Abstract
Selective autophagy is a highly regulated degradation pathway for the removal of specific damaged or unwanted cellular components and organelles such as protein aggregates. Cargo selectivity in selective autophagy relies on the action of cargo receptors and adaptors. In mammalian cells, two structurally related proteins p62 and NBR1 act as cargo receptors for selective autophagy of ubiquitinated proteins including aggregation-prone proteins in aggrephagy. Plant NBR1 is the structural and functional homolog of mammalian p62 and NBR1. Since its first reports almost ten years ago, plant NBR1 has been well established to function as a cargo receptor for selective autophagy of stress-induced protein aggregates and play an important role in plant responses to a broad spectrum of stress conditions including heat, salt and drought. Over the past several years, important progress has been made in the discovery of specific cargo proteins of plant NBR1 and their roles in the regulation of plant heat stress memory, plant-viral interaction and special protein secretion. There is also new evidence for a possible role of NBR1 in stress-induced pexophagy, sulfur nutrient responses and abscisic acid signaling. In this review, we summarize these progresses and discuss the potential significance of NBR1-mediated selective autophagy in broad plant responses to both biotic and abiotic stresses.
Keywords: autophagy, NBR1, plant stress responses, selective autophagy receptor, protein aggregates, plant heat tolerance, plant virus interaction
1. Introduction
Autophagy is an evolutionarily conserved process in eukaryotic organisms for degradation of cytoplasmic constituents including proteins and organelles in the lysosomes or vacuoles [1]. There are at least three types of autophagy, known as macroautophagy, chaperone-mediated autophagy and microautophagy. Macroautophagy is the pathway that has been most extensively characterized and is often referred to simply as autophagy [1]. Autophagy is usually induced in response to a variety of physiological and environmental stimuli and plays an important role in cell homeostasis under unfavorable and pathological conditions including starvation, extreme temperature, aging and pathogen infection [2]. Induction of autophagy is initiated by the formation of an isolation membrane called phagophore that can expand to capture cytoplasmic components and enclose the sequestered cargos to form a separate compartment, the double-membrane autophagosome [3,4]. The autophagosome is then primed to fuse with the lysosomes or vacuoles for degradation of captured materials by lysosomal/vacuolar hydrolases. Autophagosome biogenesis is a complex membrane process that requires more than 40 autophagy-related (ATG) proteins. These ATG proteins function in several physiologically continuous, but mechanistically distinct steps and are organized as several functional groups, which include (i) the ATG1–ATG13–ATG17 scaffold, formed upon activation of the ATG1 kinase during the early stage of autophagy induction, (ii) the class III phosphatidylinositol 3-kinase (PtdIns3K)–ATG14–ATG6 (Beclin 1) complex for the nucleation and assembly of the initial phagophore membrane, and iii) two interrelated ubiquitin-like conjugation systems, ATG12–ATG5–ATG16 and ATG8–PE (phosphatidylethanolamine), which are required for the regulation of membrane elongation and expansion of the forming autophagosomes [1].
The core process of autophagy and ATG proteins are highly conserved in all eukaryotic organisms including plants [1,5,6,7]. Using genetic and molecular approaches, extensive studies over the past two decades or so have demonstrated an important role of autophagy in almost all aspects of plant life, particularly in plant stress responses [5,8,9]. Autophagosome biogenesis and ATG gene expression are both induced by diverse abiotic stress conditions including nutrient starvation, heat, salt, drought and oxidative stresses [10,11,12,13,14,15]. Autophagy mutants and transgenic silencing lines are hypersensitive to nutrient starvation and compromised in tolerance to these abiotic stresses [10,11,12,13,14,15]. In addition, autophagy plays an important role in plant innate immunity. Plant mutants or transgenic silencing lines for autophagy are altered in responses to virulent and avirulent biotrophic pathogens including pathogen-induced hypersensitive responses [16,17,18,19,20,21]. Autophagy deficient mutants are compromised in resistance to necrotrophic pathogens [17,22]. As will be discussed below, autophagy is also involved in plant interaction with viral pathogens through such mechanisms as regulation of antiviral RNA silencing and targeting degradation of viral proteins. Autophagy also plays important roles in plant growth and development including root growth, leaf senescence, pollen and endosperm development [8,20,23,24,25].
Even though autophagy was initially considered to be a nonselective process of bulk degradation of intracellular contents, it has been now well established that the broad roles of autophagy are primarily mediated by selective clearance of specific cellular structures [26,27,28,29]. Ubiquitin-like ATG8, which is required for phagophore initiation, elongation and maturation, also plays a critical role in selective autophagy [27]. Upon attachment of the lipid PE to its carboxyl terminus through a conjugation pathway, ATG8 is anchored in both the inner and outer membrane of autophagosomes and provides a docking platform for the selective recruitment of cargos through selective autophagy receptors [27]. Most selective autophagy receptors recognize specific cargos and also interact with membrane-anchored ATG8 through ATG8-interacting motifs (AIMs), which have the W/Y/F-X-X-L/I/V consensus core sequence [27]. The AIM motif binds a hydrophobic patch on ATG8 known as AIM docking site [27]. Recently, a new class of selective autophagy receptors have been identified, which interact with ATG8 through ubiquitin-interacting motif (UIM)-like sequences for high-affinity binding to an ATG8 interaction site different from the AIM docking site [30,31]. Through assays with candidate UIM-containing proteins and unbiased screens, a large number of UIM-based ATG8 interactors have been identified in plants, yeast, and humans [32]. Discovery of the new class of selective autophagy receptors greatly expands the scope of selective autophagy [32].
A large number of autophagy receptors from nonplant organisms have been identified that mediate the selective autophagic degradation of a wide range of cargoes including protein aggregates, signaling complexes, mitochondria, peroxisomes, endoplasmic reticulum (ER), ribosomes and pathogens [27,29,33,34]. In plants, a substantial number of autophagy receptors have also been identified, characterized and functionally analyzed. These plant autophagy receptors include Arabidopsis protein TSPO (outer membrane tryptophan-rich sensory protein), an ATG8-interacting heme-binding protein that targets the degradation of porphyrins through autophagy [35]. TSPO also targets the aquaporin PIP2;7 for degradation in the vacuole to reduce its levels to regulate water permeability under conditions of heat and drought stress [36,37]. ATG8-interacting 1 (ATI1) and 2 (ATI2) proteins are closely related autophagy receptors partially associated with the ER under normal conditions but mainly associated with a different type of spherical compartments under carbon starvation [38,39]. ATI1 is also found on bodies associated with plastids under carbon starvation and is involved in autophagy-dependent trafficking of plastid proteins to the vacuole [39]. The three related ATI3 autophagy receptors from Arabidopsis play a critical role in plant heat tolerance and resistance to the necrotrophic fungal pathogens at least in part through interaction with ER-localized UBAC2 proteins and mediating selective autophagy of specific unknown ER components [40]. Arabidopsis DSK2 acts as a selective autophagy receptor that targets BR1-EMS suppressor 1(BES1) to modulate brassinosteroid signaling and stress tolerance [41]. Arabidopsis ORM1 and 2 proteins mediate selective autophagy of pattern recognition receptor FLS2 to negatively regulate FLS2-mediated defense [42]. More recently, Arabidopsis cytosolic protein C53 has been identified as a receptor for selective autophagy of certain ER domains (ER-phagy) [43]. C53 is also an ER-phagy receptor in mammalian cells [44]. The proteasome subunit RPN10 is a selective autophagy receptor that mediates selective autophagy of the 26S proteasome when the proteasome is inhibited chemically [31]. RPN10 interacts with ATG8 through its UIM-like sequence and is increasingly associated with the proteasome when it is ubiquitinated [30].
While many selective autophagy receptors identified in plants are plant-specific, others are evolutionarily conserved and have homologs in nonplant organisms. Plant NBR1 is structurally related to human NBR1 and SQSTM1/p62 selective autophagy receptors that act in aggrephagy [14,45,46]. Since their first reports almost ten years ago [45,46], a number of studies have revealed an important role of NBR1 in plant responses to a broad spectrum of stress conditions including heat, salt, drought and oxidative stress [14,15,47]. More recently, new discoveries have been made in the understanding the roles and mode of action of NBR1 in the modulation of plant heat stress memory, plant-viral interaction and other physiological processes associated with plant stress responses. In this review, we discuss the structure, evolution and roles of plant NBR1 in responses to a broad spectrum of biotic and abiotic stresses. We also discuss the molecular mechanisms by which NBR1-mediated selective autophagy modulates plant stress responses by highlighting the cargo proteins recognized and targeted by NBR1 and how their turnover leads to altered plant stress responses.
2. Structure, Evolution and Function of NBR1
Plant NBR1 is a structural homolog and functional hybrid of mammalian autophagy receptors NBR1 and p62, which differ in size but share similar domains and important features [45,46,48,49]. Both mammalian p62 and NBR1 proteins contain an N-terminal PB1 (Phox and Bem1p) domain followed by a ZZ-type zinc finger domain, an LC3-interacting region or LIR motif (also known as AIM motif in yeast) and a C-terminal UBA (ubiquitin-associated) domain [49]. As in many other selective autophagy receptors, the LIR motifs mediate direct interaction with ATG8, while the C-terminal UBA domain recognizes mono- and polyubiquitin. The PB1 domain of p62 mediates polymerization of p62, but the related human NBR1 protein lacks the most N-terminal basic charge cluster and, therefore, cannot polymerize through its PB1 domain [49]. However, human NBR1 can form heterodimers with p62 via its PB1 and can self-interacts via a coiled coil domain [49]. Mammalian p62 and NBR1 function as selective autophagy receptors for autophagic degradation of protein aggregates (aggrephagy), mitochondria (mitophagy), peroxisomes (pexophagy), endoplasmic reticulum (reticulophagy) and pathogens (xenophagy) [50]. p62 and NBR1 also have noncanonical role in signaling independent of autophagy [51,52,53,54]. For example, p62 promotes the expression of inflammatory genes via NF-κB through TRAF6 binding by its TRAF6-binding (TB) domain [53]. p62 can also activate mTORC1, which can upregulate c-Myc [55]. These functions are independent of the UBA or LC3-interacting region (LIR) domains of p62, which are required for its role as an autophagy receptor [53].
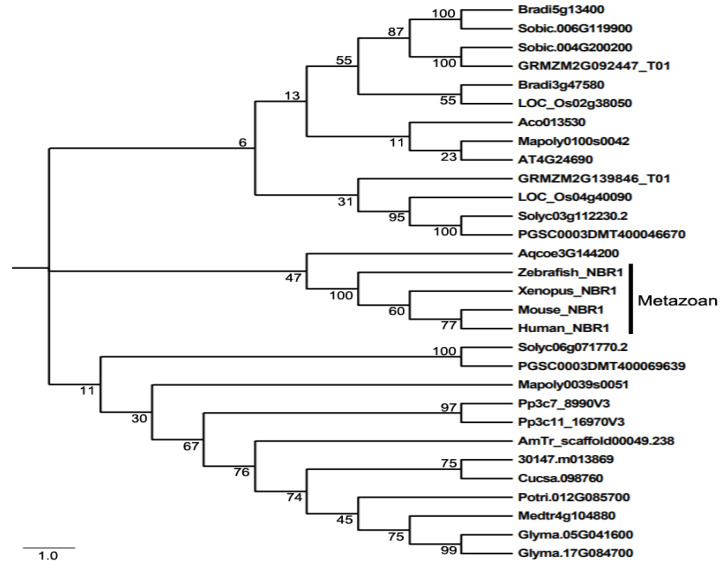
Even though NBR1 has a domain architecture similar to that of p62, it has a highly conserved globular domain characterized by the presence of 4 highly conserved tryptophan (W) residues that is absent in p62 [49]. Using this four tryptophan or FW domain to distinguish p62 and NBR1 homologs through the eukaryotic kingdom, it was discovered that only metazoans contain both p62 and NBR1 homologs, while most nonmetazonan organisms have a single NBR1 homolog [49]. Plant NBR1 homologs lack the coiled coil domain of mammalian NBR1 but have two C-terminal UBA domains [45]. In model plant Arabidopsis, there is a single gene encoding an NBR1 homolog. Like p62, Arabidopsis NBR1 can homo-oligomerizes through the N-terminal PB1 domain, indicating the plant NBR1 share some functional properties with p62 [45]. In addition, only the C-terminal UBA domain of the two UBA domains of Arabidopsis NBR1 binds ubiquitin. We have searched the sequenced genomes of representative plants along the evolutionary tree for genes encoding NBR1 homologs and identified 27 NBR1-encoding genes from 2 spore-bearing and 15 seed plants (Figure 1). As previously reported, some plants contain a single gene encoding NBR1. However, most plants including the spreading earthmoss (Physcomitrella patens), tomato, potato, rice, maize, purple false brome (Brachypodium distachyon) and soybean contain two genes encoding NBR1 proteins (Figure 1). Maize, soybean and purple false brome are known to be polyploid plants that have gone genome duplications during their evolutionary history [56,57,58], which could account for the presence of more than one NBR1 genes in their genomes. To analyze the evolutionary relationship of the conserved protein family, we performed phylogenetic analysis of NBR1 homologs from 17 plant and 4 animal species. As shown in Figure 1, there are three major clades in the phylogenetic tree. All the NBR1 proteins from the animals clustered in one clade, which intriguingly also include the NPR1 from Aquilegia coerulea, while those from plants clustered in two separate clades (Figure 1). These results indicate that the topology of phylogenetic tree for NBR1 homologs from animals and plants is in general agreement with the evolutionary tree of the organisms. However, there is no clear separation of clustering of NBR1 homologs between spore-bearing and seed plants and likewise, NBR1 proteins from monocot and dicot plants do not clustered separately in the clades of seed plants (Figure 1). Furthermore, while the twin NBR1 homologs from some plants such as soybean and P. patens clustered together, other twin NBR1 homologs from the same plant species including tomato, potato, rice, maize and purple false brome do not group together in the tree (Figure 1), indicating significant sequence divergence between these twin homologs. The evolutionary significance of the sequence variation of the NBR1 homologs is unclear but could reflect potential functional diversification of the two NBR1 homologs in these plants.
Figure 1.
The phylogenetic relationship of NBR1 homologs from plants and metazoans. The tree was inferred using the neighbor-joining method. Phylogenetic analyses were conducted in MEGA5. Bootstrap values from 1000 replicates were used to assess the robustness of the tree. NBR1 homologs in the phylogenetic analysis include those from Homo sapiens (Human NBR1), Mus musculus (Mouse NBR1), Danio rerio (Zebrafish NBR1), Xenopus tropicalis (Xenopus NBR1), Marchantia polymorpha (Mapoly0100s0042 and Mapoly0039s0051), Physcomitrella patens (Pp3c7_8990V3 and Pp3c11_16970V3), Amborella trichopoda (AmTr scaffold00049.238), Ananas comosus(Aco013530), Brachypodium distachyon (Bradi3g47580 and Bradi5g13400), Oryza sativa (LOC_Os02g38050 and LOC_Os04g40090), Sorghum bicolor (Sobic.004G200200 and Sobic.006G119900), Zea mays (GRMZM2G139846_T01 and GRMZM2G092447_T01), Aquilegia coerulea (Aqcoe3G144200), Solanum lycopersicum (Solyc03g112230.2 and Solyc06g071770.2), Solanum tuberosum (PGSC0003DMT400069639 and PGSC0003DMT400046670), Populus trichocarpa (Potri.012G085700), Ricinus communis (30147.m013869), Arabidopsis thaliana (AT4G24690), Cucumis sativus (Cucsa.098760), Glycine max (Glyma.05G041600 and Glyma.17G084700), Medicago truncatula (Medtr4g104880).
A number of groups have reported functional genetic analysis of plant NBR1 through characterization of nbr1 mutants or transgenic silencing lines [14,15,46,47,59,60,61,62]. Arabidopsis nbr1 knockout mutants grow and develop normally under normal growth conditions and is not essential for general autophagy, or for the selective clearance of peroxisomes, mitochondria, or the ER [14,60,62]. Plant NBR1 is also dispensable in age- and darkness-induced senescence but may modulates senescence under certain conditions such as short-day growth condition [14,59]. The Arabidopsis nbr1 mutants are also normal in resistance to a necrotrophic pathogen [14]. However, the nbr1 mutants are compromised in plant tolerance to heat, oxidative, salt, and drought stresses [14,47]. The role of NBR1 in plant abiotic stress tolerance is dependent on its interaction with ATG8. In Arabidopsis, NBR1 also reduces growth of bacterial pathogen Pseudomonas syringae by suppressing the establishment of an aqueous extracellular space (‘water-soaking’) [61]. Therefore, the nbr1 mutants display some but not all of the phenotypes of autophagy-deficient mutants. As will be discussed below, more recent studies have revealed specific cargo proteins of plant NBR1 and their roles in the modulation of plant heat stress memory, plant-viral interaction, senescence, reactive oxygen species-induced pexophagy and nutrient responses.
3. NBR1 in Heat Tolerance and Heat Stress Memory
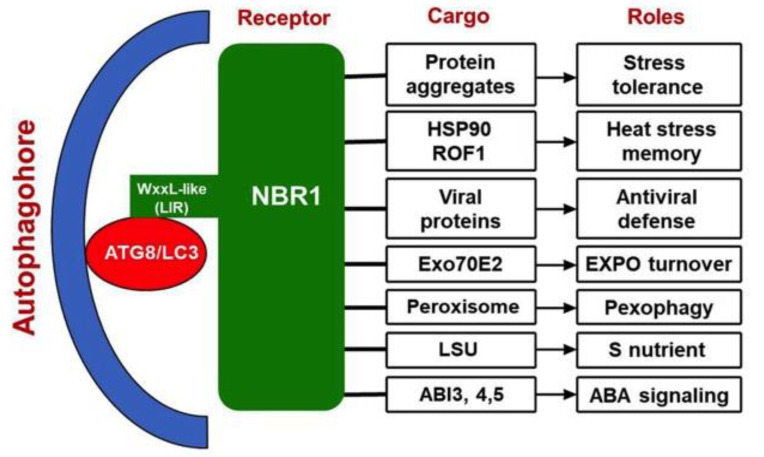
Heat stress caused by high temperature is a fundamental threat to all living organisms including plants. High temperature causes protein misfolding and misfolded proteins are highly toxic and must be efficiently removed to reduce cellular proteotoxic stress [63,64,65]. Selective autophagy functions in protein quality control by targeting degradation of misfolded and other nonnative proteins and therefore also plays an important role in plant response to heat stresses [14,15,40,47]. Arabidopsis mutants and transgenic silencing tomato plants for ATG5, ATG7 and NBR1 are all compromised in heat tolerance, associated with increased development of heat stress symptoms and compromised photosynthesis in heat-stressed leaf tissues [14,15,40,47]. The compromised heat tolerance of atg5, atg7, and nbr1 mutants was also associated with elevated accumulation of insoluble, detergent-resistant proteins that were also highly ubiquitinated under heat stress [14,15,40,47]. NBR1 also accumulated to high levels with an increasing enrichment in the insoluble protein fraction in the autophagy-deficient mutants under heat stress [14,15,40,47]. Through microscopic and biochemical analysis of nbr1 mutants expressing autophagy markers and an aggregation-prone reporter, it was found that NBR1 is required for the heat-induced formation of autophagic vesicles [56]. Moreover, cytoplasmic puncta containing aggregation-prone proteins, which were rarely observed in wild-type plants, were found to accumulate in nbr1 mutants under both control and heat stress conditions [56]. Co-localization of NBR1 with the cytoplasmic puncta suggests that Arabidopsis NBR1 is a plant aggrephagy receptor and NBR1-mediated autophagy targets ubiquitinated protein aggregates most likely derived from denatured and otherwise damaged nonnative proteins and protein aggregates generated under heat stress (Figure 2).
Figure 2.
NBR1-mediated selective autophagy in plant stress responses. The cargo proteins and biological roles of NBR1 in aggrephagy, heat stress memory, antiviral defense and Exo70E2 are well supported by biochemical, molecular and genetic evidence. Studies through analysis of protein localization, gene expression, protein-protein interactions and transgenic overexpression have also suggested possible roles of NBR1 in stress-induced pexophagy, S nutrient response and ABA signaling.
In animals, the carboxyl terminus of the Hsc70-interacting protein (CHIP) is chaperone-dependent E3 ubiquitin ligase [66,67,68,69]. CHIP plays a critical role in protein quality control by ubiquitinating Hsp70-bound misfolded proteins [66,67,68,69]. CHIP acts as both an Hsp70 co-chaperone through its N-terminal tetratricopeptide repeat (TPR) domain and an E3 ubiquitin ligase through the C-terminal U-box domain [66,67,68,69]. If chaperone-assisted refolding of denatured or damaged proteins fails, CHIP E3 ubiquitin ligase can introduce ubiquitination and thereby target denatured and damaged proteins for degradation by both UPS and autophagy [66,67,68,69]. Arabidopsis CHIP is highly similar to animal CHIPs with three tetratricopeptide repeats at the N-terminal side and a U-box domain at the C-terminal side [70,71,72]. To determine the role of CHIP in plant heat tolerance through ubiquitination of misfolded proteins and protein aggregates we have previously reported analysis of CHIP for its coordinated role with the NBR1 autophagy receptor in plant heat tolerance [47]. Two chip T-DNA insertion mutants are normal under normal growth conditions but are hypersensitive to heat, salt and oxidative stresses [47]. More importantly, the chip nbr1 double mutants were more sensitive to heat stress than the nbr1 and chip single mutants, indicating an additive role of CHIP and NBR1 in plant stress responses [47]. Proteomic profiling of stress-induced protein aggregates is consistent with CHIP and NBR1 mediating two distinct but complementary anti-proteotoxic pathways in plant stress responses [47]. More importantly, stress-induced protein aggregates were still ubiquitinated in the chip mutants [47], suggesting that other ubiquitin E3 ligases are responsible for the ubiquitination of heat-induced protein aggregates targeted by NBR1-mediated selective autophagy.
Very often, plants can be subjected repeatedly to adverse environmental conditions such as high temperature. Once a stress condition ends, plants need to orchestrate the balance between growth recovery and keeping stress memory for better survival when a second and harsher stress occurs later. Heat stress memory and recovery are complex processes regulated at multiple levels, including chromatin remodeling, transcriptional activation of heat-induced genes, regulated turnover of transcripts and proteins important for protein quality control [73,74,75]. Autophagy also helps reset cellular heat stress memory in plants [76]. Autophagy is induced in plants by moderate heat stress and remains highly active long after end of moderate heat stress [76]. The activated autophagy targets specific heat shock proteins for degradation at later stages of the thermorecovery phase [76]. Reduced levels of heat shock proteins lead to reduced heat stress memory, and consequently increased accumulation of protein aggregates after the second heat stress and a compromised heat tolerance [76]. In autophagy-deficient mutants, turnover of induced heat shock proteins is reduced, leading to improved heat stress memory and heat tolerance to the subsequent heat stress [76].
Further analysis has demonstrated that NBR1-mediated selective autophagy plays a critical role in the recovery from heat stress through reduction of heat stress memory [77]. The heat shock transcription factor HSFA2 is a key component of heat stress memory through its role in the induction of heat shock genes [78]. HSFA2 interacts with HSP90.1, a major regulator of heat tolerance [79]. Arabidopsis HSP90.1 interacts with ROF1/AFKBP62 (rotamase FKBP 1), a plant homolog of mammalianFKBP4/FKBP52, and regulates plant heat stress responses [79]. The HSP90.1-ROF1 complex remains in the cytoplasm under normal conditions but binds HSFA2 and translocates to the nucleus following exposure to heat stress [79]. Formation of this complex is probably required for enhanced transcriptional activity of HSFA2 and sustained HSP synthesis during heat stress recovery to improve plant response to an imminent recurrence of heat stress [79]. Immunoblot analysis and confocal microscopy revealed that levels of the NBR1 protein, NBR1-labeled puncta, and NBR1 activity are all induced during the heat stress recovery [77]. Co-immunoprecipitation analysis and comparative proteomic analysis identified almost 60 proteins as potential novel targets of NBR1, among which are HSP90.1 and ROF1 [77]. NBR1 mediates the degradation of HSP90.1 and ROF1 by autophagy, which attenuates the expression of heat shock protein genes regulated by the HSFA2 transcription factor, thereby repressing the response to heat stress [73] (Figure 2). In the nbr1 loss-of-function mutants, there is a stronger heat stress memory [77]. These results indicate that NBR1-mediated selective autophagy regulates plant response to recurrent heat stress. Therefore, the roles of NBR1 in plant responses to heat stress are complex. NBR1-mediated selective autophagy targets protein aggregates generated under heat stress to promote basal plant heat tolerance but also can targets degradation of protein chaperones during the recovery to weaken acquired heat tolerance in plants.
4. NBR1 in Plant-Virus Interactions
Autophagy is critical player in animal innate immunity for its role in protecting cells from diverse intracellular pathogens including viruses [80,81,82,83]. As an endolysosomal delivery system, autophagy can promote antiviral response through the transfer of viruses from the cytoplasm to the lysosome for degradation or the transfer of viral components to specific subcellular compartments for the activation of innate or adaptive antiviral immunity [80,81,82,83]. Likewise, autophagy plays an important role in plant antiviral defense. In resistant tobacco plants, silencing of genes for autophagy including ATG3 and ATG7 leads to increased spread of hypersensitive cell death and elevated accumulation of tobacco mosaic virus [18]. In these autophagy-silenced tobacco plants, the virus and viral RNA are both confined only to the infection site in the plants, indicating that the increase in TMV accumulation is not due to increased spread [18]. More likely, plant autophagy limits virus replication and/or movement through degradation of viruses in vacuoles or through effects on other antiviral mechanisms [18].
Double stranded (ds) RNA-induced RNA silencing is an important antiviral mechanism in both animal and plants. Autophagy can also modulate plant antiviral defense by targeting degradation of plant and viral proteins associated with dsRNA-induced RNA silencing. Plant antiviral RNA silencing involves the processing of viral dsRNA into small viral RNAs by the enzyme Dicer. Viral-derived small RNAs guide the degradation of viral RNAs by the RNA-induced silencing complex (RISC). To counter the antiviral defense mechanism, viruses have evolved viral suppressors that inhibit the virus-induced RNA silencing process [84]. P0, an F-box ubiquitin E3 ligase and a viral suppressor of RNA silencing from polerovirus, ubiquitinates the autophagic degradation of ARGONAUTE 1 (AGO1), a key component of RISC that binds to small interfering RNA (siRNA) and carries the RNA slicer activity [85]. Ubiquitinated AGO1 is degraded by selective autophagy instead of the 26S proteasome system based on the sensitivity of degradation to both 3-methyladenine (3-MA), an inhibitor of autophagosome formation, and E64d, a cysteine protease inhibitor of the degradation of autophagic cargo inside autolysosomes, but not to the inhibition of the proteasome [85]. In another study, it was shown that a tobacco calmodulin-like protein (rgs-CaM) functions as an antiviral factor through two distinct antiviral mechanisms, one of which is linked with autophagy [86]. First, rgs-CaM is an interacting protein of HC-Pro and structurally unrelated 2b, two RNA silencing suppressors from tobacco etch virus (TEV) and cucumber mosaic virus (CMV), respectively [86]. The tobacco rgs-CaM protein binds to the dsRNA-binding domains of the RNA silencing suppressors and sequesters them from inhibiting RNA interference (RNAi) [86]. Second, chemical inhibition or suppression of autophagy through gene silencing increased the protein levels of endogenous rgs-CaM and interacting viral RNA silencing suppressors [86]. Furthermore, accumulated endogenous rgs-CaM and interacting viral RNA silencing suppressors were associated with LysoTracker-stained autophagosomes, suggesting that they formed complexes are recruited into autophagosomes for degradation [86]. Thus, tobacco rgs-CaM binds viral RNA silencing suppressors to both decrease the suppressor activity and target their degradation by autophagy [86].
In a more recent study with cauliflower mosaic virus (CaMV), it has been shown that autophagy and NBR1-mediated selective autophagy in particular have complex roles, both anti- and proviral, in the compatible interaction of the dsDNA pararetrovirus with the model plant Arabidopsis [87]. First, NBR1-mediated selective autophagy targets nonassembled and virus particle-forming capsid proteins for degradation, thereby restricting the establishment of CaMV infection [87]. As a counter mechanism, the CaMV-induced virus factory inclusions protects capsid proteins against autophagic destruction by sequestering the viral proteins and coordinating particle assembly and storage [87]. Second, virus-triggered autophagy plays a critical role in reducing extensive senescence and tissue death of infected plants. This survival function significantly extends the timespan of virus production, thereby increasing the chances for virus particle acquisition by aphid vectors and CaMV transmission [87]. This role of autophagy is not dependent on NBR1 [87]. These results demonstrate the integration of selective autophagy into plant immunity against viruses and the potential viral strategies to evade and adapt autophagic processes for successful infection (Figure 2).
In another recently reported study, selective autophagy was identified as antiviral pathway during plant infection with turnip mosaic virus (TuMV), a positive-stranded RNA potyvirus [88]. The autophagy cargo receptor NBR1 suppresses viral accumulation by targeting the viral RNA silencing suppressor helper-component proteinase (HCpro), presumably in association with virus-induced RNA granules [88]. Again, the virus has evolved mechanisms to suppress the antiviral mechanism. TuMV seems to antagonize NBR1-dependent autophagy during infection by the activity of distinct viral proteins, thereby limiting its antiviral capacity. As found with CaMV discussed above, NBR1-independent autophagy prevents premature plant death to extend the lifespan of virus reservoirs and particle production [88]. Taken together, these results again illustrate the conserved role of selective autophagy in antiviral immunity and reveal the evolvement of viral protein functions to inhibit autophagy processes (Figure 2).
5. NBR1 in the Autophagic Degradation of Exocyst Subunit Exo70E2
As compared to endocytosis, exocytosis is a process that transport materials from inside the cell to the external part of the cell [89]. The exocyst complex directs the secretory vesicles of exocytosis from the Golgi complex to specific locations on the plasma membrane and to mediate their tethering and localization to the membrane immediately before fusion [89]. In Arabidopsis, there is a double-membrane organelle termed the exocyst-positive organelle (EXPO), which may be involved in mediating unconventional protein secretion in plants [90,91]. Arabidopsis, an exocyst complex subunit, is a marker for EXOP [90]. Upon induction of autophagy, Exo70E2-GFP positive EXPOs and autophagosome were colocalized and delivered to vacuoles for degradation in transgenic Arabidopsis plants [90]. Recently, through multiple approaches, it has been demonstrated that Arabidopsis NBR1 is a selective receptor for Exo70E2 during autophagy in Arabidopsis [59]. First, biochemical and recruitment assays demonstrated that AtNBR1 specifically interacted and recruited Exo70E2 or its EXPO to AtATG8-positive autophagosomes [59]. This interaction of NBR1 with Exo70E2 is not dependent on the UBA domains of NBR1 [59]. Second, in the nbr1 mutants, the vacuolar delivery of AtExo70E2 or EXPO upon autophagic induction was significantly reduced when compared to that in wild-type plants [59]. Taken together, these results indicate that Arabidopsis NBR1-mediated selective autophagy pathway is involved in the vacuolar delivery of Exo70E2 or EXPO in plant autophagy (Figure 2). The physiological significance of NBR1-mediated degradation of Exo70E2 or EXPO is not clear. The exocyst components are known to be involved in autophagy in yeast and mammalian cells [92,93,94]. Exocysts may also play roles in autophagy in plants [95,96]. This notion is supported by the observation that the majority of AtExo70 homologs contain ATG8-interacting motifs and Arabidopsis Exo70B1 is required for autophagy-mediated delivery of anthocyanin to the vacuole in Arabidopsis [95,96]. In addition, NBR1-mediated selective autophagy may be involved in the regulation of special protein secretion pathway through modulation of the levels of exocyst subunit proteins and the number of EXPO. It would be interesting to determine whether other exocyst complex subunits are also targeted by selective autophagy and, if so, how they are recruited to autophagosomes and what is the physiological significance for their targeted degradation in plants.
6. NBR1 in Cadmium-Induced Pexophagy
In mammalian cells, NBR1 is necessary and sufficient for selective autophagic degradation of peroxisomes (pexophagy) [33]. On the other hand, p62 is not required for pexophagy when NBR1 is in excess but its binding to NBR1 increases the efficiency of NBR1-mediated pexophagy [33]. Several reported studies, however, have shown that plant NBR1 is not required for the selective clearance of peroxisomes [60,62]. In the Arabidopsis mutants with defective LON2/At5g47040, a protease implicated in peroxisomal quality control, there is reduced responsiveness to the peroxisomally-metabolized auxin precursor indole-3-butyric acid (IBA), heightened degradation of several peroxisomal matrix proteins, and impaired processing of proteins harboring N-terminal peroxisomal targeting signals [62]. Autophagy deficiency ameliorates these defects [62]. Through comparison of peroxisome abundance between the lon2 single and lon2 nbr1 double mutants, it was established that NBR1 is not necessary for autophagy of lon2 peroxisomes [62]. Furthermore, NBR1 overexpression is not sufficient to trigger autophagy of seedling peroxisomes [62]. These results support that Arabidopsis targets peroxisomes for autophagic degradation through an NBR1-independent mechanism.
Intriguingly, another recently reported study has implicated NBR1 in heavy metal cadmium (Cd)-induced, reactive oxygen species (ROS)-dependent pexophagy in Arabidopsis [97]. Cd treatment induces transient peroxisome proliferation in Arabidopsis leaves, which is associated with upregulation of the transcript levels of ATG8 genes and ATG8 proteins [97]. The Cd-dependent induction of pexophagy was demonstrated by the accumulation of peroxisomes in Arabidopsis knockout mutants atg5 and atg7 [97]. Importantly, ATG8a colocalizes with NBR1 in the electron-dense peroxisomal core, suggesting that NBR1 may be an autophagic receptor for peroxisomes [97]. Protein carbonylation and peroxisomal redox state suggest that protein oxidation may trigger Cd-induced pexophagy [97]. Therefore, while plant NBR1 is not essential for pexophagy, it could play a role in modulating Cd-induced selective autophagy of perophagy (Figure 2). Further studies using molecular genetic approaches will be necessary to confirm the proposed role of plant NBR1 in pexophagy.
7. NBR1 in Plant Responses to Sulfur Deprivation
Sulfur (S) is an essential mineral nutrient for plants [98,99]. The plant response to sulfur deficiency includes extensive changes at multiple levels including transcriptome, proteome, metabolome even before the onset of the first visible symptoms of sulfur starvation [100,101,102]. In Arabidopsis, members of the plant-specific LSU (response to Low SUlfur) gene family are induced by sulfur deficiency [103]. Although the precise function of LSU-like proteins is still unknown, they have been identified as important stress-related hubs in plant responses to sulfur deficiency [103]. Interestingly, LSU proteins from Arabidopsis interact with CAT2 and NBR1, suggesting a possible role of NBR1 in plant S nutrient responses [103] (Figure 2). Consistent with this possibility, plants exposed to S deficit induced autophagy and the elevated transcription of NBR1, suggesting an increased demand for NBR1 during the activation of autophagy under the conditions of S deficiency [104]. Indeed, transcriptome analysis has revealed that NBR1 overexpression altered plant gene expression in response to the low S conditions [104]. One of the NBR1-regulated gene encodes RPS3, which also interacts with NBR1 in a manner independent of the UBA domains of NBR1 [104]. The interaction of NBR1 with a S deficiency-regulated ribosome protein suggests a possible role of NBR1 in ribosomes remodeling in response to S starvation. Furthermore, Arabidopsis seedling overexpressing NBR1 have significantly shorter roots than wild type when grown under S deficient conditions in the presence of TOR kinase inhibitors [104]. Thus, overexpression of NBR1 increases sensitivity to inhibition of TOR kinase under S starvation condition, both of which induce autophagy [104]. Apparently, excessive autophagy induction by the additive effect of NBR1 overexpression, starvation, and TOR inhibition cause detrimental effects on Arabidopsis growth.
8. NBR1 in Abscisic Acid (ABA) Signaling
ABA is a plant hormone that regulates a wide range of cellular and molecular processes in plant growth, development and response to both biotic and abiotic stresses [105,106]. A recently reported study has provided lines of evidence for a role of NBR1 in ABA signaling [107]. First, transcriptomic analysis of the shoots and roots of transgenic Arabidopsis NBR1-overexpressing line revealed differential gene expression in comparison with the nontransgenic wild-type plants [107]. These differentially expressed genes include those involved in responses to stimuli and stress in shoots and those related to translation and formation of ribonucleoprotein complexes in roots [107]. Analysis of interaction network of these differentially expressed genes also indicated a link between NBR1 overexpression and ABA signaling based on the observation that most hubs of this network were associated with ABA signaling [107]. Second, transgenic NBR1-overexpression lines and knockout mutants display phenotypes indicative of fine-tuning of the ABA signaling pathway [107]. These phenotypes include delayed germination, increased number of closed stomata in the NBR1-overexpression lines and increased number of lateral root initiation sites in the knockout mutants [107]. Third, despite altered gene expression associated with ABA responses in the NBR1-overexpressing lines, ABA levels were unchanged in the shoots, suggesting a possible effect of NBR1 on ABA signaling mechanisms [107]. Using bimolecular fluorescence complementation (BiFC), it was demonstrated that NBR1 interacted with three regulatory proteins of ABA pathway (ABI3, ABI4 and ABI5) in planta [107] (Figure 2). NBR1 interaction with ABI5 required its UBA domain but the interactions with ABI3 and ABI4 are independent of the ubiquitin-binding domains [107]. This observation suggests that ABI5, but not ABI3 or ABI4, requires ubiquitination prior to interaction with NBR1. Consistent with the results from BiFC, pull-down assay using recombinant proteins produced in E. coli confirmed the binding of ABI3 and ABI4, but not ABI5, to NBR1, supporting the importance of ubiquitination for ABI5 in its interaction with NBR1. It would be of interest to determine the effect of NBR1 binding on the protein levels of the regulatory proteins of ABA signaling.
9. Conclusions and Prospect
As a structural and functional homolog of mammalian p62 and NBR1, plant NBR1 also acts as selective receptor for aggrephagy that targets misfolded proteins and protein aggregates that are generated under a variety of stress conditions. The conserved role of plant NBR1 in selective autophagy likely underlies its critical roles in plant responses to a broad spectrum of biotic and abiotic stresses including heat, drought, salt and oxidative stresses. Plant NBR1 also target specific protein cargoes including heat induced protein chaperones, which paradoxically reduces heat stress memory and compromises tolerance to subsequent heat stress, probably as a mechanism to expedite the recovery of plant growth after the cessation of a heat stress. NBR1 is also increasingly recognized as a critical player in plant antiviral immunity by directly targeting specific viral proteins for degradation in the vacuole. This role of plant NBR1 in plant antiviral defense and counter-defense mechanism through exploitation of plant autophagy by viral pathogens can open new frontiers in the study of the dynamic and complex interactions between plants and microbial pathogens. Through targeting specific target proteins, NBR1-mediated selective autophagy also participates in the modulation of other important processes in plants including stress-induced pexophagy, S nutrient responses and ABA signaling. Despite the progresses, our understanding of NBR1-mediated selective autophagy is still at its infancy. The number of identified cargo proteins for plant NBR1 is still very limited and, as a result, the underlying mechanisms for the broad biological functions of plant NBR1 are still not well understood. Currently there is no comprehensive knowledge about the pathways that regulate the protein levels and activity of plant NBR1 beyond the established fact that the selective autophagy receptor is itself subjected to degradation in the vacuole during autophagy. Animal NBR1 proteins also have novel roles in signaling independent of autophagy and it remains to be determined whether plant NBR1 has similar regulatory roles in signaling in a manner independent of autophagy. A better knowledge about the broad and complex roles of NBR1 will provide new important insights into the molecular basis of plant responses to biotic and abiotic stresses.
Author Contributions
Y.Z. and Z.C. conceived the idea, wrote and evaluated the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Key Research and Development Program of Lishui (Grant No. 2020ZDYF08) and the U.S. National Science Foundation (IOS1456300).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yin Z., Pascual C., Klionsky D.J. Autophagy: Machinery and regulation. Microb. Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky D.J. Autophagy participates in, well, just about everything. Cell Death Differ. 2020;27:831–832. doi: 10.1038/s41418-020-0511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glick D., Barth S., Macleod K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 5.Bassham D.C. Plant autophagy--more than a starvation response. Curr. Opin. Plant Biol. 2007;10:587–593. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Kwon S.I., Park O.K. Autophagy in plants. J. Plant Biol. 2008;51:313–320. doi: 10.1007/BF03036132. [DOI] [Google Scholar]
- 7.Mizushima N. Autophagy. FEBS Lett. 2010;584:1279. doi: 10.1016/j.febslet.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Bassham D.C., Laporte M., Marty F., Moriyasu Y., Ohsumi Y., Olsen L.J., Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy. 2006;2:2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- 9.Signorelli S., Tarkowski L.P., Van den Ende W., Bassham D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019;24:413–430. doi: 10.1016/j.tplants.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Xiong Y., Bassham D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 12.Pu Y., Bassham D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013;8:e24297. doi: 10.4161/psb.24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J., Wang J., Cheng Y., Chi Y.J., Fan B., Yu J.Q., Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9:e1003196. doi: 10.1371/journal.pgen.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J., Wang J., Yu J.Q., Chen Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014;5:174. doi: 10.3389/fpls.2014.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayward A.P., Tsao J., Dinesh-Kumar S.P. Autophagy and plant innate immunity: Defense through degradation. Semin. Cell Dev. Biol. 2009;20:1041–1047. doi: 10.1016/j.semcdb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Lenz H.D., Haller E., Melzer E., Gust A.A., Nurnberger T. Autophagy controls plant basal immunity in a pathogenic lifestyle-dependent manner. Autophagy. 2011;7:773–774. doi: 10.4161/auto.7.7.15535. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Schiff M., Czymmek K., Talloczy Z., Levine B., Dinesh-Kumar S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Wu Y., Tang D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav. 2011;6:1408–1410. doi: 10.4161/psb.6.9.16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J., Yu J.Q., Chen Z. The perplexing role of autophagy in plant innate immune responses. Mol Plant Pathol. 2014;15:637–645. doi: 10.1111/mpp.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai Z., Wang F., Zheng Z., Fan B., Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66:953–968. doi: 10.1111/j.1365-313X.2011.04553.x. [DOI] [PubMed] [Google Scholar]
- 23.Hanamata S., Kurusu T., Kuchitsu K. Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 2014;5:457. doi: 10.3389/fpls.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison-Lowe N.J., Olsen L.J. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy. 2008;4:339–348. doi: 10.4161/auto.5629. [DOI] [PubMed] [Google Scholar]
- 25.Kurusu T., Koyano T., Hanamata S., Kubo T., Noguchi Y., Yagi C., Nagata N., Yamamoto T., Ohnishi T., Okazaki Y., et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014;10:878–888. doi: 10.4161/auto.28279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Floyd B.E., Morriss S.C., Macintosh G.C., Bassham D.C. What to eat: Evidence for selective autophagy in plants. J. Integr. Plant Biol. 2012;54:907–920. doi: 10.1111/j.1744-7909.2012.01178.x. [DOI] [PubMed] [Google Scholar]
- 27.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft C., Peter M., Hofmann K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 29.Shaid S., Brandts C.H., Serve H., Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall R.S., Hua Z., Mali S., McLoughlin F., Vierstra R.D. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell. 2019;177:766–781.e24. doi: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall R.S., Li F., Gemperline D.C., Book A.J., Vierstra R.D. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol. Cell. 2015;58:1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall M. An MAQ innovation. Med. Anthropol. Q. 2001;15:3–8. doi: 10.1525/maq.2001.15.1.3. [DOI] [PubMed] [Google Scholar]
- 33.Germain K., Kim P.K. Pexophagy: A Model for Selective Autophagy. Int. J. Mol. Sci. 2020;21:578. doi: 10.3390/ijms21020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens S., Behrends C. Molecular Mechanisms of Selective Autophagy. J. Mol. Biol. 2020;432:1–2. doi: 10.1016/j.jmb.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23:785–805. doi: 10.1105/tpc.110.081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hachez C., Veljanovski V., Reinhardt H., Guillaumot D., Vanhee C., Chaumont F., Batoko H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell. 2014;26:4974–4990. doi: 10.1105/tpc.114.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhee C., Guillon S., Masquelier D., Degand H., Deleu M., Morsomme P., Batoko H. A TSPO-related protein localizes to the early secretory pathway in Arabidopsis, but is targeted to mitochondria when expressed in yeast. J. Exp. Bot. 2011;62:497–508. doi: 10.1093/jxb/erq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avin-Wittenberg T., Michaeli S., Honig A., Galili G. ATI1, a newly identified atg8-interacting protein, binds two different Atg8 homologs. Plant Signal. Behav. 2012;7:685–687. doi: 10.4161/psb.20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaeli S., Honig A., Levanony H., Peled-Zehavi H., Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014;26:4084–4101. doi: 10.1105/tpc.114.129999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J., Wang Z., Wang X., Li X., Zhang Z., Fan B., Zhu C., Chen Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14:487–504. doi: 10.1080/15548627.2017.1422856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nolan T.M., Brennan B., Yang M., Chen J., Zhang M., Li Z., Wang X., Bassham D.C., Walley J., Yin Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell. 2017;41:33–46.e7. doi: 10.1016/j.devcel.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang F., Kimberlin A.N., Elowsky C.G., Liu Y., Gonzalez-Solis A., Cahoon E.B., Alfano J.R. A Plant Immune Receptor Degraded by Selective Autophagy. Mol. Plant. 2019;12:113–123. doi: 10.1016/j.molp.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Stephani M., Picchianti L., Gajic A., Beveridge R., Skarwan E., Sanchez de Medina Hernandez V., Mohseni A., Clavel M., Zeng Y., Naumann C., et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 2020;9 doi: 10.7554/eLife.58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerakis Y., Quintero M., Li H., Hetz C. The UFMylation System in Proteostasis and Beyond. Trends Cell Biol. 2019;29:974–986. doi: 10.1016/j.tcb.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svenning S., Lamark T., Krause K., Johansen T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011;7:993–1010. doi: 10.4161/auto.7.9.16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzynska A., Sirko A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011;7:1145–1158. doi: 10.4161/auto.7.10.16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J., Zhang Y., Qi J., Chi Y., Fan B., Yu J.Q., Chen Z. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi: 10.1371/journal.pgen.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkin V., Lamark T., Johansen T., Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 49.Lamark T., Kirkin V., Dikic I., Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 50.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puvirajesinghe T.M., Bertucci F., Jain A., Scerbo P., Belotti E., Audebert S., Sebbagh M., Lopez M., Brech A., Finetti P., et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat. Commun. 2016;7:10318. doi: 10.1038/ncomms10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim M.J., Min Y., Im J.S., Son J., Lee J.S., Lee K.Y. p62 is Negatively Implicated in the TRAF6-BECN1 Signaling Axis for Autophagy Activation and Cancer Progression by Toll-Like Receptor 4 (TLR4) Cells. 2020;9:1142. doi: 10.3390/cells9051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moscat J., Karin M., Diaz-Meco M.T. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nezis I.P., Stenmark H. p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid. Redox Signal. 2012;17:786–793. doi: 10.1089/ars.2011.4394. [DOI] [PubMed] [Google Scholar]
- 55.Duran A., Amanchy R., Linares J.F., Joshi J., Abu-Baker S., Porollo A., Hansen M., Moscat J., Diaz-Meco M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renny-Byfield S., Wendel J.F. Doubling down on genomes: Polyploidy and crop plants. Am. J. Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- 57.Udall J.A., Wendel J.F. Polyploidy and crop improvement. Crop Sci. 2006;46:S3–S14. doi: 10.2135/cropsci2006.07.0489tpg. [DOI] [Google Scholar]
- 58.Zhang K., Wang X., Cheng F. Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Hortic. Plant J. 2019;5:231–239. doi: 10.1016/j.hpj.2019.11.003. [DOI] [Google Scholar]
- 59.Ji C., Zhou J., Guo R., Lin Y., Kung C.H., Hu S., Ng W.Y., Zhuang X., Jiang L. AtNBR1 Is a Selective Autophagic Receptor for AtExo70E2 in Arabidopsis. Plant Physiol. 2020;184:777–791. doi: 10.1104/pp.20.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jung H., Lee H.N., Marshall R.S., Lomax A.W., Yoon M.J., Kim J., Kim J.H., Vierstra R.D., Chung T. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 2020;71:73–89. doi: 10.1093/jxb/erz404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ustun S., Hofius D. Anti- and pro-microbial roles of autophagy in plant-bacteria interactions. Autophagy. 2018;14:1465–1466. doi: 10.1080/15548627.2018.1475817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young P.G., Passalacqua M.J., Chappell K., Llinas R.J., Bartel B. A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy. 2019;15:941–959. doi: 10.1080/15548627.2019.1569915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kotak S., Larkindale J., Lee U., von Koskull-Doring P., Vierling E., Scharf K.D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 64.Qu A.L., Ding Y.F., Jiang Q., Zhu C. Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 2013;432:203–207. doi: 10.1016/j.bbrc.2013.01.104. [DOI] [PubMed] [Google Scholar]
- 65.von Koskull-Doring P., Scharf K.D., Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 66.Ahmed S.F., Deb S., Paul I., Chatterjee A., Mandal T., Chatterjee U., Ghosh M.K. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 2012;287:15996–16006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ballinger C.A., Connell P., Wu Y., Hu Z., Thompson L.J., Yin L.Y., Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell Biol. 1999;19:4535–4545. doi: 10.1128/MCB.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai Q., Zhang C., Wu Y., McDonough H., Whaley R.A., Godfrey V., Li H.H., Madamanchi N., Xu W., Neckers L., et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S., Wu X., Zhang J., Chen Y., Xu J., Xia X., He S., Qiang F., Li A., Shu Y., et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut. 2013;62:496–508. doi: 10.1136/gutjnl-2011-301522. [DOI] [PubMed] [Google Scholar]
- 70.Luo J., Shen G., Yan J., He C., Zhang H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 2006;46:649–657. doi: 10.1111/j.1365-313X.2006.02730.x. [DOI] [PubMed] [Google Scholar]
- 71.Shen G., Adam Z., Zhang H. The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts. Plant J. 2007;52:309–321. doi: 10.1111/j.1365-313X.2007.03239.x. [DOI] [PubMed] [Google Scholar]
- 72.Yan J., Wang J., Li Q., Hwang J.R., Patterson C., Zhang H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003;132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brzezinka K., Altmann S., Czesnick H., Nicolas P., Gorka M., Benke E., Kabelitz T., Jahne F., Graf A., Kappel C., et al. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. Elife. 2016;5 doi: 10.7554/eLife.17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar S.V., Wigge P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 75.Sedaghatmehr M., Mueller-Roeber B., Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat. Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sedaghatmehr M., Thirumalaikumar V.P., Kamranfar I., Marmagne A., Masclaux-Daubresse C., Balazadeh S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019;42:1054–1064. doi: 10.1111/pce.13426. [DOI] [PubMed] [Google Scholar]
- 77.Thirumalaikumar V.P., Gorka M., Schulz K., Masclaux-Daubresse C., Sampathkumar A., Skirycz A., Vierstra R.D., Balazadeh S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy. 2020:1–16. doi: 10.1080/15548627.2020.1820778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meiri D., Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009;59:387–399. doi: 10.1111/j.1365-313X.2009.03878.x. [DOI] [PubMed] [Google Scholar]
- 80.Deretic V., Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao L., Eissa N.T. Autophagy in Pulmonary Innate Immunity. J. Innate Immun. 2020;12:21–30. doi: 10.1159/000497414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Richetta C., Faure M. Autophagy in antiviral innate immunity. Cell. Microbiol. 2013;15:368–376. doi: 10.1111/cmi.12043. [DOI] [PubMed] [Google Scholar]
- 83.Witko-Sarsat V., Codogno P. Autophagy and innate immunity. J. Innate Immun. 2013;5:425–426. doi: 10.1159/000353153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agius C., Eamens A.L., Millar A.A., Watson J.M., Wang M.B. RNA silencing and antiviral defense in plants. Methods Mol. Biol. 2012;894:17–38. doi: 10.1007/978-1-61779-882-5_2. [DOI] [PubMed] [Google Scholar]
- 85.Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., Isono E., Schumacher K., Genschik P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA. 2012;109:15942–15946. doi: 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahara K.S., Masuta C., Yamada S., Shimura H., Kashihara Y., Wada T.S., Meguro A., Goto K., Tadamura K., Sueda K., et al. Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA. 2012;109:10113–10118. doi: 10.1073/pnas.1201628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hafren A., Macia J.L., Love A.J., Milner J.J., Drucker M., Hofius D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA. 2017;114:E2026–E2035. doi: 10.1073/pnas.1610687114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hafren A., Ustun S., Hochmuth A., Svenning S., Johansen T., Hofius D. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol. 2018;176:649–662. doi: 10.1104/pp.17.01198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu J., Guo W. The exocyst complex in exocytosis and cell migration. Protoplasma. 2012;249:587–597. doi: 10.1007/s00709-011-0330-1. [DOI] [PubMed] [Google Scholar]
- 90.Ding Y., Wang J., Chun Lai J.H., Ling Chan V.H., Wang X., Cai Y., Tan X., Bao Y., Xia J., Robinson D.G., et al. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell. 2014;25:412–426. doi: 10.1091/mbc.e13-10-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J., Ding Y., Wang J., Hillmer S., Miao Y., Lo S.W., Wang X., Robinson D.G., Jiang L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell. 2010;22:4009–4030. doi: 10.1105/tpc.110.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munz C. Autophagy Proteins in Viral Exocytosis and Anti-Viral Immune Responses. Viruses. 2017;9:288. doi: 10.3390/v9100288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Munz C. Autophagy Proteins in Phagocyte endocytosis and exocytosis. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Papandreou M.E., Tavernarakis N. Crosstalk between Endo/Exocytosis and Autophagy in Health and Disease. Biotechnol. J. 2020;15 doi: 10.1002/biot.201900267. [DOI] [PubMed] [Google Scholar]
- 95.Kulich I., Pecenkova T., Sekeres J., Smetana O., Fendrych M., Foissner I., Hoftberger M., Zarsky V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14:1155–1165. doi: 10.1111/tra.12101. [DOI] [PubMed] [Google Scholar]
- 96.Pecenkova T., Markovic V., Sabol P., Kulich I., Zarsky V. Exocyst and autophagy-related membrane trafficking in plants. J. Exp. Bot. 2018;69:47–57. doi: 10.1093/jxb/erx363. [DOI] [PubMed] [Google Scholar]
- 97.Calero-Muoz N., Exposito-Rodriguez M., Collado-Arenal A.M., Rodrguez-Serrano M., Laureano-Marn A.M., Santamara M.E., Gotor C., Daz I., Mullineaux P.M., Romero-Puertas M.C., et al. Cadmium induces reactive oxygen species-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. 2019;42:2696–2714. doi: 10.1111/pce.13597. [DOI] [PubMed] [Google Scholar]
- 98.Kopriva S., Malagoli M., Takahashi H. Sulfur nutrition: Impacts on plant development, metabolism, and stress responses. J. Exp. Bot. 2019;70:4069–4073. doi: 10.1093/jxb/erz319. [DOI] [PubMed] [Google Scholar]
- 99.Kopriva S. Plant sulfur nutrition: From Sachs to Big Data. Plant Signal. Behav. 2015;10:e1055436. doi: 10.1080/15592324.2015.1055436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang L., Kawaguchi R., Morikawa-Ichinose T., Allahham A., Kim S.J., Maruyama-Nakashita A. Sulfur Deficiency-Induced Glucosinolate Catabolism Attributed to Two beta-Glucosidases, BGLU28 and BGLU30, is Required for Plant Growth Maintenance under Sulfur Deficiency. Plant Cell Physiol. 2020;61:803–813. doi: 10.1093/pcp/pcaa006. [DOI] [PubMed] [Google Scholar]
- 101.Lewandowska M., Wawrzynska A., Moniuszko G., Lukomska J., Zientara K., Piecho M., Hodurek P., Zhukov I., Liszewska F., Nikiforova V., et al. A contribution to identification of novel regulators of plant response to sulfur deficiency: Characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol. Plant. 2010;3:347–360. doi: 10.1093/mp/ssq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lewandowska M., Sirko A. Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Pol. 2008;55:457–471. doi: 10.18388/abp.2008_3051. [DOI] [PubMed] [Google Scholar]
- 103.Niemiro A., Cysewski D., Brzywczy J., Wawrzynska A., Sienko M., Poznanski J., Sirko A. Similar but Not Identical-Binding Properties of LSU (Response to Low Sulfur) Proteins FromArabidopsis thaliana. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tarnowski L., Rodriguez M.C., Brzywczy J., Cysewski D., Wawrzynska A., Sirko A. Overexpression of the Selective Autophagy Cargo Receptor NBR1 Modifies Plant Response to Sulfur Deficit. Cells. 2020;9:669. doi: 10.3390/cells9030669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mehrotra R., Bhalothia P., Bansal P., Basantani M.K., Bharti V., Mehrotra S. Abscisic acid and abiotic stress tolerance—Different tiers of regulation. J. Plant Physiol. 2014;171:486–496. doi: 10.1016/j.jplph.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 107.Tarnowski L., Rodriguez M.C., Brzywczy J., Piecho-Kabacik M., Krckova Z., Martinec J., Wawrzynska A., Sirko A. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-64765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]