Abstract
Bloodstream infections (BSIs) are among the leading causes of morbidity and mortality worldwide, among infectious diseases. Local knowledge of the main bacteria involved in BSIs and their associated antibiotic susceptibility patterns is essential to rationalize the empiric antimicrobial therapy. The aim of this study was to define the incidence of infection and evaluate the antimicrobial resistance profile of the main pathogens involved in BSIs. This study enrolled patients of all ages and both sexes admitted to the University Hospital “San Giovanni di Dio e Ruggi d’Aragona”, Salerno, Italy between January 2015 to December 2019. Bacterial identification and antibiotic susceptibility testing were performed with Vitek 2. A number of 3.949 positive blood cultures were included out of 24,694 total blood cultures from 2015 to 2019. Coagulase-negative staphylococci (CoNS) were identified as the main bacteria that caused BSI (17.4%), followed by Staphylococcus aureus (12.3%), Escherichia coli (10.9%), and Klebsiella pneumoniae (9.4%). Gram-positive bacteria were highly resistant to Penicillin G and Oxacillin, while Gram-negative strains to Ciprofloxacin, Cefotaxime, Ceftazidime, and Amoxicillin-clavulanate. High susceptibility to Vancomycin, Linezolid, and Daptomycin was observed among Gram-positive strains. Fosfomycin showed the best performance to treatment Gram-negative BSIs. Our study found an increase in resistance to the latest generation of antibiotics over the years. This suggests an urgent need to improve antimicrobial management programs to optimize empirical therapy in BSI.
Keywords: antimicrobial sensitivity, blood culture, bloodstream infections, empiric therapy
1. Introduction
Bloodstream infections (BSIs) represent a major cause of mortality and morbidity worldwide [1]. BSIs are widely spread all over the world with direct and indirect social and economic impacts. It is estimated that BSIs affects approximately 30 million people, causing 6 million deaths each year in the world [2,3]. European reports revealed that BSI cases are more than 1.2 million each year, with a number of deaths around 157,000 patients [4]. Based on the age group, previous morbidity, and other risk factors, the mortality rate of BSIs ranges between 4.0 and 41.5% [5,6,7,8,9,10]. Healthcare costs range between $10,000 and $20,000 per hospitalized patient [11]. BSIs are caused by the presence of live bacterial and/or fungal microorganisms in the bloodstream. These events can favor a strong inflammatory response, with alteration of some clinical and hemodynamics parameters [12]. The BSIs can be divided into primary or secondary infections [13]. In primary BSIs, microorganisms are introduced directly into the bloodstream, for example, through the use of contaminated medical devices. Secondary BSI is a bloodstream infection driven by the same organism causing infection in another host tissue [14]. Moreover, BSIs are further classified in community or hospital-acquired infections. Differences among them are due to the place and duration of infection. In particular, the first one manifests in a community or within the first 48h of admission in the hospital. In the second group, BSI occurs 48 h after admission or 3 days after hospital discharge [15]. Bacteria are the leading cause of BSIs, although also the fungi may be implicated in the emergence of this infection [16]. Previous studies detected Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, and CoNS (Coagulase-negative staphylococci) strains as the most common cause of BSIs [3]. The diagnosis of BSIs is carried out through the analysis of clinical symptoms of the patient and laboratory tests [17]. The most common clinical symptoms of BSIs include (i) fever (>38 °C), (ii) chills, (iii) hypotension, and (iv) increase in white blood cell count and inflammation markers concentrations [13]. Nevertheless, the reported signs are not always present: for instance, elderly patients may develop a subdued fever or hypothermia in severe outcomes. Likewise, tachycardia and tachypnea represent the clinical signs most commonly present in critical patients. Moreover, leukopenia can be seen in severe cases, when the white blood cell is activated massively and entrapped in peripheral sites [18]. Given the complexities of clinical signs, only microbiological analysis of blood samples confirms the clinical diagnosis of sepsis [19]. Actually, blood cultures (BCs) represent the gold standard method for the diagnosis of BSIs [20]. BCs provide the identity of the pathogen and the relative pattern of antibiotic susceptibility with high sensitivity [21]. The epidemiology of BSIs differs between several countries [3,22,23,24]. These significant differences between healthcare communities require constant monitoring of local trends. The advent of antimicrobial resistance (AMR) among most bacterial pathogens causes a serious health crisis with many economic and social implications around the world [25]. AMR threatens the efficacy of antibiotics frequently used to prevent and treat BSIs. Furthermore, the lack of novel antibiotics highlights the limitations of the situation and underlines the needs of programs and actions in order to face the problem [26]. The starting point is represented by studies on bacterial etiology and antibiotic resistance profile of bacterial BSIs to improve the empirical treatment and the administration of the correct antibiotic therapy [5]. In this scenario, the current study was carried out to evaluate the bacterial pathogens involved in BSIs and their antimicrobial susceptibility pattern in patients admitted to the San Giovanni di Dio e Ruggi d’Aragona Hospital (Salerno, Italy). Knowledge about the main bacterial BSIs and related antibiotics susceptibility profile is crucial to permit the appropriate choice of antibiotic treatment, leading to a reduction in hospital stay, the cost of therapy, and mortality.
2. Results
2.1. Incidence of BSIs in Studied Patients
In the present study, 24,694 blood samples were examined. BSIs were diagnosed based on the patient’s clinical signs, biochemical parameters, and the presence of microorganisms in the blood. From 2015 to 2019, 3949 cases of BSIs were recorded (Table 1). The mean of patients with BSIs was 16.4%. Of these, 2841 (71.9%) and 1108 (28.1%) were diagnosed as primary and secondary bacteremia, respectively. Primary and secondary BSIs were included in our analysis.
Table 1.
Cases of bacteremia distributed by year.
| Year | 2015 | 2016 | 2017 | 2018 | 2019 | Total |
|---|---|---|---|---|---|---|
| Positive | 696 | 734 | 839 | 868 | 812 | 3949 |
| Negative | 4104 | 3766 | 4752 | 2924 | 5199 | 20,745 |
| Total | 4800 | 4500 | 5591 | 3792 | 6011 | 24,694 |
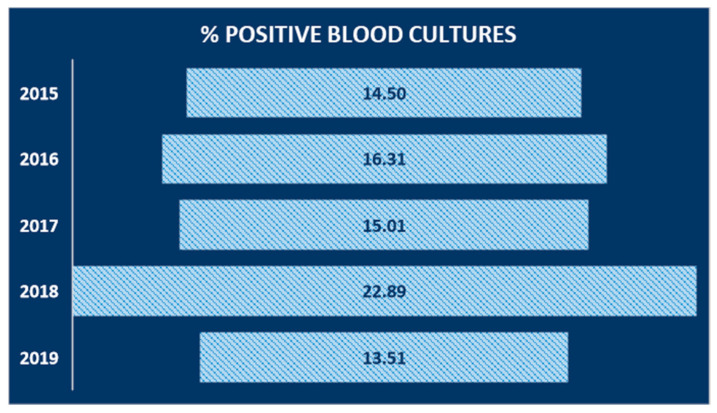
Our findings showed a rather linear trend over the years, except for the year 2018, where the number of positives exceeded 22% (Figure 1).
Figure 1.
Incidence of bloodstream infection (BSI) cases by year of study, expressed as a percentage relative to the total number of positive cases out of the total number of cases present per year of study.
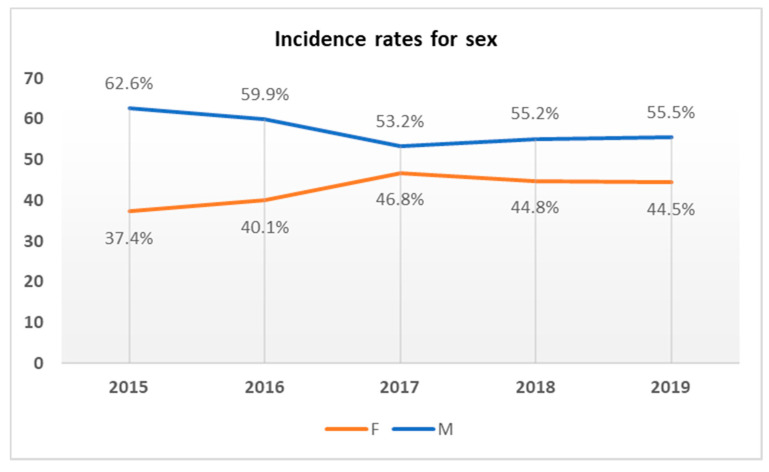
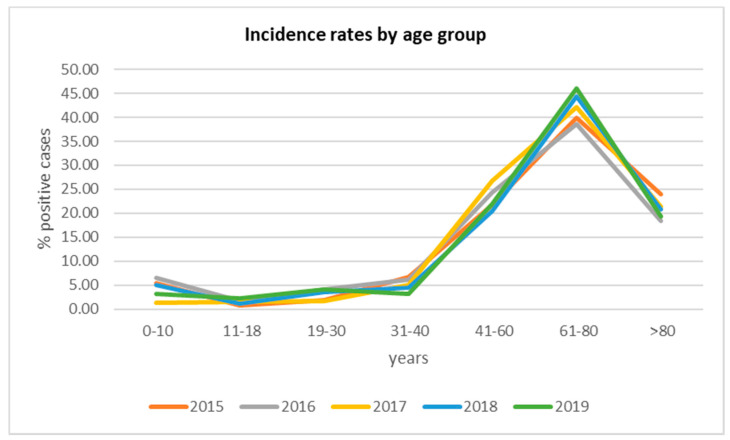
The gender and age distributions of the patients with BSIs are reported in Figure 2 and Figure 3. Regarding gender, the BSIs rate was higher in males than in females (Figure 2). Concerning age distribution, most of the positive patients were placed in the 61–80 age group (Figure 3).
Figure 2.
Incidence of BSI cases by study year associated with the sex of the patients involved.
Figure 3.
Distribution of positive cases by age group.
2.2. Isolated Bacteria
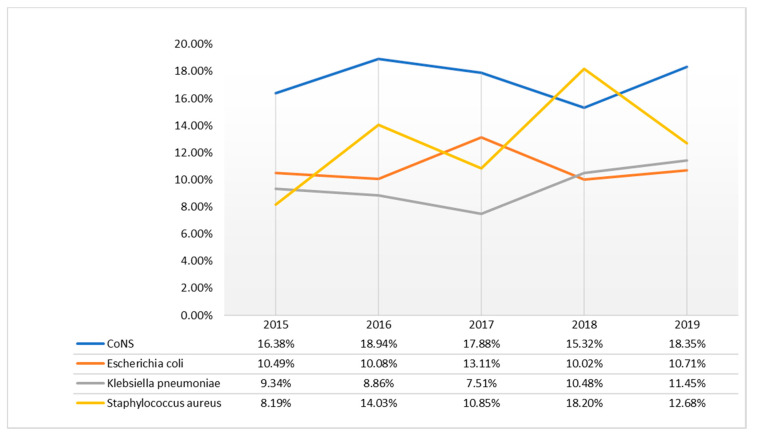
All pathogens identified over the 5 years of study, with respective incidence rates, were provided as additional data (supplementary Table S1). Our data showed that CoNS strains were the main isolated strains in the period from 2015 to 2017 and 2019 (>15.32%). Only in 2018, Staphylococcus aureus was the most identified strain (18.20%). Escherichia coli was ranked second in 2015 and 2017, while Staphylococcus aureus in 2016 and 2019. Klebsiella pneumoniae was the least represented strain. In 2018–2019, this strain was the third most isolated strain (Figure 4).
Figure 4.
Trend in percentage of batteries caused by the most frequent isolates.
2.3. Prevalence of Antimicrobial Resistance among BSI Bacteria
In the present study, the antimicrobial resistance profile of Staphylococcus aureus, CoNS strains, Escherichia coli, and Klebsiella pneumoniae had been analyzed. The antimicrobial resistance patterns are shown in Table 2, Table 3, Table 4 and Table 5. All isolated strains showed a high rate of resistance to the tested antibiotics. Among the Gram-positive bacteria analyzed, the most resistant species is represented by Staphylococcus aureus. The resistance rates for Staphylococcus aureus to Penicillin were higher than 84.6% in 2015–2018. It was relevant reduced in 2019 (68.9%). Resistance to Gentamicin in Staphylococcus aureus exhibited a relevant downward trend, ranging from 13.3 to 7.8%. The resistance to Oxacillin was detected in 229 of 515 total isolated strains of S. aureus (44%). Clindamycin, Erythromycin, Levofloxacin, Rifampicin, and Tetracycline fluctuated lightly, ranging from 36.7 to 35.9%, 43.3 to 42.7%, 33.3 to 27.8%, 7 to 7.8%, and 9.6 to 9.8%, respectively. The resistance rates for S. aureus to Vancomycin, Teicoplanin, Daptomycin, and Linezolid were lower than 7.8% but in an alarming increase (Table 2).
Table 2.
Antimicrobial resistance profile of Staphylococcus aureus from patients with BSI to commonly used antibiotics.
| St. aureus | 2015 | 2016 | 2017 | 2018 | 2019 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % |
| Fusidic acid | 60 | 0 | 103 | 1.9 | 91 | 5.5 | 158 | 4.4 | 103 | 4.9 |
| Clindamycin | 60 | 36.7 | 103 | 27.2 | 91 | 39.6 | 158 | 39.9 | 103 | 35.9 |
| Daptomycin | 60 | 0 | 103 | 1.0 | 91 | 5.5 | 158 | 5.1 | 103 | 3.9 |
| Erythromycin | 60 | 43.3 | 103 | 49.5 | 91 | 50.5 | 158 | 53.2 | 103 | 42.7 |
| Gentamicin | 60 | 13.3 | 103 | 13.6 | 91 | 9.9 | 158 | 10.8 | 103 | 7.8 |
| Levofloxacin | 57 | 33.3 | 79 | 35.4 | 77 | 53.2 | 116 | 41.4 | 90 | 27.8 |
| Linezolid | 60 | 0 | 103 | 0 | 91 | 4.4 | 158 | 4.4 | 103 | 2.9 |
| Oxacillin | 60 | 26.7 | 103 | 50.5 | 91 | 53.8 | 158 | 50.6 | 103 | 36.9 |
| Penicillin G | 60 | 86.7 | 102 | 85.3 | 91 | 84.6 | 158 | 86.7 | 103 | 68.9 |
| Rifampicin | 57 | 7.0 | 79 | 12.7 | 77 | 0 | 116 | 3.4 | 90 | 7.8 |
| Teicoplanin | 60 | 0 | 103 | 1.9 | 91 | 5.5 | 157 | 5.7 | 103 | 7.8 |
| Tetracycline | 60 | 9.6 | 103 | 9.8 | 91 | 8.8 | 158 | 3.2 | 103 | 9.8 |
| Tigecycline | 58 | 0 | 103 | 0 | 91 | 4.4 | 157 | 2.5 | 103 | 1.0 |
| Trimethoprim/ Sulfam. | 60 | 1.7 | 103 | 2.0 | 91 | 5.5 | 157 | 1.9 | 103 | 4.9 |
| Vancomycin | 60 | 0 | 103 | 1.9 | 91 | 4.4 | 158 | 4.4 | 103 | 3.9 |
Table 3.
Antimicrobial resistance profile of CoNS strains from patients with BSI to commonly used antibiotics.
| CoNS | 2015 | 2016 | 2017 | 2018 | 2019 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % |
| Fusidic acid | 212 | 28.3 | 257 | 36.6 | 274 | 27.0 | 230 | 31.7 | 242 | 33.1 |
| Clindamycin | 216 | 62.5 | 257 | 57.2 | 272 | 55.1 | 232 | 54.3 | 243 | 51.4 |
| Daptomycin | 211 | 0.5 | 257 | 1.2 | 269 | 2.2 | 229 | 0.9 | 243 | 2.9 |
| Linezolid | 200 | 0 | 237 | 0 | 258 | 0.4 | 231 | 6.9 | 243 | 3.3 |
| Oxacillin | 206 | 81.1 | 233 | 77.3 | 249 | 72.7 | 229 | 78.6 | 243 | 77.8 |
| Rifampicin | 207 | 28.0 | 233 | 31.8 | 250 | 32.4 | 201 | 37.3 | 201 | 34.3 |
| Vancomycin | 216 | 0.5 | 257 | 0.4 | 273 | 1.1 | 232 | 0.4 | 242 | 3.7 |
Table 4.
Antimicrobial resistance profile of Klebsiella pneumoniae from patients with BSI to commonly used antibiotics.
| Klebsiella pneumoniae | 2015 | 2016 | 2017 | 2018 | 2019 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % |
| Amoxicillin/A. Clav. | 67 | 77.6 | 60 | 76.7 | 53 | 79.2 | 62 | 82.3 | 116 | 94.8 |
| Cefotaxime | 67 | 85.1 | 65 | 73.8 | 63 | 87.3 | 91 | 81.3 | 92 | 92.4 |
| Ceftazidime | 67 | 79.1 | 65 | 73.8 | 63 | 85.7 | 91 | 84.6 | 93 | 92.5 |
| Ciprofloxacin | 67 | 86.6 | 65 | 73.8 | 63 | 85.7 | 91 | 76.9 | 93 | 89.2 |
| Colistin | 66 | 21.2 | 63 | 27.0 | 55 | 16.4 | 75 | 12.3 | 88 | 26.1 |
| Ertapenem | 59 | 62.7 | 65 | 64.6 | 63 | 77.8 | 91 | 54.9 | 93 | 65.6 |
| Fosfomycin | 67 | 22.4 | 60 | 28.3 | 53 | 20.7 | 63 | 14.3 | 68 | 38.2 |
| Meropenem | 67 | 64.2 | 65 | 64.6 | 63 | 74.6 | 91 | 54.9 | 93 | 65.6 |
| Piperacillin/Tazobactam | 67 | 79.1 | 65 | 70.8 | 63 | 82.5 | 91 | 81.3 | 93 | 79.6 |
| Trimethoprim/Sulf. | 67 | 77.6 | 65 | 69.2 | 63 | 88.9 | 91 | 70.3 | 93 | 58.1 |
Table 5.
Antimicrobial resistance profile of Escherichia coli from patients with BSI to commonly used antibiotics.
| Escherichia coli | 2015 | 2016 | 2017 | 2018 | 2019 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % | n. Assays | R % |
| Amoxicillin/A. Clav. | 70 | 50.0 | 66 | 56.1 | 103 | 39.8 | 84 | 51.2 | 110 | 68.2 |
| Cefotaxime | 73 | 53.5 | 74 | 54.1 | 110 | 52.7 | 87 | 57.5 | 83 | 54.2 |
| Ciprofloxacin | 73 | 61.6 | 74 | 68.9 | 110 | 66.3 | 87 | 67.8 | 87 | 71.3 |
| Ertapenem | 73 | 1.4 | 74 | 0 | 110 | 0 | 87 | 2.3 | 87 | 2.3 |
| Fosfomycin | 70 | 0 | 66 | 0 | 103 | 4.9 | 84 | 0 | 70 | 0 |
| Gentamicin | 73 | 34.2 | 74 | 40.5 | 110 | 27.3 | 87 | 27.6 | 87 | 31.0 |
| Imipenem | 73 | 0 | 74 | 0 | 110 | 0 | 74 | 0 | 23 | 0 |
| Meropenem | 73 | 0 | 74 | 0 | 110 | 0 | 87 | 2.3 | 87 | 2.2 |
| Piperacillin/tazobactam | 72 | 15.3 | 74 | 10.8 | 110 | 18.2 | 86 | 12.8 | 85 | 14.1 |
| Tigecycline | 72 | 0 | 73 | 0 | 109 | 0 | 85 | 0 | 81 | 1.2 |
| Trimethoprim/Sulf. | 73 | 41.1 | 74 | 43.2 | 110 | 45.5 | 87 | 52.9 | 87 | 55.2 |
CoNS strains represent the most frequent Gram-positive bacteria, involved in BSIs. In this study, they include the following species; Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus hominis. CoNS represent the most common blood culture contaminant. In order to differentiate contamination from real bacteremia, more than two culture series were performed per patient at the same and different time by separate venipuncture. The presence of the same isolate on multiple culture blood sets allowed us to differentiate false positives from true positives. As contamination can arise during the collection phase, having reliable factors is essential for patient management and patient surveillance [27]. The resistance rates for CoNS strains to Oxacillin were higher than 77.5%. It fluctuated slightly, ranging from 81.1 to 77.8%. Furthermore, the data analysis indicated a light variation of resistance percentage of Fusidic acid, Clindamycin, and Rifampicin, passing from 28.3 to 33.1%, 62.5 to 51.4%, and 28 to 34.3%, respectively. The resistance frequency to Daptomycin, Linezolid, and Vancomycin was less than 6.9%. In this case, the increase in resistance is worrying (Table 3).
Our findings showed that among Gram-negative bacteria, Klebsiella pneumoniae was the most resistant strain. The resistance rates for K. pneumoniae to Ertapenem, Meropenem, Piperacillin/Tazobactam, and Ciprofloxacin were above 54.9% and varies slightly over the studied period. A significant increase was observed for the resistance rates to Amoxicillin/Clavulanic acid, Cefotaxime, Ceftazidime, and Fosfomycin which passed from 77.6 to 94.8%, 85.1 to 92.4%, 79.1 to 92.5%, and 22.4 to 38.2%, respectively. A slight decrease was showed in Trimethoprim/Sulfamethoxazole resistance rate, from 77.6 to 58.1% (Table 4).
The antimicrobial susceptibility data exhibited that the resistance rate for Escherichia coli to Amoxicillin/Clavulanic acid, Ciprofloxacin, and Trimethoprim/Sulfamethoxazole was over 50%. Resistance to third-generation cephalosporins (Cefotaxime) was found in 54.1% of the E. coli strain. In contrast, Ertapenem, Fosfomycin, Imipenem, Meropenem, and Tigecycline resistance were below 5%. The resistance rates for all reported antibiotics fluctuated slightly in the studied period (Table 5).
3. Discussion
BSIs represent a global problem that needs prompt action. Timely detection, identification, and antimicrobial susceptibility testing of causative pathogens and hospital surveillance is needed to improve BSI management. This study shows the prevalence of BSI, the incidence of pathogens causing the infection, and evaluates the sensitivity profile to the main antibiotics used in the treatment, in the period between January 2015 and December 2019. In this study period, 24,694 total blood cultures were included, of which 3949 were positive (16%). For the years examined, a rather linear incidence trend (15–16%) was calculated, with the exception of 2018, where an incidence exceeds 22%. This higher value may be justified by a lower total number of blood cultures received and by a high positivity, probably caused by the S. aureus strains, more frequent in the year under review. Secondary BSIs was diagnosed in 1108 patients (28%). Our data were similar to those reported by the European Center for Disease Prevention and Control, which recorded 29% of secondary bacteremia [28]. Lower frequency of secondary BSIs was observed at University Hospital of the Canary Islands in Spain (22%) [29]. In the current study, the secondary bloodstream infections were not classified according to the district of the associated infectious process. In addition, all cases of bacteremia were included in our analysis, with the aim of evaluating the incidence and trends of antibiotic resistance of the main pathogenic bacteria that cause BSI. The average incidence of BSI cases was assessed, with a response of 16.4% per year. This isolation rate is consistent with many studies in Europe and abroad [3,30]. The incidence of bacteremia, in this study, increases rapidly with increasing age and is higher in males, in accordance with other studies [31,32]. CoNS (Coagulase-negative staphylococci) (17.4%), Staphylococcus aureus (12.8%), Escherichia coli (10.9%), and Klebsiella pneumoniae (9.4%) were the most frequently isolated species. As in other studies, the predominance of Gram-positive pathogens was documented and the bacterial distribution patterns were consistent with those reported [30,33,34,35]. Among the Gram-positive bacteria analyzed, the most resistant species is represented by Staphylococcus aureus. This species showed high resistance to Oxacillin (43.7%), Erythromycin (47.8%), and Levofloxacin (38.2%). The spread of methicillin-resistant S. aureus (MRSA) is a major public health problem [36]. To date, the most suitable treatment for resolving infections caused by MRSA is represented by to use of Glycopeptides, in particular, Vancomycin. Overuse of this antibiotic led to the emergence of resistant strains, present in this study, with an average percentage of 3.6% in the last four years, compared to 2015, where resistant strains were absent. In recent years, new antibiotics such as Linezolid and Daptomycin have been introduced into clinical practice for the treatment of MRSA infections [37]. Furthermore, for these antibiotics, as for vancomycin, there is a resistance rate of 3.9% in the last years of the study, compared to 2015 (0%). However, in this study, CoNS were the most frequent strains involved in BSI among Gram-positive bacteria. Although CoNS may act as contaminants in some cases [38], they were carefully evaluated before being included in this study, as reported in the results section. These species showed high resistance to Clindamycin (56.1%) and Oxacillin (72.7%), while low resistance was shown for Daptomycin, Linezolid and Vancomycin. CoNS are known to be the reservoir of resistance genes; therefore, the resistances shown in this study could spread among pathogenic staphylococci such as MRSA, and increase the difficulties in treating pathogen-promoted MDR infections [39]. The main Gram-negative species causing BSI, in this study, were Escherichia coli and Klebsiella pneumoniae, with a mean incidence of 10.9% and 9.5%, respectively. Isolates of K. pneumoniae showed high resistance to third-generation cephalosporins (Ceftazidime and Cefotaxime), fluoroquinolones (Ciprofloxacin), and carbapenems (Meropenem and Ertapenem), with an overall resistance range between 65% and 84%. However, they showed low resistance to Colistin and Fosfomycin, with a value of around 25%, for both. In contrast, E. coli isolates showed much lower resistance values towards carbapenems (Ertapenem, Meropenem, and Imipenem) around 1%, although they share high resistance values to third-generation cephalosporins and fluoroquinolones. These values were higher than the European average and the national average [40]. By using broad-spectrum antibiotics, such as cephalosporin and fluoroquinolones, they have favored the colonization and spread of resistant Enterobacteriaceae, including E. coli [41]. In 2019 there was a relevant increase in the percentage of K. pneumoniae isolates to penicillin-resistant (Amoxicillin/Clavulanic Acid) from 77.6% in 2015 to 94.8% in 2019. Furthermore, the study showed an increase in the percentage of resistance to third-generation both around 92% in 2019. While resistance to carbapenems from 64.2% in 2015 to 65.6% in 2019, and to fluoroquinolones, from 86. 6% in 2015 to 89.2% in 2019, has remained fairly stable, even if very high values are recorded, compared to the European average (among 25–50%) [42,43].
4. Materials and Methods
4.1. Samples Collection
A total of 24,694 blood samples were collected from patients admitted to the University Hospital “San Giovanni di Dio e Ruggi d’Aragona” in the period between January 2015 and December 2019. Blood draw was performed in accordance with the hospital’s aseptic guidelines. The protocol required the disinfection at the collection site with 2% chlorhexidine.
4.2. Isolation, Identification and Antimicrobial Susceptibility Test for BSI Pathogens
A volume of 5–10 mL and 2–3 mL were inoculated in blood culture bottles for adult and pediatric patients, respectively. For pediatric patients, the survey was performed only on the aerobic bottle, while an aerobic bottle and an anaerobic bottle were used for adult patients. Blood culture samples were delivered to the Microbiology Laboratory for testing. Blood culture bottles were incubated in the automated blood culture monitoring BACTEC 9240 blood culture system (Becton Dickinson Diagnostic Instrument Systems) system. The most common incubation time for bacteria was 5 days, it was increased for slow-growing organisms. When a positive alarm occurred in the blood culture instrument, 1 drop from each bottle was plated on standard bacteriology media: Chocolate agar, blood agar, MacConkey, and Sabouraud Glucose agar medium (Oxoid, Hampshire, UK). All plates were incubated overnight at 37 °C. The Chocolate agar was maintained in the presence of CO2. After 24–48 h of incubation, each plate was examined and bacterial identification and antimicrobial susceptibility test were performed. The bacterial identification and antimicrobial susceptibility test were performed via technology Vitek 2 (bioMe’rieux, Marcy l’Etoile, France), following the manufacturer’s recommendations. The results of antimicrobial susceptibility were interpreted as “susceptible”, “resistant”, or “intermediate” according to EUCAST guidelines.
4.3. Ethical Consideration Statement
Ethical approval by the Human Research Ethics Committee was not requested. The present study used laboratory management data, collected from database. This is a retrospective study and not directly associated with patients.
4.4. Statistical Analysis
Demographic data of patients, including age, gender, isolated strain(s), and drug sensitivity results, were used for the analysis. The crude incidence and age- and sex-standardized incidence were calculated. The chi-framework test was used to compare the differences in the incidence of bacteria in hospitalized patients and the differences among antibiotic sensitivities over the range of years considered in the study. The p-value was calculated using the chi-squared test for a row-by-column contingency table with appropriate degrees of freedom. p < 0.05 was considered statistically significant. The IBM Statistical Package for Social Sciences Version 22.00 (SPSS Inc, Chicago, USA (http://www.spss.com)) was used for data analysis.
5. Conclusions
Hospital surveillance studies of blood infections allow for a deeper understanding of BSI-causing microorganisms and their pattern of antibiotic susceptibility to improve empirical antibiotic therapy [44]. It is essential to evaluate the etiological agents, the results of microbial culture, and antimicrobial susceptibility, in order to be able to follow the trend of resistance to the most frequently administered antibiotics [45,46]. Efficient control methods are needed to decrease resistance to antibiotic drugs and to ensure that patients receive effective treatment [47,48]. Therefore, programs should be implemented to improve the quality of empirical therapy in patients with suspected BSI and the optimization of definitive therapy, improving the antimicrobial management program in our university hospital [49].
Acknowledgments
The authors would like to thank the staff of the U.O.C University Hospital of Campania “Luigi Vanvitelli” in Naples, Italy and the staff of University Hospital “San Giovanni di Dio e Ruggi d’Aragona”, for their contributions.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/12/851/s1, Table S1: Percentage trend of all bacterial isolates that caused bloodstream infections.
Author Contributions
Conceptualization, B.S.; writing—review and editing, B.S. and V.F.; supervision, V.F., G.F. and M.G.; funding acquisition, G.B. and G.F.; Data curation, G.M.P. and P.P.; Visualization, O.M., E.S. (Enrica Serretiello), G.M., E.S. (Emanuela Santoro), C.Z., F.D.C., M.C. and G.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Timsit J.-F., Ruppé E., Barbier F., Tabah A., Bassetti M. Bloodstream infections in critically ill patients: An expert statement. Intensiv. Care Med. 2020;46:266–284. doi: 10.1007/s00134-020-05950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fleischmann M.C., Scherag A., Adhikari N.K.J., Hartog C., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 3.Wisplinghoff H., Bischoff T., Tallent S.M., Seifert H., Wenzel R.P., Edmond M.B. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin. Infect. Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 4.Wu J.-N., Gan T.-E., Zhu Y.-X., Cao J.-M., Ji C.-H., Wu Y.-H., Lu B. Epidemiology and microbiology of nosocomial bloodstream infections: Analysis of 482 cases from a retrospective surveillance study. J. Zhejiang Univ. Sci. B. 2015;16:70–77. doi: 10.1631/jzus.B1400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed D., Nahid A., Sami A.B., Halim F., Akter N., Sadique T., Rana S., Bin Elahi S., Rahman M. Bacterial etiology of bloodstream infections and antimicrobial resistance in Dhaka, Bangladesh, 2005–2014. Antimicrob. Resist. Infect. Control. 2017;6:2. doi: 10.1186/s13756-016-0162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanoksil M., Jatapai A., Peacock S.J., Limmathurotsakul D. Epidemiology, microbiology and mortality associated with community-acquired bacteremia in northeast Thailand: A multicenter surveillance study. PLoS ONE. 2013;8:e54714. doi: 10.1371/annotation/e199ebcc-0bc1-4be1-ad91-ad2a8c0c9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallés J., Palomar M., Alvárez-Lerma F., Rello J., Blanco A., Garnacho-Montero J., Martín-Loeches I. Evolution Over a 15-Year Period of Clinical Characteristics and Outcomes of Critically Ill Patients With Community-Acquired Bacteremia. Crit. Care Med. 2013;41:76–83. doi: 10.1097/CCM.0b013e3182676698. [DOI] [PubMed] [Google Scholar]
- 8.Kollef M.H., Zilberberg M.D., Shorr A.F., Vo L., Schein J., Micek S.T., Kim M. Epidemiology, microbiology and outcomes of healthcare-associated and community-acquired bacteremia: A multicenter cohort study. J. Infect. 2011;62:130–135. doi: 10.1016/j.jinf.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Søgaard M., Nørgaard M., Dethlefsen C., Schønheyder H.C. Temporal Changes in the Incidence and 30-Day Mortality associated with Bacteremia in Hospitalized Patients from 1992 through 2006: A Population-based Cohort Study. Clin. Infect. Dis. 2011;52:61–69. doi: 10.1093/cid/ciq069. [DOI] [PubMed] [Google Scholar]
- 10.Valles J., Rello J., Ochagavia A., Garnacho J., Alcala M.A. Community-acquired bloodstream infection in critically ill adult patients: Impact of shock and inappropriate antibiotic therapy on survival. Chest. 2003;123:1615–1624. doi: 10.1378/chest.123.5.1615. [DOI] [PubMed] [Google Scholar]
- 11.Kilgore M.L., Brossette S. Cost of bloodstream infections. Am. J. Infect. Control. 2008;36:S172.e1–S172.e3. doi: 10.1016/j.ajic.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Hotchkiss R.S., Moldawer L.L., Opal S.M., Reinhart K., Turnbull I.R., Vincent J.L. Sepsis and septic shock. Nat. Rev. Dis. Primers. 2016;2:16045. doi: 10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathur P., Varghese P., Tak V., Gunjiyal J., Lalwani S., Kumar S., Misra M.C. Epidemiology of Blood Stream Infections at a Level-1 Trauma Care Center of India. J. Lab. Physicians. 2014;6:022–027. doi: 10.4103/0974-2727.129086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwiecińska-Piróg J., Skowron K., Gospodarek-Komkowska E. Primary and Secondary Bacteremia Caused by Proteus spp.: Epidemiology, Strains Susceptibility and Biofilm Formation. Pol. J. Microbiol. 2018;67:471–478. doi: 10.21307/pjm-2018-055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegman-Igra Y., Fourer B., Orni-Wasserlauf R., Golan Y., Noy A., Schwartz D., Giladi M., Yardena S.-I., Boaz F., Ruth O.-W., et al. Reappraisal of Community-Acquired Bacteremia: A Proposal of a New Classification for the Spectrum of Acquisition of Bacteremia. Clin. Infect. Dis. 2002;34:1431–1439. doi: 10.1086/339809. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Z., Wang Q., Zhu F., An Y. Epidemiology, species distribution, antifungal susceptibility and mortality risk factors of candidemia among critically ill patients: A retrospective study from 2011 to 2017 in a teaching hospital in China. Antimicrob. Resist. Infect. Control. 2019;8:89. doi: 10.1186/s13756-019-0534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian L., Zhang Z., Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: A 20-year surveillance study (1998-2017) Antimicrob. Resist. Infect. Control. 2019;8:86. doi: 10.1186/s13756-019-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent J.-L. The Clinical Challenge of Sepsis Identification and Monitoring. PLoS Med. 2016;13:e1002022. doi: 10.1371/journal.pmed.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seifert H. The Clinical Importance of Microbiological Findings in the Diagnosis and Management of Bloodstream Infections. Clin. Infect. Dis. 2009;48:238–245. doi: 10.1086/598188. [DOI] [PubMed] [Google Scholar]
- 20.Dailey P.J., Osborn J., Ashley E.A., Baron E.J., Dance D.A.B., Fusco D., Fanello C., Manabe Y.C., Mokomane M., Newton P.N., et al. Defining System Requirements for Simplified Blood Culture to Enable Widespread Use in Resource-Limited Settings. Diagnostics. 2019;9:10. doi: 10.3390/diagnostics9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peker N., Couto N., Sinha B., Rossen J.W.A. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: Recent developments in molecular approaches. Clin. Microbiol. Infect. 2018;24:944–955. doi: 10.1016/j.cmi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Sader H.S., Castanheira M., Streit J.M., Carvalhaes C.G., Mendes R.E. Frequency and antimicrobial susceptibility of bacteria causing bloodstream infections in pediatric patients from United States (US) medical centers (2014–2018): Therapeutic options for multidrug-resistant bacteria. Diagn. Microbiol. Infect. Dis. 2020;98:115108. doi: 10.1016/j.diagmicrobio.2020.115108. [DOI] [PubMed] [Google Scholar]
- 23.Hattori H., Maeda M., Nagatomo Y., Takuma T., Niki Y., Naito Y., Sasaki T., Ishino K. Epidemiology and risk factors for mortality in bloodstream infections: A single-center retrospective study in Japan. Am. J. Infect. Control. 2018;46:e75–e79. doi: 10.1016/j.ajic.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Buetti N., Atkinson A., Marschall J., Kronenberg A., Anresis T.S.C.F.A.R. Incidence of bloodstream infections: A nationwide surveillance of acute care hospitals in Switzerland 2008–2014. BMJ Open. 2017;7:e013665. doi: 10.1136/bmjopen-2016-013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llor C., Bjerrum L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014;5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall K.K., Lyman J.A. Updated Review of Blood Culture Contamination. Clin. Microbiol. Rev. 2006;19:788–802. doi: 10.1128/CMR.00062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. [(accessed on 4 July 2013)]; Available online: https://www.ecdc.europa.eu/en/publications-data/point-prevalence-survey-healthcare-associated-infections-and-antimicrobial-use-0.
- 29.Sante L., Aguirre-Jaime A., Miguel M.A., Ramos M.J., Pedroso Y., Lecuona M. Epidemiological study of secondary bloodstream infections: The forgotten issue. J. Infect. Public Heal. 2019;12:37–42. doi: 10.1016/j.jiph.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Panday R.S.N., Wang S., Van De Ven P.M., Hekker T.A.M., Alam N., Nanayakkara P.W.B. Evaluation of blood culture epidemiology and efficiency in a large European teaching hospital. PLoS ONE. 2019;14:e0214052. doi: 10.1371/journal.pone.0214052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gubbels S., Nielsen J., Voldstedlund M., Kristensen B., Schonheyder H.C., Vandenbroucke-Grauls C., Arpi M., Björnsdóttir M., Knudsen J.D., Dessau R.B., et al. Utilization of blood cultures in Danish hospitals: A population-based descriptive analysis. Clin. Microbiol. Infect. 2015;21:344.e13–344.e21. doi: 10.1016/j.cmi.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Uslan D.Z., Crane S.J., Steckelberg J.M., Cockerill F.R., 3rd, St Sauver J.L., Wilson W.R., Baddour L.M. Age- and sex-associated trends in bloodstream infection: A population-based study in Olmsted County, Minnesota. Arch. Intern. Med. 2007;167:834–839. doi: 10.1001/archinte.167.8.834. [DOI] [PubMed] [Google Scholar]
- 33.Karlowsky J.A., Jones M.E., Draghi D.C., Thornsberry C., Sahm D.F., Volturo G.A. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 2004;3:7. doi: 10.1186/1476-0711-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thakur S., Thakur K., Sood A., Chaudhary S. Bacteriological profile and antibiotic sensitivity pattern of neonatal septicaemia in a rural tertiary care hospital in North India. Indian J. Med Microbiol. 2016;34:67–71. doi: 10.4103/0255-0857.174108. [DOI] [PubMed] [Google Scholar]
- 35.Gohel K., Jojera A., Soni S., Gang S., Sabnis R., Desai M. Bacteriological Profile and Drug Resistance Patterns of Blood Culture Isolates in a Tertiary Care Nephrourology Teaching Institute. BioMed Res. Int. 2014;2014:1–5. doi: 10.1155/2014/153747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pignataro D., Foglia F., Della Rocca M.T., Melardo C., Santella B., Folliero V., Shinde S., Pafundi P.C., Sasso F., Iovene M.R., et al. Methicillin-resistant Staphylococcus aureus: Epidemiology and antimicrobial susceptibility experiences from the University Hospital ‘Luigi Vanvitelli’ of Naples. Pathog. Glob. Heal. 2020;2020:1–6. doi: 10.1080/20477724.2020.1827197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choo E.J., Chambers H.G. Treatment of Methicillin-Resistant Staphylococcus aureus Bacteremia. Infect. Chemother. 2016;48:267–273. doi: 10.3947/ic.2016.48.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sidhu S.K., Malhotra S., Devi P., Tuli A.K. Significance of coagulase negative Staphylococcus from blood cultures: Persisting problems and partial progress in resource constrained settings. Iran. J. Microbiol. 2016;8:366–371. [PMC free article] [PubMed] [Google Scholar]
- 39.Saber H., Jasni A.S., Jamaluddin T.Z.M.T., Ibrahim R. A Review of Staphylococcal Cassette Chromosome mec (SCCmec) Types in Coagulase-Negative Staphylococci (CoNS) Species. Malays. J. Med Sci. 2017;24:7–18. doi: 10.21315/mjms2017.24.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camins B.C., Marschall J., De Vader S.R., Maker D.E., Hoffman M.W., Fraser V.J. The clinical impact of fluoroquinolone resistance in patients with E coli bacteremia. J. Hosp. Med. 2011;6:344–349. doi: 10.1002/jhm.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawat D., Nair D. Extended-spectrum beta-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010;2:263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apondi O.E., Oduor O.C., Gye B.K., Kipkoech M.K. High Prevalence of Multi-Drug Resistant Klebsiella Pneumoniae in A Tertiary Teaching Hospital in Western Kenya. Afr. J. Infect. Dis. 2016;10:89–95. doi: 10.21010/ajid.v10i2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuner E.A., Yeh J.-Y., Hall G.S., Sekeres J., Endimiani A., Bonomo R.A., Shrestha N.K., Fraser T.G., Van Duin D. Treatment and outcomes in carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Diagn. Microbiol. Infect. Dis. 2011;69:357–362. doi: 10.1016/j.diagmicrobio.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sydnor E.R.M., Perl T.M. Hospital Epidemiology and Infection Control in Acute-Care Settings. Clin. Microbiol. Rev. 2011;24:141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Belkum A., Bachmann T.T., Ludke G., Lisby J.G., Kahlmeter G., Mohess A., Becker K., Hays J.P., Woodford N., Mitsakakis K., et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2019;17:51–62. doi: 10.1038/s41579-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fair R.J., Tor Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Med. Chem. 2014;6:25–46. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee C.-R., Cho I.H., Jeong B.C., Lee S.H. Strategies to Minimize Antibiotic Resistance. Int. J. Environ. Res. Public Heal. 2013;10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zannella C., Shinde S., Vitiello M., Falanga A., Galdiero E., Fahmi A., Santella B., Nucci L., Gasparro R., Galdiero M., et al. Antibacterial Activity of Indolicidin-Coated Silver Nanoparticles in Oral Disease. Appl. Sci. 2020;10:1837. doi: 10.3390/app10051837. [DOI] [Google Scholar]
- 49.Retamar P., Portillo M.M., López-Prieto M.D., Rodríguez-López F., De Cueto M., García M.V., Gómez M.J., Del Arco A., Muñoz A., Sánchez-Porto A., et al. Impact of Inadequate Empirical Therapy on the Mortality of Patients with Bloodstream Infections: A Propensity Score-Based Analysis. Antimicrob. Agents Chemother. 2012;56:472–478. doi: 10.1128/AAC.00462-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.