Abstract
The fates of RNA species in a cell are controlled by ribonucleases, which degrade them by exploiting the universal structural 2′-OH group. This phenomenon plays a key role in numerous transformative technologies, for example, RNA interference and CRISPR/Cas13-based RNA editing systems. These approaches, however, are genetic or oligomer-based and so have inherent limitations. This has led to interest in the development of small molecules capable of degrading nucleic acids in a targeted manner. Here we describe click-degraders, small molecules that can be covalently attached to RNA species through click-chemistry and can degrade them, that are akin to ribonucleases. By using these molecules, we have developed the meCLICK-Seq (methylation CLICK-degradation Sequencing) a method to identify RNA modification substrates with high resolution at intronic and intergenic regions. The method hijacks RNA methyltransferase activity to introduce an alkyne, instead of a methyl, moiety on RNA. Subsequent copper(I)-catalyzed azide–alkyne cycloaddition reaction with the click-degrader leads to RNA cleavage and degradation exploiting a mechanism used by endogenous ribonucleases. Focusing on N6-methyladenosine (m6A), meCLICK-Seq identifies methylated transcripts, determines RNA methylase specificity, and reliably maps modification sites in intronic and intergenic regions. Importantly, we show that METTL16 deposits m6A to intronic polyadenylation (IPA) sites, which suggests a potential role for METTL16 in IPA and, in turn, splicing. Unlike other methods, the readout of meCLICK-Seq is depletion, not enrichment, of modified RNA species, which allows a comprehensive and dynamic study of RNA modifications throughout the transcriptome, including regions of low abundance. The click-degraders are highly modular and so may be exploited to study any RNA modification and design new technologies that rely on RNA degradation.
Short abstract
RNA methylation controls many biological processes. We describe a small molecule-based platform to hijack RNA methylation by guided degradation for high resolution profiling of modified substrates.
Targeted cutting and degradation of RNA plays key roles in a wide array of cellular mechanisms, such as RNA turnover,1,2 processing,3 and innate immune responses.4 These mechanisms are harnessed in technologies, such as RNA interference5 and CRISPR/Cas13 systems.6 However, these platforms are genetically encoded or oligomer-based, which limits their applicability. Small molecules that target and degrade RNA circumvent many of these limitations and expand the scope for RNA degradation approaches.7,8 So far, their availability is limited and their platforms lack the modularity of genetically encoded approaches. Thus, a strategy that retains the modularity and scope of small-molecule targeting of RNA is highly desirable.
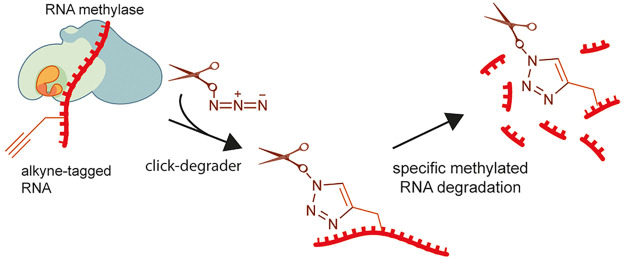
We have come up with a concept of click-degraders, small molecules that can be covalently attached to any RNA species by click chemistry, degrading them in a similar way to ribonucleases. To demonstrate the power of this concept, we have developed meCLICK-Seq (methylation–CLICK-degradation sequencing), a small-molecule-based transcriptome-editing platform that hijacks endogenous RNA methylation pathways9 to induce specific cleavage and degradation of methylated RNAs (Figure 1a), and used it to carry out a transcriptome-wide sequencing of methylated RNA writer substrates to determine the specificity of m6A writer enzymes. What makes meCLICK-Seq unique among RNA modification sequencing methods is that its readout is depletion, not enrichment, of methylated RNA species, which allows reliable high-throughput sequencing in a wider range of RNA species than before. meCLICK-Seq has numerous other advantages over other available RNA sequencing methods. Unlike antibody-based methods,2,10−12 meCLICK-Seq does not require large quantities of RNA (so is ideal for large parallel studies and for use with rare cellular populations), does not rely on availability of an antibody against a particular modification and is catalytically dependent, and so captures a dynamic picture of RNA modifications at a given time. Unlike antibody-free RNA methylation mapping methods,13−15 meCLICK-Seq does not involve enzymatic or any other kind of RNA processing besides the regular sequencing pipelines. RNA degradation is achieved by direct treatment of cultured cells with small molecules; this avoids enzymatic biases. Importantly, the meCLICK-Seq platform is highly modular, and the click-degrader can be used with different probes to map different RNA modifications, for example, acetylation or isopentenylation, and with suitable cellular models, the approach described here can be used to study any type of RNA methylation.
Figure 1.
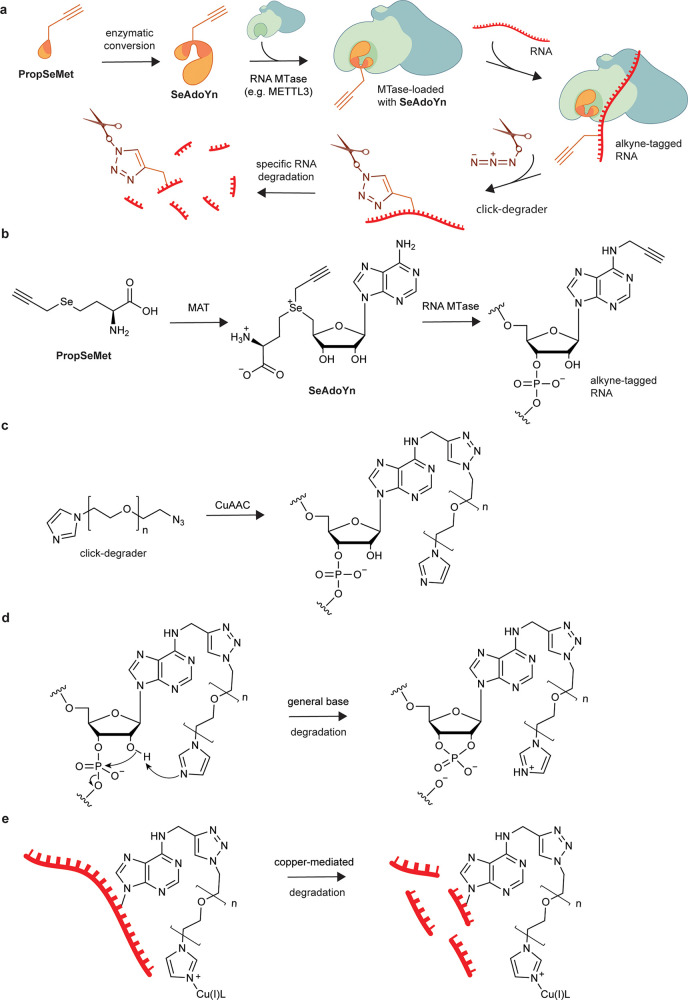
meCLICK-Seq, a small molecule-based methylated RNA editing platform. (a) Proposed mechanism of action of meCLICK-Seq. (b) Conversion of PropSeMet into SeAdoYn and subsequent introduction of propargyl groups into RNA. (c) Functionalization of propargylated RNA with the click-degrader. (d) Proposed mechanism of the general base RNA degradation. (e) Copper-mediated RNA degradation.
Despite the complex mechanism, meCLICK-Seq involves just two cell culture treatment steps. In the first step, cells are incubated with methionine surrogate PropSeMet (PSM) after 30 min of methionine starvation. Cells take up the methionine surrogate, and native methionine adenosyltransferases transform it into SeAdoYn, an S-adenosyl methionine (SAM) surrogate. RNA methyltransferases (MTases), including METTL3 and METTL16, use SeAdoYn as a cofactor, instead of SAM, which leads to the addition of propargyl, instead of methyl, groups onto RNA (Figure 1b).16 A Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) reaction is then carried out directly on cultured cells to tag RNA with a click-degrader, which acts as a functional artificial RNA modification that catalyzes the cleavage of RNA and leads to its degradation (Figure 1c). This in situ step prevents further RNA processing and minimizes treatment biases. The imidazole-based click-degrader has a dual mechanism. It acts as a general base that abstracts a proton from the 2′-O position on RNA that leads to cleavage, similarly to ribonucleases and other artificial nuclease mimics (Figure 1d).17−19 It also cleaves RNA in a copper-dependent manner, which mimics several nucleic acid-cleaving natural products (Figure 1e20). As a result, this cleavage leads to decreased levels of methylated substrates, which can be directly quantified to provide information about the methylation status of any RNA transcript in real time.
Results and Discussion
Click-Degraders Function through a Dual Chemical Mechanism
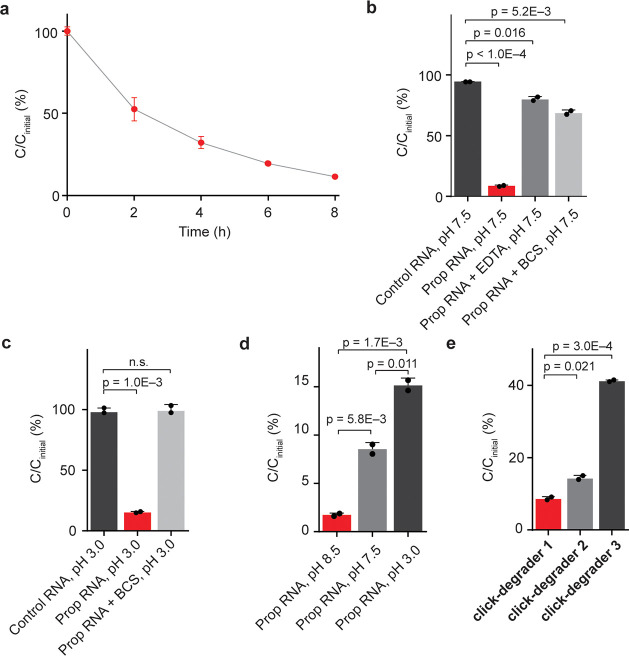
To demonstrate the validity of our RNA degradation strategy and to probe the chemical mechanism of click-degraders, we have carried out in vitro experiments on a synthetic RNA 11-mer functionalized with a propargyl moiety on one of its 6A positions. An unmodified 11-mer was used as a control. We went on to attach the click-degrader onto alkynylated RNA so as to observe what effect it has on RNA stability. Under optimal conditions, CuAAC RNA functionalization was complete in approximately 10 min (Figure S1a–c). Furthermore, we found that upon incubation at 37 °C, the functionalized RNA gets gradually degraded (Figure 2a). Although the kinetic profile of RNA degradation may be complex, first-order half-lives are often used as an estimate of RNA stability.21 By using a first-order approximation, we have calculated the functionalized RNA 11-mer to have a half-life of 2.6 h (Figure S1d). Additionally, we carried out the click-degrader functionalization and degradation on two longer oligomers (Figure S2a–c). In both cases, longer oligomers got degraded more extensively than the 11-mer (Figure S1e). This might be because longer oligomers have more sites susceptible to degradation. We have also compared the effects of click-chemistry components (copper sulfate, THPTA, sodium ascorbate, and the click-degrader) on alkynylated and nonalkynylated control oligomer. Only 9% of the alkynylated click-degrader-functionalized RNA remained after 14 h of incubation, whereas almost no degradation was observed when an identical click-degrader treatment was applied to a control RNA oligomer that lacks the propargyl handle and the ability to be click-functionalized (Figure 2b). To obtain further evidence that the click-components only affect the stability of functionalized RNA, we have carried out the click-functionalization and degradation on a mixture of control and propargylated 11-mers (both at 100 μM). Again, extensive degradation was observed exclusively for the click-degrader functionalized oligomer (Figure S1f,g). These results demonstrate that only the covalently imidazole-functionalized RNA is degraded, so the observed effect is specific to modified RNA and not the result of nonspecific degradation, for example from the click-degrader, copper, and ascorbate.
Figure 2.
Study of the chemical mechanism of click-degraders. (a) Time-dependent degradation of click-degrader 1 functionalized RNA 11-mer at 37 °C. n = 2. (b) Extent of RNA degradation after 14 h at 37 °C, pH 7.5, n = 2. (c) Extent of RNA degradation after 14 h at 37 °C, pH 3.0, n = 2. (d) Extent of RNA degradation in neutral and acidic conditions, n = 2. (e) Extent of RNA degradation after 14 h at 37 °C, pH 7.5, with PEG linkers of differing lengths. Cinitial corresponds to initial concentration; C corresponds to concentration at a specified time point. Click-degraders 1, 2, and 3 have linkers with 6, 4, and 2 PEG subunits, respectively, n = 2. Error bars represent SD.
To gain insight into the mechanism of click-degraders, we carried out in vitro degradation reactions under conditions that suppress certain mechanisms. To investigate whether copper plays a role, we ran reactions quenched with either the copper chelator bathocuproinedisulfonic acid (BCS) or the general metal chelator ethylenediaminetetraacetic acid (EDTA). In the presence of these chelators, 68% and 79% of RNA remained intact, respectively (Figure 2b), which suggests that copper plays a key role in the degradation process. To investigate the role of our click-degrader as a general base, we carried out the reaction under acidic conditions (pH 3), in which imidazole on the click-degrader is protonated and nonbasic but intrinsic RNA stability is comparable to neutral conditions,22 as well as under basic conditions (pH 8.5), in which the imidazole is nearly completely deprotonated. In the case of pH 3.0, 15% of functionalized RNA was retained, which is almost twice as much as under pH 7.5, whereas the nonpropargylated oligomer was not significantly affected; at pH 8.5 only 2% of functionalized RNA remained intact (Figures 2c,d and S1h). Similar trends were observed when the functionalized 11-mer was degraded at pH 3.0, 7.5, and 8.5 for 2 h (Figure S1i). Importantly, no significant RNA degradation was observed in the presence of BCS at pH 3 when both copper and general base mechanisms are blocked (Figure 2c). These findings show that both copper-dependent and general base mechanisms are relevant and sufficient to explain the chemical mechanism of our click-degrader. We were also interested whether other transition metals as well as noncoordinated copper in tandem with the click-degrader are able to facilitate degradation. To look into this, we used the minimum amount of copper–THPTA complex (100 μM) for the CuAAC reaction, followed by quenching of copper with BCS (200 μM) and the addition of other transition metals (1 mM of corresponding chloride). Interestingly, we found that Zn(II) facilitates the degradation but Fe(II) does not (Figure S1j). This result suggests that in the cellular environment, cofactors other than copper can facilitate the degradation of functionalized RNAs. In addition, we also investigated how the length of the click-degrader linker affects the efficiency of degradation. We tested three different linker lengths [6, 4, and 2 poly(ethylene glycol) (PEG) subunits] and found that the longest linker led to the most efficient degradation, possibly because it is more flexible and provides a better reach for imidazole (Figure 2e). It was difficult to isolate and test click-degraders with even longer linkers, so we used click-degrader 1 (6 PEG subunits) for downstream experiments.
To gain further insight into the mechanism of the click-degrader, we have carried out liquid chromatography–mass spectrometry (MS) and RNA gel analyses on the degradation products of the click-degrader functionalized 11-mer. The MS analysis was carried out on LCMS traces of degraded RNA products of the functionalized 11-mer. In all the degraded samples many peaks corresponding to different masses were observed, suggesting that the degradation results in the formation of many different RNA products (Figure S3a–c, Supporting Table S1). An RNA degradation reaction quenched with BCS also revealed formation of numerous RNA products, however there was a different pattern of observed peaks, suggesting that the click-degrader can cleave RNA on many different sites through the general base mechanism (Figure S3d, Supporting Table S2). The RNA gels were in agreement with LCMS results: formation of a smear was observed upon prolonged degradation, further suggesting formation of multiple RNA species (Figure S3e,f). Altogether, these results have implications for detection of 2′-O-methylated RNA species (e.g., m6Am). As the degradation can take place on many different positions, the 2′-O-Me should not protect RNA from degradation (with potential exceptions where several such modifications are nested together); thus meCLICK-Seq should be adaptable for detection of modifications such as m6Am.
Elucidation of mRNA Substrates of m6A Writers
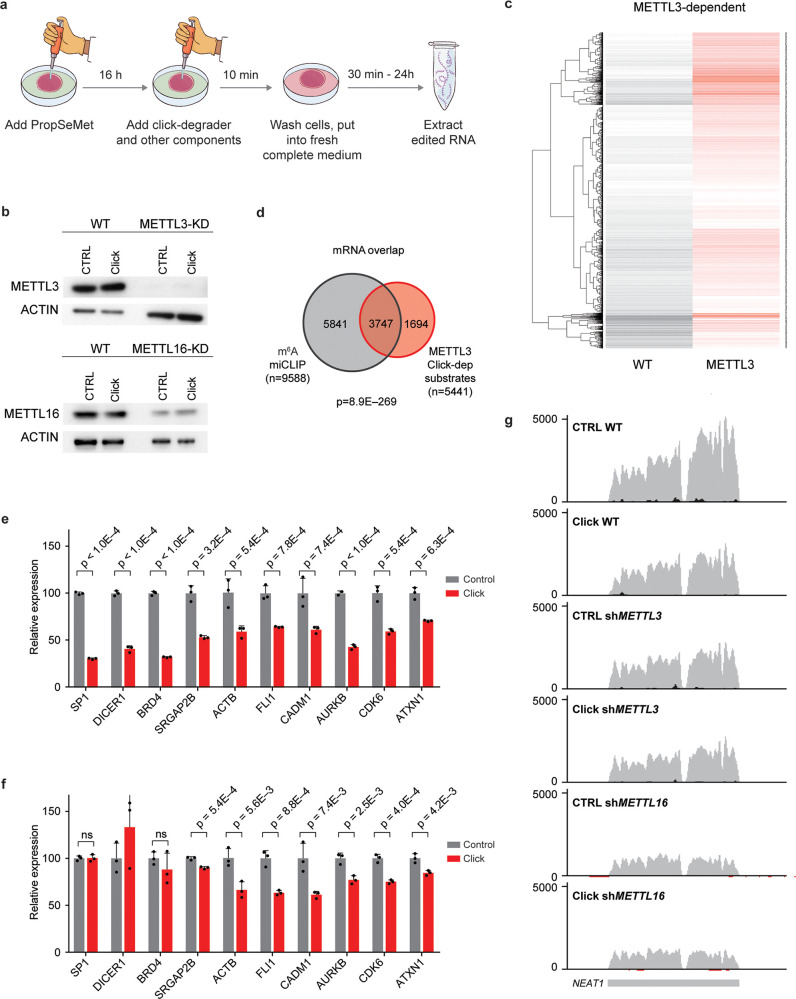
Upregulation of RNA methylation was shown to play a pivotal role in the maintenance of cancers, including acute myeloid leukemia (AML) cells.23,24 We thus went on to investigate whether PropSeMet treatment results in formation of propargyl groups on a human acute myeloid leukemia cell line, MOLM13, as well as a kidney cancer cell line, HEK293T, focusing on the propargyl version (Pr6A) of the RNA modification m6A, the most abundant type of mRNA methylation. Indeed, having analyzed via mass spectrometry the RNA of these cells after PropSeMet treatment, we observed formation of the propargylated adenosine on polyA-enriched RNA (Figure S4a,b). Having demonstrated that PropSeMet is a probe capable of inducing RNA propargylation, we went on to apply meCLICK-Seq to the human AML cell line MOLM-13 (Figure 3a). We used isogenic MOLM-13 cells stably transduced with conditional shRNAs against METTL3 or METTL16, both known to be m6A writers,25,26 or scrambled shRNA as control (Figure 3b).27 As our platform is catalysis-dependent, the loss or down-regulation of an RNA methyltransferase is predicted to eliminate degradation of its RNA substrates. Initially, we observed that the abundance of many mRNAs was reduced in clicked but not control (PropSeMet-fed but not clicked) cells, which suggests successful intracellular degradation of methylated transcripts (Figure 3c and Figure S5a). Strikingly, levels of many of these transcripts were restored in cells with either METTL3 or METT16 downregulation, which suggests that methylation of the relevant RNAs was mediated by the catalytic activity of the corresponding MTase. In particular, we identified 5441 METTL3- and 7656 METTL16-dependent mRNA substrates (Supporting Table S3). We then cross-compared our findings to results of an m6A individual-nucleotide resolution cross-linking and immunoprecipitation (miCLIP)12 sequencing experiment carried out on MOLM-13 cells (Supporting Table S4).28 We observed a consistent overlap for both enzymes with 69% of METTL3 and 67% of METTL16 mRNA substrates identified through meCLICK-Seq reported to contain m6A sites by miCLIP (Figure 3d and Figure S5b). Furthermore, we found that the m6A methylation of 5159 mRNAs depends on both MTases and that the majority of the identified methylated substrates are dependent on METTL16, which is in line with the fact that METTL16 is the modulator of cellular cofactor SAM and its levels (Figure S5c).25,29 It is thus likely that many of the overlapping substrates are methylated predominantly by METTL3, whose catalytic activity in part depends on METTL16. In a similar fashion, METTL16- but not METTL3-dependent m6A-containing substrates are most likely directly methylated by METTL16. To check whether these results can be observed by independent methods, we used RT-qPCR to validate a panel of m6A-containing transcripts (Figure 3e,f and Figure S5d). Notably, this validation mirrored the results of the RNA-seq highlighting the specificity of our platform. Furthermore, to show that the click components, on their own, have little effect on the individual transcript levels, we carried out qPCR on a panel of m6A substrates using RNA extracted from starved MOLM13 cells treated with the same click conditions as PropSeMet-fed cells. We observed no significant differences in substrate abundances between treated and nontreated cells, showing that the observed effects result from the click-degrader functionalization and not the individual click components (Figure S5e). Thus, we have demonstrated that meCLICK-Seq can specifically identify the mRNA substrates of RNA MTases and the results agree closely with the results of antibody-based approaches.
Figure 3.
meCLICK-Seq elucidates the relationship between m6A writers and methylation of mRNAs and lncRNAs. (a) meCLICK-Seq workflow. (b) Western blots demonstrate the extent of METTL3 and METTL16 depletion in conditional knock-down MOLM-13 cells treated with PropSeMet. Application of click-degrader does not significantly alter the levels of MTases. (c) Heat map showing decrease of methylated mRNA levels upon clicking and rescue of METTL3-dependent transcripts upon METTL3 depletion. (d) Overlap of m6A-containing mRNAs determined through m6A miCLIP and METTL3 mRNA substrates determined by meCLICK-Seq; significance indicated by Fisher’s exact test. (e) RT-qPCR-based meCLICK-Seq validation of a panel of genes, n = 3. (f) RT-qPCR-based meCLICK-Seq validation of a panel of genes in METTL3 depleted cells, n = 3. (g) Genome browser snapshot of NEAT1. Applying click machinery diminishes the WT but not METTL3 or METTL16 levels. p values determined with one-tailed t test. ns = not significant (p ≥ 0.05). Error bars represent SD.
Many lncRNAs Are METTL3 or METTL16 Substrates
Another group of RNAs reported to be heavily methylated are long noncoding RNAs (lncRNAs). We therefore investigated whether our meCLICK-Seq platform could identify methylation changes on this RNA subgroup focusing again on METTL3- and METTL16-dependent m6A events.30 For example, NEAT1 was shown to be a target of m6A demethylase ALKBH5, although the MTases that deposit the modifications were not identified.31 Through meCLICK-Seq, we demonstrate that the methylation of NEAT1 depends on both METTL3 and METTL16 (Figure 3g). In total, we identified 689 METTL3-dependent and 889 METTL16-dependent lncRNAs, out of which 77 and 104, respectively, significantly overlapped with lncRNA substrates shown to contain m6A sites by miCLIP sequencing (Figure S5f,g and Supporting Table S5). Interestingly, we observed that the overlap between meCLICK-Seq and m6A miCLIP, albeit significant, is much smaller for lncRNAs than for mRNAs. Furthermore, meCLICK-Seq was able to identify a greater number of lncRNA substrates than m6A miCLIP, the opposite to what was observed for mRNAs, which suggests that our platform efficiently probes lncRNA methylation without antibody biases. Similar to mRNAs, the majority of lncRNAs are promiscuous m6A substrates because methylation of 562 lncRNAs (56% of total identified) appeared to be dependent on both METTL3 and METTL16 (Figure S5h). We were also able to validate a number of these findings by RT-qPCR (Figure S5i). Furthermore, to investigate whether meCLICK-Seq is able to degrade and therefore identify RNA species that contain methylation on known secondary structures, we validated using qPCR the lncRNA MALAT1 with primers designed to target the m6A-containing loop (Figure S6a).32 We observed extensive degradation, showing that our method could indeed target methylation on secondary structures. Overall, using meCLICK-Seq, we have thus revealed m6A methylation on lncRNAs in a MTase-dependent fashion.
To provide additional evidence that PropSeMet treatment does indeed result in formation of propargylated adenosine (Pr6A) on natively m6A-containing species and to show that CuAAC click reaction can effectively take place on these sites, we carried out a biotin–streptavidin pull-down experiment. Briefly, we treated MOLM13 cells with PropSeMet and reacted the propargylated RNA with biotin-azide probe, followed by streptavidin pull-down and qPCR analysis of a panel of known m6A-containing species. As expected, these RNA species were successfully enriched after full PropSeMet–biotin treatment but not in any of the controls that omit one of the components, indicating that these RNA species were indeed labeled with PropSeMet as well as providing an additional method to validate sufficiently abundant methylated RNA species (Figure S6b). Altogether, these experiments provide further evidence that PropSeMet treatment indeed results in propargylation of mRNA.
meCLICK-Seq Reveals That m6A Is Widespread in Intronic and Intergenic Regions
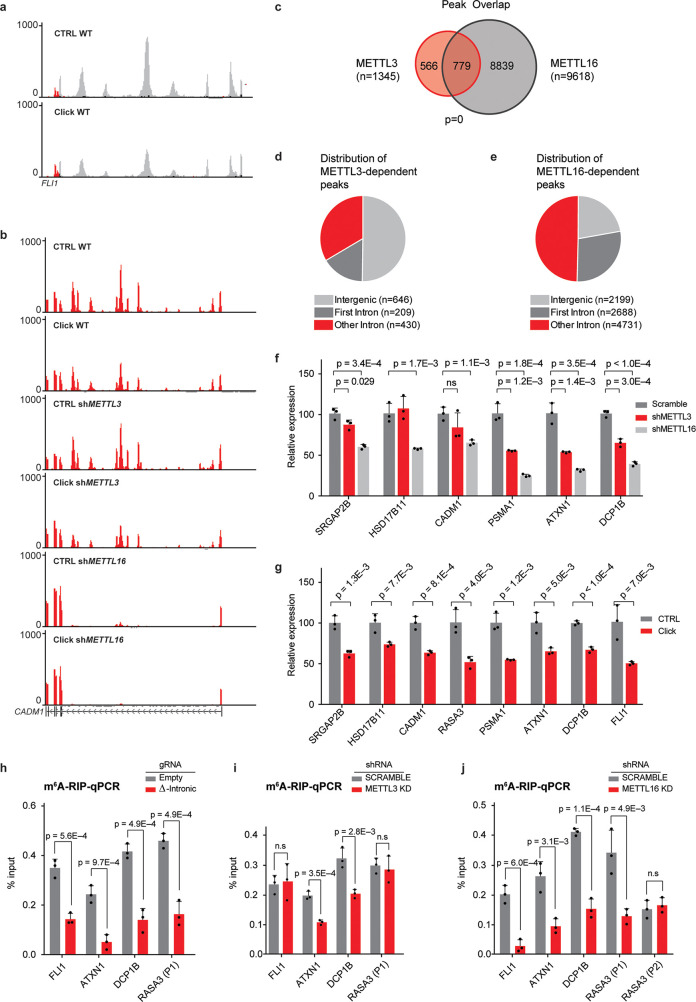
Rather unexpectedly, further interrogation of our RNAseq data with the meCLICK-Seq platform revealed high-resolution peaks in intronic and intergenic regions. The majority of these peaks were highly sensitive to functionalization with the click-degrader, which indicates that these regions were indeed modified by MTases (Figure 4a). These peaks might be a result of the artificial propargyl modification interfering with RNA recycling pathways and perhaps leading to increased stability. Another interesting observation was the pronounced loss of abundance of many identified peaks in cells depleted of either METTL3 or METTL16, which implies that these MTases regulate methylation at the corresponding peaks (Figure 4b and Figure S7a–d). In total, we have found 1345 METTL3- and 9618 METTL16-dependent intronic and intergenic peaks that lost abundance upon clicking and hence had the modification (Figure 4c) with the distribution of these peaks appearing different for the two MTases (Supporting Table S6). In line with recent literature,23,33,34 50% of METTL3-dependent peaks were found in intergenic regions (relative to 23% for METTL16) further highlighting the involvement of METTL3 in chromatin associated or cotranscriptional pathways (Figure 4d,e). Moreover, more than 77% of METTL16-dependent peaks were located in intronic regions with 36% of those found in the first intron, unexpected for a 3′-biased method. This highlights the different roles these MTases play in mRNA modification, with METTL16 regulating the modification in many more intronic regions than METTL3. This is in line with previous reports about the cotranscriptional role of METTL3 and METTL16 in depositing m6A in introns.25,35 meCLICK-Seq is thus the first method to capture widespread methylation in low-abundance transcriptomic regions at such high resolution and low RNA input. Unlike antibody-based intronic analysis, meCLICK-Seq does not require enrichment of nascent RNA to observe intronic methylation.
Figure 4.
meCLICK-Seq reveals widespread m6A mark in introns and intergenic regions. (a) Intronic peaks in the first intron of FLI1. (b) Intronic peaks in the first intron of CADM1 in three isogenic cell models. Intronic peaks are abolished specifically in METTL16-KD cells. (c) Overlap between METTL3- and METTL16-dependent intronic peaks. (d) Distribution of METTL3-dependent peaks in intronic and intergenic regions. (e) Distribution of METTL16-dependent peaks in intronic and intergenic regions. (f) RT-qPCR-based validation of a panel intronic peaks, n = 3. (g) Validation of dependence of intronic peaks on RNA methylases METTL3 and METTL16, n = 3. (h) Results of m6A-RIP in cells with methylated introns removed by dual gRNA system, n = 3. (i) Results of m6A-RIP in cells with depleted METTL3, n = 3. (j) Results of m6A-RIP in cells with depleted METTL16. RASA3 peak 2 is not affected by the knock-down illustrating that the observed effect is enzyme-specific, n = 3. ns = not significant (p ≥ 0.05). Error bars represent SD.
To further demonstrate that the peaks we found contain bona fide m6A sites, we performed motif analysis. We find that variations of the DRACH and TACAG motifs, the respective consensus sequences for METTL3 and METTL16, are dramatically overrepresented in METTL3- and METTL16-dependent peaks, respectively (Figure S8a,b).29,36,37 We also compared the overlap between these peaks and m6A sites found in introns and exons by m6A miCLIP. A significant overlap was observed for both METTL3- and METTL16-dependent peaks, which provides further evidence that the observed peaks have m6A sites (Figure S8c,d). Furthermore, a centered distribution of distances between miCLIP sites and meCLICK-Seq peaks was observed (Figure S8e,f). We also validated some of these peaks by RT-qPCR. The results echoed the findings of RNA-seq (Figure 4f). Furthermore, we observed a decrease in the intensity of meCLICK-Seq peaks in the click group, identical to the results obtained by RNA-Seq (Figure 4g). To validate the intronic results with additional, independent methods, we derived cell lines with deletions in intronic regions that correspond to METTL3- or METTL16-dependent meCLICK-Seq peaks. This was done by using CRISPR-Cas9, with dual gRNAs targeting the flanks of selected meCLICK-Seq peaks. Using m6A-RIP-qPCR, we found that cells with introns removed via CRISPR had significantly less m6A compared to cells containing empty CRISPR-Cas9 vectors (Figure 4h). Additionally, in MOLM-13 cells with either METTL3 or METTL16 knock-down, the m6A-RIP signal was selectively decreased at peaks dependent on the relevant RNA MTase, further validating the results of meCLICK-Seq (Figure 4i,j). Combined, this shows that the meCLICK-Seq peaks contain m6A sites and hold information about MTase specificity.
meCLICK-Seq Can Determine Substrates for Other RNA Methylations
Through further interrogation of meCLICK-Seq data sets we demonstrated that the method can be used for study of different types of methylation. For this aim, we compared our findings with a published data set for 7-methyl guanosine (m7G), obtained through m7G-Seq.38 We found a significant overlap between modified mRNAs determined by the two methods, with 378 overlapping genes (Figure S9a and Supporting Table S7). Strikingly, 82 click-sensitive intronic and intergenic peaks were found to be in vicinity of m7G-Seq-determined sites, a number significantly higher than expected from a random distribution (Figure S9b,c and Supporting Table S8). Thus, meCLICK-Seq is able to probe various types of RNA methylation.
METTL16 Is Linked to Intronic Polyadenylation Sites
To better understand the presence of intronic peaks in our analysis, we decided to investigate the connection to intronic polyadenylation (IPA) given the fact that we used polyA-enriched RNA for RNA-seq analysis. IPA has been recently reported to be widespread in cancers and to inactivate certain tumor suppressors in leukemia.39,40 To determine whether IPA is associated with the presence of m6A, we compared the positions of meCLICK-Seq peaks to IPA sites of leukemia cells identified in a published study by 3′-Seq.39 For additional confidence, we applied very strict parameters and only considered meCLICK-Seq peaks positioned on the exact IPA site. Strikingly, significant overlap was found between METTL16- but not METTL3-dependent peaks and the IPA sites (Figure S10a,b and Supporting Table S4). Moreover, the distribution of METTL16 peaks around the IPA sites suggested a link between the two, with a large number of the peaks overlapping or being close to the IPA sites (Figure S10c). These data suggest that METTL16 has a possible role in IPA and, in turn, splicing, and also demonstrate that meCLICK-Seq can be used to interrogate methylation of low-abundance transcripts.
Conclusions
We have designed click-degraders, small molecules that are clicked onto RNA to induce its degradation, and by using them we developed meCLICK-Seq, a powerful small-molecule-based method for the study of diverse aspects of cellular RNA methylation, including high-confidence mapping of methylation in low abundance transcripts transcribed from introns and intergenic regions. Unlike many RNA methylation sequencing methods, meCLICK-Seq is unbiased and depends strictly on the catalytic activity of RNA MTases, such that it can determine their transcript and locus specificity. The method offers the reproducibility characteristics of small-molecule-based methods, is easy to perform, and has low RNA input requirements, which make it ideal for parallel characterization of different cell types, rare purified populations, and tissues. meCLICK-Seq is perhaps the first non-microscopic method to apply click chemistry directly on cells with a quantifiable output. Antibody-based methods provide a biased picture of methylation at any given time, whereas meCLICK-Seq reports a dynamic state of methylation in a cell. Unlike most antibody-free RNA methylation mapping methods, meCLICK-Seq does not involve complex postextraction RNA modification or processing. Furthermore, this is one of the first studies that exploit functional artificial RNA modifications for a measurable biological output.
We used meCLICK-Seq to define the transcript substrates of m6A writers METTL3 and METTL16 and their overlap. We have also demonstrated that m6A is widespread in lncRNAs and intronic and intergenic regions, where it is deposited primarily by METTL16. Furthermore, we demonstrate for the first time the prevalence of m6A modification in polyadenylated introns, which have been shown to be oncogenic contributions in cancer.39,40
meCLICK-Seq is highly modular, and by use of suitable models and metabolic labeling it can be adapted to study of other RNA methyltransferases and demethylases as well as other RNA modifications. With a modified workflow this method could also be adapted to investigate and characterize DNA modifications. The majority of studies of RNA methylation focus on high-abundance RNA species, whereas the role of methylation in low-abundance species is largely uncharacterized owing to a lack of molecular tools. We foresee meCLICK-Seq enabling the study of RNA modifications in previously inaccessible regions of the transcriptome. Defects in intron-related mechanisms are implicated in a wide range of pathologies,41 and much remains to be learned about the role of noncoding RNAs in disease.42 We expect that our click-degradation platform will facilitate both fundamental and translational discoveries about the role of RNA modifications throughout the transcriptome and, in parallel, pave the way for the development of new technologies aimed to expand the capabilities of targeted nucleic acid degradation.
Experimental Section
In Vitro Click-Degrader Functionalization Reactions
In a standard reaction, CuSO4 (final concentration 1.0 mM), tris(3-hydroxypropyltriazolylmethyl)amine (THPTA, 3.0 mM), click-degrader (2.0 mM), and the RNA oligomer (200 μM) were added to pH 7.5 or pH 3 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 20 mM) or pH 8.5 TRIS (100 mM) buffer supplemented with 10 mM MgCl2 and 100 mM KCl. CuAAC was initiated by adding sodium ascorbate (NaAsc, 50 mM). The reaction mixture was then incubated at 37 °C. Reactions to generate calibration curves were quenched with EDTA (12 mM) 20 min after adding NaAsc to allow complete functionalization of the RNA oligomer. Reactions involving prequenching were quenched with EDTA (12 mM) or bathocuproinedisulfonic acid (BCS, 3 mM) 20 min after adding NaAsc. After quenching, the reaction mixtures were analyzed using LCMS. For all samples, an initial concentration was calculated from analysis of an appropriate oligomer treated under click conditions for 20 min, ensuring full functionalization of propargylated oligomers. Identities of RNA species present were determined by their mass (Figure S6–S10). For nonstandard reactions, the conditions are specified in the text.
LCMS Analysis of Oligonucleotides
Oligomers were analyzed using a Xevo G2-S TOF mass spectrometer coupled to an Acquity UPLC system using an Acquity UPLC BEH C18 1.7 μm column. The system utilizes electrospray ionization (ESI). Two mobile phases were used, 16.3 mM TEA, 400 mM HFIP in H2O and 16.3 mM TEA, 400 mM HFIP in 80:20 v/v MeCN and H2O, with a flow rate of 0.200 mL/min. Calibration curves for the RNA species were based either on A260 or intensities of specified negative m/z signals (Figure S1e–h). Intensities of integrated peaks were calculated using native modules of KNIME software platform.43 Total mass spectra were reconstructed from the ion series using the MaxEnt algorithm preinstalled on MassLynx software (v. 4.1 from Waters) according to the manufacturer’s instructions. To obtain the negative ion series described, the oligomer peak in the chromatogram was selected for integration and further analysis.
RNA Degradation Gel Electrophoresis
In vitro RNA degradation reactions were carried out as described above. The quenched reaction mixture was mixed in 1:1 ratio with a loading buffer (95% formamide, 0.025% SDS, 0.025% bromophenol blue (BPB), 0.025% xylene cyanol FF, 0.025% ethidium bromide, 0.5 mM EDTA), heated at 70 °C for 5 min, and cooled to 0 °C. PAGE was performed on NovexTM TBE-Urea Gels, containing 15% polyacrylamide under 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 180 V for 60 min. Gel staining was performed using SYBR Green II RNA Gel Stain (Invitrogen) in 1× TBE buffer. The stained RNA was visualized with ChemiDoc MP (Bio-Rad, United Kingdom).
Cell Culture
MOLM13 cells were cultured in RPMI1640 (Gibco) supplemented with 10% v/v FBS and 1% v/v penicillin/streptomycin/l-glutamine. 293T cells were cultured in DMEM (Gibco) supplemented with 10% v/v FBS and 1% v/v penicillin/streptomycin/l-glutamine.
Lentiviral Vector Production, Infection, and Transfection
For virus production, 293T cells were transfected with lentiviral vector pLKO.1 together with the packaging plasmids PAX2 and VSVg at a 1:1.5:0.5 ratio. Supernatant was harvested 48 and 72 h after transfection. Cells (1 × 106) and viral supernatant were mixed in 2 mL of culture medium supplemented with 8 μg mL–1 Polybrene (Millipore), followed by spinfection (60 min, 900g, 32 °C), and further incubated overnight at 37 °C. The medium was refreshed on the following day, and the transduced cells were further cultured.
Generation of Conditional Knock-down and Intronic Deletion-Containing Cells
MOLM-13 cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions using pLKO-TETon-Puro lentiviral vectors expressing shRNAs against the coding sequence of human METTL3, METTL16, or a scrambled control. Twenty-four hours after spinfection, the cells were replated in fresh medium containing 1 μg mL–1 of puromycin and kept in selection medium for 7 days. Anti-METTL3 and scrambled shRNAs were induced by treating the cells with 200 ng mL–1 tetracycline for 3 days, anti-METTL16 was induced by identical treatment for 2 days.
gRNA assays were performed using dual gRNA vectors as reported previously.44 Viral supernatants were collected 48 h after transfection. All transfections and viral collections were performed in 15 cm plates as mentioned below. For virus production, 5 μg of the above plasmids and 5 μg of psi-Eco packaging vector were transfected dropwise into the 293T cells using 47.5 μL TransIT LT1 (Mirus) and 600 μL of Opti-MEM (Invitrogen). The resulting viral supernatant was harvested, and transduction of cells was performed in 6-well plates. After transduction, transduced cells were sorted for BFP (for gRNA). The gRNA sequences are listed in Table S12.
Cellular RNA Degradation Reactions (meCLICK-Seq)
MOLM13 cells were suspended in methionine-free RPMI-1640 media (Gibco) supplemented with 10% v/v FBS and 1% v/v penicillin/streptomycin/l-glutamine at a density of 1 000 000 cells mL–1. The cells were incubated for 30 min at 37 °C followed by addition of PropSeMet at a final concentration of 150 μM. Treated cells were incubated for further 16 h at 37 °C. Aqueous solutions of premixed CuSO4 and THPTA were added at final concentrations of 100 and 300 μM, respectively, followed by the click-degrader 1 at 400 μM and NaAsc at 5 mM. Treated cells were incubated for 10 min at 37 °C and resuspended in complete RPMI-1640 medium. Afterward, the cells were again incubated at 37 °C and harvested after 5 h for RNA extraction.
m6A RNA Immunoprecipitation
Total RNA was isolated from MOLM-13 control, Δintronic, METTL3-KD, or METTL16-KD cells (two independent biological replicates for each shRNA) 8 days after doxycycline administration using the RNAeasy midi kit (Qiagen). Successively polyA enriched RNA was purified from 300 μg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). For each immunoprecipitation reaction, 500 ng of polyA+ purified RNA was used. m6A RNA immunoprecipitation was performed using the Magna MeRIP m6A kit (Millipore) according to the manufacturer’s instructions. Immunoprecipitated RNA was analyzed via RT-qPCR.
RNA Extraction, Reverse Transcription, and RT-qPCR
Total RNA was extracted from pelleted cells using RNAeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Twenty micrograms of total RNA was enriched for polyA-containing sequences using Dynabeads mRNA purification kit (Invitrogen) according to the manufacturer’s instructions. One microgram of total RNA and all of the polyA-enriched RNA from the previous step were retrotranscribed using SuperScript Vilo Master Mix (Invitrogen) according the to manufacturer’s instructions. Levels of specific RNAs were measured using fast mode of StepOnePlus Real-Time PCR System (Applied Biosystems) and Fast SYBR Green Master Mix (Applied Biosystems) according to the manufacturer’s instructions. For reactions where total RNA was used, RNA levels were normalized to 18S subunit of the ribosome. For reactions where polyA-enriched RNA was used, RNA levels were normalized to RPL32 mRNA. These housekeeping genes were chosen as their levels fluctuated the least under various conditions tested. Primer sequences are listed below.
Statistical Analysis
General statistical analyses were carried out using a one-sided (in cases where we were interested only in a decrease of signal in the treated group) or two-sided Student’s t test at a confidence interval of 95%.
Protein Extraction and Immunoblotting
The cells were lysed in whole-cell lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% NP-40, and 1 mM EDTA) supplemented with 1 mM DTT, protease inhibitors (Sigma), and phosphatase inhibitors (Sigma). Protein quantities were estimated with Bradford assays (Bio-Rad). The protein samples were supplemented with SDS–PAGE sample buffer, and DTT was added to each sample. Protein (10–40 μg) was separated on a 4–12% Bis-Tris SDS–PAGE gel (Invitrogen) with a same amount of protein added to each track of a gel and blotted onto poly(vinylidene difluoride) membranes (Millipore). Visualization was performed using LumiGLO Chemiluminescent Substrate (KPL, 54-61-00) and X-ray film (GE Healthcare). The following antibodies were used: anti-METTL3 from Bethyl Laboratories (A301-568A), anti-METTL16 from Abcam (ab185990), anti-β-actin from Abcam (ab8227), goat anti-rabbit IgG H&L (HRP) from Abcam (ab205718).
HPLC-MS/MS Analysis of RNA
Up to 10 μg of poly(A) RNA was decapped with Cap-Clip Acid Pyrophosphatase (Cellscript) for 90 min at 37 °C, then digested with 100 U of P1 nuclease (New England Biolabs) at 37 °C overnight and dephosphorylated with 1 U of rSAP (New England Biolabs) at 37 °C for 1 h. The 100 μL samples were filtered with Millex-GV 0.22 μm filters (Millipore Sigma). Five to ten microliters from each sample was injected into the Agilent 6470 Triple Quad LC/MS instrument with Agilent Zorbax Eclipse C18 reverse phase HPLC column. The samples were run at 500 μL/min flow rate in mobile phase buffer A (water with 0.1% formic acid) and 0–20% gradient of buffer B (methanol with 0.1% formic Acid). MRM transitions were measured for adenosine (268.1 to 136.1), guanosine (284.1 to 152.1), and Pr6A (306.1 to 174.1). Standard compound for Pr6A was run on HLPC-MS/MS to optimize the HPLC method and determine retention times. For LC-MS/MS data collection and analysis, Agilent Mass Hunter LC/MS Data Acquisition, version B.08.00, and Quantitative Analysis, version B.07.01, software were used.
Biotin Pull-down Experiment
RNA was extracted from PropSeMet treated MOLM13 cells as described above. CuAAC reaction was then performed in 20 mM HEPES (pH 7.5) on 7 μg of RNA, 20 μM azide-PEG3–biotin conjugate (Sigma-Aldrich), 5 mM CuSO4, 15 mM THPTA, and 5 mM NaAsc, to a final volume of 100 μL. After initiating the reaction with NaAsc, the mixture was stirred at 37 °C for 30 min, followed by quenching of the reaction with 6 mM EDTA. Biotinylated RNA was then precipitated with EtOH and redissolved in 30 μL of nuclease-free water. Streptavidin pull-down was carried out using streptavidin beads (Invitrogen) according to the manufacturer’s instructions. RNA was then analyzed via RT-qPCR as described above with abundance of each transcript in the pulled-down RNA compared to corresponding input RNA.
Bioinformatic Analysis
Total RNA was extracted from pelleted cells using RNAeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNaseq data from all experiments consisting of 75 bp paired-end Illumina reads were mapped to the human genome assembly GRCh38 by STAR 2.7.1b,45 using arguments “--outSAMunmapped Within KeepPairs --outFilterIntronMotifs RemoveNoncanonicalUnannotated --chimSegmentMin 0 --chimJunctionOverhangMin 20”. BigWig files were produced by deepTools 3.3.246 bamCoverage using RPKM normalization. Exonic reads were removed using the intersect function in Bedtools, v2.29.0,47 and exon regions derived from the Ensembl GRCh38, version 93. Reads on the forward strand were extracted using Samtools 1.9,48 with “view -f 128 -F 16” and “-f 80” and merged into one file. Reads on the reverse strand were extracted using “view -f 144” and “-f 64 -F 16” and merged. Broad peaks were called by MACS2 2.1.4,49 using WT as treatment and METTL16/METTL3 as control, with arguments “callpeak --broad --extsize = 300 --keep-dup 20 --nomodel -g hs --broad-cutoff 0.9 --max-gap 500”. Intersection of peaks between the two replicates was taken. Peaks that show reduced signals in meCLICK-Seq compared to WT were selected as the final set.
To search for motifs, DNA sequences of peaks were extracted from the GRCh38 genomic FASTA file. Motifs were discovered by MEME-chip 5.0.550 using parameters “-meme-nmotifs 30”. For gene quantification, exonic regions were obtained from Ensembl GRCh38, version 93. Number of reads in each exon was calculated by customized code and normalized by RPKM. To counter the 3′ bias, the level of the most 3′ exon was used as expression of the gene.
m6A sites were analyzed by first mapping Reads from the miCLIP experiments to the GRCh38 assembly using STAR 2.7.1a45 with arguments “--outFilterMismatchNoverReadLmax 0.05 --alignIntronMax 0”. Forward and reverse genomic-coverage tracks were subsequently produced by the bamCoverage function in deepTools 3.3.246 with arguments “--normalizeUsing None --filterRNAstrand forward” and “--normalizeUsing None --filterRNAstrand reverse” respectively. Regions with nonzero coverage were extracted by customized codes. Sites called from 4 miCLIP runs were merged into a union set, and the ones also present in the input data were removed.
Acknowledgments
We thank UKRI (BBSRC DTP scholarships to S.M. and H.K.C) and the Jardine Foundation and Cambridge Trust (Ph.D. scholarship to M.E.H.). K.T. was funded by a Wellcome Trust Sir Henry Wellcome Fellowship (grant reference RG94424). G.S.V. is funded by a Cancer Research UK Senior Cancer Fellowship (C22324/A23015). Research in the Shi lab is supported by Ludwig Institute for Cancer Research. Y.S. is American Cancer Society Research Professor. G.J.L.B. is a Royal Society University Research Fellow (URF\R\180019) and an FCT Investigator (IF/00624/2015). We thank Dr. Vikki Cantrill for her help with the editing of this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01094.
Putative mass spectra peaks for the 11-mer after 2 hour degradation and for the functionalized and BCS-quenched 11-mer after 14 hour degradation, detected RNA species, oligomers used for the in vitro study, list of primers and gRNAs, analysis of propargylated and non-propargylated RNA oligomers degradation by click-degrader, predicted structures of oligomers used, analyses of CTRL and Prop oligomers, analysis of mRNA and lncRNA species, examples of intronic peak snapshots, analyses of METTL3- and METTL16-dependent peaks, comparison of methylated mRNAs determined via meCLICK-Seq and m7G-Seq, analysis of MeCLICK-Seq peaks and IPA sites, mass spectra of functionalized 11-mer species, and chemical synthesis and characterization including NMR spectra (PDF)
METTL3 and METTL16 mRNA substrates determined via meCLICK-Seq (TXT)
Results of a miCLIP study of m6A in MOLM13 cells (XLSX)
METTL3 and METTL16 lncRNA substrates determined via meCLICK-Seq (TXT)
METTL13 and METTL16-dependent intronic/intergenic peaks determined via meCLICK-Seq (XLSX)
Methylated mRNA species determined via meCLICK-Seq with an overlap with m7G-Seq study (XLSX)
Intronic and intergenic click-degrader sensitive RNA peaks with an overlap with m7G-Seq study (XLSX)
Accession Codes
The accession number for the RNA-seq data generated in this study is GSE147238.
The authors declare the following competing financial interest(s): S.M., K.T., and G.J.L.B. are co-inventors on a patent application that incorporates discoveries described in this manuscript. All other authors declare no conflict of interests.
Notes
All data and methods are available in the manuscript or the Supporting Information. All cell lines are available upon request through a material transfer agreement with the University of Cambridge.
Supplementary Material
References
- Meyer S.; Temme C.; Wahle E. Messenger RNA Turnover in Eukaryotes: Pathways and Enzymes. Crit. Rev. Biochem. Mol. Biol. 2004, 39 (4), 197–216. 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Shu X.; Cao J.; Cheng M.; Xiang S.; Gao M.; Li T.; Ying X.; Wang F.; Yue Y.; Lu Z.; Dai Q.; Cui X.; Ma L.; Wang Y.; He C.; Feng X.; Liu J. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 2020, 16 (8), 887–895. 10.1038/s41589-020-0526-9. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18 (11), 655–670. 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A.; Jha B. K.; Silverman R. H. New Insights into the Role of RNase L in Innate Immunity. J. Interferon Cytokine Res. 2011, 31 (1), 49–57. 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y.; Wang C. C.; Choy K. W.; Du Q.; Chen J.; Wang Q.; Li L.; Chung T. K. H.; Tang T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538 (2), 217–227. 10.1016/j.gene.2013.12.019. [DOI] [PubMed] [Google Scholar]
- Cox D. B. T.; Gootenberg J. S.; Abudayyeh O. O.; Franklin B.; Kellner M. J.; Joung J.; Zhang F. RNA editing with CRISPR-Cas13. Science 2017, 358 (6366), 1019. 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales M. G.; Matsumoto Y.; Velagapudi S. P.; Disney M. D. Small Molecule Targeted Recruitment of a Nuclease to RNA. J. Am. Chem. Soc. 2018, 140 (22), 6741–6744. 10.1021/jacs.8b01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Disney M. D. Precise Small Molecule Degradation of a Noncoding RNA Identifies Cellular Binding Sites and Modulates an Oncogenic Phenotype. ACS Chem. Biol. 2018, 13 (11), 3065–3071. 10.1021/acschembio.8b00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkuviene M.; Clouet-d’Orval B.; Cerniauskas I.; Weinhold E.; Klimasauskas S. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases. Nucleic Acids Res. 2012, 40 (14), 6765–6773. 10.1093/nar/gks381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D.; Saletore Y.; Zumbo P.; Elemento O.; Mason C. E.; Jaffrey S. R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149 (7), 1635–1646. 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; Sorek R.; Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485 (7397), 201–206. 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Linder B.; Grozhik A. V.; Olarerin-George A. O.; Meydan C.; Mason C. E.; Jaffrey S. R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12 (8), 767–772. 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Chen L.-Q.; Zhao Y.-L.; Yang C.-G.; Roundtree I. A.; Zhang Z.; Ren J.; Xie W.; He C.; Luo G.-Z. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 2019, 5 (7), eaax0250. 10.1126/sciadv.aax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D. DART-seq: an antibody-free method for global m6A detection. Nat. Methods 2019, 16 (12), 1275–1280. 10.1038/s41592-019-0570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Xiao Y.; Dong S.; Yu Q.; Jia G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol. 2020, 16 (8), 896–903. 10.1038/s41589-020-0525-x. [DOI] [PubMed] [Google Scholar]
- Hartstock K.; Nilges B. S.; Ovcharenko A.; Cornelissen N. V.; Püllen N.; Lawrence-Dörner A.-M.; Leidel S. A.; Rentmeister A. Enzymatic or In Vivo Installation of Propargyl Groups in Combination with Click Chemistry for the Enrichment and Detection of Methyltransferase Target Sites in RNA. Angew. Chem., Int. Ed. 2018, 57 (21), 6342–6346. 10.1002/anie.201800188. [DOI] [PubMed] [Google Scholar]
- Raines R. T. Ribonuclease A. Chem. Rev. 1998, 98 (3), 1045–1066. 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Cheng L.; Abhilash K. G.; Breslow R. Binding and biomimetic cleavage of the RNA poly(U) by synthetic polyimidazoles. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (32), 12884. 10.1073/pnas.1210846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N. G.; Fabani M. M.; Zenkova M. A.; Bichenkova E. V.; Polushin N. N.; Sil’nikov V. V.; Douglas K. T.; Vlassov V. V. Sequence-specific artificial ribonucleases. I. Bis -imidazole-containing oligonucleotide conjugates prepared using precursor-based strategy. Nucleic Acids Res. 2004, 32 (13), 3887–3897. 10.1093/nar/gkh702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.-R.; Li J.; Cai W.; Lai J. Y. H.; McKinnie S. M. K.; Zhang W.-P.; Moore B. S.; Zhang W.; Qian P.-Y. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat. Chem. 2019, 11 (10), 880–889. 10.1038/s41557-019-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. R.; Thor S.; Kohler L.; Kohler P. R. A.; Metcalf W. W.; Luthey-Schulten Z. Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans. BMC Genomics 2016, 17, 924. 10.1186/s12864-016-3219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oivanen M.; Kuusela S.; Lönnberg H. Kinetics and Mechanisms for the Cleavage and Isomerization of the Phosphodiester Bonds of RNA by Brønsted Acids and Bases. Chem. Rev. 1998, 98 (3), 961–990. 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- Barbieri I.; Tzelepis K.; Pandolfini L.; Shi J.; Millán-Zambrano G.; Robson S. C.; Aspris D.; Migliori V.; Bannister A. J.; Han N.; De Braekeleer E.; Ponstingl H.; Hendrick A.; Vakoc C. R.; Vassiliou G. S.; Kouzarides T. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552 (7683), 126–131. 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L. P.; Pickering B. F.; Cheng Y.; Zaccara S.; Nguyen D.; Minuesa G.; Chou T.; Chow A.; Saletore Y.; MacKay M.; Schulman J.; Famulare C.; Patel M.; Klimek V. M.; Garrett-Bakelman F. E.; Melnick A.; Carroll M.; Mason C. E.; Jaffrey S. R.; Kharas M. G. The N6-methyladenosine m6A-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23 (11), 1369–1376. 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton K. E.; Chen B.; Liu K.; Hunter O. V.; Xie Y.; Tu B. P.; Conrad N. K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169 (5), 824–835. 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J. A.; Shambaugh M. E.; Polayes D.; Matera A. G.; Rottman F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3 (11), 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Wiederschain D.; Wee S.; Chen L.; Loo A.; Yang G.; Huang A.; Chen Y.; Caponigro G.; Yao Y.-M.; Lengauer C.; Sellers W. R.; Benson J. D. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 2009, 8 (3), 498–504. 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- Leger A.; Amaral P. P.; Pandolfini L.; Capitanchik C.; Capraro F.; Barbieri I.; Migliori V.; Luscombe N. M.; Enright A. J.; Tzelepis K.; Ule J.; Fitzgerald T.; Birney E.; Leonardi T.; Kouzarides T.. RNA modifications detection by comparative Nanopore direct RNA sequencing. bioRxiv 2019, 843136, https://www.biorxiv.org/content/10.1101/843136v1. [DOI] [PMC free article] [PubMed]
- Doxtader K. A.; Wang P.; Scarborough A. M.; Seo D.; Conrad N. K.; Nam Y. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol. Cell 2018, 71 (6), 1001–1011. 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.; Qiao J.; Wang G.; Lan Y.; Li G.; Guo X.; Xi J.; Ye D.; Zhu S.; Chen W.; Jia W.; Leng Y.; Wan X.; Kang J. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46 (8), 3906–3920. 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Guo S.; Piao H.-Y.; Wang Y.; Wu Y.; Meng X.-Y.; Yang D.; Zheng Z.-C.; Zhao Y. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J. Physiol. Biochem. 2019, 75 (3), 379–389. 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. I.; Parisien M.; Dai Q.; Liu N.; Diatchenko L.; Sachleben J. R.; Pan T. N6-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J. Mol. Biol. 2016, 428, 822–833. 10.1016/j.jmb.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.; Weng H.; Zhou K.; Wu T.; Zhao B. S.; Sun M.; Chen Z.; Deng X.; Xiao G.; Auer F.; Klemm L.; Wu H.; Zuo Z.; Qin X.; Dong Y.; Zhou Y.; Qin H.; Tao S.; Du J.; Liu J.; Lu Z.; Yin H.; Mesquita A.; Yuan C. L.; Hu Y.-C.; Sun W.; Su R.; Dong L.; Shen C.; Li C.; Qing Y.; Jiang X.; Wu X.; Sun M.; Guan J.-L.; Qu L.; Wei M.; Müschen M.; Huang G.; He C.; Yang J.; Chen J. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567 (7748), 414–419. 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Dou X.; Chen C.; Chen C.; Liu C.; Xu M. M.; Zhao S.; Shen B.; Gao Y.; Han D.; He C. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 2020, 367 (6477), 580–586. 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louloupi A.; Ntini E.; Conrad T.; Ørom U. A. V. Transient N6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell Rep. 2018, 23 (12), 3429–3437. 10.1016/j.celrep.2018.05.077. [DOI] [PubMed] [Google Scholar]
- Mendel M.; Chen K.-M.; Homolka D.; Gos P.; Pandey R. R.; McCarthy A. A.; Pillai R. S. Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell 2018, 71 (6), 986–1000. 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H.; Matsumoto M.; Ishigami Y.; Ebina M.; Muto A.; Sato Y.; Kumagai S.; Ochiai K.; Suzuki T.; Igarashi K. S-Adenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 2017, 21 (12), 3354–3363. 10.1016/j.celrep.2017.11.092. [DOI] [PubMed] [Google Scholar]
- Zhang L.-S.; Liu C.; Ma H.; Dai Q.; Sun H.-L.; Luo G.; Zhang Z.; Zhang L.; Hu L.; Dong X.; He C. Transcriptome-wide Mapping of Internal N7-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell 2019, 74 (6), 1304–1316. 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-H.; Singh I.; Tisdale S.; Abdel-Wahab O.; Leslie C. S.; Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 2018, 561 (7721), 127–131. 10.1038/s41586-018-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh I.; Lee S.-H.; Sperling A. S.; Samur M. K.; Tai Y.-T.; Fulciniti M.; Munshi N. C.; Mayr C.; Leslie C. S. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat. Commun. 2018, 9 (1), 1716. 10.1038/s41467-018-04112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.; Lee D.; Lee J.; Park D.; Kim Y. J.; Park W.-Y.; Hong D.; Park P. J.; Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet. 2015, 47 (11), 1242–1248. 10.1038/ng.3414. [DOI] [PubMed] [Google Scholar]
- Kopp F.; Mendell J. T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172 (3), 393–407. 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas A. A. Comprehensible classification models: a position paper. SIGKDD Explor. Newsl. 2014, 15 (1), 1–10. 10.1145/2594473.2594475. [DOI] [Google Scholar]
- Tzelepis K.; Koike-Yusa H.; De Braekeleer E.; Li Y.; Metzakopian E.; Dovey O. M.; Mupo A.; Grinkevich V.; Li M.; Mazan M.; Gozdecka M.; Ohnishi S.; Cooper J.; Patel M.; McKerrell T.; Chen B.; Domingues A. F.; Gallipoli P.; Teichmann S.; Ponstingl H.; McDermott U.; Saez-Rodriguez J.; Huntly B. J. P.; Iorio F.; Pina C.; Vassiliou G. S.; Yusa K. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016, 17 (4), 1193–1205. 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A.; Davis C. A.; Schlesinger F.; Drenkow J.; Zaleski C.; Jha S.; Batut P.; Chaisson M.; Gingeras T. R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29 (1), 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F.; Ryan D. P.; Grüning B.; Bhardwaj V.; Kilpert F.; Richter A. S.; Heyne S.; Dündar F.; Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44 (W1), W160–W165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R.; Hall I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26 (6), 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Handsaker B.; Wysoker A.; Fennell T.; Ruan J.; Homer N.; Marth G.; Abecasis G.; Durbin R.; The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25 (16), 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Liu T.; Meyer C. A.; Eeckhoute J.; Johnson D. S.; Bernstein B. E.; Nussbaum C.; Myers R. M.; Brown M.; Li W.; Liu X. S. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9 (9), R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P.; Bailey T. L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011, 27 (12), 1696–1697. 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.