Typically, there is a large initial nonspecific adhesion of the hMSC to the hydrogel, followed by smaller force events after the bulk of the hMSC has detached as it moves away from the surface. The integrated area under the curve (blue) represents the total adhesion energy binding the single hMSC to the hydrogel surface. Inset illustrates ramp-like change in force (dotted line) preceding a force step event (between solid lines). (b) Total adhesion energy for hMSCs bound to 100%, 10%, and 0% RGD hydrogels. n = 7–13 per condition. Parametric one-way ANOVA with Tukey multiple comparison test. Box plots represent median ± IQR, whiskers represent minimum and maximum. ns = not significant. (c) Histogram of total registered force events during unbinding of hMSCs from 100% and 10% RGD hydrogels. N = 3 cells measured at n = 5 locations per n = 3 hydrogel replicates per condition. (d) Average total and RGD specific rupture events occurring on 100% and 10% RGD hydrogels, as defined through controlled interaction experiments (see Figure S5). N = 3 cells measured at n = 5 locations per n = 3 hydrogel replicates per condition. (e) Percentage of RGD rupture events per force displacement curve occurring on 100% and 10% RGD hydrogels.
Further analysis of the individual rupture events revealed the sensitivity of SCFS to the availability of RGD in the hydrogels. The average number of total rupture events (NT) detected in a single force–distance curve for the 100% RGD hydrogel (Figure 3d) was almost twice (178%) the NT for the 10% RGD hydrogel. Comparatively, the Au-RGD surface generated a nearly 10-fold increase in NT relative to the 100% RGD hydrogel (Figure S5d), attributed to the higher density of available RGD when chemisorbed directly onto the gold surface.44 To characterize the proportion of RGD-hMSC single unbinding events compared to nonspecific events, the number of rupture events occurring within the range of 63 ± 10 pN (accounting for baseline noise in the AFM) were quantified. The average number of RGD-hMSC unbinding events in a single force–distance curve (NRGD) followed a similar pattern; there were almost twice the number of NRGD on the 100% RGD hydrogel compared to the 10% RGD hydrogel, and again a 10-fold increase in the NRGD on the Au-RGD compared to the 100% RGD hydrogel (Figures 3d and S5d). Curiously, the NT measured for the 0% and 10% RGD hydrogel were similar (Figure 3d and S5d), but the total number of events on the plain gold surface was greatly reduced as previously described (Figure S5c). This indicates a baseline level of nonspecific binding between the hMSC and the PEG hydrogel; as observed in the cell culture experiments, this nonspecific binding was not sufficient to facilitate adhesion of the hMSC to the 0% RGD hydrogel.
Interestingly, the proportion of RGD interaction rupture events on the RGD hydrogels remained very similar; specifically, we found that 30% and 36% of all rupture events on 100% and 10% RGD hydrogels, respectively, occurred at 63 ± 10 pN (Figure 3e), which was comparable to the Au-RGD surface (30%) (Figure S5e). Hence even as the quantity of rupture events (NT) increased with increasing RGD availability, the proportion of these forces identified to be specific hMSC-RGD interactions remained consistent. This suggests that SCFS is sensitive to the saturation of available RGD binding sites on the hMSC, and that even at “low” concentrations the RGD density available is more than sufficient for initial cell adhesion.45 More complex analysis of the possibility of multiple binding events and clustering behavior is difficult to accurately determine using SCFS, hence applying spatial molecular quantification methods such as dSTORM can further elucidate the hMSC-RGD hydrogel interaction.
Characterization of Integrin α5β1 hMSC Surface Clustering Using dSTORM
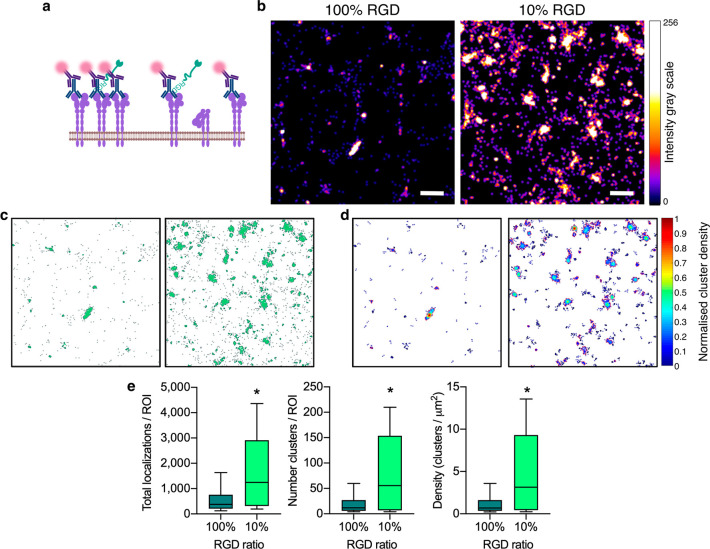
We proceeded to quantify the number and arrangement of α5β1 integrins on the plasma membrane surface of hMSCs adhered to RGD-functionalized hydrogels. Immunolabeling α5β1 integrins in their active, extended conformation enabled assessment of the surface availability of the integrin for ligand binding (Figure 4a). dSTORM image acquisition was carried out via temporally separated blinking events of AlexaFluor647. The individual detections were Gaussian fitted, and super-resolved images were reconstructed. The localization precision of the individual AlexaFluor647 detections was measured to be 16.7 nm (σ) (Figure S7a), while the cluster sizes had a measured diameter of 140–160 nm (based on the full width half-maximum, FWHM, of their fluorescent intensity) (Figure S7b). This is similar to the previously reported localization precision of AlexaFluor647 using dSTORM,46 and the reported size of integrin clusters.47
Figure 4.
dSTORM imaging of integrin α5β1. (a) Schematic representing immunolabeling of active integrin α5β1. (b) Representative dSTORM reconstructed images of integrin α5β1 clusters on the surface of hMSCs bound to 100% and 10% RGD hydrogels. Scale bar = 500 nm. (c) Representative DBSCAN cluster maps of images in panel b. Maps are 4 × 4 μm. (d) Representative cluster density maps of images in panel b. Maps are 4 × 4 μm. (e) Analysis of total number of surface localizations, number of clusters and density of clusters of integrin α5β1 per ROI for hMSCs in contact with 100% and 10% RGD hydrogels. N = 10–15 ROIs from n = 3 hydrogel replicates per condition. Welch’s unequal variances unpaired two-tailed t test. Box plots represent median ± IQR; whiskers represent minimum and maximum. *p < 0.05.
The monoclonal JBS5 clone primary antibody to integrin α5β1 was used as it has been extensively characterized in the literature to bind only the active, extended conformation of the integrin and thus specifically represents the integrins that are available for binding at the cell surface.19,48,49 Further, these studies also tested various RGD peptide sequences, and found that short RGD peptides, which do not include the synergy fibronectin sequence (as used in the present study), permit antibody binding. To ensure that the RGD-integrin interactions did not obscure α5β1 labeling in our setup, a “saturated” labeling control was used where hMSCs were seeded on untreated glass and dSTORM was performed (Figure S8). No difference in the total number of localizations, number of clusters or density of clusters was seen between the hMSCs on glass or on 100% RGD hydrogels, suggesting the RGD peptide indeed does not obscure antibody labeling of the integrin, and thus the differences measured are attributed to cell surface availability of the integrin.
There was a dramatic increase in α5β1 availability at the cell-material interface on the 10% RGD hydrogel compared to the 100% RGD hydrogel (Figure 4b). Using the integrated density-based spatial clustering of applications with noise (DBSCAN) function of the cluster analysis algorithm Clus-DoC,50 the α5β1 clusters were identified and maps of the clusters and their densities were generated within regions of interest (ROI) of 4 × 4 μm (Figure 4c and d). Upon analysis of the clusters, we measured a significant increase in the median number of surface localizations by 232%, number of clusters by 363% and density of clusters by 363%, for 10% hydrogels compared to 100% (Figure 4e). It was previously established that increased cell traction forces are due to local activation of α5β1 integrins,51 and hence, the increased receptor numbers and clustering on 10% RGD hydrogels likely underpins the increased migration speed measured at the macro scale. However, to date, no study has demonstrated the effect of RGD concentration in a hydrogel on integrin α5β1 presentation. We concluded that the availability and clustering of α5β1 on the hMSC membrane corresponds inversely to available RGD binding sites on the hydrogel surface. Hence dSTORM provides molecular sensitivity to differences in cell surface localization of integrin α5β1, driven by the availability of RGD on the surface of the hydrogel.
Conclusions
To explore the correlative relationship between the multiscale interfacing measurements, we indexed the various parameters between 0 and 1 (Figure 5). While this indexing simplifies the complexity of cell-material interactions, it enables the comparison and combination of the various cellular processes underlying cell adhesion and migration across several length scales. Plotting these indexed values side-by-side revealed stark differences between unbinding events and receptor localization that help to explain the macroscopic cell migration observations. On high binding (100% RGD) hydrogels, strong interactions between the hMSC and RGD, illustrated by a higher quantity of RGD-hMSC unbinding events, enabled stable cell adhesion to the substrate and slower migration. On low binding (10% RGD) hydrogels, fewer specific interactions between the hMSC and the hydrogel, illustrated by reduced RGD-hMSC unbinding events, correlated with increased α5β1 surface localizations and clustering, likely as a cellular response to attempt to improve cell adhesion, leading to faster migration.
Figure 5.
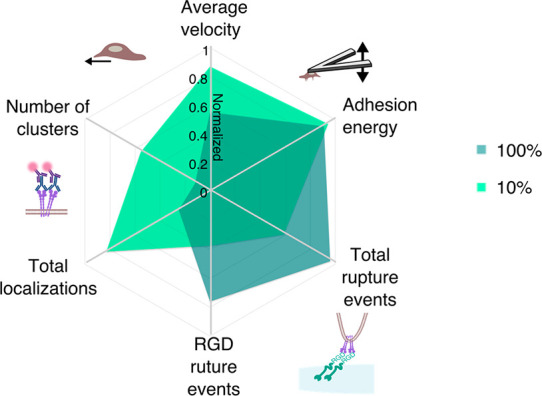
Correlation of multiscale interfacing parameters. 2D Radar plot of median measurements on 100% and 10% RGD hydrogels, with normalized values indexed between 0 and 1 on each axis. Migration velocity was indexed between 0 and 3 μm/min; adhesion energy was indexed between 0 and 600 nJ. Total rupture events were indexed between 0 and 12, and RGD rupture events were indexed between 0 and 5. Total localizations were indexed between 0 and 1500 per ROI, and the number of clusters were indexed between 0 and 100 per ROI.
Taken together our measurements reveal a relationship between the availability of RGD binding sites and the subsequent cell response. Furthermore, although it appears counterintuitive at first, increased RGD availability and proportion of RGD-hMSC unbinding events did not correlate with increased receptor localizations. While we do not rule out that there were likely changes occurring in other RGD-binding receptors that we did not measure, a clear difference in α5β1 surface availability was seen in the low versus high binding conditions, which agrees with a recent study where increased RGD ligand spacing (which correlates with reduced surface concentration) promoted growth of total α5β1-containing focal adhesions on epithelial cells.52 Our data also agree with studies on other motile cell types, such as cancer cells, where increased migration and invasion were associated with increased surface α5β1 protein levels, as measured by Western blots, and thus spatial organization of the receptors could not be appreciated.53,54 Thus, our dSTORM results fully support this increase in measured hMSC migration velocity on low binding RGD hydrogels through increased integrin surface localizations and clustering, suggesting that ligand density partly determines hMSC adhesion stability and migratory behavior. Our mechanistic insights thus provide a key consideration for peptide concentration when designing materials to support cell adhesion and migration. Moreover, our technical approach also highlights the insights that can be provided by such complementary and quantitative techniques to precisely characterize cell-material interactions. Both SCFS and dSTORM are relatively recent techniques that are not yet widely utilized in biomaterial characterization. More traditional methods for analyzing cell-material interactions include measuring bulk material stiffness and surface topography by AFM combined with conventional fluorescence microscopy of cell shape.55 However, cell behavior can now be assessed in greater detail via examining nanoscale interactions with the immediate microenvironment. SCFS and dSTORM are complementary to traditional techniques, such as quartz crystal microbalance with dissipation (QCM-D), where individual unbinding events cannot be measured,56,57 and diffraction-limited fluorescence microscopy where individual receptors cannot be visualized or quantified due to the diffraction limit of 200 nm.18 As integrins direct cell adhesion and migration, the relationship between surface receptor organization and unbinding events, with respect to macroscale migration, could not be previously quantified.7,17 While SCFS and dSTORM have enabled measurable advances in our understanding of cell binding and receptor distribution in isolated cell culture systems, we believe that our demonstration of these techniques used in parallel offers important insights into biomaterial interfacing that could not be quantified using traditional techniques. We successfully demonstrated the capacity to elucidate previously unquantified nanoscale observations within an accessible biomaterials system with two concentrations of available RGD peptide, providing a solid methodological foundation that we hope will be built on in the future. We limited our study to investigating a single peptide-integrin pair; however, this system lends itself to the study of many other receptor–peptide interactions, and therefore, further interesting insights could be gained by studying different types and conformations of adhesive ligands at a variety of concentrations. Additionally, our system was limited to 20 kPa hydrogels, which are generally considered to be stiff. Previous work demonstrated that hMSCs can respond very differently to hydrogel substrates with varied stiffness and presentation of adhesive ligands58−62 therefore, future studies should aim to expand upon our work for tissue specific regimens.
Even more precise exploration of how peptide-functionalized hydrogels interact with cells at the molecular scale appears to be in reach and would establish a basis for a priori determination of peptide amount, as well as its spatial and temporal presentation. This setup is envisaged to be straightforward for use by many others as AFM is often used for material characterization, in addition to recent advances in single molecule interaction studies, specifically the super-resolution technique dSTORM, which can be carried out on a standard widefield microscope with the addition of a powerful laser and careful sample preparation. As these techniques become more widely available to researchers at the intersection of biology and biomaterials, we foresee attaining even more critical insights in the future that will drive discovery forward in interdisciplinary research.
Methods/Experimental Section
Hydrogel Synthesis and Fabrication
Hydrogels were formed from a macromer solution prepared with 10% w/w eight-arm PEG-norbornene (MW 20 000)63 in Dulbecco’s phosphate buffered saline (DPBS; Life Technologies), with 13.7 mM PEG-dithiol cross-linker (MW 1000, Sigma-Aldrich), 6.8 mM peptide (cell-adhesive CGGRGDSP or scrambled CGGRDGSP), and 0.05% w/w Irgacure 2959 (Sigma-Aldrich). These amounts of cross-linker and peptide were used to react with all available norbornenes. Peptides were manually synthesized by Fmoc solid-phase peptide synthesis, followed by HPLC purification (water/acetonitrile gradient with 0.1% v/v TFA) and pure peptide molecular weight verification by MALDI-ToF. Hydrogels were formed by pipetting the macromer solution into disc-shaped silicone molds (6 mm diameter, 0.5 mm height) between glass coverslips and cured 7 min with 365 nm light (5 mW cm–2), where the upper coverslip was coated with nonadhesive Rain-X, and the lower 13 mm diameter glass coverslip was thiol-functionalized using 3-mercaptopropyl trimethoxysilane (Sigma-Aldrich). To thiol-functionalize, coverslips were rinsed 1× with acetone, soaked 10 min in 4% v/v 3-mercaptopropyl trimethoxysilane in acetone, rinsed 3× with acetone, cured 10 min at 80 °C, then cooled and stored at −20 °C until use. Following gelation, the upper coverslip was removed and the hydrogel remained covalently bound to the lower coverslip. The hydrogels were rinsed in PBS-antis (DPBS supplemented with 1% v/v antibiotic/antimycotic (A/A; Invitrogen) and 1% v/v penicillin–streptomycin (P/S; Invitrogen)), transferred to individual wells of a 24-well tissue culture plastic (TCP) plate (Corning), covered with fresh PBS-antis, sterilized by exposing to 365 nm light (5 mW cm–2) for 15 min with the plate cover on, sealed with parafilm, and stored at 4 °C until use.
Fluoraldehyde Peptide Assay
Peptide functionalization to PEG hydrogels was confirmed using fluoraldehyde o-pthaldialdehyde (OPA) reagent solution (Thermo Fisher Scientific). Free-standing 6 mm diameter hydrogels (formed between two Rain-X coated glass coverslips) were equilibrated in PBS-antis and transferred to individual wells in a black 96-well plate. PBS-antis (50 μL) and 50 μL of OPA were added to each well, and fluorescence was measured immediately in scanning mode on a PerkinElmer EnVision plate reader, according to OPA reagent specifications.
Gold-RGD Self-Assembled Monolayer Surface
Gold-coated silicon wafer substrates were treated with O2 plasma (Plasma Prep 5, Gala Instruments) for 6 min and then soaked in ethanol for 10 min. One hundred microliters of 10 mmol RGD in DPBS solution was pipetted onto the substrate and incubated for 30 min; then, it was rinsed 3× with DPBS and used for the SCFS measurements in serum-free Leibovitz’s L-15 CO2 independent media (L-15; Life Technologies).
Cell Culture
Primary human bone marrow-derived mesenchymal stem cells (hMSCs; Lonza) were cultured under standard cell culture conditions (37 °C, humidified atmosphere, 5% CO2). hMSCs were expanded in mesenchymal stem cell basal medium (Lonza), supplemented with mesenchymal stem cell growth medium (Lonza) and 1% v/v A/A. Cells were grown to 80–90% confluency in T175 cell culture flasks (Corning) before use in each experiment.
Live Cell Imaging and Migration Analysis
Autoclaved silicone o-rings (9.19 mm inner diameter, RS PRO) were placed on each hydrogel in a 24-well TCP plate (Corning). hMSCs were labeled with 5 μM CellTracker Orange CMRA (Thermo Fisher Scientific) in MesenPRO RS medium (Thermo Fisher Scientific) for 30 min, washed with DPBS, trypsinized (0.05% v/v Trypsin-EDTA (1×), Life Technologies) and seeded onto hydrogels at 20 000 cells/cm2 or empty TCP wells (10 000 cells/cm2, a lower cell density was used for these controls to avoid confluency) in alpha minimal essential medium Glutamax–1 (αMEM; Life Technologies) supplemented with 10% v/v mesenchymal stem cell grade fetal bovine serum (MSC-FBS; Life Technologies) and 1% v/v P/S, and incubated 24 h to attach. The culture medium was changed to MesenPRO RS supplemented with 20 ng mL–1 bFGF and 10 ng mL–1 TGF-β1 (both recombinant human growth factors, Peprotech), and the plate was transferred to the sample chamber on a Zeiss Axio Observer live imaging microscope and equilibrated 1 h (37 °C, 5% CO2) before beginning measurements. 3× ROIs per well were imaged under bright field and fluorescent illumination (45 HQ TexasRed filter) every 10 min for 6 h using a 10× objective, at 20% stage speed to minimize drift. The migration data was analyzed using Icy (https://icy.bioimageanalysis.org)64 with the Spot Detector65 and Spot Tracking66 plugins. The tracking data was analyzed and exported with the Motion Profiler plugin.
Confocal Microscopy and Cell Number Analysis
Following live imaging, sample wells were washed with DPBS and fixed with 4% v/v paraformaldehyde (PFA; Electron Microcopy Sciences) for 20 min at room temperature. All indicated dilutions were in DPBS. O-rings were removed from gels, and all following immunolabeling steps were performed on an orbital shaker (75 rpm) at room temperature, with DPBS washes between each step. Samples were treated with 0.25% v/v Triton X-100 (Sigma-Aldrich) for 30 min, blocked 30 min with 5% v/v normal goat serum (Sigma-Aldrich), immunolabeled 20 min in 0.1% v/v bovine serum albumin (BSA) supplemented DPBS with 1:500 AlexaFluor 488 Phalloidin (Thermo Fisher Scientific), and 10 min with 1:1000 DAPI. Immunolabeled samples were stored at 4 °C in PBS-antis up to 1 week before imaging. The coverslip-bound gels were inverted on a 35 mm glass bottom μ-dish (Ibidi GmbH) in DPBS and imaged on a Leica SP8 inverted confocal microscope. Images were taken with a 10× objective and 63× oil immersion objective. The DAPI channel was extracted from the confocal microscopy images using FIJI. Images were transformed into 8-bit and an automatic threshold based on intensity gray levels applied to the image. The “analyze particles” function was then utilized to extract the number of nuclei per image.
Atomic Force Microscopy (AFM)
All AFM measurements were performed using a Keysight 5500 AFM.
Indentation for Young’s Modulus
Colloidal cantilevers (sQUBE) with a nominal tip radius of 20 μm and calibrated with a spring constant of 3.7 N/m. For each hydrogel sample, 5 force–indentation measurements were performed at 3 individual locations. The force–indentation measurements were modeled using a spherical Hertz model to quantify the Young’s modulus.
Single-Cell Force Spectroscopy (SCFS)
Probe Functionalization
Tipless cantilevers (Nanoworld PNP-TR) were first O2 plasma treated for 20 min. The cantilevers were then incubated in 0.5 mg/mL biotinylated BSA (Thermo Fisher Scientific) for 12 h at 4 °C. The cantilevers were then rinsed with DPBS; then, they were incubated in 0.5 mg/mL streptavidin (Sigma-Aldrich) for 1 h at room temperature. The cantilevers were rinsed with DPBS, then finally incubated in 0.1 mg/mL biotinylated concanavalin A (Vector Laboratories) for 1 h at room temperature. The cantilevers were rinsed and then stored in DPBS at 4 °C until use or up to 2 weeks.
Single-Cell Attachment and SCFS Measurements
The SCFS was performed with a Keysight temperature-controlled stage to enable local temperature control at 37 °C in a method similarly described in ref (67). The hydrogel samples were placed into a TCP Petri dish (Corning) and incubated in L-15 media 1 h prior to SCFS measurements at 37 °C. The hMSCs were cultured in αMEM supplemented with 10% v/v MSC-FBS and 1% A/A with a density of 1000 cells/cm2 for 24 h prior to detachment. The hMSCs were incubated in 0.5 mM EDTA for 45 min to detach from the culture plate to produce a solution of hMSC in suspension. 50 μL of the hMSC suspension was added to the hydrogel sample in the Petri dish and L-15 media and allowed to settle for 10 min. The functionalized cantilever was then introduced to the media during this 10 min period to allow the cantilever to reach thermal equilibrium. Using the optical system of the AFM, the cantilever was positioned above a single cell and brought into contact with an applied force of 0.5 nN for 30 s. The cantilever was then retracted with the cell attached and was, subsequently, left to adhere for 15 min prior to SCFS measurements.
For the SCFS measurements, the hMSC-functionalized cantilever was positioned over a cell-free region of the hydrogel and force–distance curves performed. In five locations across the hydrogel, five force–distance curves were performed sequentially with 2 min rest period between each measurement and performed with a rate of 1 μ m/s, a 2 nN set-point, and a 30 s dwell period when engaged with the hydrogel surface. For the loading rate measurements, the retraction velocity of the cantilever was varied with a range of 0.1, 1, 5, 10, 15, and 20 μm/s. The loading rate was calculated as the product of the retraction velocity and the calibrated spring constant of the cantilever. The contact time was set to 50 ms, and for each velocity, 10 force curves were performed, except for 0.1 μm/s, which were performed 5 times.
On each hydrogel sample (100%, 10%, and 0% RGD), these two force–distance curve parameters were repeated 3 times with a freshly functionalized hMSC cantilever. The resulting force–distance curves were analyzed using custom MATLAB code to quantify force jumps within the retraction curve; these force jumps are identified as rupture events, which occur when a bond formed between the hMSC integrins and the hydrogel detaches.
AFM Blocking
Detached cells were incubated in a solution of 10 mM RGD in L-15 media for 15 min prior to being attached to the functionalized cantilever, and then, the SCFS protocol described above was performed on a 100% RGD hydrogel.
Direct Stochastic Optical Reconstruction Microscopy (dSTORM)
Preparation of dSTORM Samples
Cells were trypsinized in 0.05% v/v trypsin-EDTA (1×) and seeded at a concentration of 20 000 cells/cm2 in αMEM supplemented with 10% v/v MSC-FBS and 1% v/v A/A onto the hydrogels in 12-well TCP plates (Corning). After 24 h, cells were fixed in 0.3% v/v glutaraldehyde (GA; Electron Microscopy Sciences) in cytoskeleton stabilization buffer (10 mM MES buffer pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2),68 with 0.25% v/v Triton X-100 for 5 min; then, they were fixed in 3% v/v GA in cytoskeleton stabilization buffer for 10 min. The cells were then treated with 0.1% w/v NaBH4 in DPBS for 10 min, rinsed 1× in DPBS, followed by two more washes for 10 min each. Cells were then blocked in 3% w/v BSA in DPBS for 2 h at room temperature, incubated with α5β1 primary antibody (mouse monoclonal, clone JBS5, 1:2000; Millipore) in 3% w/v BSA in DPBS for 1 h 30 min at room temperature, washed three times in DPBS, and then incubated with AlexaFluor647 secondary antibody (goat antimouse IgG, 1:2000; Life Technologies) 1 h 30 min in 3% w/v BSA in DPBS. The cells were washed a further three times in DPBS followed by post fixation with 2% v/v PFA in DPBS for 10 min followed by a further three washes with DPBS. The samples were stored overnight at 4 °C for imaging the following day.
dSTORM Image Acquisition
The cells were imaged in 25% v/v VectaShield (Vector Laboratories) in glycerol (Sigma-Aldrich).69 First, a drop of the 25% v/v VectaShield in glycerol was placed on a 35 mm high μ-dish with a #1.5H glass coverslip (Ibidi-GmbH), and the hydrogel was placed upside-down onto the imaging solution, with a small weight of 8.6 g placed on top, bringing the cells into contact with the glass coverslip. Imaging was carried out on a Zeiss Elyra PS.1 AxioObserver Z1 motorized inverted microscope with an electron-multiplying charge-coupled device (EMCCD) camera (Andor iXon DU 897), an alpha Plan-Apochromat 100x/1.46 NA immersion oil DIC VIS Elyra objective and a 640 nm solid-state laser (150 mW). ZEN black image software v.2012 was utilized to acquire the movies. Images were captured with EPI illumination, in ultrahigh-power mode (PALM_uHP), in 16-Bit depth with a pixel size of 100 nm and an image size of 24.8 μm × 24.8 μm. AlexaFluor647 was excited at 640 nm with an exposure time of 50 ms per frame at 80% laser power with an EMCCD gain of 10%, and the fluorescence emission was acquired over 15 000 frames. As hMSCs are much bigger than the field of view with a 100× objective, images were taken with as much of the cell in as possible.
Cluster Analysis
The single-molecule localization data were analyzed using the versatile, open source software ThunderSTORM plug-in for FIJI,70 which has previously been used to reconstruct dSTORM images of varying cellular structures.32,71−74 Camera parameters were input (pixel size = 100 nm, photoelectrons per A/D count = 8.6, base level = 414, EM gain = 10). Default fitting parameters were used (wavelet-based filter, local maximum detection of single molecules, and integrated two-dimensional Gaussian fitting). Multiemitter fitting was selected for reconstruction to correct for overblinking as each secondary antibody was conjugated with 5 fluorophores. Postprocessing involved drift correction by cross-correlation, followed by filtering for an uncertainty ≤15 nm and frames merged (maximum = 10) to remove localizations blinking continuously across several frames from the same molecule, thus avoiding overcounting. Images were reconstructed as 2D average shifted histograms with a bin size of 20 nm corresponding to a 5× magnification. Cluster analysis was carried out using the Clus-DoC script50 with MATLAB v.2017. The exported data from ThunderSTORM were uploaded into Clus-DoC and 5 regions of interest (ROI) of 4 μm × 4 μm were randomly selected for each image. This was done to ensure no background was analyzed but only the flat surface of the hMSCs. DBSCAN was then used with a minimum number of neighbors (MinPts) of 3 for cluster propagation within a radius (epsilon) of 30 nm, with a cluster defined as having 10 localizations or more.
Quantification and Statistical Analysis
All graphing and statistical analysis were carried out using the software GraphPad Prism v.8. Data were tested for normality of distribution using D’Agostino-Pearson and Kolmogorov–Smirnov tests. Parametric data were analyzed with either an unpaired two-tailed t test or an unpaired one-way ANOVA. Parametric data that displayed unequal variances were analyzed with Welch’s unpaired t test or Brown-Forsythe and Welch unpaired one-way ANOVA with Dunnett’s T3 multiple comparison test. Nonparametric data were analyzed using a two-tailed unpaired t test with Mann–Whitney post hoc or a Kruskal–Wallis ANOVA with Dunn’s multiple comparison test. Data is represented as violin plots, box and whisker plots or dot plots with median ± the interquartile range (IQR). *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant.
Acknowledgments
S.A.M. was supported by a PhD studentship in Biomedicine and Bioengineering in Osteoarthritis, Imperial College London. S.C.S. acknowledges support from Horizon 2020 through the Marie Skłodowska-Curie Individual Fellowship “RADoTE” (701664). I.J.P. acknowledges support from an individual fellowship from the Whitaker International Program, Institute of International Education, United States of America. C.L-R. acknowledges funding from a studentship from an EPSRC doctoral training grant (1975740, EP/R512540/1). T.E.W. and M.M.S. acknowledge support from the Rosetrees Trust. S.C.S and M.M.S. acknowledge support from the UK regenerative medicine platform “Acellular/Smart Materials—3D architecture” (MR/R015651/1). A.G., J.E.S., T.E.W., and M.M.S. acknowledge support from a Wellcome Trust Senior Investigator Award (098411/Z/12/Z). J.E.S. and M.M.S. acknowledge support from the Engineering and Physical Science Research Council (EPSRC) programme grant “Advanced Functional Materials” (EP/M020398/1). A.G. acknowledges support from the European Union’s Horizon 2020 Research and Innovation Programme through the Marie Skłodowska-Curie Individual Fellowship “RAISED” (660757). The authors acknowledge the use of the Facility for Imaging by Light Microscopy (FILM) at Imperial College London partly supported by funding from the BBSRC (BB/L015129/1). The authors thank Sebastian Kuhn (Imperial College London) for synthesizing the RGD peptides. The authors also thank Professor Phil Williams (University of Nottingham) for critical reviewing and insightful feedback on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c07428.
Characterization of RGD peptide-functionalized hydrogel stiffness and total peptide content, analysis of number of cells bound to hydrogels of different RGD concentration, average migration velocity of hMSCs on RGD hydrogels, AFM force curve controls on different substrates, AFM event forces for controls, AFM loading rate, dSTORM localization precision and cluster size, and dSTORM cluster analysis for controls on untreated glass (PDF)
Author Present Address
† A.G.: Applied Chemistry and Environmental Science, School of Science, RMIT University, Melbourne, VIC 3001, Australia.
Author Present Address
‡ J.E.S.: University of Bath, Claverton Down, Bath, BA2 7AY, United Kingdom.
Author Contributions
Δ S.A.M. and A.G. contributed equally. Conception and experimental design were performed by S.A.M., S.C.S., and A.G. Experiments were carried out by S.A.M., S.C.S., C.L-R., and A.G. The migration setup was carried out by J.E.S. and T.E.W. Data analysis was carried out by S.A.M., I.J.P., and A.G. M.M.S. contributed to study design and supervised the project. The manuscript was written by S.A.M., S.C.S., and A.G. with feedback from all the authors.
The authors declare no competing financial interest.
Notes
Raw data is available online at DOI: 10.5281/zenodo.4281240.
Supplementary Material
References
- Lam J.; Segura T. The Modulation of MSC Integrin Expression by RGD Presentation. Biomaterials 2013, 34, 3938–3947. 10.1016/j.biomaterials.2013.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A.; Anseth K. S. Advances in Bioactive Hydrogels to Probe and Direct Cell Fate. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 421–444. 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- Neves S. C.; Pereira R. F.; Araújo M.; Barrias C. C.. Bioengineered Peptide-Functionalized Hydrogels for Tissue Regeneration and Repair. Peptides and Proteins as Biomaterials for Tissue Regeneration and Repair, 1st ed.; Elsevier Ltd.: United Kingdom, 2018; pp 101–125. [Google Scholar]
- Kyburz K. A.; Anseth K. S. Three-Dimensional HMSC Motility within Peptide-Functionalized PEG-Based Hydrogels of Varying Adhesivity and Crosslinking Density. Acta Biomater. 2013, 9, 6381–6392. 10.1016/j.actbio.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahal A. S.; Schweikle M.; Heyward C. A.; Tiainen H. Attachment and Spatial Organisation of Human Mesenchymal Stem Cells on Poly(ethylene Glycol) Hydrogels. J. Mech. Behav. Biomed. Mater. 2018, 84, 46–53. 10.1016/j.jmbbm.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Salinas C. N.; Anseth K. S. The Influence of the RGD Peptide Motif and Its Contextual Presentation in PEG Gels on Human Mesenchymal Stem Cell Viability. J. Tissue Eng. Regener. Med. 2008, 2, 296–304. 10.1002/term.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M.; Cavalcanti-Adam E. A.; Glass R.; Blümmel J.; Eck W.; Kantlehner M.; Kessler H.; Spatz J. P. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. ChemPhysChem 2004, 5, 383–388. 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3, 793–795. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger A. V.; Hutmacher D. W.; Muller D. J. Single-Cell Force Spectroscopy, an Emerging Tool to Quantify Cell Adhesion to Biomaterials. Tissue Eng., Part B 2014, 20, 40–55. 10.1089/ten.teb.2013.0125. [DOI] [PubMed] [Google Scholar]
- Ma S.; Xie N.; Li W.; Yuan B.; Shi Y.; Wang Y. Immunobiology of Mesenchymal Stem Cells. Cell Death Differ. 2014, 21, 216–225. 10.1038/cdd.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q.; Ren H.; Han Z. Mesenchymal Stem Cells: Immunomodulatory Capability and Clinical Potential in Immune Diseases. J. Cell. Immunother. 2016, 2, 3–20. 10.1016/j.jocit.2014.12.001. [DOI] [Google Scholar]
- Mohammadi M.; Jaafari M. R.; Mirzaei H. R.; Mirzaei H. Mesenchymal Stem Cell: A New Horizon in Cancer Gene Therapy. Cancer Gene Ther. 2016, 23, 285–286. 10.1038/cgt.2016.35. [DOI] [PubMed] [Google Scholar]
- Dang Y.; Zhang C.; Zhu Y. Stem Cell Therapies for Age-Related Macular Degeneration: The Past, Present, and Future. Clin. Interventions Aging 2015, 10, 255–264. 10.2147/CIA.S73705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia Maria Ferri A.; Bersano A.; Lisini D.; Boncoraglio G.; Frigerio S.; Parati E. Mesenchymal Stem Cells for Ischemic Stroke: Progress and Possibilities. Curr. Med. Chem. 2016, 23, 1598–1608. 10.2174/0929867323666160222113702. [DOI] [PubMed] [Google Scholar]
- Richardson S. M.; Kalamegam G.; Pushparaj P. N.; Matta C.; Memic A.; Khademhosseini A.; Mobasheri R.; Poletti F. L.; Hoyland J. A.; Mobasheri A. Mesenchymal Stem Cells in Regenerative Medicine: Focus on Articular Cartilage and Intervertebral Disc Regeneration. Methods 2016, 99, 69–80. 10.1016/j.ymeth.2015.09.015. [DOI] [PubMed] [Google Scholar]
- Zachar L.; Bačenková D.; Rosocha J. Activation, Homing, and Role of the Mesenchymal Stem Cells in the Inflammatory Environment. J. Inflammation Res. 2016, 9, 231–240. 10.2147/JIR.S121994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti-Adam E. A.; Volberg T.; Micoulet A.; Kessler H.; Geiger B.; Spatz J. P. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophys. J. 2007, 92 (8), 2964–2974. 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veevers-Lowe J.; Ball S. G.; Shuttleworth A.; Kielty C. M. Mesenchymal Stem Cell Migration Is Regulated by Fibronectin through Alpha5beta1-Integrin-Mediated Activation of PDGFR- and Potentiation of Growth Factor Signals. J. Cell Sci. 2011, 124, 1288–1300. 10.1242/jcs.076935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould A. P.; Askari J. A.; Humphries M. J. Molecular Basis of Ligand Recognition by Integrin Alpha5beta1: I. Specificity of Ligand Binding Is Determined by Amino Acid Sequences in the Second and Third NH2-Terminal Repeats of the Subunit. J. Biol. Chem. 2000, 275, 20324–20336. 10.1074/jbc.M000572200. [DOI] [PubMed] [Google Scholar]
- Takagi J.; Strokovich K.; Springer T. A.; Walz T. Structure of Integrin α5β1 in Complex with Fibronectin. EMBO J. 2003, 22, 4607–4615. 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P.; Lauer-Fields J. L.; Schmoekel H. G.; Metters A. T.; Weber F. E.; Fields G. B.; Hubbell J. A. Synthetic Matrix Metalloproteinase-Sensitive Hydrogels for the Conduction of Tissue Regeneration: Engineering Cell-Invasion Characteristics. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 5413–5418. 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis S. L. Advantages of RGD Peptides for Directing Cell Association with Biomaterials. Biomaterials 2011, 32, 4205–4210. 10.1016/j.biomaterials.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersel U.; Dahmen C.; Kessler H. RGD Modified Polymers: Biomaterials for Stimulated Cell Adhesion and Beyond. Biomaterials 2003, 24, 4385–4415. 10.1016/S0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- Kapp T. G.; Rechenmacher F.; Neubauer S.; Maltsev O. V.; Cavalcanti-Adam E. A.; Zarka R.; Reuning U.; Notni J.; Wester H. J.; Mas-Moruno C.; Spatz J.; Geiger B.; Kessler H. A Comprehensive Evaluation of the Activity and Selectivity Profile of Ligands for RGD-Binding Integrins. Sci. Rep. 2017, 7, 39805. 10.1038/srep39805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-C.; Ki C. S.; Shih H. Thiol-Norbornene Photoclick Hydrogels for Tissue Engineering Applications. J. Appl. Polym. Sci. 2015, 132, 41563. 10.1002/app.41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J.; Heisenberg C. P.; Gaub H. E.; Muller D. J. Single-Cell Force Spectroscopy. J. Cell Sci. 2008, 121, 1785–1791. 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- Puckert C.; Tomaskovic-Crook E.; Gambhir S.; Wallace G. G.; Crook J. M.; Higgins M. J. Molecular Interactions and Forces of Adhesion between Single Human Neural Stem Cells and Gelatin Methacrylate Hydrogels of Varying Stiffness. Acta Biomater. 2020, 106, 156–169. 10.1016/j.actbio.2020.02.023. [DOI] [PubMed] [Google Scholar]
- Friedrichs J.; Legate K. R.; Schubert R.; Bharadwaj M.; Werner C.; Müller D. J.; Benoit M. A Practical Guide to Quantify Cell Adhesion Using Single-Cell Force Spectroscopy. Methods 2013, 60, 169–178. 10.1016/j.ymeth.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Selhuber-Unkel C.; López-García M.; Kessler H.; Spatz J. P. Cooperativity in Adhesion Cluster Formation during Initial Cell Adhesion. Biophys. J. 2008, 95, 5424–5431. 10.1529/biophysj.108.139584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M.; Van De Linde S.; Schüttpelz M.; Kasper R.; Seefeldt B.; Mukherjee A.; Tinnefeld P.; Sauer M. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chem., Int. Ed. 2008, 47, 6172–6176. 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Reinhard B. M. Characterizing Large-Scale Receptor Clustering on the Single Cell Level: A Comparative Plasmon Coupling and Fluorescence Superresolution Microscopy Study. J. Phys. Chem. B 2019, 123, 5494–5505. 10.1021/acs.jpcb.9b05176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäldchen F.; Schlegel J.; Götz R.; Luciano M.; Schnermann M.; Doose S.; Sauer M. Whole-Cell Imaging of Plasma Membrane Receptors by 3D Lattice Light-Sheet DSTORM. Nat. Commun. 2020, 11, 887. 10.1038/s41467-020-14731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddig S.; Aufmkolk S.; Doose S.; Jobin M. L.; Werner C.; Sauer M.; Calebiro D. Super-Resolution Imaging Reveals the Nanoscale Organization of Metabotropic Glutamate Receptors at Presynaptic Active Zones. Sci. Adv. 2020, 6, eaay7193 10.1126/sciadv.aay7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q.; Cai M.; Zhou L.; Xu H.; Shi Y.; Sun J.; Jiang J.; Gao J.; Wang H. Using an RNA Aptamer Probe for Super-Resolution Imaging of Native EGFR. Nanoscale Adv. 2019, 1, 291–298. 10.1039/C8NA00143J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A.; Devarasetty M.; Kang H. W.; Mead I.; Bishop C.; Shupe T.; Lee S. J.; Jackson J.; Yoo J.; Soker S.; Atala A. A Hydrogel Bioink Toolkit for Mimicking Native Tissue Biochemical and Mechanical Properties in Bioprinted Tissue Constructs. Acta Biomater. 2015, 25, 24–34. 10.1016/j.actbio.2015.07.030. [DOI] [PubMed] [Google Scholar]
- Chien H. W.; Fu S. W.; Shih A. Y.; Tsai W. B. Modulation of the Stemness and Osteogenic Differentiation of Human Mesenchymal Stem Cells by Controlling RGD Concentrations of Poly(carboxybetaine) Hydrogel. Biotechnol. J. 2014, 9, 1613–1623. 10.1002/biot.201300433. [DOI] [PubMed] [Google Scholar]
- Kidd S.; Spaeth E.; Dembinski J. L.; Dietrich M.; Watson K.; Klopp A.; Battula V. L.; Weil M.; Andreeff M.; Marini F. C. Direct Evidence of Mesenchymal Stem Cell Tropism for Tumor and Wounding Microenvironments Using in Vivo Bioluminescent Imaging. Stem Cells 2009, 27, 2614–2623. 10.1002/stem.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D. J.; Krieg M.; Alsteens D.; Dufrêne Y. F. New Frontiers in Atomic Force Microscopy: Analyzing Interactions from Single-Molecules to Cells. Curr. Opin. Biotechnol. 2009, 20, 4–13. 10.1016/j.copbio.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Taubenberger A.; Cisneros D. A.; Friedrichs J.; Puech P. H.; Muller D. J.; Franz C. M. Revealing Early Steps of α2β1 Integrin-Mediated Adhesion to Collagen Type I by Using Single-Cell Force Spectroscopy. Mol. Biol. Cell 2007, 18, 1634–1644. 10.1091/mbc.e06-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tees D. F. J.; Waugh R. E.; Hammer D. A. A Microcantilever Device to Assess the Effect of Force on the Lifetime of Selectin-Carbohydrate Bonds. Biophys. J. 2001, 80, 668–682. 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R.; Nassoy P.; Leung A.; Ritchie K.; Evans E. Energy Landscapes of Receptor-Ligand Bonds Explored with Dynamic Force Spectroscopy. Nature 1999, 397, 50–53. 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Strohmeyer N.; Bharadwaj M.; Costell M.; Fässler R.; Müller D. J. Fibronectin-Bound α5β1 Integrins Sense Load and Signal to Reinforce Adhesion in Less than a Second. Nat. Mater. 2017, 16, 1262–1270. 10.1038/nmat5023. [DOI] [PubMed] [Google Scholar]
- Li F.; Redick S. D.; Erickson H. P.; Moy V. T. Force Measurements of the α5β1 Integrin-Fibronectin Interaction. Biophys. J. 2003, 84, 1252–1262. 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Yan C.; Ye K.; He Y.; Li Z.; Ding J. Effect of RGD Nanospacing on Differentiation of Stem Cells. Biomaterials 2013, 34, 2865–2874. 10.1016/j.biomaterials.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Sankaran S.; Jaatinen L.; Brinkmann J.; Zambelli T.; Vörös J.; Jonkheijm P. Cell Adhesion on Dynamic Supramolecular Surfaces Probed by Fluid Force Microscopy-Based Single-Cell Force Spectroscopy. ACS Nano 2017, 11, 3867–3874. 10.1021/acsnano.7b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T.; Tinnefeld P. Photophysics of Fluorescent Probes for Single-Molecule Biophysics and Super-Resolution Imaging. Annu. Rev. Phys. Chem. 2012, 63, 595–617. 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changede R.; Xu X.; Margadant F.; Sheetz M. Nascent Integrin Adhesions Form on All Matrix Rigidities after Integrin Activation. Dev. Cell 2015, 35, 614–621. 10.1016/j.devcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Mould A. P.; Askari J. A.; Aota S. I.; Yamada K. M.; Irie A.; Takada Y.; Mardon H. J.; Humphries M. J. Defining the Topology of Integrin Alpha5beta1-Fibronectin Interactions Using Inhibitory Anti-Alpha5 and Anti-Beta1Monoclonal Antibodies. Evidence That the Synergy Sequence of Fibronectin Is Recognized by the Amino-Terminal Repeats of the Alpha5 Subunit. J. Biol. Chem. 1997, 272, 17283–17292. 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- Mould A. P.; Askari J. A.; Byron A.; Takada Y.; Jowitt T. A.; Humphries M. J. Ligand-Induced Epitope Masking: Dissociation of Integrin α5β1-Fibronectin Complexes Only by Monoclonal Antibodies with an Allosteric Mode of Action. J. Biol. Chem. 2016, 291, 20993–21007. 10.1074/jbc.M116.736942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pageon S. V.; Nicovich P. R.; Mollazade M.; Tabarin T.; Gaus K. Clus-DoC: A Combined Cluster Detection and Colocalization Analysis for Single-Molecule Localization Microscopy Data. Mol. Biol. Cell 2016, 27, 3627–3636. 10.1091/mbc.e16-07-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G. L.; Cohen D. M.; Desai R. A.; Breckenridge M. T.; Gao L.; Humphries M. J.; Chen C. S. Activation of Beta 1 but Not Beta 3 Integrin Increases Cell Traction Forces. FEBS Lett. 2013, 587, 763–769. 10.1016/j.febslet.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria R.; Wiegand T.; Escribano J.; Elosegui-Artola A.; Uriarte J. J.; Moreno-Pulido C.; Platzman I.; Delcanale P.; Albertazzi L.; Navajas D.; Trepat X.; García-Aznar J. M.; Cavalcanti-Adam E. A.; Roca-Cusachs P. Force Loading Explains Spatial Sensing of Ligands by Cells. Nature 2017, 552, 219–224. 10.1038/nature24662. [DOI] [PubMed] [Google Scholar]
- Maschler S. M.; Wirl G.; Spring H.; Bredow D. V.; Sordat I.; Beug H.; Reichmann E. Tumor Cell Invasiveness Correlates with Changes in Integrin Expression and Localization. Oncogene 2005, 24, 2032–2041. 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- Mierke C. T.; Frey B.; Fellner M.; Herrmann M.; Fabry B. Integrin α5β1 Facilitates Cancer Cell Invasion through Enhanced Contractile Forces. J. Cell Sci. 2011, 124, 369–383. 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxboim A.; Rajagopal K.; Brown A. E. X.; Discher D. E. How Deeply Cells Feel: Methods for Thin Gels. J. Phys.: Condens. Matter 2010, 22, 194116. 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin L.; Thakar D.; Goyer C.; Labbé P.; Boturyn D.; Coche-Guérente L. Controlled Surface Density of RGD Ligands for Cell Adhesion: Evidence for Ligand Specificity by Using QCM-D. J. Mater. Chem. B 2015, 3, 5577–5587. 10.1039/C5TB00420A. [DOI] [PubMed] [Google Scholar]
- Iturri J.; García-Fernández L.; Reuning U.; García A. J.; del Campo A.; Salierno M. J. Synchronized Cell Attachment Triggered by Photo-Activatable Adhesive Ligands Allows QCM-Based Detection of Early Integrin Binding. Sci. Rep. 2015, 5, 9533. 10.1038/srep09533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove B. D.; Mui K. L.; Driscoll T. P.; Caliari S. R.; Mehta K. D.; Assoian R. K.; Burdick J. A.; Mauck R. L. N-Cadherin Adhesive Interactions Modulate Matrix Mechanosensing and Fate Commitment of Mesenchymal Stem Cells. Nat. Mater. 2016, 15, 1297–1306. 10.1038/nmat4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N.; Arany P. R.; Mao A. S.; Shvartsman D.; Ali O. A.; Bencherif S. A.; Rivera-Feliciano J.; Mooney D. J. Harnessing Traction-Mediated Manipulation of the Cell/Matrix Interface to Control Stem-Cell Fate. Nat. Mater. 2010, 9, 518–526. 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. Y.; Martin K. E.; García J. R.; Johnson C. T.; Theriault H. S.; Han W. M.; Zhou D. W.; Botchwey E. A.; García A. J. Integrin-Specific Hydrogels Modulate Transplanted Human Bone Marrow-Derived Mesenchymal Stem Cell Survival, Engraftment, and Reparative Activities. Nat. Commun. 2020, 11, 114. 10.1038/s41467-019-14000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Hou Y.; Xie W.; Camacho J. L. C.; Cheng C.; Holle A.; Young J.; Trappmann B.; Zhao W.; Melzig M. F.; Cavalcanti-Adam E. A.; Zhao C.; Spatz J. P.; Wei Q.; Haag R. Ligand Diffusion Enables Force-Independent Cell Adhesion via Activating α5β1 Integrin and Initiating Rac and RhoA Signaling. Adv. Mater. 2020, 32, 2002566. 10.1002/adma.202002566. [DOI] [PubMed] [Google Scholar]
- Tan S. J.; Chang A. C.; Anderson S. M.; Miller C. M.; Prahl L. S.; Odde D. J.; Dunn A. R. Regulation and Dynamics of Force Transmission at Individual Cell-Matrix Adhesion Bonds. Sci. Adv. 2020, 6, eaax0317 10.1126/sciadv.aax0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaalure S. C.; Chu S.; Bryant S. J. An Enzyme-Sensitive PEG Hydrogel Based on Aggrecan Catabolism for Cartilage Tissue Engineering. Adv. Healthcare Mater. 2015, 4, 420–431. 10.1002/adhm.201400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chaumont F.; Dallongeville S.; Chenouard N.; Hervé N.; Pop S.; Provoost T.; Meas-Yedid V.; Pankajakshan P.; Lecomte T.; Le Montagner Y.; Lagache T.; Dufour A.; Olivo-Marin J. C. Icy: An Open Bioimage Informatics Platform for Extended Reproducible Research. Nat. Methods 2012, 9, 690–696. 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- Olivo-Marin J. C. Extraction of Spots in Biological Images Using Multiscale Products. Pattern Recognit. 2002, 35, 1989–1996. 10.1016/S0031-3203(01)00127-3. [DOI] [Google Scholar]
- Chenouard N.; Bloch I.; Olivo-Marin J. C. Multiple Hypothesis Tracking for Cluttered Biological Image Sequences. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2736–2750. 10.1109/TPAMI.2013.97. [DOI] [PubMed] [Google Scholar]
- Friedrichs J.; Helenius J.; Muller D. J. Quantifying Cellular Adhesion to Extracellular Matrix Components by Single-Cell Force Spectroscopy. Nat. Protoc. 2010, 5, 1353–1361. 10.1038/nprot.2010.89. [DOI] [PubMed] [Google Scholar]
- Xu K.; Babcock H. P.; Zhuang X. Dual-Objective STORM Reveals Three-Dimensional Filament Organization in the Actin Cytoskeleton. Nat. Methods 2012, 9, 185–188. 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N.; Keller D.; Rajan V. S.; Gönczy P.; Manley S. Simple Buffers for 3D STORM Microscopy. Biomed. Opt. Express 2013, 4, 885–899. 10.1364/BOE.4.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesný M.; Křížek P.; Borkovec J.; Švindrych Z.; Hagen G. M. ThunderSTORM: A Comprehensive ImageJ Plug-in for PALM and STORM Data Analysis and Super-Resolution Imaging. Bioinformatics 2014, 30, 2389–2390. 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel J.; Peters S.; Doose S.; Schubert-Unkmeir A.; Sauer M. Super-Resolution Microscopy Reveals Local Accumulation of Plasma Membrane Gangliosides at Neisseria Meningitidis Invasion Sites. Front. Cell Dev. Biol. 2019, 7, 194. 10.3389/fcell.2019.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckl L.; Pedram K.; Roy A. R.; Krishnan V.; Gustavsson A. K.; Dorigo O.; Bertozzi C. R.; Moerner W. E. Quantitative Super-Resolution Microscopy of the Mammalian Glycocalyx. Dev. Cell 2019, 50, 57–72. 10.1016/j.devcel.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.; Tong Z.; Ye Q.; Sun T.; Hong Z.; Zhang L.; Bortnick A.; Cho S.; Beuzer P.; Axelrod J.; Hu Q.; Wang M.; Evans S. M.; Murre C.; Lu L. F.; Sun S.; Corbett K. D.; Cang H. Molecular Organization of Mammalian Meiotic Chromosome Axis Revealed by Expansion STORM Microscopy. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 18423–18428. 10.1073/pnas.1902440116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Linde S. Single-Molecule Localization Microscopy Analysis with ImageJ. J. Phys. D: Appl. Phys. 2019, 52, 203002. 10.1088/1361-6463/ab092f. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.