Abstract
Understanding how emerging influenza viruses recognize host cells is critical in evaluating their zoonotic potential, pathogenicity, and transmissibility between humans. The surface of the influenza virus is covered with hemagglutinin (HA) proteins that can form multiple interactions with sialic acid-terminated glycans on the host cell surface. This multivalent binding affects the selectivity of the virus in ways that cannot be predicted from the individual receptor–ligand interactions alone. Here, we show that the intrinsic structural and energetic differences between the interactions of avian- or human-type receptors with influenza HA translate from individual site affinity and orientation through receptor length and density on the surface into virus avidity and specificity. We introduce a method to measure virus avidity using receptor density gradients. We found that influenza viruses attached stably to a surface at receptor densities that correspond to a minimum number of approximately 8 HA–glycan interactions, but more interactions were required if the receptors were short and human-type. Thus, the avidity and specificity of influenza viruses for a host cell depend not on the sialic acid linkage alone but on a combination of linkage and the length and density of receptors on the cell surface. Our findings suggest that threshold receptor densities play a key role in virus tropism, which is a predicting factor for both their virulence and zoonotic potential.
Short abstract
Virus binding to receptor density gradients at cell surface mimics shows that the avidity and selectivity of influenza A virus are governed by receptor structure and density.
Introduction
Cross-species (zoonotic) transmission of viruses such as influenza A and coronaviruses is a continuous threat to humans.1,2 However, the determinants governing the ability of these viruses to switch host species and potentially trigger pandemics remain poorly understood.3 A clearer understanding of the mechanisms underlying virus–host interactions at the molecular level allows for better predictions of the threats of emerging viruses.4,5 These insights can also shed light on other unexplained virus phenotypes, such as restricted host ranges and varying degrees of pathogenicity.
The hemagglutinin (HA) of influenza A virus (IAV) initiates infection by binding to sialic acid-terminated glycan receptors on the host cell glycocalyx. Mutations in the receptor-binding domain (RBD) of HA can lead to changes in both affinity and specificity. These changes are associated with influenza zoonoses,6−8 antigenic drift,9 transmissibility,10,11 and pathogenicity.4,12 Mutations outside the RBD may influence sialoglycan binding indirectly by adjustment of HA thermostability or virus morphology.13,14
While the HA of avian IAV preferentially binds to sialic acids linked via an α2,3-linkage to a penultimate galactose, those adapted to humans favor the α2,6-linkage.6 The impact of RBD mutations on receptor binding is reflected in the orientation of these receptors in their complex with HA.15 With sialic acid in the binding pocket, the remaining glycan residues extend in different topologies based on the α2,3- or α2,6-linkage and interactions between the glycan and the surface of HA.16 In avian-adapted HA subtypes, the α2,3-glycans extend out from the RBD perpendicular to the HA protein. In human-adapted HA subtypes, the α2,6-glycans extend over the HA head domain.17,18
Although the monomeric solution affinity (Kd) for HA–sialoglycan binding is typically in the millimolar range, intact IAV can bind to cells at picomolar virus concentrations.19−22 The binding affinity is dependent on receptor density,19,20 but the exact number of HA–sialoglycan contributions remains unclear. Quantitative assessment of influenza interactions with the host cell glycocalyx requires a method that deconvolutes affinity, specificity, density dependence, and structural aspects of influenza–glycan interactions in a representative environment, as well as a physical–mathematical model that translates the individual molecular determinants of the affinity and contact area into the overall affinity and specificity. Previously, a numerical model was applied to show that multivalent particles that form weak receptor–ligand interactions bind to areas of varying receptor density in a nonlinear fashion, characterized by a threshold receptor density at which binding becomes efficient.23
We show how intrinsic structural differences in binding of HA to α2,3 and α2,6 receptors transfer through the length and density of sialoglycans at the cell surface into virus avidity and specificity. We developed a multivalent affinity platform to probe these individual contributions in an environment that approximates the molecular architecture of the cell membrane. We accomplished this by creating sialoglycan receptor density gradients on supported lipid bilayers to obtain virus threshold receptor densities. We combine information from multiple platforms to study the combined effects of receptor type, density, and length on the virus binding. We developed an analytical model to quantitatively describe binding profiles of IAV to assess multivalent binding at the threshold density. Finally, the molecular mechanism is further explored by molecular dynamics simulations to provide detailed insights into structural aspects of the binding process.
Results and Discussion
Multivalent Affinity Profiling
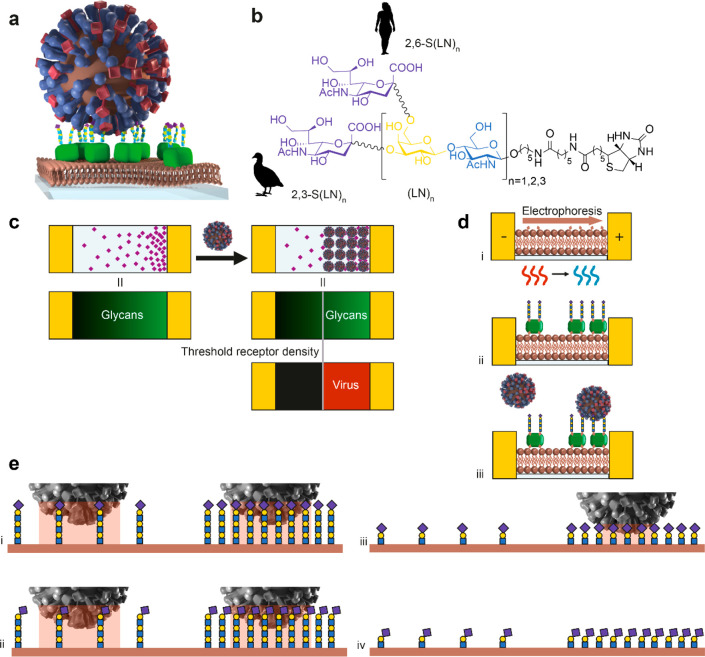
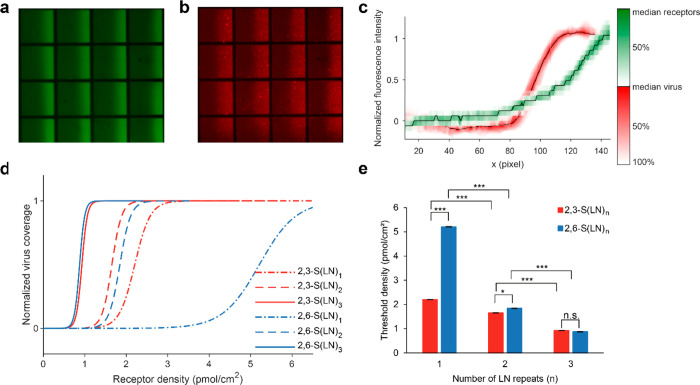
To study receptor density dependence of IAV binding, we developed multivalent affinity profiling (MAP), a method based on the colocalization of fluorescently labeled viruses and receptors on broad-range and continuous surface gradients. We applied this method to study the binding of IAV as a function of receptor density using glycans with different lengths and linkages. To mimic the presentation of glycans on a cell surface, we use biotinylated glycans immobilized by streptavidin (SA) on a supported lipid bilayer (SLB) (Figure 1a). The glycans consist of sialic acid α2,3- or α2,6-linked to one, two, or three N-acetyl lactosamine repeats (2,3- or 2,6-S(LN)n, Figure 1b). They are presented in a density gradient to visualize the threshold receptor density, above which viruses bind with maximum coverage, and with negligible coverage below (Figure 1c).23,24 This threshold receptor density is the key indicator of virus binding specificity in the MAP platform.
Figure 1.
The MAP chip is a platform to quantitate IAV binding to a host cell-mimicking surface. (a) Cartoon showing how glycans immobilized by streptavidin onto supported lipid bilayers (SLBs) were used to mimic the host cell architecture (not to scale). (b) Structure of the biotinylated glycans. 2,3-S(LN)n are avian-type receptors and 2,6-S(LN)n human-type. (LN)2 has no sialic acid and was used to control receptor density on QCM and BLI platforms. (c) Schematic representation of the MAP platform, where viruses only bind above the threshold receptor density for a particular virus–sialoglycan combination. (d) Methodology to create the gradient platform and to study virus binding on receptor gradients. Electrophoretic gradients of biotinylated lipids are formed in SLBs in a microfluidic device at elevated temperature and subsequently cooled down to lock the gradients (i). The biotin gradient is modified with fluorescently labeled streptavidin (SA) and biotinylated sialoglycans (ii). The fluorescently labeled influenza virus is passed over the SLBs with the sialoglycans acting as receptors (iii). (e) Cartoons showing how length and linkage of the sialoglycans can affect virus binding in the case of PR8. The contact area is indicated in peach. If both receptor length and linkage (2,3 or 2,6) are favorable, the virus binds at high and low receptor density (i). If linkage is unfavorable (ii), PR8 may still bind at high and low densities. Other influenza viruses may show a stronger preference for one receptor type and bind only at high densities. If length is unfavorable (iii), the virus binds only at high receptor density. If both are unfavorable, the virus may not bind at all (iv).
The MAP chip gradients are formed in a microfluidic device that we developed for this purpose.25,26 First, we comixed a charged biotinylated lipid and a zwitterionic base lipid (MPPC) to form the SLB and subsequently subjected the SLB to an electrical field at elevated temperature, yielding stable surface–biotin gradients at room temperature (Figure 1d). We then functionalized the gradients of biotinylated lipids with fluorescently labeled SA and S(LN)n and subsequently passed fluorescently labeled IAV over these receptor gradients and mapped the colocalization of virus and receptors to determine the threshold receptor density. This allows us to study the combined effect of the glycan length and 2,3- or 2,6-linkage on the binding of influenza as a function of the receptor density (Figure 1e).
Threshold Density Dependence on Receptor Type and Length
To study the density-dependent binding of IAV on different glycan types, we used UV-inactivated influenza A/Puerto Rico/8/34 (PR8) Mount Sinai strain with fluorescent label. The colocalization of this virus with S(LN)n receptors on green-labeled streptavidin was imaged with fluorescence microscopy (Figure 2a,b). While the receptors formed a gradient with an exponential curve, the fluorescent signal of the virus showed the characteristic sigmoidal profile (Figure 2c). Because the average receptor density of the gradient is known, the sigmoidal binding profile of the virus can be related to the absolute receptor densities of 2,3- and 2,6-S(LN)n (Figure 2d). Virus binding data from multiple pairs of micrographs were combined to afford these binding profiles (Figures S1 and S2).
Figure 2.
Multivalent affinity measurements using different glycan receptors. (a) Fluorescence micrograph of receptor gradients with labeled SA. (b) Fluorescence micrograph of adsorbed labeled virus, binding selectively at high receptor densities. (c) Fluorescence intensity profiles of labeled SA (green) and virus (red) as a function of distance along the horizontal direction. The black lines show the median along the vertical direction. The variance is given by the red and green contours which indicate the percentage of data points between each contour line and the median. (d) Multivalent affinity profiles of influenza PR8 virus as a function of receptor density. The data to which the curves were fitted are shown in Figure S1. (e) Threshold receptor densities for S(LN)n. Error bars show the 95% confidence interval of the fit. n.s., not significant; *, p < 0.05; ***, p < 0.001.
We observe for PR8 that the threshold receptor densities for all glycans lie within the range 0.9–5.2 pmol/cm2. The longest 2,3- and 2,6-S(LN)3 glycan receptors display the lowest binding thresholds, which are the same for both glycans (Figure 2e). The threshold density becomes higher with shorter glycans, but the effect is more profound for 2,6-S(LN)n than for 2,3-S(LN)n. Analyses with quartz crystal microbalance (QCM, Figure S3a) and biolayer interferometry (BLI, Figure S3b) show approximately the same threshold density values, but the binding for 2,6-S(LN)1 was too low to detect using these techniques (Figure S3c).
Similarly, we compared the data with the binding of PR8 on a traditional glycan array (Figure S3d). Linear S(LN)3 and branched glycans of different lengths, each with 2,3-, 2,6-, or without sialic acid, were immobilized on a microarray and incubated with labeled PR8. The relative fluorescence intensity of bound PR8 was measured. As observed for MAP, the microarray data show that PR8 can bind to both 2,3- and 2,6-linkages, and the binding to shorter glycans is lower, in particular for 2,6-linked glycans. Interestingly, PR8 is also more sensitive to the branching of 2,6- than of 2,3-linked glycans.
Theoretical Model to Describe the Multivalent Binding of Influenza Virus
To understand how glycan type and length affect the position of the threshold receptor density, we developed a theoretical binding model, based on the statistical thermodynamics of multivalent adsorption.23,27 In the Supporting Discussion 1 section of the Supporting Information, we derive a theoretical expression for the multivalent equilibrium binding constant Kav of virus particles on a receptor surface and the average number of possible simultaneous virus–glycan receptor interactions Ñ. The resulting expression depends on several fundamental properties of the virus, glycans, and surface receptor chemistry (shown in Figure 3a,b):
| 1 |
| 2 |
Figure 3.
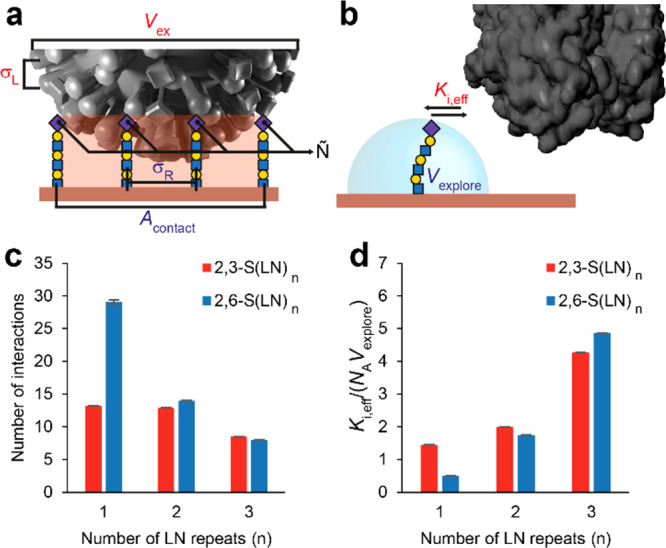
Theoretical model for multivalent binding of influenza. (a) Cartoon of a virus bound to a glycan-coated surface with model parameters indicated. σL is the density of RBDs on the virus. Vex is the excluded volume of a bound virus, which is a function of its diameter. Ñ is the average number of HA–glycan interactions that can form simultaneously. σR is the density of receptors on the surface. Acontact is the receptor surface area that can be reached by RBDs on the virus. (b) zoom-in of a single HA–glycan interaction. Ki,eff is the monovalent receptor–ligand equilibrium of a surface-bound glycan and virus-bound RBD. Vexplore is the volume accessible to a glycan. (c) Values of Ñthreshold for different glycan types and lengths, calculated from the threshold receptor density and maximum length of each glycan. (d) Fitted values of Ki,eff/NAVexplore, the contribution of individual interactions to the avidity.
The following parameters are virus characteristics: the excluded volume Vex of a virus particle, the effective equilibrium constant of an individual HA–glycan interaction Ki,eff (the inverse of Kd), and the RBD density on the virus surface σL. Vexplore, Acontact, and σR are surface characteristics: the receptor density σR, the volume accessible to a glycan Vexplore, and the surface area that can be reached by RBDs when the virus is bound Acontact. The quantity (1/NAVexplore), where NA is Avogadro’s number, is the “effective molarity” of glycan receptors on the surface, from the perspective of a single HA monomer on a bound virus. The effective equilibrium constant Ki,eff can be higher than the equilibrium constant in solution due to mass-transfer effects associated with rebinding to the surface,28 or lower due to additional free energy costs such as from steric repulsion between HA and the surface and any configurational entropy cost for forming the bond.29,30 Although no significant nonspecific binding was observed in a binding study without glycans in QCM-D (Figure S4a), the possible effect of a nonspecific free energy contribution on the virus binding profiles is also discussed in the Supporting Discussion 1 section of the Supporting Information.
We applied the theoretical model to fit the binding profiles of IAV (Figure 2d) and determine the number of interactions needed at the threshold and Ki,eff (Supporting Discussion 1 section in the Supporting Information). Molecular dynamics (MD) simulations were used to determine the minimum and maximum length of each glycan and to derive Acontact and Vexplore (Figure S5). The number of interactions at the threshold Ñthreshold is calculated using eq 2. Because σL is on average 3.8 pmol/cm231 and can locally be as high as 7.7 pmol/cm2 for close-packed HA,32 σR is generally lower than σL at the threshold density (Figure 2e). This applies even to 2,6-S(LN)1 if we assume that viruses that bound at the threshold density used their areas with the highest HA density. Ñthreshold is therefore the product of the threshold receptor density and Acontact (Figure 3c). Ñthreshold is approximately 8 for S(LN)3 and up to 29 for 2,6-S(LN)1 as shorter glycans contribute less to the overall avidity than longer glycans (Figure 3d), thus requiring a higher number of interactions to bind.
Molecular Modeling of HA–Glycan Binding
To understand the influence of glycan length, structure, and presentation on the virus binding, MD simulations were employed to explore the possible shapes S(LN)n glycan–linker–biotin molecules can adopt when bound to the SA tetramer. To take into account the orientational constraints of the HA and SA molecules that arise from their immobilization on the virus and array surfaces, respectively, the angle between the center lines of the HA and SA was determined (Figure 4a). Based on a simple geometric model (Figure 4c) for a spherical virus interacting with glycans projecting from a flat SA surface, the allowable angular range for the glycan relative to the surface depends on the glycan length and the virion radius. The MD data were used to provide estimates of the average glycan length for each type of glycan bound to SA. With these values, the allowable angles were determined (Figure 4b; Supporting Discussion 2 section in the Supporting Information).
Figure 4.
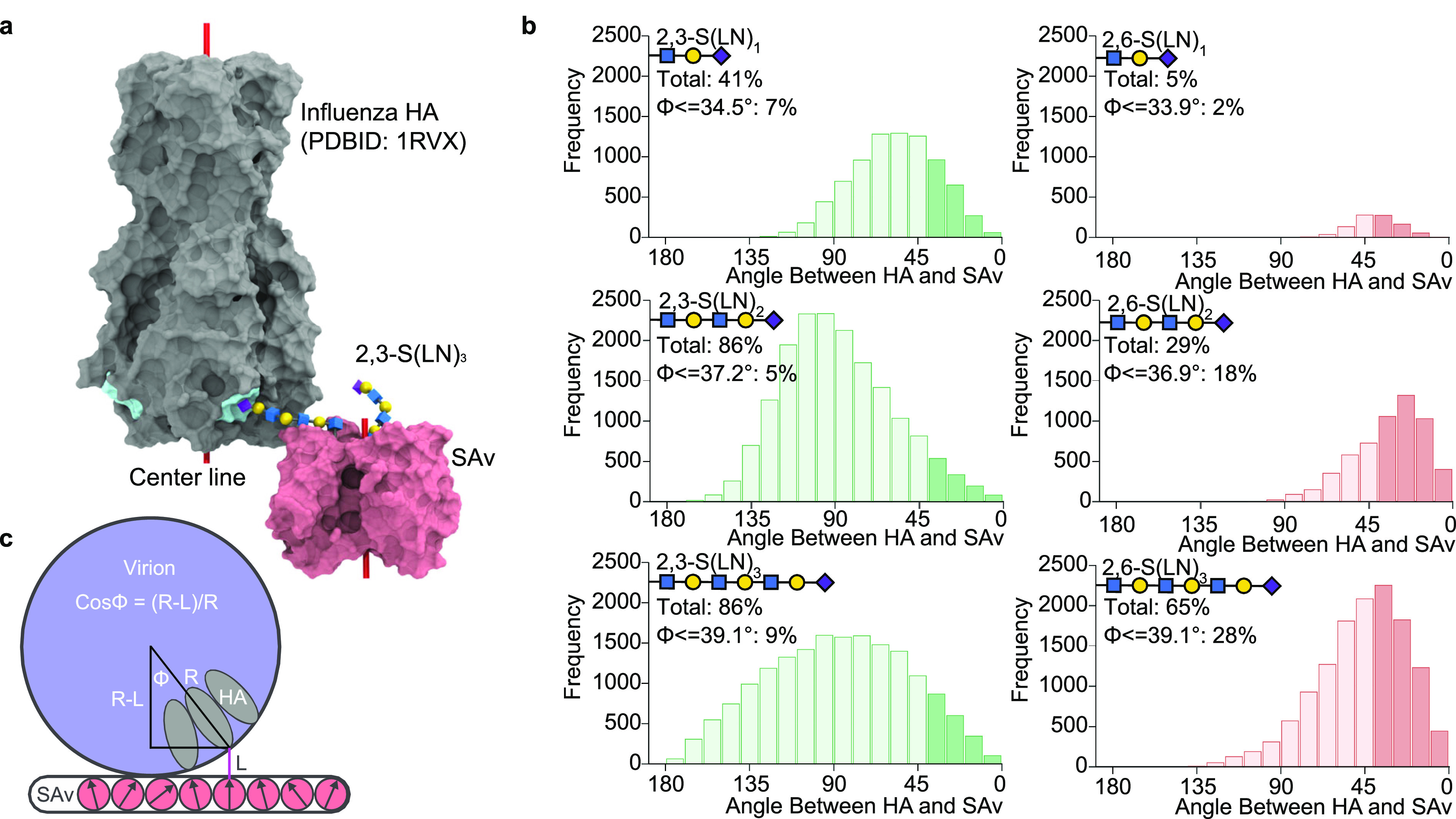
Glycan shapes and HA–SA angles that lead to binding in molecular dynamics. (a) The crystal structure of influenza HA in complex with 2,3-S(LN)1 (gray surface, binding sites in cyan) superimposed onto the biotinylated 2,3-S(LN)3 bound by SA via the penultimate Gal residue of the glycan (shown as 3D-SNFG). This superimposition was repeated for each of the four biotinylated glycans in each glycan system, and for each snapshot taken from the MD simulations. Any structures with atomic overlaps between the HA and SA were removed from consideration. The angle between the two center lines of the HA and SA was measured (180° for the structure shown here). (b) Histogram plots of the frequency of shapes that did not result in atomic overlap between HA and SA as binned every 10° of the angle between the center lines of SA and HA. The total percentage of glycan shapes without atomic overlap and the percentage of shapes accessible if the rotations of HA and SA are restricted are shown under each glycan structure (n = 20 000 for each system). (c) Range of acceptable angles (ϕ) between the HA axis and the normal to the flat surface as a function of virion radius (R) and glycan length (L).
The percentage of shapes for each SA-immobilized glycan type that could allow HA binding were determined (Figure 4b). In general, as the glycan becomes shorter, it displays fewer shapes compatible with HA binding, particularly in the case of 2,6-S(LN)1, which correlates with the observed avidity data (Figure 2e). The decrease in shapes compatible with HA binding corresponds to the decrease in conformational entropy of the glycans but ignores interactions between the HA head and LN repeats.17 For each glycan, the total number of orientations with allowable angles correlates well with the observed dependence on receptor densities (Figure 2); the shorter glycans, especially 2,6-S(LN)1, have fewer acceptable glycan poses and therefore require a higher receptor density to bind the virus.
Because the modes of binding that are accessible are unequally affected by glycan length, we investigated (Supporting Discussion 2 section in the Supporting Information) the conditions under which (A) two glycans on different SAs could bind to different RBDs in the same HA trimer (2 SA–1 HA), (B) two glycans on the same SA could bind to two different HA trimers (1 SA–2 HA), or (C) two glycans on the same SA could bind to the same HA trimer (1 SA–1 HA). Both the 2 SA–1 HA mode and the 1 SA–2 HA mode are more accessible for 2,3-S(LN)n glycans than for 2,6-S(LN)n and are more strongly affected by glycan length for 2,6-S(LN)n. The bidentate 1 SA–1 HA mode was only accessible to 2,6-S(LN)3 and not to shorter or 2,3-linked glycans, which is consistent with the findings of Peng et al.33 and Nemanichvili et al.18 for branched glycans.
Discussion
Glycan structure and presentation have a profound influence on both the monovalent and multivalent surface affinities. The values for Ki,eff that we found for S(LN)3 (Figure S5f) are comparable to the solution equilibrium constants that were measured for X-31, which has HA and NA of Aichi/2/68 (H3N2) and other genes from PR8 and has highly similar binding profiles in BLI.19,34 For S(LN)1, Ki,eff is an order of magnitude lower, which is consistent with the fewer number of shapes of the immobilized S(LN)1 glycan that are able to bind to the HA. This is consistent with the increased entropic cost of forming an S(LN)1–RBD interaction that is suggested by MD (Figure 4b). In addition, the entropy of the multivalent virus–surface interaction is affected by differences in the availability of multivalent binding modes (Supporting Discussion 2 section in the Supporting Information). The 1 SA–2 HA and 2 SA–1 HA modes are both more accessible to 2,3- than to 2,6-S(LN)n. The equally high avidity observed for 2,6-S(LN)3 could therefore be a result of its unique ability to form a bidentate interaction between two SA-bound glycans and a single HA. Because all modes are less available to the shorter 2,6-S(LN)1 and 2,6-S(LN)2, their avidity is lower.
Through the length and density of glycans, their effects on virus binding translate into different multivalent affinities and selectivities. This is especially evident for PR8, which binds readily to both α2,3- and α2,6-linked sialoglycans, but likely applies also to other IAVs possessing clear specificity. The type, length, and density dependence of these complex virus–receptor interactions may explain why some IAVs infect certain cell types, and others do not. For example, duck intestinal mucosa is rich in α2,3-sialosides whereas human upper airway tissue expresses high levels of α2,6-sialosides.35 The IAVs circulating in humans are of avian origin and acquired the ability to use α2,6-linked sialosides. It is becoming increasingly clear that the glycocalyx varies considerably between (host) species,6,36 and between different cell types.37,38 Not only differences in α2,3- or α2,6-linkage of sialoglycans but also their branching, the length of their branches, and the density of sialoglycans determine to which cells IAV binds. Although glycomic studies of human lung tissue have shown the presence of sialosides presented on extended LN moieties, the density of functional receptors is not known, and glycan arrays that present a variety of α2,3- and α2,6-linked sialoglycans of varied length and branching are unable to predict infection of human respiratory tissue.38
The total density of sialic acid in the glycocalyx of cells is typically 10-fold higher than the threshold density on our sensor.39,40 However, as the glycocalyx presents a dense brush that extends approximately 1 μm from the cell surface,41 only a small fraction of these moieties can be involved in the initial binding of a virus so that only the density variations near the outer surface of the glycocalyx may be involved in host cell recognition. That the accessibility of sialoglycans is more important than their number is supported by the observations on human alveolar epithelial cells that IAVs associate more strongly with the mucin MUC1 than with the much larger MUC1642 as well as in synthetic systems where IAVs responded to density variations of sialic acid-modified polymers in a mucuslike brush where each polymer displayed on average 90 sialic acid groups.43
For weaker-binding IAVs than PR8, where the threshold receptor density would be higher than their density of RBDs, the 3D presentation and flexibility of receptors may play a major role in their specificity by forming sufficient interactions only when the glycocalyx shapes itself to the virus. Most human IAVs bind less strongly to α2,3-linked glycans than PR8 but more strongly to α2,6-linked glycans of the same length, whereas the opposite is true for most avian IAVs, and some such as Anhui/1/2013 (H7N9) bind less strongly to glycans of both linkages.19,20,34,43,44 It is therefore likely that variations in both the density and structure of sialoglycans in the glycocalyx play a role in the host cell specificity of IAV. The preference of IAV to bind one cell type over another was further associated with increased airborne transmissibility11 and A/H5N1 virus pathogenicity.37
The MAP chip and supporting model offer a method to quantitatively characterize the multivalent binding of any virus that binds multivalently to cellular glycan receptors. This study shows that IAV needs a specific minimum number of RBD–glycan pairs to form. Not only the structure of the virus but also that of the glycans determines the density at which this number of interactions is achieved. This way, intrinsic structural (and energetic) differences of an α2,3- or α2,6-linked glycan in the RBD are translated into different multivalent binding profiles. MAP may allow studying not only how individual RBD receptor mutations influence the Ki but also how changes in virus morphology affect Kav. We expect that it is through multivalency that such mutations are associated with virus pathogenicity and transmissibility. MAP may therefore be used in concert with quantitative glycomics to further elucidate the connection between the multivalency of viruses and these phenotypes in order to predict the tropism, transmissibility, and pathogenicity of novel zoonotic viruses.
Acknowledgments
We thank Wouter Vijselaar for the cleanroom fabrication of the gradient chips. This study was supported by the Volkswagen Foundation (FlapChips project to E.v.d.V. and J.H.), by The Netherlands Organization for Scientific Research (NWO, TOP 715.015.001 to J.H., and TOP-PUNT 718.015.003 to G.-J.P.H.B.), by the European Commission (Marie Curie Innovative Training Network MULTI-APP, 642793 to J.H.), by the Dutch 4TU High-Tech Materials program (New Horizons in Designer Materials grant to N.B.T.), and by the National Institutes of Health (NIH, grants U01 CA207824 and P41 GM103390 to R.J.W.). R.P.d.V. is a recipient of an ERC Starting Grant from the European Commission (802780) and a Beijerinck Premium of the Royal Dutch Academy of Sciences.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01175.
MATLAB files (ZIP)
Methods; discussion of the derivation and fitting of the theoretical binding model; discussion of the possible modes of divalent HA–glycan complexation; and additional figures including binding profiles, threshold receptor densities, fluorescence intensities, virus contact areas, and structure illustrations (PDF)
Author Present Address
¶ M.C.B.: INFIQC, Departamento de Química Orgánica, Facultad de Ciencias Químicas, Universidad Nacional de Córdoba, Ciudad Universitaria (X5000HUA), Córdoba, Argentina
Author Contributions
E.v.d.V. and J.H. conceived and designed the study. N.J.O. was responsible for the MAP experiments. D.D.I. and M.C.B. performed the QCM studies. M.T. and E.v.d.V. produced and characterized the viruses and performed BLI studies. Z.L., G.V., and G.-J.P.H.B. were responsible for the synthesis of the glycans. R.P.d.V. was responsible for the glycan array studies. O.C.G. and R.J.W. were responsible for the MD simulations. N.B.T. was responsible for the analytical model. P.H.E.H. and N.J.O. were responsible for the image analysis and the fitting of the analytical model to experimental data. N.J.O. wrote the paper with input from all authors.
The authors declare no competing financial interest.
Notes
Data availability: The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
References
- Schaapveld M.; Aleman B. M. P.; van Eggermond A. M.; Janus C. P. M.; Krol A. D. G.; van der Maazen R. W. M.; Roesink J.; Raemaekers J. M. M.; de Boer J. P.; Zijlstra J. M.; et al. Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N. Engl. J. Med. 2009, 360 (25), 2605–2615. 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- Wu F.; Zhao S.; Yu B.; Chen Y.-M.; Wang W.; Song Z.-G.; Hu Y.; Tao Z.-W.; Tian J.-H.; Pei Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579 (7798), 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S.; Fouchier R. Epidemiological and Genetic Investigations of Human-to-Human Transmission of Zoonotic Influenza Viruses. Eurosurveillance 2014, 19 (25), 20840. 10.2807/1560-7917.ES2014.19.25.20840. [DOI] [PubMed] [Google Scholar]
- Maines T. R.; Chen L. M.; Van Hoeven N.; Tumpey T. M.; Blixt O.; Belser J. A.; Gustin K. M.; Pearce M. B.; Pappas C.; Stevens J.; et al. Effect of Receptor Binding Domain Mutations on Receptor Binding and Transmissibility of Avian Influenza H5N1 Viruses. Virology 2011, 413 (1), 139–147. 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Iorio D.; Verheijden M. L.; van der Vries E.; Jonkheijm P.; Huskens J. Weak Multivalent Binding of Influenza Hemagglutinin Nanoparticles at a Sialoglycan-Functionalized Supported Lipid Bilayer. ACS Nano 2019, 13 (3), 3413–3423. 10.1021/acsnano.8b09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M.; Fouchier R. A. M. Role of Receptor Binding Specificity in Influenza A Virus Transmission and Pathogenesis. EMBO J. 2014, 33 (8), 823–841. 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson J. C.; de Vries R. P. H5N1 Receptor Specificity as a Factor in Pandemic Risk. Virus Res. 2013, 178 (1), 99–113. 10.1016/j.virusres.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M.; Tuzikov A.; Bovin N.; Gambaryan A.; Klimov A.; Castrucci M. R.; Donatelli I.; Kawaoka Y. Early Alterations of the Receptor-Binding Properties of H1, H2, and H3 Avian Influenza Virus Hemagglutinins after Their Introduction into Mammals. J. Virol. 2000, 74 (18), 8502–8512. 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel B. F.; Burke D. F.; Bestebroer T. M.; van der Vliet S.; Zondag G. C. M.; Vervaet G.; Skepner E.; Lewis N. S.; Spronken M. I. J.; Russell C. A.; et al. Substitutions Near the Receptor Binding Site Determine Major Antigenic Change During Influenza Virus Evolution. Science 2013, 342, 976–979. 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- Yamada S.; Suzuki Y.; Suzuki T.; Le M. Q.; Nidom C. A.; Sakai-Tagawa Y.; Muramoto Y.; Ito M.; Kiso M.; Horimoto T.; et al. Haemagglutinin Mutations Responsible for the Binding of H5N1 Influenza A Viruses to Human-Type Receptors. Nature 2006, 444 (7117), 378–382. 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- Richard M.; van den Brand J. M. A.; Bestebroer T. M.; Lexmond P.; de Meulder D.; Fouchier R. A. M.; Lowen A. C.; Herfst S. Influenza A Viruses Are Transmitted via the Air from the Nasal Respiratory Epithelium of Ferrets. Nat. Commun. 2020, 11 (1), 766. 10.1038/s41467-020-14626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M. J.; Bristol T.; Proudfoot K. E.; Sally Davis A.; Dunham E. J.; Taubenberger J. K. In Vivo Evaluation of Pathogenicity and Transmissibility of Influenza A(H1N1)Pdm09 Hemagglutinin Receptor Binding Domain 222 Intrahost Variants Isolated from a Single Immunocompromised Patient. Virology 2012, 428 (1), 21–29. 10.1016/j.virol.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C. J.; Hu M.; Okda F. A. Influenza Hemagglutinin Protein Stability, Activation, and Pandemic Risk. Trends Microbiol. 2018, 26 (10), 841–853. 10.1016/j.tim.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahey M. D.; Fletcher D. A. Influenza A Virus Surface Proteins Are Organized to Help Penetrate Host Mucus. eLife 2019, 8, 1–24. 10.7554/eLife.43764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A.; Srinivasan A.; Raman R.; Viswanathan K.; Raguram S.; Tumpey T. M.; Sasisekharan V.; Sasisekharan R. Glycan Topology Determines Human Adaptation of Avian H5N1 Virus Hemagglutinin. Nat. Biotechnol. 2008, 26 (1), 107–113. 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Shi Y.; Lu X.; Shu Y.; Qi J.; Gao G. F. An Airborne Transmissible Avian Influenza H5 Hemagglutinin Seen at the Atomic Level. Science 2013, 340 (6139), 1463–1467. 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- Ji Y.; White Y. J.; Hadden J. A.; Grant O. C.; Woods R. J. New Insights into Influenza A Specificity: An Evolution of Paradigms. Curr. Opin. Struct. Biol. 2017, 44, 219–231. 10.1016/j.sbi.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemanichvili N.; Tomris I.; Turner H. L.; McBride R.; Grant O. C.; van der Woude R.; Aldosari M. H.; Pieters R. J.; Woods R. J.; Paulson J. C.; et al. Fluorescent Trimeric Hemagglutinins Reveal Multivalent Receptor Binding Properties. J. Mol. Biol. 2019, 431 (4), 842–856. 10.1016/j.jmb.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X.; Coombs P. J.; Martin S. R.; Liu J.; Xiao H.; McCauley J. W.; Locher K.; Walker P. A.; Collins P. J.; Kawaoka Y.; et al. Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus. Nature 2013, 497 (7449), 392–396. 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- Vachieri S. G.; Xiong X.; Collins P. J.; Walker P. a; Martin S. R.; Haire L. F.; Zhang Y.; McCauley J. W.; Gamblin S. J.; Skehel J. J. Receptor Binding by H10 Influenza Viruses. Nature 2014, 511 (7510), 475–477. 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- Müller M.; Lauster D.; Wildenauer H. H. K.; Herrmann A.; Block S. Mobility-Based Quantification of Multivalent Virus-Receptor Interactions: New Insights into Influenza A Virus Binding Mode. Nano Lett. 2019, 19 (3), 1875–1882. 10.1021/acs.nanolett.8b04969. [DOI] [PubMed] [Google Scholar]
- Benton D. J.; Martin S. R.; Wharton S. A.; McCauley J. W. Biophysical Measurement of the Balance of Influenza A Hemagglutinin and Neuraminidase Activities. J. Biol. Chem. 2015, 290 (10), 6516–6521. 10.1074/jbc.M114.622308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Veracoechea F. J.; Frenkel D. Designing Super Selectivity in Multivalent Nano-Particle Binding. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (27), 10963–10968. 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacheva G. V.; Curk T.; Auzély-Velty R.; Frenkel D.; Richter R. P. Designing Multivalent Probes for Tunable Superselective Targeting. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (18), 5579–5584. 10.1073/pnas.1500622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weerd J.; Krabbenborg S. O.; Eijkel J.; Karperien M.; Huskens J.; Jonkheijm P. On-Chip Electrophoresis in Supported Lipid Bilayer Membranes Achieved Using Low Potentials. J. Am. Chem. Soc. 2014, 136 (1), 100–103. 10.1021/ja411287u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbenborg S. O.; van Weerd J.; Karperien M.; Jonkheijm P.; Huskens J. Locked-in Biomimetic Surface Gradients That Are Tunable in Size, Density and Functionalization. ChemPhysChem 2014, 15 (16), 3460–3465. 10.1002/cphc.201402509. [DOI] [PubMed] [Google Scholar]
- Curk T.; Bren U.; Dobnikar J. Bonding Interactions between Ligand-Decorated Colloidal Particles. Mol. Phys. 2018, 116 (21–22), 3392–3400. 10.1080/00268976.2018.1503354. [DOI] [Google Scholar]
- Schuck P.; Zhao H.. The Role of Mass Transport Limitation and Surface Heterogeneity in the Biophysical Characterization of Macromolecular Binding Processes by SPR Biosensing. In Life Sciences; Mol N. J., Fischer M. J. E., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, 2010; Vol. 627, pp 15–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Veracoechea F. J.; Leunissen M. E. The Entropic Impact of Tethering, Multivalency and Dynamic Recruitment in Systems with Specific Binding Groups. Soft Matter 2013, 9 (12), 3213. 10.1039/c3sm27766f. [DOI] [Google Scholar]
- Varilly P.; Angioletti-Uberti S.; Mognetti B. M.; Frenkel D. A General Theory of DNA-Mediated and Other Valence-Limited Colloidal Interactions. J. Chem. Phys. 2012, 137 (9), 094108. 10.1063/1.4748100. [DOI] [PubMed] [Google Scholar]
- Vahey M. D.; Fletcher D. A. Low-Fidelity Assembly of Influenza A Virus Promotes Escape from Host. Cell 2019, 176 (1–2), 281–294. 10.1016/j.cell.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.; Cardone G.; Winkler D. C.; Heymann J. B.; Brecher M.; White J. M.; Steven A. C. Influenza Virus Pleiomorphy Characterized by Cryoelectron Tomography. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (50), 19123–19127. 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; de Vries R. P.; Grant O. C.; Thompson A. J.; McBride R.; Tsogtbaatar B.; Lee P. S.; Razi N.; Wilson I. A.; Woods R. J.; et al. Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-Type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 2017, 21 (1), 23–34. 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D. J.; Wharton S. A.; Martin S. R.; McCauley J. W. Role of Neuraminidase in Influenza A(H7N9) Virus Receptor Binding. J. Virol. 2017, 91 (11), 1–10. 10.1128/JVI.02293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J.; Blixt O.; Tumpey T. M.; Taubenberger J. K.; Paulson J. C.; Wilson I. A. Structure and Receptor Specificity of the Hemagglutinin from an H5N1 Influenza Virus. Science 2006, 312 (5772), 404–410. 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Long J. S.; Mistry B.; Haslam S. M.; Barclay W. S. Host and Viral Determinants of Influenza A Virus Species Specificity. Nat. Rev. Microbiol. 2019, 17 (2), 67–81. 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- van Riel D.; Munster V. J.; de Wit E.; Rimmelzwaan G. F.; Fouchier R. A. M.; Osterhaus A. D. M. E.; Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science 2006, 312 (5772), 399–399. 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- Walther T.; Karamanska R.; Chan R. W. Y.; Chan M. C. W.; Jia N.; Air G.; Hopton C.; Wong M. P.; Dell A.; Malik Peiris J. S.; et al. Glycomic Analysis of Human Respiratory Tract Tissues and Correlation with Influenza Virus Infection. PLoS Pathog. 2013, 9 (3), e1003223. 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman G.; Knox R.; Nordt F.; Regan D. Red Cell Agins. I. Surface Charge Density and Sialic Acid Content of Density-Fractionated Human Erythrocytes. Blood 1977, 50 (6), 1001–1011. 10.1182/blood.V50.6.1001.bloodjournal5061001. [DOI] [PubMed] [Google Scholar]
- Mammen M.; Dahmann G.; Whitesides G. M. Effective Inhibitors of Hemagglutination by Influenza Virus Synthesized from Polymers Having Active Ester Groups. Insight into Mechanism of Inhibition. J. Med. Chem. 1995, 38 (21), 4179–4190. 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- Kesimer M.; Ehre C.; Burns K. A.; Davis C. W.; Sheehan J. K.; Pickles R. J. Molecular Organization of the Mucins and Glycocalyx Underlying Mucus Transport over Mucosal Surfaces of the Airways. Mucosal Immunol. 2013, 6 (2), 379–392. 10.1038/mi.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J. L.; Corcilius L.; Tan H.-X.; Payne R. J.; McGuckin M. A.; Brown L. E. The Cell Surface Mucin MUC1 Limits the Severity of Influenza A Virus Infection. Mucosal Immunol. 2017, 10 (6), 1581–1593. 10.1038/mi.2017.16. [DOI] [PubMed] [Google Scholar]
- Huang M. L.; Cohen M.; Fisher C. J.; Schooley R. T.; Gagneux P.; Godula K. Determination of Receptor Specificities for Whole Influenza Viruses Using Multivalent Glycan Arrays. Chem. Commun. 2015, 51 (25), 5326–5329. 10.1039/C4CC08613A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen A. J. G.; Spaan T.; Low H. Z.; Di Iorio D.; van den Brand J.; Tieke M.; Barendrecht A.; Rohn K.; van Amerongen G.; Stittelaar K.; et al. Influenza-Induced Thrombocytopenia Is Dependent on the Subtype and Sialoglycan Receptor and Increases with Virus Pathogenicity. Blood Adv. 2020, 4 (13), 2967–2978. 10.1182/bloodadvances.2020001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.