Abstract
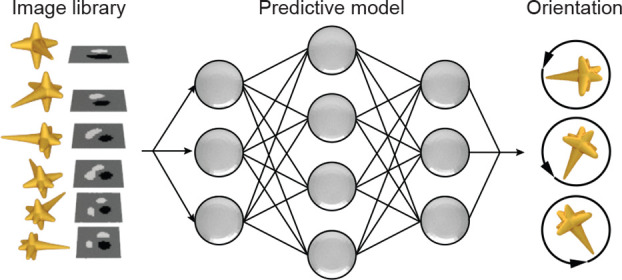
This paper describes a computational imaging platform to determine the orientation of anisotropic optical probes under differential interference contrast (DIC) microscopy. We established a deep-learning model based on data sets of DIC images collected from metal nanoparticle optical probes at different orientations. This model predicted the in-plane angle of gold nanorods with an error below 20°, the inherent limit of the DIC method. Using low-symmetry gold nanostars as optical probes, we demonstrated the detection of in-plane particle orientation in the full 0–360° range. We also showed that orientation predictions of the same particle were consistent even with variations in the imaging background. Finally, the deep-learning model was extended to enable simultaneous prediction of in-plane and out-of-plane rotation angles for a multibranched nanostar by concurrent analysis of DIC images measured at multiple wavelengths.
Short abstract
We describe a deep-learning platform for identifying the three-dimensional orientation of anisotropic optical probes used in differential interference contrast microscopy.
Automated single-particle tracking techniques1−3 have played key roles in studying biological processes ranging from cellular motion4 to targeted drug delivery.5−7 These methods analyze cellular dynamics by first recording a video of an optical probe (fluorescent molecules8−10 or semiconductor quantum dots11,12) and then extracting the translational trajectories by analysis algorithms.13,14 Although rotational dynamics has not been studied in such tracking processes, they can provide additional molecular level information on cellular activities such as protein diffusion and cytoskeleton formation.15−18 One major drawback for visualizing rotational motion is that common fluorescence probes either have orientation-invariant emission or limited signal intensity.19 Also, existing algorithms using Gaussian fitting or cross-correlation cannot identify probe orientation.13,14,20 The development of next-generation, particle-tracking platforms for rotational dynamics requires simultaneous advances in both the optical probe and the analysis method.
Optical imaging approaches using anisotropic metal nanoparticle probes can resolve rotational motion because of their polarization-dependent optical responses.21−23 For example, gold nanorods show a different scattering intensity under polarized dark-field microscopy based on orientation24,25 but cannot be used in cellular environments with strong background scattering.26 Alternatively, differential interference contrast (DIC) microscopy27,28 can generate orientation-dependent patterns (bright and dark pixels) from anisotropic plasmonic nanoparticles.23 However, because of the symmetry of gold nanorods, their orientation can only be tracked during in-plane rotation because DIC image signals decrease dramatically with out-of-plane rotation.29 Gold nanostars30−32 (AuNS) are three-dimensional (3D) optical probes that have multiple branches oriented in different directions. Compared to nanorods, AuNS show more complicated DIC patterns33,34 that can be correlated with their 3D nanostructure and orientation.35
Data-driven machine-learning algorithms can assist single-particle tracking in physiological environments when fast and accurate pattern analyses are needed.36−39 Statistical-learning approaches determine particle trajectories from optical microscopy images by locating the Haar-like features36,37,40 (developed for object detection) that identify particle position but not orientation. A different set of features must be established between DIC image pattern and particle orientation to solve the inverse problem. Deep convolutional networks41,42 are universal machine learning tools that can automatically identify robust features associated with a response or category in an image data set.43,44 To reduce data acquisition time, data-augmentation strategies45,46 have been developed so that data sets of sufficient sizes (>104 labeled images) can be obtained from only a small set of original images. Therefore, deep-learning models may become an effective tool to predict the orientation of nanoparticle probes from their diffraction-limited microscopy images.
Here we show a deep-learning platform that can identify the 3D orientation of optical probes used in DIC microscopy. We constructed DIC image data sets of both Au nanorods and AuNS with labeled orientations to establish the deep-learning models. The optimized model predicted the in-plane orientation of nanorods from their DIC images with an accuracy limited only by the inherent angular resolution of the imaging-and-probe system. The angular range of in-plane orientation sensing was expanded from 0–180° to 0–360° using anisotropic AuNS probes with a lower structural symmetry than the nanorods. We further confirmed the robustness of our deep-learning model by showing that predictions were accurate even with different backgrounds. Finally, we determined the 3D orientation of a multibranched AuNS by the simultaneous detection of in-plane and out-of-plane angles.
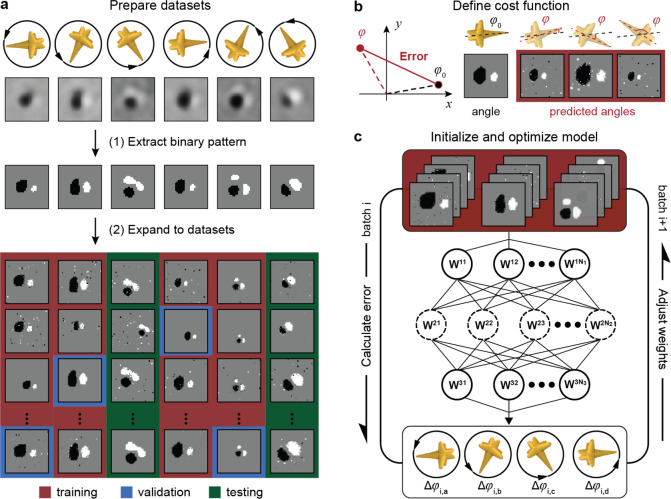
Figure 1 depicts a scheme of the prediction process for nanoparticle orientations based on DIC microscopy in the de Sénarmont configuration47,48 and convolutional neural networks. An unpolarized light source is converted into elliptically polarized light by a linear polarizer and a quarter-wave plate, which is then split into two orthogonally polarized beams that are spatially separated by 120 nm at the first Nomarski prism (Figure 1a). The beams experience different phase changes at the nanoparticle before being combined by a second Nomarski prism to form either bright or dark pixels in the DIC images. At each in-plane orientation of the nanoparticle probe, we collected raw DIC images to produce a data set labeled by the corresponding angle (Figure 1b). Convolutional neural networks were then constructed by the PyTorch toolbox49,50 based on the library of DIC images (Figure 1c).
Figure 1.
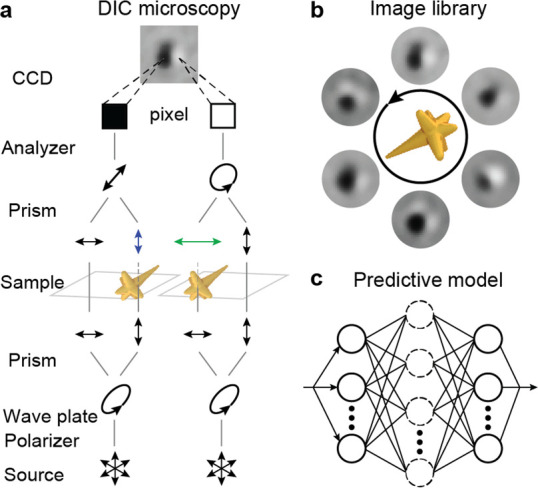
Intelligent orientation sensing by differential interference contrast (DIC) microscopy images and deep learning. Schemes depicting (a) DIC imaging setups, (b) libraries of DIC images, and (c) convolutional networks for deep learning.
Our deep-learning model was optimized iteratively using a training data set to minimize the prediction errors defined by a cost function.51,52Figure 2a summarizes our procedure to prepare DIC data sets for model training and testing. We processed raw DIC images by our multistep thresholding method that produced clean black–white patterns from both calculated and measured DIC data (Figures S1–S2). By a data-augmentation process,45,46 the images corresponding to each particle orientation were converted to a data set class (∼1500 images) labeled by angle (Figure S3). In this step, the DIC patterns in the original images were randomly resized and spatially shifted to produce image copies in the class. The image-scaling process ensured that the optimized models are insensitive to changes in the DIC pattern size so that different imaging setups can use this deep-learning approach. Random noise was also added to the images to improve the tolerance of the model to pixel-level imaging defects. A fraction of classes with evenly distributed orientations was reserved as the testing data set, while the remaining classes were split into training and validation data sets with a 4:1 ratio. The training data set was used to optimize the weights in the convolutional network during model training, and the validation data set was used to monitor overfitting errors. The testing data set evaluated the accuracy of the optimized models.
Figure 2.
Deep-learning models for orientation prediction of anisotropic gold nanoparticles based on DIC microscopy images. (a) Scheme of data set preparation from raw DIC images with labeled nanoparticle orientations. (b) Definition of cost function based on labeled and predicted angles. (c) Training and testing of the artificial neural networks in the deep learning model by batches of images in the training data set. Data set preparation steps in (a) include (1) extracting DIC patterns from raw images by computer vision methods (Python Scikit-Image Package) and (2) expanding images in each orientation into a data set by tailoring randomly the pattern size, position, and noise level. In each epoch described in (c), the artificial neural network calculated the predicted angle for batches of images in the training data sets, evaluated the corresponding error based on (b), and adjusted the weights in the model in a back-propagation process for the next batch.
Figure 2b shows the cost function to be minimized by the deep-learning model. For a general prediction model of in-plane rotations in the range 0–360°, the prediction error was defined based on polar coordinates as
| 1 |
where φ and φ0 are the predicted and labeled angles corresponding to each image in the training set, respectively.Figure 2c depicts the training process of the deep-learning model starting from a convolutional network with randomly initialized weights (Code section S1). In each epoch of the optimization, the model-training function (1) predicts the particle orientations for all images in the training data set with the model; (2) calculates the corresponding errors by eq 1; and (3) adjusts the model weights to reduce the total error (Figures S4–S5). After each training epoch, the model also makes orientation predictions for images in the validation data set, and the average validation error (per image) is compared to the training error. If the validation error was below 10% of the error at the initial epoch, the convergence criterion was satisfied, and the model was evaluated using the testing data set.
We first tested the deep-learning method for tracking the in-plane rotation of a gold nanorod (length l = 90 nm, width w = 40 nm) with a localized surface plasmon (LSP) resonance at λ = 620 nm (Figure S6). Because of contrast inversion33 when the imaging wavelength is tuned across the LSP, DIC images are typically collected at a wavelength shorter or longer than this resonance. We collected the raw images at λ = 700 nm and constructed a library of DIC patterns of the nanorod with 36 in-plane angles (relative to the x axis) φ0 = 0°, 5°, ..., 175° by the data augmentation process. Among the 36 angles, images at φ0 = 0°, 20°, ..., 160° were selected as the testing data set, and the remaining angles were split randomly into training (∼70% images) and validation (∼30% images) data sets. Because of the 2-fold symmetry of the nanorod, the cost function was modified from eq 1 to
| 2 |
so that the error was a periodic function of φ between 0 and 180°.
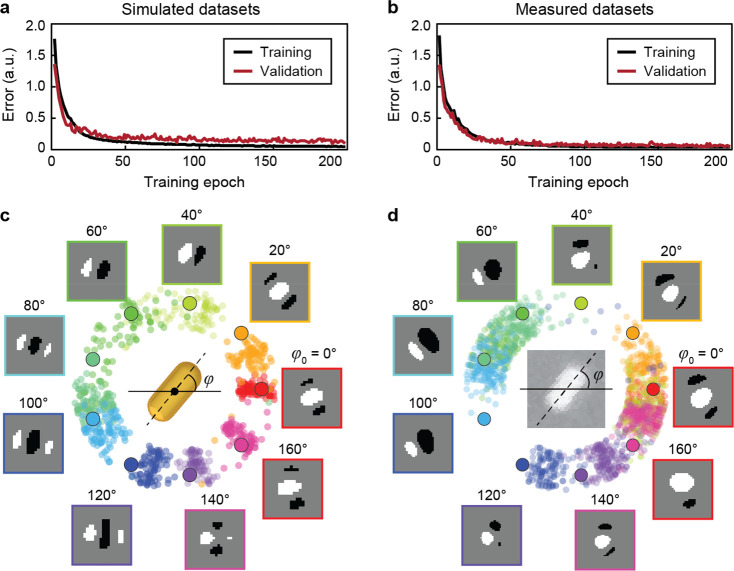
Figure 3a–b shows the separate training processes of the neural network for calculated and measured DIC images, respectively, which are consistent in bright–dark contrast but can have differences in the patterns.33 A model using a three-layer convolutional network can reach convergence in 50 epochs for the simulated data sets (Figure S7). In comparison, training using the (noisy) measured data required four convolutional layers to converge within the same time (Figure S8). Figure 3c–d shows the performance of these optimized networks on predicting in-plane particle orientation from images in their corresponding testing data set (φ0 = 0°, 20°, ..., 160°). Without learning directly from DIC patterns acquired at these angles, the model could determine particle orientations with errors below ±20° for the simulated data set, which is the experimental angular resolution of DIC and plasmonic optical probes (Figures S9–S10). These results indicate that convolutional networks can predict the in-plane orientation of nanoparticles.
Figure 3.
Deep learning models can predict in-plane angles of gold nanorods. Mean-square errors of the neural networks during the training process based on (a) simulated and (b) measured DIC images. Prediction of nanorod in-plane orientations by the deep learning model based on (c) calculated images (d) experimental results. The image library consisted of DIC images calculated by finite-difference time-domain (FDTD) simulations or measured experimentally for a nanorod (length l = 90 nm, width w = 40 nm) with in-plane angles φ0 = 0°, 5°, ..., 175° at wavelength λ = 700 nm. DIC images at φ0 = 0°, 20°, ..., 160° were used for testing the model, and the images at remaining angles were used for training and validation. Scanning electron microscopy (SEM) in (d) shows the structure of the nanorod.
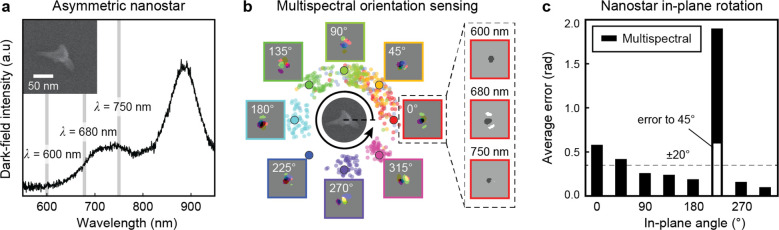
To realize in-plane orientation tracking in the full 0–360° range, we tested our deep-learning approach with a low-symmetry AuNS having two long branches positioned in the imaging plane. Compared to nanorods, the angular separation in the raw data set was increased to 15° to reduce the data collection time. Figure 4a shows our selected AuNS, where DIC images were collected at three wavelengths (λ = 600, 680, 750 nm) around the LSP resonance at λ = 730 nm. Imaging was not conducted around the other LSP resonance at λ = 880 nm because the silicon detector has poor sensitivity. To increase the accuracy of the orientation predictions, we developed a model to account for the three wavelengths simultaneously based on three convolutional layers (Figures S11–S12). Figure 4b shows the performance of the optimized neural network on predicting in-plane orientation from images at φ0 = 0°, 45°, ..., 315°. The predictions were accurate at all angles except for φ0 = 225°, where the DIC pattern was similar to φ0 = 45°. Figure 4c shows the accuracy of the multiwavelength model averaged over ∼600 test images at each angle. At all angles except φ0 = 45° and 225°, the multiwavelength model showed reduced average errors compared to the single-wavelength models that used only the images at λ = 680 nm (Figures S13–S14). At most angles, both models showed an average error below 20°, which indicates that the orientation of appropriately shaped anisotropic probes can be predicted over the 0–360° range. The prediction accuracy of the model can be improved further by accounting for DIC images at multiple wavelengths simultaneously.
Figure 4.
Low-symmetry imaging probe enables full 0–360° orientation sensing. (a) SEM image of the gold nanostar (AuNS) probes and the scattering spectrum measured by dark-field microscopy. (b) The predicted angles based on the testing data sets collected at three wavelengths λ = 600, 680, 750 nm. (c) Prediction errors of the deep-learning models at each angle. The image library consisted of DIC images for the AuNS with in plane angles φ0 = 0°, 15°, ..., 360°. DIC images at angles φ0 = 0°, 45°, ..., 315° were used for testing the model, and the images at remaining angles were used for training and validation.
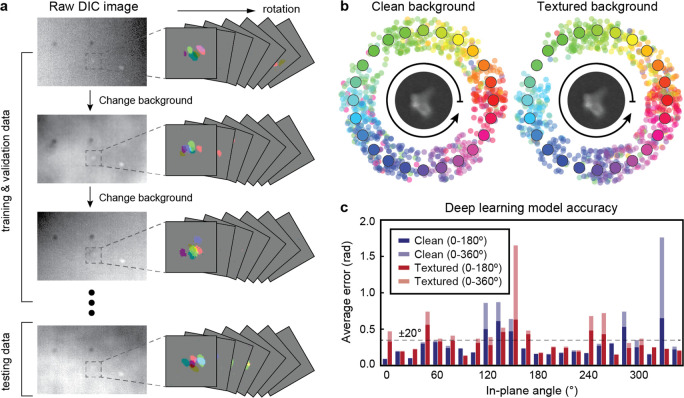
We tested the robustness of the orientation prediction platform under different background imaging conditions. Figure 5a shows the raw DIC images of four AuNS, including our selected optical probe (dashed box) with two LSP resonances at λ = 770 and 910 nm (Figure S15) and its extracted patterns under different background conditions. We prepared microscale patterns in 5 nm Cr films on replaceable coverslips that produced background noises in the imaging field of view (Figure S16). We trained the multiwavelength deep-learning model by four data sets prepared from images with clean or randomly textured background. Figure 5b–c shows the test results of the optimized model for orientations of the AuNS based on DIC images collected with and without backgrounds. For both tested data sets, the model exhibited average errors below ±20° for 75% of the selected angles; large discrepancies at some angles were observed because of difficulties in distinguishing between ±180° orientations. These consistent predictions of in-plane angles with tolerance to imaging conditions will be important for imaging in live cells.
Figure 5.
Robust orientation sensing under complex imaging backgrounds. (a) Examples of (left) raw DIC images and (right) extracted multiwavelength patterns. (b) The predicted angles from the testing data sets collected at three wavelengths λ = 660, 700, and 750 nm and (c) the corresponding error analysis for testing data sets with clean and random backgrounds.
Finally, we demonstrated that out-of-plane orientation sensing is possible using an anisotropic, multispectral optical probe based on calculated DIC images. Figure 6a–b shows the structure and optical responses of our AuNS probe. This AuNS showed three LSP resonances (λ = 725, 785, 815 nm), each corresponding to a branch at a different spatial orientation (Figure S17). We prepared data sets of calculated DIC images at all combinations of in-plane angles φ0 = 0°, 5°, ..., 355° and out-of-plane angles θ0 = −90°, −85°, ..., 90° for four wavelengths (λ = 710, 750, 800, 825 nm) that spanned the range of the LSP resonances. To account for both rotation angles (φ0 and θ0), the cost function was defined based on a spherical coordinate as
| 3 |
where θ and θ0 are the predicted and labeled out-of-plane angles corresponding to each image in the training set, respectively. Images at φ0 = 0°, 20°, ..., 340° were selected as the testing data set, and the remaining angles were used for training and validation of the model. Figure 6c shows the performance of the deep-learning model for all combinations of φ0 and θ0. For 80% of the orientations in the testing data set, the average prediction errors were below 20° (Figure S18). We believe that our multispectral optical probe can be realized experimentally with developments in synthesis32 and sorting methods53 that improve control over AuNS shape and homogeneity.
Figure 6.
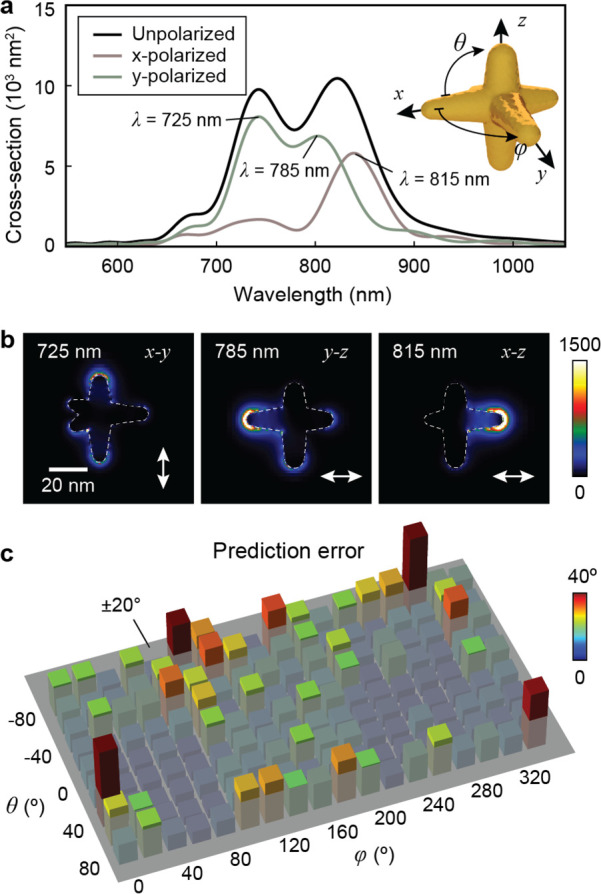
Multispectral AuNS enables three-dimensional (3D) orientation sensing. (a) Scattering spectra and (b) near-field electric-field intensity map of the selected AuNS with LSP resonances at λ = 725, 785, and 815 nm. (c) Prediction errors evaluated with a test data set consisting of DIC images at all combinations of in-plane angles φ0 = 0°, 20°, ..., 340° and out-of-plane rotation θ0 = −80°, −60°, ..., 80°.
In summary, we demonstrated a deep-learning approach to determine the orientation of optical nanoparticle probes from their microscope images. Innovations in anisotropic probe design enabled the sensing of in-plane orientations in the full 0–360° range with an accuracy at the intrinsic limit of the DIC technique with a AuNS probe. We also showed the prediction of the out-of-plane orientation for a multispectral AuNS by imaging simultaneously at multiple wavelengths. The model is robust against noise in imaging background and has the potential to achieve fast, fully automated tracking of particle rotations during live-cell interactions. We expect that this deep-learning platform can resolve cellular interactions involving 3D rotational dynamics that are not accessible by existing imaging techniques but are critical for understanding and optimizing next-generation therapeutic systems.
Acknowledgments
The work related to discovering optically responsive nanomaterials by machine learning was supported by the National Science Foundation (NSF) under NSF Award NSF CMMI-1848613 (J.H. and T.W.O.). The optical imaging development was supported by the National Institutes of Health (NIH) Grant 5 R01 GM131421-02 (T.L. and P.C.). This work was also partially supported by CONIX center of STARnet Semiconductor Research Corporation program, which was sponsored by MARCO and DARPA (S.W.). This work made use of the EPIC and NUFAB facility of Northwestern University’s NUANCE Center, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205); the MRSEC program (NSF DMR-1720139) at the Materials Research Center; the International Institute for Nanotechnology (IIN); the Keck Foundation; and the State of Illinois, through the IIN. This research was supported in part through the computational resources and staff contributions provided for the Quest high performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation Grant No. ACI-1548562.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c01252.
Methods; data set preparation from differential interference contrast microscopy images; layer units in convolutional neural networks for orientation prediction; deep-learning models for predicting in-plane nanorod orientation; deep-learning models for predicting in-plane nanostar orientation; orientation prediction in different imaging background; out-of-plane orientation tracking by multispectral nanostars (PDF)
Author Contributions
J.H. and T.W.O. conceived the idea of using machine-learning models for the orientation prediction of anisotropic nanoparticle probes. J.H., T.L., and P.C. collected the raw experimental DIC images to prepare the data sets and performed the dark-field spectroscopy for the nanoparticles. J.H. and A.D.S. calculated the simulated DIC images based on FDTD calculations. J.H. and S.W. developed the data-processing pipeline to produce the training, validation, and testing data sets. J.H. and S.W. implemented and constructed the deep-learning models based on PyTorch. T.L. and P.C. synthesized the nanoparticles imaging probes. J.H. and T.R. fabricated the chromium patterns used as imaging backgrounds.
The authors declare no competing financial interest.
Supplementary Material
References
- Kusumi A.; Sako Y.; Yamamoto M. Confined Lateral Diffusion of Membrane Receptors as Studied by Single Particle Tracking (Nanovid Microscopy). Effects of Calcium-Induced Differentiation in Cultured Epithelial Cells. Biophys. J. 1993, 65, 2021–2040. 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason T. G.; Ganesan K.; van Zanten J. H.; Wirtz D.; Kuo S. C. Particle Tracking Microrheology of Complex Fluids. Phys. Rev. Lett. 1997, 79, 3282–3285. 10.1103/PhysRevLett.79.3282. [DOI] [Google Scholar]
- Saxton M. J.; Jacobson K. Single-Particle Tracking:Applications to Membrane Dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 373–399. 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- Stone J. W.; Sisco P. N.; Goldsmith E. C.; Baxter S. C.; Murphy C. J. Using Gold Nanorods to Probe Cell-Induced Collagen Deformation. Nano Lett. 2007, 7, 116–119. 10.1021/nl062248d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.; Cui Y.; Levenson R. M.; Chung L. W. K.; Nie S. In Vivo Cancer Targeting and Imaging with Semiconductor Quantum Dots. Nat. Biotechnol. 2004, 22, 969–976. 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- Tada H.; Higuchi H.; Wanatabe T. M.; Ohuchi N. In Vivo Real-Time Tracking of Single Quantum Dots Conjugated with Monoclonal Anti-Her2 Antibody in Tumors of Mice. Cancer Res. 2007, 67, 1138. 10.1158/0008-5472.CAN-06-1185. [DOI] [PubMed] [Google Scholar]
- Suh J.; Dawson M.; Hanes J. Real-Time Multiple-Particle Tracking: Applications to Drug and Gene Delivery. Adv. Drug Delivery Rev. 2005, 57, 63–78. 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Giepmans B. N. G.; Adams S. R.; Ellisman M. H.; Tsien R. Y. The Fluorescent Toolbox for Assessing Protein Location and Function. Science 2006, 312, 217. 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Thompson R. E.; Larson D. R.; Webb W. W. Precise Nanometer Localization Analysis for Individual Fluorescent Probes. Biophys. J. 2002, 82, 2775–2783. 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A.; Nakada C.; Ritchie K.; Murase K.; Suzuki K.; Murakoshi H.; Kasai R. S.; Kondo J.; Fujiwara T. Paradigm Shift of the Plasma Membrane Concept from the Two-Dimensional Continuum Fluid to the Partitioned Fluid: High-Speed Single-Molecule Tracking of Membrane Molecules. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 351–378. 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- Medintz I. L.; Uyeda H. T.; Goldman E. R.; Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater. 2005, 4, 435–446. 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Michalet X.; Pinaud F. F.; Bentolila L. A.; Tsay J. M.; Doose S.; Li J. J.; Sundaresan G.; Wu A. M.; Gambhir S. S.; Weiss S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science 2005, 307, 538. 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheezum M. K.; Walker W. F.; Guilford W. H. Quantitative Comparison of Algorithms for Tracking Single Fluorescent Particles. Biophys. J. 2001, 81, 2378–2388. 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini I. F.; Koumoutsakos P. Feature Point Tracking and Trajectory Analysis for Video Imaging in Cell Biology. J. Struct. Biol. 2005, 151, 182–195. 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Gu Y.; Sun W.; Wang G.; Fang N. Single Particle Orientation and Rotation Tracking Discloses Distinctive Rotational Dynamics of Drug Delivery Vectors on Live Cell Membranes. J. Am. Chem. Soc. 2011, 133, 5720–5723. 10.1021/ja200603x. [DOI] [PubMed] [Google Scholar]
- Roosen-Runge F.; Hennig M.; Zhang F.; Jacobs R. M. J.; Sztucki M.; Schober H.; Seydel T.; Schreiber F. Protein Self-Diffusion in Crowded Solutions. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 11815. 10.1073/pnas.1107287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H.; Higashida C.; Yuan Y.; Ishizaki T.; Narumiya S.; Watanabe N. Rotational Movement of the Formin Mdia1 Along the Double Helical Strand of an Actin Filament. Science 2011, 331, 80. 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- Macias-Romero C.; Didier M. E. P.; Zubkovs V.; Delannoy L.; Dutto F.; Radenovic A.; Roke S. Probing Rotational and Translational Diffusion of Nanodoublers in Living Cells on Microsecond Time Scales. Nano Lett. 2014, 14, 2552–2557. 10.1021/nl500356u. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R.Fluorescence Polarization. In Principles of Fluorescence Spectroscopy; Lakowicz J. R., Ed.; Springer US: Boston, MA, 1983; pp 111–153. [Google Scholar]
- Jaqaman K.; Loerke D.; Mettlen M.; Kuwata H.; Grinstein S.; Schmid S. L.; Danuser G. Robust Single-Particle Tracking in Live-Cell Time-Lapse Sequences. Nat. Methods 2008, 5, 695–702. 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen C.; Alivisatos A. P. Gold Nanorods as Novel Nonbleaching Plasmon-Based Orientation Sensors for Polarized Single-Particle Microscopy. Nano Lett. 2005, 5, 301–304. 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- Chang W.-S.; Ha J. W.; Slaughter L. S.; Link S. Plasmonic Nanorod Absorbers as Orientation Sensors. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 2781. 10.1073/pnas.0910127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won Ha J.; Sun W.; Wang G.; Fang N. Differential Interference Contrast Polarization Anisotropy for Tracking Rotational Dynamics of Gold Nanorods. Chem. Commun. 2011, 47, 7743–7745. 10.1039/c1cc11679g. [DOI] [PubMed] [Google Scholar]
- Kaplan L.; Ierokomos A.; Chowdary P.; Bryant Z.; Cui B. Rotation of Endosomes Demonstrates Coordination of Molecular Motors During Axonal Transport. Science Advances 2018, 4, e1602170. 10.1126/sciadv.1602170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L.; Qiao Y.; He Y.; Yeung E. S. Three Dimensional Orientational Imaging of Nanoparticles with Darkfield Microscopy. Anal. Chem. 2010, 82, 5268–5274. 10.1021/ac1006848. [DOI] [PubMed] [Google Scholar]
- Hu R.; Yong K.-T.; Roy I.; Ding H.; He S.; Prasad P. N. Metallic Nanostructures as Localized Plasmon Resonance Enhanced Scattering Probes for Multiplex Dark-Field Targeted Imaging of Cancer Cells. J. Phys. Chem. C 2009, 113, 2676–2684. 10.1021/jp8076672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta M.Advanced Light Microscopy; Elsevier, PWN: Amsterdam, Warsaw, 1989; Vol. 2.. [Google Scholar]
- Wang G.; Sun W.; Luo Y.; Fang N. Resolving Rotational Motions of Nano-Objects in Engineered Environments and Live Cells with Gold Nanorods and Differential Interference Contrast Microscopy. J. Am. Chem. Soc. 2010, 132, 16417–16422. 10.1021/ja106506k. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Ha J. W.; Wei L.; Wang G.; Fang N. Determining the Full Three-Dimensional Orientation of Single Anisotropic Nanoparticles by Differential Interference Contrast Microscopy. Angew. Chem., Int. Ed. 2012, 51, 7734–7738. 10.1002/anie.201202340. [DOI] [PubMed] [Google Scholar]
- Dam D. H. M.; Lee J. H.; Sisco P. N.; Co D. T.; Zhang M.; Wasielewski M. R.; Odom T. W. Direct Observation of Nanoparticle–Cancer Cell Nucleus Interactions. ACS Nano 2012, 6, 3318–3326. 10.1021/nn300296p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Dam D. H. M.; Ha J. W.; Yue J.; Odom T. W. Enhanced Human Epidermal Growth Factor Receptor 2 Degradation in Breast Cancer Cells by Lysosome-Targeting Gold Nanoconstructs. ACS Nano 2015, 9, 9859–9867. 10.1021/acsnano.5b05138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra K.; Culver K. S. B.; Werner S. E.; Lee R. C.; Odom T. W. Manipulating the Anisotropic Structure of Gold Nanostars Using Good’s Buffers. Chem. Mater. 2016, 28, 6763–6769. 10.1021/acs.chemmater.6b03242. [DOI] [Google Scholar]
- Choo P.; Hryn A. J.; Culver K. S.; Bhowmik D.; Hu J.; Odom T. W. Wavelength-Dependent Differential Interference Contrast Inversion of Anisotropic Gold Nanoparticles. J. Phys. Chem. C 2018, 122, 27024–27031. 10.1021/acs.jpcc.8b08995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik D.; Culver K. S. B.; Liu T.; Odom T. W. Resolving Single-Nanoconstruct Dynamics During Targeting and Nontargeting Live-Cell Membrane Interactions. ACS Nano 2019, 13, 13637–13644. 10.1021/acsnano.9b03144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver K. S. B.; Liu T.; Hryn A. J.; Fang N.; Odom T. W. In Situ Identification of Nanoparticle Structural Information Using Optical Microscopy. J. Phys. Chem. Lett. 2018, 9, 2886–2892. 10.1021/acs.jpclett.8b01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.; Zhou X.; Kirchhausen T.; Wong S. T. C. Detection of Molecular Particles in Live Cells Via Machine Learning. Cytometry, Part A 2007, 71A, 563–575. 10.1002/cyto.a.20404. [DOI] [PubMed] [Google Scholar]
- Bruijning M.; Visser M. D.; Hallmann C. A.; Jongejans E. Trackdem: Automated Particle Tracking to Obtain Population Counts and Size Distributions from Videos in R. Methods in Ecology and Evolution 2018, 9, 965–973. 10.1111/2041-210X.12975. [DOI] [Google Scholar]
- Kowalek P.; Loch-Olszewska H.; Szwabiński J. Classification of Diffusion Modes in Single-Particle Tracking Data: Feature-Based Versus Deep-Learning Approach. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2019, 100, 032410. 10.1103/PhysRevE.100.032410. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gil G.; Garcia-March M. A.; Manzo C.; Martín-Guerrero J. D.; Lewenstein M. Single Trajectory Characterization Via Machine Learning. New J. Phys. 2020, 22, 013010. 10.1088/1367-2630/ab6065. [DOI] [Google Scholar]
- Lienhart R.; Maydt J.. An Extended Set of Haar-Like Features for Rapid Object Detection. In Proceedings of the International Conference on Image Processing; IEEE, 2002. [Google Scholar]
- Zhong Y.; Li C.; Zhou H.; Wang G. Developing Noise-Resistant Three-Dimensional Single Particle Tracking Using Deep Neural Networks. Anal. Chem. 2018, 90, 10748–10757. 10.1021/acs.analchem.8b01334. [DOI] [PubMed] [Google Scholar]
- Newby J. M.; Schaefer A. M.; Lee P. T.; Forest M. G.; Lai S. K. Convolutional Neural Networks Automate Detection for Tracking of Submicron-Scale Particles in 2d and 3d. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 9026. 10.1073/pnas.1804420115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietterich T. G.Ensemble Methods in Machine Learning. In International Workshop on Multiple Classifier Systems; Springer: 2000; pp 1–15. [Google Scholar]
- Welling M.; Teh Y. W.. Bayesian Learning Via Stochastic Gradient Langevin Dynamics. In Proceedings of the 28th International Conference on Machine Learning (ICML-11), 2011; pp 681–688. [Google Scholar]
- Perez L.; Wang J.. The Effectiveness of Data Augmentation in Image Classification Using Deep Learning. arXiv preprint, arXiv: 1712.04621, 2017.
- Ronneberger O.; Fischer P.; Brox T. In U-Net: Convolutional Networks for Biomedical Image Segmentation, Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015; Navab N.; Hornegger J.; Wells W. M.; Frangi A. F., Eds.; Springer International Publishing: Cham, 2015; pp 234–241. [Google Scholar]
- Chen J.; Xu Y.; Lv X.; Lai X.; Zeng S. Super-Resolution Differential Interference Contrast Microscopy by Structured Illumination. Opt. Express 2013, 21, 112–121. 10.1364/OE.21.000112. [DOI] [PubMed] [Google Scholar]
- Gu Y.; Wang G.; Fang N. Simultaneous Single-Particle Superlocalization and Rotational Tracking. ACS Nano 2013, 7, 1658–1665. 10.1021/nn305640y. [DOI] [PubMed] [Google Scholar]
- Ketkar N., Introduction to Pytorch. In Deep Learning with Python; Springer: 2017; pp 195–208. [Google Scholar]
- Paszke A.; Gross S.; Chintala S.; Chanan G.; Yang E.; DeVito Z.; Lin Z.; Desmaison A.; Antiga L.; Lerer A.. Automatic Differentiation in Pytorch. NIPS 2017 Workshop Autodiff, 2017.
- LeCun Y.; Bengio Y.; Hinton G. Deep Learning. Nature 2015, 521, 436. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Di W.; Bhardwaj A.; Wei J.. Deep Learning Essentials: Your Hands-on Guide to the Fundamentals of Deep Learning and Neural Network Modeling. Packt Publishing, 2018; p 284. [Google Scholar]
- Chandra K.; Kumar V.; Werner S. E.; Odom T. W. Separation of Stabilized Mops Gold Nanostars by Density Gradient Centrifugation. ACS Omega 2017, 2, 4878–4884. 10.1021/acsomega.7b00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.