Abstract
Simple Summary
Long non-coding RNAs (lncRNA) have been associated with a number of diseases including cancer. A well-studied lncRNA called XIST (X-inactive specific transcript) acts as a major effector of the X-inactivation process. It is expressed on the inactive X chromosome providing a dosage equivalence between males and females. Recently XIST has been implicated in the development of lung cancer. Using a bioinformatics approach, we demonstrate the XIST is over-expressed in female patients compared to males. When XIST gene was silenced in two different cell lines (of male and female origin), a number of genes were differentially expressed; playing a role in signal transduction pathways, energy balance and metabolism, thus providing a better insight of the role of this lncRNA in cancer. Finally, we showed that expression of XIST with another 4 genes provided a strong diagnostic potential to discriminate lung cancer from healthy controls.
Abstract
Long non-coding RNAs (lncRNAs) perform a wide functional repertoire of roles in cell biology, ranging from RNA editing to gene regulation, as well as tumour genesis and tumour progression. The lncRNA X-inactive specific transcript (XIST) is involved in the aetiopathogenesis of non-small cell lung cancer (NSCLC). However, its role at the molecular level is not fully elucidated. The expression of XIST and co-regulated genes TSIX, hnRNPu, Bcl-2, and BRCA1 analyses in lung cancer (LC) and controls were performed in silico. Differentially expressed genes (DEGs) were determined using RNA-seq in H1975 and A549 NSCLC cell lines following siRNA for XIST. XIST exhibited sexual dimorphism, being up-regulated in females compared to males in both control and LC patient cohorts. RNA-seq revealed 944 and 751 DEGs for A549 and H1975 cell lines, respectively. These DEGs are involved in signal transduction, cell communication, energy pathways, and nucleic acid metabolism. XIST expression associated with TSIX, hnRNPu, Bcl-2, and BRCA1 provided a strong collective feature to discriminate between controls and LC, implying a diagnostic potential. There is a much more complex role for XIST in lung cancer. Further studies should concentrate on sex-specific changes and investigate the signalling pathways of the DEGs following silencing of this lncRNA.
Keywords: XIST, x-inactivation center, NSCLC, lncRNA, TCGA, GTEX, RNA-seq, lung cancer, bioinformatics
1. Introduction
The large-scale genome sequencing studies of the last decade have documented the pervasive transcription of almost 90% of the human genome [1], with 98% of the transcriptome consisting of long non-coding RNAs (lncRNAs) [2]. However, despite these efforts, our current understanding of lncRNAs’ complex biology is still limited. Long non-coding RNAs are a class of RNA molecules that do not encode proteins and range in length from 200 nucleotides to 100 kilobases [3]. Previous analyses have pointed to a wide range of functions for lncRNAs in developmental and cellular processes, including gene expression, chromatin remodeling and modification, splicing, editing, translation and degradation of the RNA, and gene silencing with endogenous small interfering RNA (siRNA) [4,5,6].
LncRNAs were found to be expressed in a variety of diseases including cancer suggesting potential roles as biomarkers or even therapeutic targets. Studies have identified key lncRNAs as regulators of oncogenes and tumour suppressors such as PTEN and KRAS [7,8,9]. Other lncRNAs were shown to be important for genomic imprinting, and for regulating epigenetic procedures. This is the case for lncRNA X-inactive specific transcript (XIST) that inactivates one of the two X chromosomes in females [10,11,12].
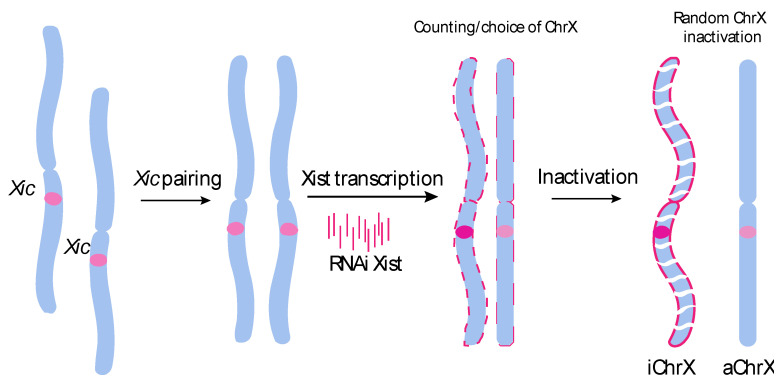
In literature, XIST is described interchangeably as either a lncRNA or a pseudogene and acts as a major effector in the X chromosome inactivation process. It is expressed only on the inactive X chromosome providing a dosage equivalence between males and females (reviewed in [10,13]). XIST was the first non-coding gene identified in the X inactivation center (XIC) region [14]. The expression of this lncRNA is the essential step for the initiation of the X inactivation. XIST is transcribed, spliced, and polyadenylated resulting in an mRNA. However, no protein products have ever been observed. XIST is coating and inactivates one of the X chromosomes (Figure 1). However, the lack of XIST activity leads to the failed inactivation and duplication of the gene on the second X chromosome, resulting in its activation [14,15].
Figure 1.
Process of X chromosome inactivation (Xi). XIST is triggering an X chromosome inactivation while the other X chromosome remains active. XIST is spreading along one of the X chromosomes (coating) and its binding triggers the chromatin inactivation. Xic: X-inactivation center, iChrX: inactive Xchr, aChrX: active Xchr [14].
Another key lncRNA that mediates the X chromosome inactivation is TSIX. TSIX acts as a XIST repressor. TSIX is the antisense RNA of XIST and their differential expression patterns define the activation or inactivation of the X chromosome. Apart from the XIST-TSIX mechanism, XIST RNA and the nuclear matrix protein heterogeneous nuclear ribonucleoprotein U (hnRNPu) interact and, upon the depletion of hnRNPu, XIST is detached from the inactive X chromosome (Xi) and diffusely localized into the nucleoplasm [10,15].
Recent studies have shown that XIST has an aberrant expression pattern in breast cancer [16], cervical squamous cell carcinoma [17], colorectal cancer [18], gastric cancer [19], glioma [20], hepatocellular carcinoma [21], nasopharyngeal carcinoma [22], non-small cell lung cancer (NSCLC) [23], pancreatic cancer [24], osteosarcoma [25], and ovarian cancer [26]. Furthermore, XIST was shown to regulate tumour cell migration, proliferation, and invasion, in NSCLC [6,27]. Wang et al. showed that cell lines and patient samples of NSCLC overexpressed XIST and shown that XIST knockdown inhibits tumour growth in vivo. Moreover, XIST exhibits oncogenic properties by regulating the miR-449a and B-cell lymphoma 2 (Bcl-2) gene in NSCLC [5]. The same study has shown that BRCA1 also influences the concentration of XIST on the Xi. Specifically, RNAi of BRCA1 decreases the concentration of XIST on Xi, and the reduction of BRCA1 by Cre-mediated excision also decreases XIST concentration on Xi [28]. Collectively these data indicate that XIST could be an important novel biomarker for the detection of NSCLC [23,29]. We hypothesize that there is a higher order of complexity in the regulation of XIST and its impact in multiple signaling pathways. In this study, we investigate changes in the transcriptional landscape of cell lines from male and female patients with NSCLC where XIST was downregulated and studied the expression and correlation of associated genes in lung cancer.
2. Results
2.1. Expression Level of XIST, TSIX, hnRNPu, Bcl-2, and BRCA1 in NSCLC
Leveraging the available expression data from TCGA and GTEX, as tumour and respectively normal control lung samples (referring as ‘Lung’ in figures), we have investigated differential expression patterns of XIST, TSIX, hnRNPu, Bcl-2, and BRAC1 in lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) (Supplementary Figure S1).
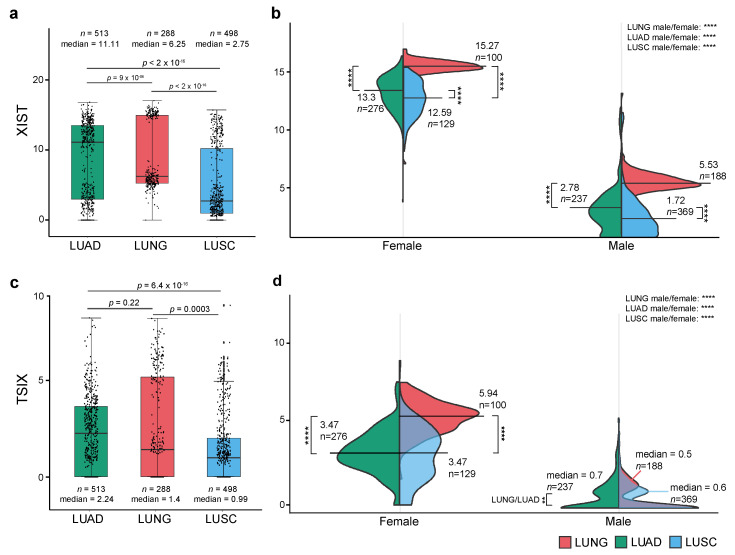
Overall, similar to previously published results where XIST is upregulated in various cancers [6,30], we found a similar trend, in LUAD compared to normal lung cohort (LUNG), whereas in LUSC a downregulation was observed (Figure 2a). However, given the involvement of XIST in X chromosome inactivation, we dissected these data further by measuring the expression of XIST in males and females. Here, a different picture emerges, suggesting a gender-specific downregulation in tumour samples compared to control Figure 2b. For TSIX, a modest overall upregulation in LUAD was observed, whereas in LUSC a significant downregulation when compared to normal lung (LUNG) was noted (Figure 2c). Further gender stratification showed the emergence of a similar distribution as noted earlier for XIST (Figure 2d).
Figure 2.
XIST and TSIX expression. (a) Boxplots of the overall XIST expression in LUAD, LUSC and normal lung (LUNG), (b) Violin plots of the gender-specific expression of XIST in LUAD, LUSC and LUNG, (c) Boxplots of the overall TSIX expression in LUAD, LUSC, and LUNG (d) Violin plots of the gender-specific expression of TSIX in LUAD, LUSC, and normal lung. All expression data is presented in units of log2(norm_count + 1). Median expression and total sample size (count, n) are shown. p-values were calculated the Wilcoxon test in R, statistically, significant represented as ****: p ≤ 0.0001, non-significant results are not labelled in the graph.
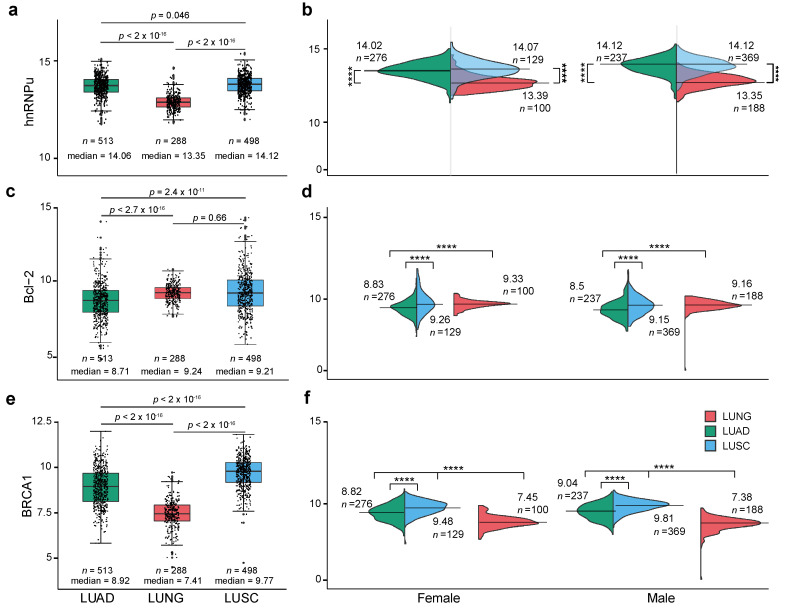
Next, we investigated the expression patterns of hnRNPu, Bcl-2, and BRCA1 in the same cohorts of patients (Figure 3). Overall, we found that the expression of hnRNPu and BRCA1 in LUAD and LUSC is significantly upregulated compared to the normal lung samples in both males and females. On the other hand, Bcl-2 appears to be downregulated only in LUAD when compared to controls. Moreover, the three genes did not display any gender-specific expression patterns as observed earlier for XIST and TSIX suggesting that the transcriptional landscape of hnRNPu, Bcl-2, and BRCA1 is gender agnostic.
Figure 3.
Gene expression patterns for (a,b) hnRNPu, (c,d) Bcl-2 and (e,f) BRCA1 in units of log2(norm_count + 1) for normal lung, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Median expression and sample size were labelled. Median expression and total sample size (count, n) are shown. p-values were calculated the Wilcoxon test in R, statistically, significant represented as ****: p ≤ 0.0001, non-significant results are not labelled in the graph.
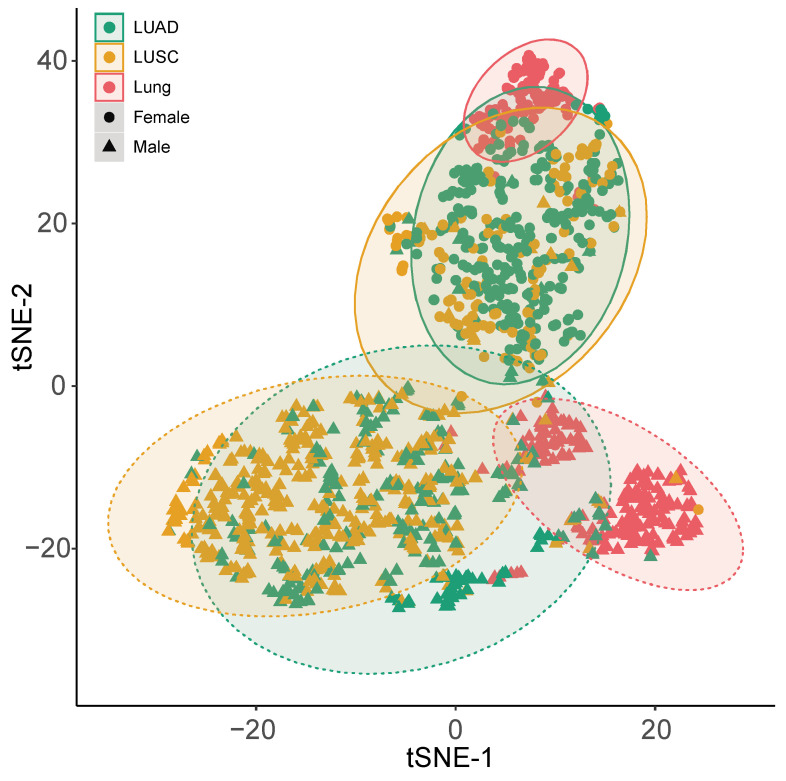
Following the sample and gender-specific expression analysis, we used T-distributed stochastic neighbor embedding (t-SNE) to evaluate the discriminatory power of the gene expression patterns to differentiate between normal and tumour samples (Figure 4 and Supplementary Figure S2). Visual inspection of the t-SNE plot showed a clear and distinct clustering of male and female samples. Furthermore, by combining the expression information, we were able to discriminate between normal and cancer samples, suggesting a good collective diagnosis biomarker potential for the five genes. Notably, some cancer samples are overlapping healthy samples, suggesting that some healthy individual’s expression pattern of the five genes is similar to the ones observed commonly in tumour samples, hinting at the potential existence of cancer precursors in healthy individuals.
Figure 4.
Clustering of tumour and healthy lung samples in a 2D map using t-SNE algorithm based on the normalized expression levels of XIST, TSIX, hnRNPu, Bcl-2, and BRCA1. tSNE-1 and tSNE-2 are the tSNE projection axes.
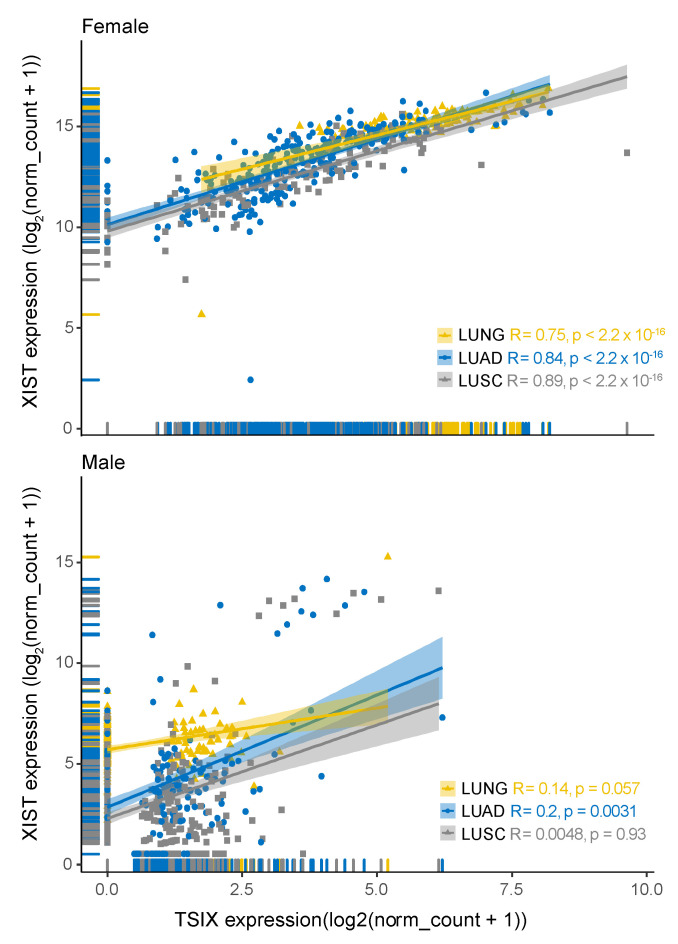
Next, we investigated whether the XIST and the four associated genes have correlated expression patterns in NSCLC (Supplementary Figure S3). For this, we used Spearman’s rank correlation tests evaluating male and female samples individually. We found that XIST has an independent expression pattern with respect to hnRNPu, Bcl-2, and BRCA1. However, as expected we observed a strong correlation between XIST and TSIX expressions in samples derived from female patients (R = 0.75 for LUNG cohort, 0.84 for LUAD and 0.89 for LUSC) (Figure 5).
Figure 5.
Correlation with regression line for gene expression (in units if log2(norm_count + 1)) for XIST vs. TSIX, Spearman’s rank correlation test was used to generate the correlation coefficient represented as R in the analyses, p represents p-value. Healthy lung, LUAD, and LUSC were compared separately in both sexes. Total sample number is 1299, 794 derived from male patients and 505 from female patients. There are 116 samples from male patients with 0 expression in XIST, 461 in TSIX. In female patients derived samples, 17 show zero expression for TSIX gene. All samples with zero expression in both genes were removed from the analysis.
2.2. Functional Analysis
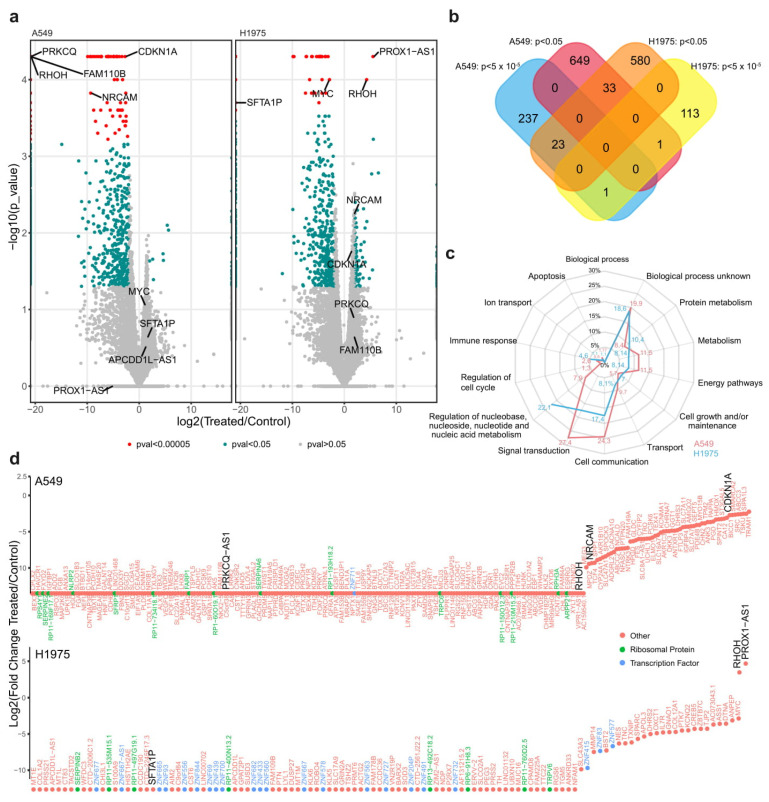
Following the sexual dimorphism of XIST, we investigated the change in the genome transcriptional landscape A549 (derived from male patient) and H1975 (derived from female patient) NSCLC cell lines upon silencing XIST using siRNA (Figure 6). The differential expressed genes were identified using RNAseq analysis. The results are summarized in the Vulcan plots. Overall, we distinguished three types of genes (Figure 6a). First, there are genes (shown in red) that show a statistically significant (p-value < 10−4) differential transcription pattern, with a large absolute value of log2FoldChange > 2, between the two analyzed conditions. Second, there are genes (seen in green) that show a moderately significant change in expression (p-value < 0.05) and an absolute value of log2FoldChange > 2. Finally, some genes do not show any significant change in the transcription levels between the analyzed states (shown here in grey).
Figure 6.
(a) Vulcan plots for the A549 and H1975 gene expression. Data shows mostly up-regulation or down-regulation of genes in the siRNA XIST treated samples in comparison to non-treated ones (left of center = down-regulation, right of center = up-regulation), with a handful of genes shown to be very significantly up or down-regulated. (b) Venn diagram showing the total genes significantly dysregulated for both cell lines. (c) The biological processes where the down-regulated genes are found. Most of the dysregulated genes in both cells lines are playing a significant role in signal transduction (27.4% for the A549), cell-to-cell communication (24.3% for the A549 and 17.4% for the H1975) and regulation of nucleic acid metabolism (22.1% for the H1975 and 7.9% for the A549), (d) Ranking of all the highly significantly differentially expressed genes (DEGs) in both cell lines (p < 5 × 10−5).
Overall, 944 genes were significantly dysregulated after the treatment with the siRNA XIST for the A549 cell line. From those genes, the 683 were downregulated at p < 0.05, and 261 were highly-significantly down-regulated at p < 5 × 10−5. In the H1975, 751 genes were significantly dysregulated after the treatment with the siRNA XIST. 536 genes were downregulated at p < 0.05 and 113 significantly down-regulated p < 5 × 10−5 with two genes to present and highly significant upregulation after the transfection. The DEGs are presented as supplementary table. The two cell lines had 34 downregulated genes in common with p < 0.05, 24 downregulated genes with p < 5 × 10−5, and one gene (RHOH) that was up-regulated in the H1975 and down-regulated for the A549 (Figure 6b). Subsequent analysis of the differentially expressed genes (DEGs) using FunRich revealed distinct biological processes for the two cell lines, including signal transduction, cell communication, metabolism, and energy pathways (Figure 6c).
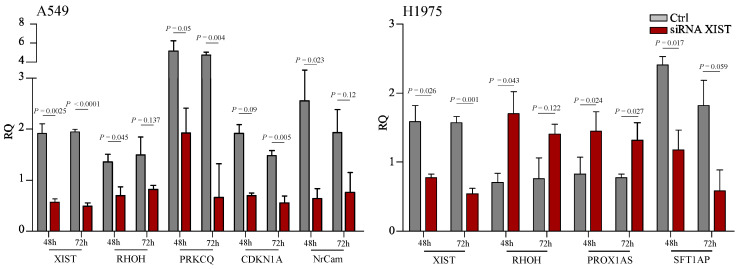
A summary of the top DEG in the A549 and H1975 cell lines respectively is shown in Figure 6d. In both cell lines, XIST was downregulated in agreement with previous studies that siRNA was used [6,27,31,32]. PROX1AS1 (antisense RNA) and RHOH (a negative regulator of cell growth and survival) were markedly upregulated in H1975 cells in agreement with RNAseq data. We expanded on these observations in A549 cells where RHOH, PRKCQ (required for the activation of the transcription factors NF-kB and AP-1), NrCAM (required for normal responses to cell-cell contacts in the brain and peripheral nervous system), and CDKN1A (involved in p53 mediated inhibition of cellular proliferation to DNA damage) were downregulated in siRNA treated cells in agreement with the general trend observed in RNAseq for this cell line (Figure 7).
Figure 7.
Relative expression of selected targets validating RNAseq data. For A549 cell line, XIST, RHOH, PRKCQ, CDKN1A and NrCam were validated using qRT-PCR. Significant downregulation corroborated the RNA-seq data. Similarly, XIST, RHOH, PROX1AS and SFT1AP were also assessed. Downregulation of XIST and SFT1AP was noted and upregulation of RHOH and PROX1AS following siRNA transfection. These data also corroborate the trends seen in RNA-seq for this cell line.
3. Discussion
In this study, we provide a novel insight into the expression of XIST and key co-expressed (?) genes. Leveraging the available large-scale sequence data from TCGA and GTEX, we demonstrate that XIST, TSIX, hnRNPu, Bcl-2, and BRCA1 are differentially expressed in two different types of lung cancer when compared to controls. Initial observations suggested that XIST was significantly upregulated in LUAD compared to controls. This finding corroborates previous studies where XIST has been shown to be over-expressed in lung cancer and other cancers [23,31,33,34,35,36,37].
However, subsequent stratification by sex revealed that XIST is highly expressed in females and downregulated in LUAD and LUSC when compared to normal controls. Although this might appear as a discrepancy, it is evident that there is a vast range in XIST’s gene expression when all samples are measured independently of gender. Thus, depending on data preparation and processing a potentially different picture can emerge. For example, in all earlier studies, XIST expression was not stratified in male and female lung cancer patients and numbers were considerably lower to the ones assessed in this study. Of note, our detailed analysis revealed the presence of two groups in control and LUAD cohorts (Figure 3a). Apart from issues relating to sex-specific expression, the preparation of RNA and subsequent analyses of data might also be contributing factors for the detection of lncRNAs. It has been recommended -when using sequencing for lncRNAs- that rRNA depletion (instead of polyA selection) should be performed [38]. If RNA-seq is polyA+ enriched, it will bias the analysis. It has also been suggested that 50 million paired reads are needed for tissues, as there are so many and low abundant lncRNAs. Another factor that can potentially induce different results is the method used for the quantification of lncRNAs [39]. Our data provides a deeper insight into the differential expression of XIST and highlights the need of standardization of RNA preparation protocols that would ultimately increase the transcriptomic comparability between different RNA-seq datasets, thus enabling a better mapping of lncRNAs [40,41].
Clustering of tumour and healthy lung samples in a 2D map using t-SNE algorithm based on the normalized expression levels of XIST, TSIX, hnRNPu, Bcl-2, and BRCA1, revealed that collectively these five genes -when assessed together- can have a diagnostic potential in both LUAD and LUSC. This observation warrants further investigation in both tissue and liquid biopsies from NSCLC patients using a much larger cohort especially for the healthy control group. Further, a strong correlation between XIST and TSIX was seen. TSIX is the antisense of XIST. It is the complementary sequence to XIST that is 40 kilobases long as is transcribed in the opposite direction across the XIST gene. Much like XIST, TSIX only acts on the chromosome it is produced by. The relationship between TSIX and XIST is inverse meaning that when the expression of TSIX is increased, XIST expression is reduced therefore it blocks inactivation in the cis or same X chromosome. When TSIX expression is reduced, XIST expression is increased and causes the inactivation of the X chromosome [4].
Silencing XIST in vitro resulted in 944 DEGs for A549 and 751 DEGs for H1975 cell lines respectively. A common trend for both cell lines was that the majority of DEGs were downregulated compared to controls, providing further evidence for a critical role of XIST in cell proliferation as it has been shown in previous studies [6].
For example, in A549 cells, genes like protein kinase C theta (PRKCQ/PKC-θ), cyclin-dependent kinase inhibitor 1A (CDKN1A/p21), neuro-glia-related cell-adhesion molecule (NrCAM) and ras homolog family member H (RHOH) were amongst the most downregulated genes. PKC-θ enhances anchorage-independent survival, growth-factor-independent proliferation, and migration and its down-regulation enhances anoikis [42]. In addition PKC-θ regulates cell cycle checkpoint pathways in lung cancer [43]. Cyclin-dependent kinase inhibitor 1A (p21) can exert multiple roles including progression of the cell cycle, DNA repair, apoptosis, and can also function as an oncogene which promoting tumour growth by inhibiting apoptosis. Furthermore, another role of p21 is that it can be used as a predictive biomarker of response to therapy in TP53 and KRAS mutated NSCLC [44,45,46]. NrCAM is upregulated in a series of carcinomas like papillary thyroid carcinomas [47,48] and significant overexpression of the NrCAM in SCLC was also noted in comparison to normal lungs [49]. Of note, stable expression of the NrCAM ectodomain in NIH3T3 cells induced tumorigenesis in mice [50]. RHOH on the other hand, is a negative regulator of cell growth and survival. Rho GTPases regulate cell migration, proliferation, survival, and death. All these cellular processes are crucial for the maintenance of normal tissues, but also contribute to cancer progression [51]. For example, RHOH expression levels correlate with prostate cancer progression [52].
In H1975 cells silencing XIST lead to a highly significant down-regulation of 113 genes. One of these genes was MYC, a family of regulator genes and proto-oncogenes that code for transcription factors. It is well known that c-myc oncogene is frequently amplified in lung tumours and has been linked to their malignancy [53]. In a conditional model for the metastasis of NSCLC, c-myc played a crucial role in this process [54], and is overexpressed in 40–75% of NSCLC cases [55]. Inhibiting MYC on NSCLC in mice, lead to rapid regression of tumours with mild reversible side effects [56]. It is possible therefore that MYC can be targeted via silencing XIST. In contrast to A549 cells, RHOH was upregulated in H1975 cells following downregulation of XIST. This warrants further research as it might be a case of a differential expression in a cell- or sex-specific manner. Future studies should involve further research into the role of XIST using a wider repertoire of in vitro models. Our current cell lines have an EGFR mutant phenotype harbouring a T790M resistance mutation and a L858R sensitizing mutation (H1975), whereas the A549 cell line is KRAS mutant. Of note, mutations of the inactive X chromosome constitute a feature of tumorigenesis [57].
Gene enrichment analyses revealed that for A549 cells the main biological processes (in terms of percentage of genes involved) are signal transduction, cell communication, energy pathways, and metabolism; with the latter two having a number of same genes involved. For example, CYP24A1 has oncogenic properties in lung adenocarcinoma [58,59] and AKR1B10 has been shown to induce the metastatic potential of lung cancer cells to the brain in vitro. MicroRNA-1304 inhibited the growth of an NSCLC cell line by targeting HMOX1 [60]. Similarly, enrichment analysis in siRNA-treated H1975 cells, revealed that a number of processes including cell communication and signal transduction. These involve genes like MUC16 (CA125) whose levels relate to different stages of NSCLC [61], and PTK7 that is associated with lymph node metastasis as well as ALK and EGFR mutations in lung cancer [62]. In addition, PTN another gene effected by XIST is a heparin-binding growth factor that is involved with tumour progression [63]. In the same cell line, XIST affected the expression of 13 different zinc finger proteins. This is of increasing importance since they constitute the largest transcription factor family in the human genome and have been implicated in cancer progression [64].
Collectively, our study raises issues regarding detection of lncRNA using RNA-seq and provides a novel insight into a sex-specific role of XIST that warrants further investigation that can be of translational value in terms of the development of biomarkers and new therapeutic approaches targeting XIST and/or components of the signaling pathways it regulates in NSCLC.
4. Materials and Methods
4.1. Cell Lines
Two cell lines were used as in vitro models for lung cancer, A549 (ATCC® CCL-185™, Gaithersburg, MD, USA) and H1975 (ATCC® CRL-5908™, Gaithersburg, MD, USA). A549 cells were grown in complete DMEM (Dulbecco’s Modified Eagle’s Medium, Gibco, Fisher Scientific, Loughborough, UK) with 10% fetal bovine serum (FBS, Gibco, Fisher Scientific, Loughborough, UK), 1% penicillin/streptomycin (Gibco, Fisher Scientific, Loughborough, UK) and 1% L-Glutamine (Gibco, Fisher Scientific, Loughborough, UK). H1975 cells were grown in RPMI + 1% L-Glutamine (Gibco, Fisher Scientific, Loughborough, UK), 10% FBS and 1% Pen/strep. Cell lines were cultured at 37°C, in a 5% CO2 environment.
4.2. siRNA for XIST
We used siRNA targeted to XIST (SMARTpool: ON-TARGETplus, Dharmacon, CO, USA) to suppress XIST gene expression in A549 and H1975 cells. Both cell lines were seeded at 2 × 104 per well in 6-well plates. The siRNA transfection was carried out once at a concentration of 25 nmol/L per well/dish using Dharmafect 1 transfection reagent and serum-free media, a scrambled control was also included (ON-TARGETplus Non-targeting Pool; Horizon Discovery, UK). Cells were cultured in 3 biological replicates for 48 and 72 h. Then, 24 h post-transfection the medium was changed and replaced with fresh antibiotic-free medium [31].
4.3. RNA Isolation, cDNA Synthesis, and qPCR
RNA was extracted from cell lines, using the GenElute mRNA MiniPrep Kit (Merck, Feltham, UK), a silica membrane/spin column method, and stored at −80 °C until further use. cDNA was synthesized from mRNA using Superscript II (Fisher Scientific, Loughborough, UK). cDNA concentration was normalized using RNA concentrations determined by NanoDrop (Fisher Scientific, Loughborough, UK) and was synthesized to a concentration of 1000 ng according to manufacturer’s instructions.
4.4. RT-qPCR
Relative expression of the genes of interest was assessed by quantitative PCR using the SYBR green PCR master mix ROX (ABI, Fisher Scientific, Loughborough, UK) (Table 1). 18S RNA was used as the endogenous control and the targets investigated were manufactured by Sigma-Aldrich. Then, 1 μL from the cDNA (1000 ng/mL) and 0.5 μL of the primers (10 μM) were used for each reaction. All reactions were run in triplicate and a non-template control (NTC) was included in every run.
Table 1.
Primers for targets selected.
| Primer | Sequence 5′–3′ |
|---|---|
| 18S RNA-Forward | ATGGCCGTTCTGAGTTGGTG |
| 18S RNA-Reverse | CGCTGAGCCAGTCAGTGTAG |
| PRKCQ-Forward | CTTGTGGCAGCTTTGGATGT |
| PRKCQ-Reverse | CGTTTCTGACGCACATGTTT |
| NrCAM-Forward | TTGTGCAAAGAGGGAGCATG |
| NrCAM-Reverse | GGGCAGTTCCCTGTTGTCCT |
| CDKN1A-Forward | GCAGACCAGCATGACAGATTT |
| CDKN1A-Reverse | GGATTAGGGCTTCCTCTTGGA |
| RHOH Forward | GAGAAGTAACATTCTGCAAATCGC |
| RHOH Reverse | AGCACACGCCATTCAGCAAG |
| XIST-Forward | AGGTCAGGCAGAGGAAGTCA |
| XIST-Reverse | AGGTCAGGCAGAGGAAGTCA |
| ROX1-AS1 Forward | CTAGTTAGCAGGGGCAGCAC |
| PROX1-AS1 Reverse | AACAGAGAGGCGTGGAAGAA |
| MYC-Forward | TGGACATCCGCAAAGACCTGTAC |
| MYC-Reverse | TCAGGAGGAGCAATGATCTTGA |
| SFTA1P-Forward | CAGCATTCCAGGTGGGCTTT |
| SFTA1P-Reverse | CCTTGTTTGGCTTACTCGTGC |
qPCR data of relative gene expression was analyzed using the ΔΔCt method whereby the endogenous control Ct is subtracted from the gene of interest Ct to calculate ΔCt, and a relative quantity value was calculated by finding 2−ΔΔCt. A Student’s unpaired t-test was used to calculate statistical significance (p < 0.05).
4.5. RNAseq
RNA sequencing was carried out on lung cancer cell lines treated with siRNA XIST for 48 h. All samples were sequenced at 75bp paired-end according to Illumina specifications. The data were analyzed using open-source software from the Tuxedo suite. Reads were mapped to the human reference genome hg19 using the GENCODE 24 annotations [6], with TopHat2 [23] (Bowtie 2) under standard conditions. The alignments were filtered for high-quality hits with a minimum selection threshold score of 30. The mapped reads were assembled into transcripts and quantified using the Cufflinks package [34]. Functional enrichment analyses and Venn diagrams were performed in the open software FunRich. The statistical cut-off of functional enrichment analyses using this stand-alone software was kept at default setting with a p-value < 0.05 after Bonferroni correction.
4.6. Bioinformatic Analysis
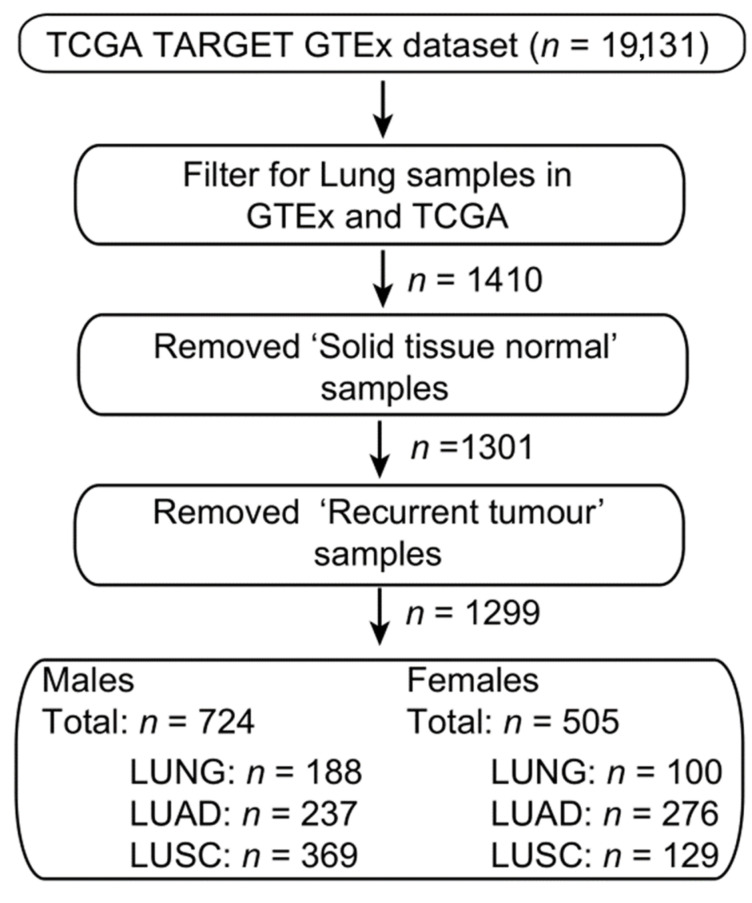
The XIST expression data and associated sample survival information were extracted from the UCSC Xenabrowser (xenabrowser.net), cohort “TCGA TARGET GTEx”. Samples from the publicly available datasets from The Cancer Genome Atlas project (TCGA), and Genotype-Tissue Expression project (GTEx) labelled as “primary tumour” and respectively “normal tissue” were selected for further processing (Figure 8). All expression data were cross-study normalized by UCSC using TOIL [35]. All expression data is presented in units of log2(norm_count + 1). Disease-specific phenotype data related to cancer stage, patient age, gender, and smoking status were extracted from the UCSC Xenabrowser TCGA Lung Adenocarcinoma (LUAD) and TCGA Lung Squamous Cell Carcinoma (LUSC) cohorts.
Figure 8.
Workflow for the extraction and pre-processing of TCGA and GTEX samples from the UCSC Xenabrowser.
The Wilcoxon test was used to compare the gene expression level between two cohorts and Spearman’s rank test was used to determine the correlation coefficient between two gene expression patterns. All statistical tests performed in R v3.5.0. A summary of the data pre-processing steps is shown in Figure 2. T-SNE analysis was performed using the RtSNE v0.15 package in R v3.5.0. (www.rstudio.com/)
5. Conclusions
Our study provides a novel insight into the role of XIST in lung cancer. Apart from exhibiting sexual dimorphism in terms of expression, the downregulation of this lncRNA in NSCLC appears to influence differential signalling cascades and genes in male versus female cancer cell lines. The co-expression of XIST, TSIX, hnRNPu, Bcl-2, and BRCA1 provided a strong collective feature to discriminate between controls and LC, implying a diagnostic potential.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/12/3499/s1, Figure S1: Heatmap of gene expression of XIST, TSIX, HNRNPU, BCL-2 and BRCA1 for each individual sample collected from TCGA and GTEx. Sexes and tissue types (healthy lung, lung adenocarcinoma/LUAD and lung squamous cell carcinoma/LUSC) are labelled as two additional columns. Gene expressions have units of log2(norm_count + 1), normalized by upper quartile method. Individual sample IDs were labeled on the left side of the heatmap. Total number of samples is 1299, data directly obtained from Xenabrowser cohort, “TCGA TARGET GTEx”, Figure S2: tSNE clustering of various gene combinations differentiating for gender and sample type: (a) XIST/TSIX, (b) XIST/HNRNPU, (c) XIST/BCL2, (d) XIST/BRCA1, (e) XIST/TSIX/HNRNPU, (f) XIST/HNRNPU/BCL2, (g) XIST/TSIX/BRCA1, (h) XIST/TSIX/BCL2, (i) XIST/BCL2/BRCA1, (j) XIST/HNRNPU/BRCA1, (k) XIST/TSIX/HNRNPU/BRCA1, (l) XIST/BCL2/BRCA1/HNRNPU, (m) XIST/TSIX/BCL2/BRCA1, (n) XIST/TSIX/BCL2/HNRNPU. Figure S3: Correlation with regression line for gene expression (in units if log2(norm_count + 1)) for all pairwise combinations of genes from XIST, TSIX, hnRNPu, TSIX, Bcl-2 and BRCA1. Spearman’s rank correlation test was used to generate the correlation coefficient represented as R in the graph, p represents p-value. Healthy lung, LUAD, and LUSC were compared separately in both sexes. Total sample number is 1299, 794 derived from male patients and 505 from female patients. There are 116 samples from male patients with 0 expression in XIST, 461 in TSIX, 1 in hnRNPu, Bcl-2, and BRCA1. In female patients derived samples, 17 show zero expression for TSIX gene. All samples with zero expression in both genes investigated were removed the analysis. Table S1: Spreadsheet with the differentially expressed genes in A549 and H1975 cell lines following siRNA of XIST.
Author Contributions
Conceptualization, P.K., V.A., C.S., and E.K.; methodology, P.K., Q.D., H.H., C.I.F., and C.S.; software, P.K., C.I.F., Q.D., and C.S.; validation, P.K., Q.S., and C.S.; formal analysis, P.K., H.H., Q.D., C.S., and E.K.; investigation, P.K., A.P., V.A., C.S., and E.K.; resources, V.A. and A.P.; data curation, C.S. and E.K.; writing—original draft preparation, P.K., A.P., V.A., C.S., and E.K.; writing—review and editing, P.K., H.H., A.P., V.A., C.S., and E.K.; visualization, P.K., Q.D., and C.S.; supervision, E.K. and C.S.; project administration, E.K., C.S., and V.A.; funding acquisition, V.A. and A.P.; E.K. and C.S. should be considered joint last co-authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Cancer Treatment and Research Trust (CTRT), Mount Vernon Hospital, Rickmansworth Road, Northwood, Middlesex, HA6 2RN, UK; grant number 11016100.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Delás M.J., Hannon G.J. lncRNAs in development and disease: From functions to mechanisms. Open Biol. 2017;7:170121. doi: 10.1098/rsob.170121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y.-L., Li X.-B., Hou Y.-X., Fang N.-Z., You J.-C., Zhou Q.-H. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancerThis article has been corrected since Advanced Online Publication, and an erratum is also printed in this issue. Acta Pharmacol. Sin. 2017;38:371–381. doi: 10.1038/aps.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H., Shen Q., Zhang X., Yang C., Cui S., Sun Y., Wang L., Fan X., Xu S. The Long Non-Coding RNA XIST Controls Non-Small Cell Lung Cancer Proliferation and Invasion by Modulating miR-186-5p. Cell. Physiol. Biochem. 2017;41:2221–2229. doi: 10.1159/000475637. [DOI] [PubMed] [Google Scholar]
- 7.Inamura K. Major Tumor Suppressor and Oncogenic Non-Coding RNAs: Clinical Relevance in Lung Cancer. Cells. 2017;6:12. doi: 10.3390/cells6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do H., Kim W. Roles of Oncogenic Long Non-coding RNAs in Cancer Development. Genom. Inform. 2018;16:e18. doi: 10.5808/GI.2018.16.4.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Z., Sun T., Hacisuleyman E., Fei T., Wang X., Brown M., Rinn J.L., Lee M.G.-S., Chen Y., Kantoff P.W., et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 2016;7:10982. doi: 10.1038/ncomms10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gendrel A.V., Heard E. Fifty years of x-inactivation research. Development. 2011;138:5049–5055. doi: 10.1242/dev.068320. [DOI] [PubMed] [Google Scholar]
- 11.Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 12.Jonkers I., Barakat T.S., Achame E.M., Monkhorst K., Kenter A., Rentmeester E., Grosveld F., Grootegoed J.A., Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z., Jiang X., Jiang X., Zhao H. X-inactive-specific transcript: A long noncoding RNA with complex roles in human cancers. Gene. 2018;679:28–35. doi: 10.1016/j.gene.2018.08.071. [DOI] [PubMed] [Google Scholar]
- 14.Pintacuda G., Young A.N., Cerase A. Function by Structure: Spotlights on Xist Long Non-coding RNA. Front. Mol. Biosci. 2017;4:1–11. doi: 10.3389/fmolb.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerase A., Pintacuda G., Tattermusch A., Avner P. Xist localization and function: New insights from multiple levels. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salvador M.A., Wicinski J., Cabaud O., Toiron Y., Finetti P., Josselin E., Lelièvre H., Kraus-Berthier L., Depil S., Bertucci F., et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin. Cancer Res. 2013;19:6520–6531. doi: 10.1158/1078-0432.CCR-13-0877. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi R., Miyagawa R., Yamashita H., Morikawa T., Okuma K., Fukayama M., Ohtomo K., Nakagawa K. Increased expression of long non-coding RNA XIST predicts favorable prognosis of cervical squamous cell carcinoma subsequent to definitive chemoradiation therapy. Oncol. Lett. 2016;12:3066–3074. doi: 10.3892/ol.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D.-L., Chen L.-Z., Lu Y.-X., Zhang D.-S., Zeng Z.-L., Pan Z.-Z., Huang P., Wang F.-H., Li Y.-H., Ju H.-Q., et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. doi: 10.1038/cddis.2017.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D., Ren J., Wang Y., Li B., Gu Y. Intraoperative monitoring of blood perfusion in port wine stains by laser Doppler imaging during vascular targeted photodynamic therapy: A preliminary study. Photodiagn. Photodyn. Ther. 2016;14:142–151. doi: 10.1016/j.pdpdt.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Du P., Zhao H., Peng R., Liu Q., Yuan J., Peng G., Liao Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo Y., Lu Y., Wang P., Huang S., He L., Li D., Li F., Huang J., Lin X., Li X., et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017;39 doi: 10.1177/1010428317690999. [DOI] [PubMed] [Google Scholar]
- 22.Song P., Ye L.-F., Zhang C., Peng T., Zhou X.-H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 23.Fang J., Sun C.-C., Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem. Biophys. Res. Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Wei W., Liu Y., Lu Y., Yang B., Tang L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J. Cell. Biochem. 2017;118:3349–3358. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 25.Li G.-L., Wu Y.-X., Li Y.-M., Li J. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2829–2834. [PubMed] [Google Scholar]
- 26.Huang K.-C., Rao P.H., Lau C.C., Heard E., Ng S.-K., Brown C., Mok S.C., Berkowitz R.S., Ng S.-W. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol. Cancer Ther. 2002;1:769–776. [PubMed] [Google Scholar]
- 27.Yao Y., Ma J., Xue Y., Wang P., Li Z.Z., Liu J., Chen L., Xi Z., Teng H., Wang Z., et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 28.Silver D.P., Dimitrov S.D., Feunteun J., Gelman R., Drapkin R., Lu S.D., Shestakova E., Velmurugan S., Denunzio N., Dragomir S., et al. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128:991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Wang P., Chen D., Ma H., Li Y. Long non-coding RNA MEG3 regulates proliferation and apoptosis in non-small cell lung cancer: Via the miR-205-5p/LRP1 pathway. RSC Adv. 2017;7:49710–49719. doi: 10.1039/C7RA08057C. [DOI] [Google Scholar]
- 30.Shen L., Li C., Liu M., Wei D., Chang Q., Cui J. Prognostic and clinicopathological roles of long non-coding RNA XIST in human cancers: A meta-analysis. Transl. Cancer Res. 2018;7:1624–1633. doi: 10.21037/tcr.2018.12.13. [DOI] [Google Scholar]
- 31.Sun W., Zu Y., Fu X., Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol. Rep. 2017;38:3347–3354. doi: 10.3892/or.2017.6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhang G., Cheng Z., Dai L., Jia L., Jing X., Wang H., Zhang R., Liu M., Jiang T., et al. Knockdown of LncRNA-XIST Suppresses Proliferation and TGF-β1-Induced EMT in NSCLC Through the Notch-1 Pathway by Regulation of miR-137. Genet. Test. Mol. Biomark. 2018;22:333–342. doi: 10.1089/gtmb.2018.0026. [DOI] [PubMed] [Google Scholar]
- 33.Hu B., Shi G., Li Q., Li W., Zhou H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J. Cell. Biochem. 2018 doi: 10.1002/jcb.27920. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q., Hu W., Zhu W., Zhang F., Lin-lin L., Liu C., Songyang Y. Clinica Chimica Acta Long non coding RNA XIST as a prognostic cancer marker—A meta-analysis. Clin. Chim. Acta. 2018;482:1–7. doi: 10.1016/j.cca.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Simpkins F., Jang K., Yoon H., Hew K.E., Kim M., Azzam D.J., Sun J., Zhao D., Ince T.A., Liu W., et al. Dual Src and MEK Inhibition Decreases Ovarian Cancer Growth and Targets Tumor Initiating Stem-Like Cells. Clin. Cancer Res. 2018;24:4874–4886. doi: 10.1158/1078-0432.CCR-17-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y., Wang J., Wang J. Long noncoding RNA XIST promotes proliferation and invasion by targeting miR-141 in papillary thyroid carcinoma. OncoTargets Ther. 2018;11:5035–5043. doi: 10.2147/OTT.S170439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tantai J., Hu D., Yang Y., Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8:7887–7895. doi: 10.1287/mnsc.1060.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Y., Zhao S., Sheng Q., Guo M., Lehmann B., Pietenpol J., Samuels D.C., Shyr Y. RNAseq by Total RNA Library Identifies Additional RNAs Compared to Poly(A) RNA Library. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/862130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng H., Brennan K., Hernaez M., Gevaert O. Benchmark of long non-coding RNA quantification for RNA sequencing of cancer samples. Gigascience. 2019;8 doi: 10.1093/gigascience/giz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang R., Jaritz M., Guenzl P., Vlatkovic I., Sommer A., Tamir I.M., Marks H., Klampfl T., Kralovics R., Stunnenberg H.G., et al. An RNA-Seq Strategy to Detect the Complete Coding and Non-Coding Transcriptome Including Full-Length Imprinted Macro ncRNAs. PLoS ONE. 2011;6:e27288. doi: 10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilott N.E., Ponting C.P. Predicting long non-coding RNAs using RNA sequencing. Methods. 2013;63:50–59. doi: 10.1016/j.ymeth.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Byerly J., Halstead-Nussloch G., Ito K., Katsyv I., Irie H.Y. PRKCQ promotes oncogenic growth and anoikis resistance of a subset of triple-negative breast cancer cells. Breast Cancer Res. 2016;18:95. doi: 10.1186/s13058-016-0749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deeds L., Teodorescu S., Chu M., Yu Q., Chen C.-Y. A p53-independent G1 cell cycle checkpoint induced by the suppression of protein kinase C alpha and theta isoforms. J. Biol. Chem. 2003;278:39782–39793. doi: 10.1074/jbc.M306854200. [DOI] [PubMed] [Google Scholar]
- 44.Zamagni A., Pasini A., Pirini F., Ravaioli S., Giordano E., Tesei A., Calistri D., Ulivi P., Fabbri F., Foca F., et al. CDKN1A upregulation and cisplatin-pemetrexed resistance in non-small cell lung cancer cells. Int. J. Oncol. 2020;56:1574–1584. doi: 10.3892/ijo.2020.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoji T., Tanaka F., Takata T., Yanagihara K., Otake Y., Hanaoka N., Miyahara R., Nakagawa T., Kawano Y., Ishikawa S., et al. Clinical significance of p21 expression in non-small-cell lung cancer. J. Clin. Oncol. 2002;20:3865–3871. doi: 10.1200/JCO.2002.09.147. [DOI] [PubMed] [Google Scholar]
- 46.Groeger A.M., Caputi M., Esposito V., Baldi A., Rossiello R., Santini D., Mancini A., Kaiser H.E., Baldi F. Expression of p21 in non small cell lung cancer relationship with PCNA. Anticancer Res. 2000;20:3301–3305. [PubMed] [Google Scholar]
- 47.Górka B., Skubis-Zegadło J., Mikula M., Bardadin K., Paliczka E., Czarnocka B. NrCAM, a neuronal system cell-adhesion molecule, is induced in papillary thyroid carcinomas. Br. J. Cancer. 2007;97:531–538. doi: 10.1038/sj.bjc.6603915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Sui F., Ma J., Ren X., Guan H., Yang Q., Shi J., Ji M., Shi B., Sun Y., et al. Positive Feedback Loops Between NrCAM and Major Signaling Pathways Contribute to Thyroid Tumorigenesis. J. Clin. Endocrinol. Metab. 2017;102:613–624. doi: 10.1210/jc.2016-1677. [DOI] [PubMed] [Google Scholar]
- 49.Rohrbeck A., Neukirchen J., Rosskopf M., Pardillos G.G., Geddert H., Schwalen A., Gabbert H.E., von Haeseler A., Pitschke G., Schott M., et al. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J. Transl. Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conacci-Sorrell M., Kaplan A., Raveh S., Gavert N., Sakurai T., Ben-Ze’ev A. The shed ectodomain of Nr-CAM stimulates cell proliferation and motility, and confers cell transformation. Cancer Res. 2005;65:11605–11612. doi: 10.1158/0008-5472.CAN-05-2647. [DOI] [PubMed] [Google Scholar]
- 51.Haga R.B., Ridley A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases. 2016;7:207–221. doi: 10.1080/21541248.2016.1232583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tajadura-Ortega V., Garg R., Allen R., Owczarek C., Bright M.D., Kean S., Mohd-Noor A., Grigoriadis A., Elston T.C., Hahn K.M., et al. An RNAi screen of Rho signalling networks identifies RhoH as a regulator of Rac1 in prostate cancer cell migration. BMC Biol. 2018;16:29. doi: 10.1186/s12915-018-0489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barr L.F., Campbell S.E., Diette G.B., Gabrielson E.W., Kim S., Shim H., Dang C.V. c-Myc suppresses the tumorigenicity of lung cancer cells and down-regulates vascular endothelial growth factor expression. Cancer Res. 2000;60:143–149. [PubMed] [Google Scholar]
- 54.Rapp U.R., Korn C., Ceteci F., Karreman C., Luetkenhaus K., Serafin V., Zanucco E., Castro I., Potapenko T. MYC is a metastasis gene for non-small-cell lung cancer. PLoS ONE. 2009;4:e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H., Liu H., Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soucek L., Whitfield J.R., Sodir N.M., Massó-Vallés D., Serrano E., Karnezis A.N., Swigart L.B., Evan G.I. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jäger N., Schlesner M., Jones D.T.W., Raffel S., Mallm J.P., Junge K.M., Weichenhan D., Bauer T., Ishaque N., Kool M., et al. XHypermutation of the inactive X chromosome is a frequent event in cancer. Cell. 2013;155:567. doi: 10.1016/j.cell.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiratsuchi H., Wang Z., Chen G., Ray P., Lin J., Zhang Z., Zhao L., Beer D., Ray D., Ramnath N. Oncogenic Potential of CYP24A1 in Lung Adenocarcinoma. J. Thorac. Oncol. 2017;12:269–280. doi: 10.1016/j.jtho.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Liu W., Song J., Du X., Zhou Y., Li Y., Li R., Lyu L., He Y., Hao J., Ben J., et al. AKR1B10 (Aldo-keto reductase family 1 B10) promotes brain metastasis of lung cancer cells in a multi-organ microfluidic chip model. Acta Biomater. 2019;91:195–208. doi: 10.1016/j.actbio.2019.04.053. [DOI] [PubMed] [Google Scholar]
- 60.Li C.-G., Pu M.-F., Li C.-Z., Gao M., Liu M.-X., Yu C.-Z., Yan H., Peng C., Zhao Y., Li Y., et al. MicroRNA-1304 suppresses human non-small cell lung cancer cell growth in vitro by targeting heme oxygenase-1. Acta Pharmacol. Sin. 2017;38:110–119. doi: 10.1038/aps.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Wang D., Zheng G., Yang Y., Du L., Dong Z., Zhang X., Wang C. Clinical evaluation and therapeutic monitoring value of serum tumor markers in lung cancer. Int. J. Biol. Markers. 2016;31:80–87. doi: 10.5301/jbm.5000177. [DOI] [PubMed] [Google Scholar]
- 62.Jiang W., He J., Lv B., Xi X., He G., He J. PTK7 expression is associated with lymph node metastasis, ALK and EGFR mutations in lung adenocarcinomas. Histol. Histopathol. 2020;35:489–495. doi: 10.14670/HH-18-183. [DOI] [PubMed] [Google Scholar]
- 63.Wang H.Q., Wang J. Expression of pleiotrophin in small cell lung cancer. J. Biol. Regul. Homeost. Agents. 2015;29:175–179. [PubMed] [Google Scholar]
- 64.Jen J., Wang Y.-C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.