Abstract
Dagger nematodes of the genus Xiphinema include a remarkable group of invertebrates of the phylum Nematoda comprising ectoparasitic animals of many wild and cultivated plants. Damage is caused by direct feeding on root cells and by vectoring nepoviruses that cause diseases on several crops. Precise identification of Xiphinema species is critical for launching appropriate control measures. We deciphered the cryptic diversity of the Xiphinema hispanum-species complex applying integrative taxonomical approaches that allowed us to verify a paradigmatic example of the morphostatic speciation and the description of a new species, Xiphinema malaka sp. nov. Detailed morphological, morphometrical, multivariate and genetic studies were carried out, and mitochondrial and nuclear haploweb analyses were used for species delimitation of this group. The new species belongs to morphospecies Group 5 from the Xiphinema nonamericanum-group species. D2-D3, ITS1, partial 18S, and partial coxI regions were used for inferring the phylogenetic relationships of X. malaka sp. nov. with other species within the genus Xiphinema. Molecular analyses showed a clear species differentiation not paralleled in morphology and morphometry, reflecting a clear morphostatic speciation. These results support the hypothesis that the biodiversity of dagger nematodes in southern Europe is greater than previously assumed.
Keywords: Bayesian inference, cryptic species, coxI, D2-D3 expansion domains of 28S rRNA-gene, integrative taxonomy, principal component analysis
1. Introduction
Plant-parasitic nematodes (PPN) are characterized by the presence of a stylet used for root tissue penetration, comprise about 15% of the total number of nematode species currently known, of which over 4100 species have been identified as PPN [1,2]. Annual crop losses caused by PPN are estimated to be about 8–15% of total crop production worldwide [3,4]. Accurate identification of PPN is essential for the selection of appropriate control measures against plant pathogenic species, as well as for a reliable method allowing distinction between species under quarantine or regulatory strategies and a better understanding of their implications in pest control and soil ecology [5,6]. PPN species have been defined historically based on morphological characteristics [7,8]. However, the adoption of molecular techniques in nematode taxonomy has revealed unexpected genetic diversity within species throughout the phylum Nematoda [9]. This has been especially accurate for the family Longidoridae, a large group of ectoparasitic nematodes feeding from the root tip zone to the hairy root region, and characterized by a substantial intra and interspecific homogeneity of the morphometric characters used for species discrimination [1,6,10,11]. Use of molecular data in species identification of dagger and needle nematodes over the last three decades has indicated that many widespread species actually comprise multiple genetically divergent and morphologically similar cryptic species [6,11,12,13]. Complexes of cryptic species often result from nonecological speciation in which diversification is not accompanied by apparent ecological or morphological separation in traditional quantitative traits [14].
The genus Xiphinema is one of the most diversified group species of longidorid nematodes with more than 280 valid species [5,6,11,15]. The ecological and phytopathological importance of this group of nematodes lies in its wide range of host plants and cosmopolitan distribution [5,11], but some species of this genus are vectors of several important plant viruses (genus Nepovirus, family Comoviridae) that cause significant damage to a wide range of crops [10,16]. Considering the great diversity of this group, the genus Xiphinema was divided into two different species groups [5,17,18]: (i) the Xiphinema americanum-group comprising a complex of about 60 species [15,17]; and (ii) the Xiphinema nonamericanum-group which comprises a complex of more than 220 species [5,6,19]. Later, this group was divided into eight morphospecies groups for helping identification [18]. However, some cryptic species and species complexes within Xiphinema have been recently revealed based on integrative taxonomical approaches, including morphometric multivariate methods, genetic analyses based on ribosomal and mitochondrial DNA (rDNA and mtDNA, respectively) and species delimitation (haplonet tools) [6,11,20,21]. A paradigmatic example of these species complexes comprises the Xiphinema hispanum-complex, viz. didelphic Xiphinema species from the Iberian Peninsula characterized by a rounded tail in females with or without an inconspicuous bulge projecting slightly ventrally and a uterus showing spiniform structures [22]. The cryptic diversity of this species complex has been deciphered by our team over the last ten years applying integrative taxonomical approaches that allowed us to verify these species as valid, and the recent description of a new species, X. subbaetense [11,20]. Recent studies on this species complex clearly separated three species (X. adenohystherum, X. hispanum and X. subbaetense) revealing high levels of genetic diversity within them that showed little morphological differentiation [11]. In new nematode surveys carried out in natural areas in the provinces of Málaga and Almería, Andalusia, southern Spain, we have detected nine unidentified Xiphinema isolates resembling X. hispanum-complex morphology. Detailed morphological and morphometrical observations using light microscopy indicated that these isolates appeared undistinguishable from X. hispanum complex species, a fact which prompted us to undertake comprehensive multivariate and genetic analyses, compared with previous reported data, to decipher this taxonomic conundrum.
Morphostatic evolution can be defined as genetic modifications, and even complete speciation events, which are not reflected in morphology, often being a result of nonadaptive radiation marked by the rapid proliferation of species without ecological differentiation [23,24]. Although no data have yet been specifically mentioned in Nematoda, morphostatic evolution seems not to be a rare phenomenon in longidorids based on the numerous complexes and cryptic species documented [6,11,12,13,15,20,25]. In Longidoridae, it is very common that molecular divergences among species are not reflected in morphological or morphometric traits, which conforms a morphostatic model of evolution with numerous cryptic species within this group [6,11,13,15,20,21,25,26].
In this context, we investigated (1) the existence of a new cryptic species within the X. hispanum-complex confirming a morphostatic speciation in this group using an integrative species delineation approach based on multivariate morphometric analysis and haplonet mitochondrial and nuclear haploweb tools; (2) a new species of the genus Xiphinema (Xiphinema malaka sp. nov.) described through integrative methods based on the combination of morphological, morphometric and molecular data; and (3) phylogenetic analyses based on D2-D3 expansion domains of the 28S rRNA gene, ITS1, the partial 18S rRNA gene, and the partial mitochondrial coxI gene sequences to clarify the relationships of the new Xiphinema species.
2. Results
Species boundaries within the Xiphinema complex included in this research (Figure 1) were based on the integrative application of morphological, morphometric and molecular methods to unravel potential cryptic species diversity (Table 1). Species delimitation was carried out using two independent approaches based on morphometric (multivariate analysis) and molecular data using ribosomal and mitochondrial sequences (haplonet). Multivariate morphometric and haplonet methods were performed on the nine studied isolates including previous isolates from the X. hispanum-complex to verify species identifications. The integration of this procedure with the analysis of nematode morphology allowed us to verify Xiphinema malaka sp. nov. as a valid new species within the X. hispanum cryptic complex. Additionally, we maintained a consensus approach for the different species delimitation methods, including concordant results in phylogenetic trees inferred from nuclear and mitochondrial markers and/or different morphological or morphometric characteristics.
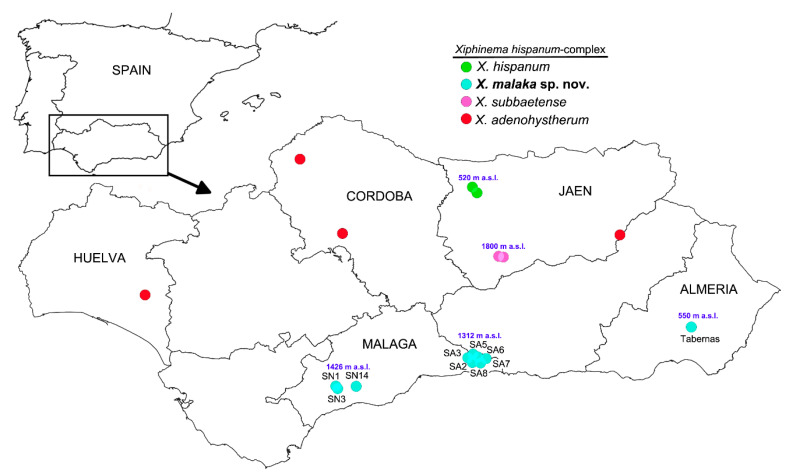
Figure 1.
Geographic distribution of Xiphinema hispanum-complex species and locations of sampling sites of which the recovered isolates of the new species were characterized morphometrically and molecularly. Arrow indicates the location of Andalusia in the Iberian Peninsula.
Table 1.
Isolates sampled for Xiphinema malaka sp. nov. from several localities of Málaga and Almería provinces (Southern Spain), and Xiphinema adenohystherum sequences used in this study.
| Sample Code—Host-Plant Locality | D2-D3 Haplotype | coxI Haplotype | D2-D3 | ITS1 | 18S | coxI | |
|---|---|---|---|---|---|---|---|
| Xiphinema malaka sp. nov. | |||||||
| Maritime Pine (Pinus pinaster Aiton) | |||||||
| 1. SA03-DF91 | Canillas de Albaida (Málaga) a | D2-D3-Hm3 | coxI-Hm2 | MT584052 | MT584088 | MT584086 | MT580263 |
| 2. SA03-DF92 | Canillas de Albaida (Málaga) a | D2-D3-Hm10 | coxI-Hm2 | MT584053 | - | - | MT580264 |
| 3. SA03-DF15 | Canillas de Albaida (Málaga) a | D2-D3-Hm10 | coxI-Hm2 | MT584054 | - | - | MT580265 |
| 4. SA03-DF90 | Canillas de Albaida (Málaga) a | D2-D3-Hm10 | - | MT584055 | - | - | - |
| 5. SA03-DF93 | Canillas de Albaida (Málaga) a | D2-D3-Hm10 | - | MT584056 | - | - | - |
| 6. SA03-DF94 | Canillas de Albaida (Málaga) a | D2-D3-Hm10 | - | MT584057 | - | - | - |
| 7. SA02-DF42 | Canillas de Albaida (Málaga) | D2-D3-Hm12 | - | MT584058 | MT584089 | - | |
| 8. SA02-AU62 | Canillas de Albaida (Málaga) | D2-D3-Hm12 | - | MT584059 | - | - | |
| 9. SA05_DF16 | Canillas de Albaida (Málaga) | D2-D3-Hm11 | - | MT584060 | MT584090 | - | |
| 10. SA05_DH96 | Canillas de Albaida (Málaga) | D2-D3-Hm1 | - | MT584061 | - | - | - |
| 11. SA05_DH97 | Canillas de Albaida (Málaga) | D2-D3-Hm2 | - | MT584062 | - | - | - |
| 12. SA05_DH98 | Canillas de Albaida (Málaga) | D2-D3-Hm3 | - | MT584063 | - | - | - |
| 13. SA05_DH99 | Canillas de Albaida (Málaga) | D2-D3-Hm3 | - | MT584064 | - | - | - |
| 14. SA06-DG12 | Canillas de Albaida (Málaga) | D2-D3-Hm12 | - | MT584065 | MT584091 | - | - |
| 15. SA06-DG13 | Canillas de Albaida (Málaga) | D2-D3-Hm5 | - | MT584066 | MT584092 | - | - |
| 16. SA06-DI01 | Canillas de Albaida (Málaga) | D2-D3-Hm4 | - | MT584067 | - | - | - |
| 17. SA06-DI02 | Canillas de Albaida (Málaga) | D2-D3-Hm5 | - | MT584068 | - | - | - |
| 18. SA06-DI03 | Canillas de Albaida (Málaga) | D2-D3-Hm6 | - | MT584069 | - | - | - |
| 19. SA06-DI04 | Canillas de Albaida (Málaga) | D2-D3-Hm4 | - | MT584070 | - | - | - |
| 20. SA07-AU46 | Canillas de Albaida (Málaga) | D2-D3-Hm9 | coxI-Hm3 | MT584071 | MT584093 | MT584087 | MT580266 |
| 21. SA07-AU47 | Canillas de Albaida (Málaga) | D2-D3-Hm9 | - | MT584072 | MT584094 | - | - |
| 22. SA07-AU48 | Canillas de Albaida (Málaga) | D2-D3-Hm9 | - | MT584073 | - | - | - |
| 23. SA08-DF19 | Canillas de Albaida (Málaga) | D2-D3-Hm2 | coxI-Hm1 | MT584074 | MT584095 | - | MT580267 |
| Black pine (Pinus nigraArnold) | - | ||||||
| 24. SN01-DI10 | Igualeja (Málaga) | D2-D3-Hm7 | coxI-Hm5 | MT584075 | MT584096 | - | MT580268 |
| 25. SN01-DE88 | Igualeja (Málaga) | D2-D3-Hm3 | coxI-Hm5 | MT584076 | - | - | MT580269 |
| 26. SN01-DF46 | Igualeja (Málaga) | D2-D3-Hm3 | coxI-Hm6 | MT584077 | - | - | MT580270 |
| 27. SN03-DE93 | Igualeja (Málaga) | D2-D3-Hm7 | coxI-Hm5 | MT584078 | MT584097 | - | MT580271 |
| Cork oak (Quercus suber L.) | |||||||
| 28. SN14-DF11 | Monda (Málaga) | D2-D3-Hm8 | - | MT584079 | MT584098 | - | - |
| 29. SN14-DI12 | Monda (Málaga) | D2-D3-Hm8 | - | MT584080 | - | - | - |
| 30. SN14-DI13 | Monda (Málaga) | D2-D3-Hm8 | - | MT584081 | - | - | - |
| 31. SN14-DI14 | Monda (Málaga) | D2-D3-Hm8 | - | MT584082 | - | - | - |
| 32. SN14-DI15 | Monda (Málaga) | D2-D3-Hm8 | coxI-Hm4 | MT584083 | - | - | MT580272 |
| 33. SN14-DI16 | Monda (Málaga) | D2-D3-Hm8 | coxI-Hm4 | MT584084 | - | - | MT580273 |
| Yellow broom (Cytisus scoparius (L.) Link) | |||||||
| 34. RAMB-AO44 | Tabernas, Almería | D2-D3-Hm13 | coxI-Hm1 | MT584085 | MT584099 | - | MT580274 |
| Xiphinema adenohystherum | |||||||
| Grapevine (Vitis vinifera L.) | |||||||
| 406f | Villalba del Alcor, Huelva | - | - | - | MT584100 | - | - |
| Wild olive (Olea europaea L. subsp europaea var. sylvestris) | |||||||
| AR139 | Aroche, Huelva | - | - | - | MT584101 | - | - |
| European holly (Ilex aquifolium L.) | |||||||
| Z137f | Arevalo de la Sierra, Soria | - | - | - | MT584102 | - | - |
a Type locality (paratype specimens); (-) Not obtained or not performed.
2.1. Multivariate Morphometric Analysis
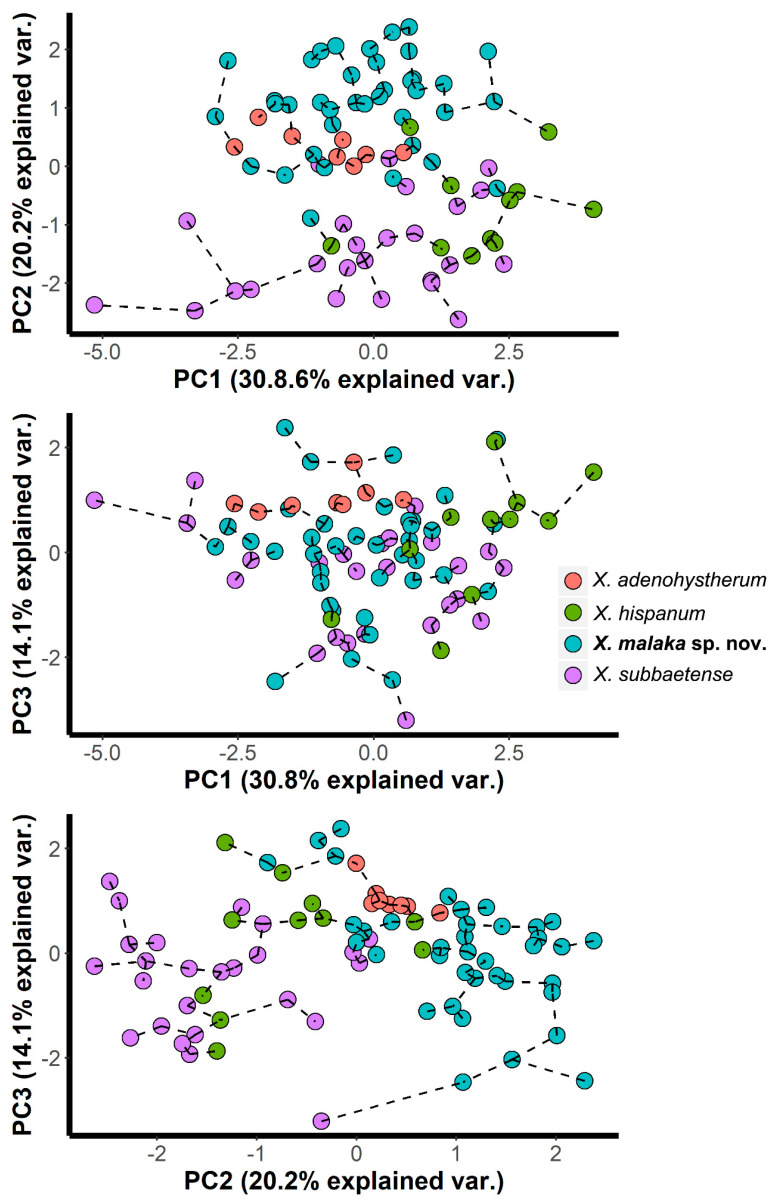
In principal component analysis (PCA), the first three components (sum of squares (SS) loadings > 1) accounted for 65.1% of the total variance in the morphometric characteristics of the X. hispanum-complex (Table 2). The eigenvalues for each character were used to interpret the biological meaning of the factors. First, the principal component 1 (PC1) was mainly dominated by a stylet with a high positive correlation (eigenvalue = 0.523). PC2 was mainly dominated by high negative correlation for the vulva position (eigenvalue = −0.547) as well as a high positive correlation for the a ratio (eigenvalue = 0.482) (Table 2). This component was, therefore, related with the overall nematode size and shape. Finally, PC3 was mainly dominated by the highest positive correlation found for the c’ ratio and lower, but also high, positive correlation for the hyaline region length (eigenvalues = 0.774 and 0.458, respectively). This component was then related with tail shape. Overall, these results suggest that all of the extracted PCs were related to the overall size and shape of nematode isolates. The results of the PCA were represented graphically in Cartesian plots in which isolates of the X. hispanum-complex were projected on the plane of the x- and y-axes, respectively, as pairwise combinations of components 1 to 3 (Figure 2). In the graphic representation of the X. hispanum-complex, and with the exception of X. adenohystherum, we observed that the specimens of all species were projected showing an expanded distribution along the plane for all the projected combinations of the components. One reason might be the wide morphometric variation detected in these species (Table 3 and Table 4) [6,11]. As a consequence, we did not detect a clear separation amongst species within the X. hispanum-complex, all the specimens being projected at random for all the projected combinations. These patterns suggest a clear example of morphostatic speciation within the X. hispanum-complex. However, it should be noted that when projected on the plane of the combinations of PC1-2 and PC2-3, almost all specimens of X. malaka sp. nov. and X. subbaetense were separated among them (Figure 2). This graphical separation was shown by the projection of PC2 (dominated by the V and a ratios). This graphical separation is due to the variation found in the ratio a among these species, as pointed out below. A minimum spanning tree (MST) superimposed on the plot of the first three principal components showed the same patterns observed with PCA, that is, not clear separation amongst species within the X. hispanum-complex (Figure 2).
Table 2.
Eigenvectors and SS loadings of factors derived from nematode morphometric characters for Xiphinema hispanum-complex (Xiphinema malaka sp. nov., Xiphinema adenohystherum, Xiphinema hispanum, Xiphinema subbaetense).
|
Xiphinema hispanum-Complex Principal Components a |
|||
|---|---|---|---|
| Character b | PC1 | PC2 | PC3 |
| Body length (L) | −0.382 | 0.032 | −0.003 |
| a | −0.081 | 0.483 | 0.262 |
| c’ | 0.009 | 0.137 | 0.774 |
| d | −0.371 | 0.444 | −0.100 |
| d’ | −0.318 | −0.384 | 0.086 |
| V | −0.170 | −0.550 | −0.046 |
| Stylet | −0.523 | −0.001 | −0.015 |
| Oa-gr | −0.440 | 0.198 | −0.320 |
| Hyaline region length | −0.334 | −0.256 | 0.458 |
| SS loadings | 1.67 | 1.35 | 1.13 |
| % of total variance | 30.80 | 20.21 | 14.10 |
| Cumulative % of total variance | 30.80 | 51.01 | 65.1 |
a Based on 41 female specimens of Xiphinema malaka sp. nov. from seven isolate samples, 25 female specimens of Xiphinema subbaetense from two isolate samples, eight female specimens of Xiphinema adenohystherum from a isolate sample, and 11 female specimens of Xiphinema hispanum from a isolate sample. Values of morphometric variables 1 to 3 (eigenvector > 0.458) are underlined. All isolates were molecularly identified and located at southern Spain. The c’ ratio was excluded by the multicollinearity test and then, it was not included in the multivariate analysis for the Xiphinema hispanum-complex; b Morphological and diagnostic characters according to Jairajpuri and Ahmad [7] with some inclusions. a = body length/maximum body width; c’ = tail length/body width at anus; d = anterior to guiding ring/body diam. at lip region; d’ = body diameter at guiding ring/body diameter at lip region; Oa-gr = Oral aperture-guiding ring distance; V = (distance from anterior end to vulva/body length) × 100.
Figure 2.
Principal component analysis on morphometric characters to characterize Xiphinema hispanum-complex with a superimposed minimum spanning tree (based on Euclidean distance).
Table 3.
Morphometrics of paratypes for Xiphinema malaka sp. nov. from maritime pine (Pinus pinaster Aiton) at Canillas de Albaida (Málaga province) southern Spain a.
| Paratypes | ||||||
|---|---|---|---|---|---|---|
| Characters-Ratios b | Holotype | Females | J1 | J2 | J3 | J4 |
| n | 1 | 20 | 7 | 4 | 4 | 6 |
| L (mm) | 4.3 | 4.2 ± 0.36 (3.51–4.90) |
1.25 ± 0.58 (1.13–1.31) |
1.81 ± 0.11 (1.66–1.92) |
2.19 ± 0.88 (2.11–2.33) |
3.11 ± 0.16 (2.86–3.31) |
| a | 73.6 | 74.2 ± 5.4 (65.6–82.2) |
48.8 ± 2.1 (45.3–51.5) |
43.2 ± 3.3 (40.8–48.1) |
56.9 ± 2.0 (53.5–58.7) |
64.8 ± 3.4 (62.1–70.0) |
| b | 8.8 | 8.0 ± 0.7 (6.7–9.3) |
5.1 ± 0.7 (4.4–6.1) |
6.0 ± 0.8 (5.3–7.0) |
5.9 ± 0.4 (5.5–6.3) |
6.6 ± 0.5 (6.0–7.2) |
| c | 117.4 | 110.6 ± 7.8 (97.3–126.3) |
20.5 ± 1.1 (18.6–22.4) |
28.5 ± 1.8 (26.8–31.0) |
39.9 ± 1.6 (37.4–41.5) |
67.4 ± 5.8 (60.2–75.4) |
| c’ | 0.9 | 0.9 ± 0.04 (0.9–1.0) |
3.5 ± 0.2 (3.2–3.8) |
2.4 ± 0.1 (2.3–2.6) |
2.0 ± 0.1 (1.8–2.1) |
1.3 ± 0.1 (1.2–1.4) |
| d | 8.9 | 8.4 ± 0.7 (6.9–9.3) |
5.4 ± 0.1 (5.3–5.6) |
7.6 ± 0.1 (7.5–7.6) |
7.3 ± 0.04 (7.3–7.4) |
8.4 ± 0.5 (8.1–9.2) |
| d’ | 2.7 | 2.6 ± 0.1 (2.4–2.9) |
2.0 ± 0.1 (1.9–2.1) |
2.68 ± 0.02 (2.67–2.70) |
2.4 ± 0.1 (2.3–2.5) |
2.7 ± 0.1 (2.6–2.8) |
| V | 50.1 | 50.2 ± 0.7 (49.2–51.5) |
- | - | - | - |
| G1 | 13.7 | 13.6 ± 1.4 (11.1–15.8) |
- | - | - | - |
| G2 | 12.5 | 11.6 ± 1.1 (9.9–13.5) |
- | - | - | - |
| Odontostyle length | 143.0 | 140.4 ± 4.7 (131.0–148.5) |
64.1 ± 0.6 (63.5–65.0) |
76.3 ± 1.5 (75.0–78.0) |
93.6 ± 5.6 (84.0–98.0) |
117.8 ± 4.0 (115.0–125.5) |
| Odontophore length | 77.5 | 79.5 ± 1.9 (75.0–83.0) |
40.6 ± 0.7 (40.0–42.0) |
56.1 ± 1.5 (54.0–57.5) |
59.9 ± 2.2 (57.5–63.0) |
71.2 ± 2.0 (67.0–72.5) |
| Total stylet | 220.5 | 219.9 ± 5.8 (206.0–229.0) |
104.7 ± 1.1 (104.0–107.0) |
135.8 ± 3.2 (132.5–140.0) |
153.5 ± 5.9 (145.0–161.0) |
189.0 ± 3.8 (187.0–197.0) |
| Replacement odontostyle | - | - | 76.7 ± 2.4 (74.0–80.0) |
100.9 ± 4.5 (95.0–105.5) |
117.8 ± 8.8 (105.0–127.0) |
143.9 ± 2.9 (140.0–148.0) |
| Lip region width | 14.5 | 14.6 ± 0.4 (14.0–15.0) |
9.4 ± 0.3 (9.0–9.5) |
10.3 ± 0.4 (10.0–10.5) |
11.1 ± 0.3 (11.0–11.5) |
12.5 ± 0.4 (12.0–13.0) |
| Oral aperture-guiding ring | 129.0 | 124.9 ± 8.8 (96.0–135.0) |
51.6 ± 1.6 (50.0–54.0) |
75.1 ± 3.6 (71.5–80.0) |
81.80 ± 2.0 (80.0–84.0) |
103.6 ± 4.5 (98.0–110.0) |
| Tail length | 37.0 | 38.0 ± 2.3 (34.0–43.0) |
60.8 ± 1.9 (57.5–64.0) |
63.5 ± 1.7 (62.0–65.0) |
54.8 ± 2.2 (53.0–58.0) |
46.3 ± 2.6 (43.0–49.0) |
| J | 12.0 | 11.9 ± 1.1 (10.0–13.5) |
9.1 ± 0.6 (8.5–10.5) |
13.8 ± 1.3 (12.5–15.0) |
12.3 ± 0.3 (12.0–12.5) |
10.5 ± 0.9 (9.5–11.5) |
a Measurements are in µm and in the form: mean ± standard deviation (range); b a = body length/maximum body width; b = body length/pharyngeal length; c = body length/tail length; c’ = tail length/body width at anus; d = anterior to guiding ring/body diam. at lip region; d’= body diam. at guiding ring/body diam. at lip region; V = (distance from anterior end to vulva/body length) × 100; J = hyaline tail region length; G1 = (anterior genital branch length/body length) × 100; G2 = (posterior genital branch length/body length) × 100.
Table 4.
Morphometrics of Xiphinema malaka sp. nov. from several sampling points and localities at Málaga and Almería provinces, southern Spaina.
| Málaga Province | Almería Province | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Host-plant Locality Sample Code |
Maritime Pine (Canillas de Albaida) SA2 |
Maritime Pine (Canillas de Albaida) SA5 |
Maritime Pine (Canillas de Albaida) SA6 |
Maritime Pine (Canillas de Albaida) SA8 |
Black Pine (Iguaneja) SN1 |
Black Pine (Iguaneja) SN3 |
Cork oak (Monda) SN14 |
Yellow Broom Tabernas AO44 |
|
| Characters-ratios b | Females | Females | Females | Male | Female | Female | Female | Females | J4 |
| n | 1 | 4 | 9 | 1 | 1 | 1 | 1 | 2 | 1 |
| L (mm) | 4.6 | 4.5 ± 0.4 (4.0–5.0) |
4.8 ± 0.2 (4.5–5.2) |
4.8 | 4.3 | 4.3 | 4.2 | (4.85, 4.92) | 3.08 |
| a | 82.8 | 85.4 ± 11.0 (76.0–99.8) |
87.2 ± 5.2 (79.6–96.5) |
109.5 | 76.1 | 72.1 | 70.9 | (82.0, 82.2) | 70.0 |
| b | 9.9 | 9.0 ± 1.1 (8.0–10.0) |
9.5 ± 0.7 (8.6–11.0) |
9.2 | 9.0 | 12.2 | 8.1 | (9.8, 10.3) | 7.0 |
| c | 123.1 | 132.8 ± 15.2 (115.3–151.2) |
164.7 ± 16.3 (133.2–178.6) |
170.2 | 133.5 | 98.3 | 113.9 | (103.2, 105.8) | 65.5 |
| c’ | 0.91 | 0.91 ± 0.03 (0.87–0.95) |
0.78 ± 0.06 (0.72–0.89) |
0.8 | 0.8 | 1.0 | 0.9 | (1.0, 1.1) | 1.3 |
| d | 8.5 | 8.2 ± 0.2 (8.0–8.4) |
8.3 ± 0.3 (7.9–8.9) |
7.6 | 7.6 | 8.1 | 9.4 | (7.4, 7.9) | 9.2 |
| d’ | 2.6 | 2.48 ± 0.01 (2.47–2.50) |
2.54 ± 0.13 (2.33–2.75) |
2.3 | 2.5 | 2.6 | 2.8 | (2.6, 2.7) | 2.8 |
| V or T | 49.2 | 49.4 ± 0.6 (48.5–50.0) |
49.8 ± 1.1 (47.8–51.5) |
62.0 | 47.1 | 52.2 | 50.0 | (51.8, 53.8) | - |
| G1 | 9.4 | 11.5 ± 0.8 (10.7–12.3) |
12.1 ± 2.0 (9.9–13.6) |
- | 13.5 | 12.8 | 10.5 | (9.2, 10.7) | - |
| G2 | 8.9 | 10.0 ± 0.7 (9.5–10.8) |
11.4 ± 1.2 (10.1–12.3) |
- | 13.0 | 11.7 | 10.2 | (8.1, 10.2) | - |
| Odontostyle length | 135.5 | 137.0 ± 4.2 (132.0–141.0) |
143.1 ± 2.2 (138.5–145.5) |
144.5 | 131.0 | 134.5 | 136.0 | (143.0, 149.0) | 125.5 |
| Odontophore length | 79 | 80.9 ± 0.9 (80.0–82.0) |
79.1 ± 2.7 (76.0–84.0) |
78.0 | 83.0 | 80.0 | 80.0 | (82.0, 88.0) | 71.5 |
| Total stylet | 215 | 217.9 ± 4.9 (212.5–222.0) |
222.2 ± 3.5 (215.5–227.5) |
225.5 | 214.0 | 215.0 | 216.0 | (231.0) | 197.0 |
| Replacement odontostyle | - | - | - | - | - | - | - | - | 146.0 |
| Lip region width | 14.5 | 14.5 ± 0.4 (14.0–15.0) |
14.7 ± 0.5 (14.0–15.5) |
16.0 | 15.0 | 14.0 | 14.0 | (15.0, 15.5) | 14.0 |
| Oral aperture-guiding ring | 115 | 118.5 ± 2.1 (116.0–121.0) |
122.5 ± 3.8 (119.0–129.0) |
121.5 | 114.0 | 114.0 | 122.0 | (115.0, 118.0) | 110.0 |
| Tail length | 37 | 33.6 ± 1.1 (32.5–35.0) |
29.2 ± 2.9 (26.0–34.5) |
28.0 | 32.5 | 44.0 | 37.0 | (46.5, 47.0) | 47.0 |
| J | 12.0 | 10.3 ± 0.4 (10.0–10.5) |
10.5 ± 2.0 (8.0–13.0) |
8.0 | 8.5 | 12.0 | 11.0 | (13.0) | 12.0 |
| Spicules | - | - | - | 64.0 | - | - | - | - | - |
| Lateral guiding pieces | - | - | - | 26.5 | - | - | - | - | - |
a Measurements are in µm and in the form: mean ± standard deviation (range); b a = body length/maximum body width; b = body length/pharyngeal length; c = body length/tail length; c’ = tail length/body width at anus; d = anterior to guiding ring/body diam. at lip region; d’= body diam. at guiding ring/body diam. at lip region; V = (distance from anterior end to vulva/body length) × 100; J = hyaline tail region length; G1 = (anterior genital branch length/body length) × 100; G2 = (posterior genital branch length/body length) × 100.
2.2. Mitochondrial Haplonet and Nuclear Haploweb Networks
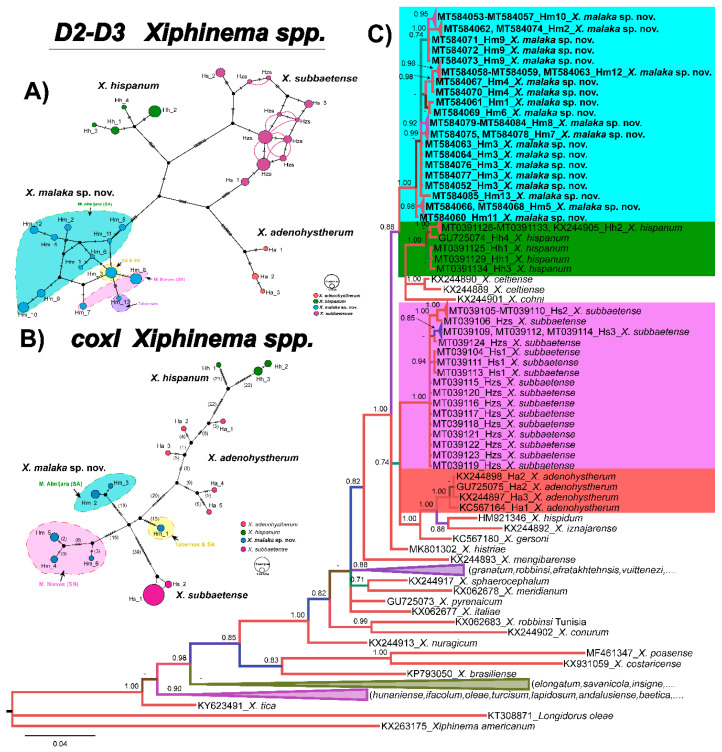
Species delimitation using haplonet methods in X. hispanum-complex species contained 75 sequences (35 sequences from X. malaka sp. nov., four sequences from X. adenohystherum, 13 sequences from X. hispanum, and 23 sequences from X. subbaetense) with 13, 3, 4, and 3 different haplotypes and several heterozygous individuals, respectively (Table 1, Figure 3A). The TCS haplotype analysis inferred from the D2-D3 region showed four well-differentiated haplogroups corresponding to four different main lineages (X. adenohystherum, X. hispanum, X. malaka sp. nov., and X. subbaetense) (Figure 3A). Xiphinema malaka sp. nov. comprised a higher diversity in Mountain Almijara (SA, with nine haplotypes) than that detected in Mountain Nieves (SN 2 haplotypes), one haplotype in Tabernas, and one haplotype (Hm3) jointly detected in SA and SN (Figure 3A).
Figure 3.
(A). Construction of D2-D3 haploweb of Xiphinema malaka sp. nov. (B). coxI haplonet of Xiphinema malaka sp. nov. Coloured circles represent haplotypes and their diameter are proportional to the number of individuals sharing the same haplotype. Black short lines on the branches indicate the number of mutated positions in the alignment that separate each haplotype. Co-occurring haplotypes are enclosed in black dashes. (C). Phylogenetic relationships within the genus Xiphinema. Bayesian 50% majority rule consensus tree as inferred from D2 and D3 expansion domains of 28S rRNA sequence alignment under the general time-reversible model of sequence evolution with correction for invariable sites and a gamma-shaped distribution (GTR + I + G) +. Posterior probabilities more than 0.70 are given for appropriate clades. Newly obtained sequences in this study are shown in bold. Scale bar = expected changes per site. Some branches were collapsed for improving readability of Xiphinema species. Abbreviations: Ha = X. adenohystherum haplotypes; Hh = X. hispanum haplotypes; Hm = X. malaka sp. nov. haplotypes; Hs = X. subbaetense haplotypes; Hzs = X. subbaetense heterozygous specimens. SA = Mountain of Almijara and Tejeda; SN = Mountain of Nieves.
However, in coxI haplonet (Figure 3B), six different haplotypes of X. malaka sp. nov. were detected, three in SN and three in SA. One from SA shared the same haplotype with the Tabernas isolate, and this haplotype kept a far molecular distance with the other two haplotypes from SA. It was worth noting that the number of D2-D3 haplotypes of X. malaka sp. nov. was higher than coxI haplotypes (13 vs. 6), but there were more mutations between these coxI haplotypes than D2-D3 haplotypes (Figure 3A,B). Besides, X. subbaetense also comprised more haplotypes in the D2-D3 haplonet than the coxI haplonet (11 vs. 2); the situation of X. hispanum, X. adenohystherum were the same as previously described by Cai et al. [11].
2.3. Molecular Characterization
The amplification of D2-D3 expansion domains of 28S rRNA, ITS1 rRNA, the partial 18S rRNA, and partial coxI genes, yielded single fragments of ~900 bp, 1100 bp, 1800 bp, and 500 bp, respectively, based on gel electrophoresis. D2-D3 for X. malaka sp. nov. (MT584052–MT584085) showed a low intraspecific variability with 1–7 different nucleotides and 0 indels (99% similarity). The molecular diversity of this marker within SA (1–7 nucleotides, 0 indels) and SN (2–3 nucleotides, 0 indels) isolates was similar among them and differed from the closest related species, X. hispanum (KX244905, MT039125–MT039134) by 20–21 different nucleotides and 1–2 indels (97% similarity), X. subbaetense (MT039104–MT039124) by 22–25 different nucleotides and 2–3 indels (97% similarity), and from X. adenohystherum (KC567164, KX244898, GU725075, KX244897) by 29–42 different nucleotides and 3 indels (96% similarity).
The ITS1 region for X. malaka sp. nov. showed an intraspecific variability with 26–39 different nucleotides and 4–10 indels (96%–98% similarity). The molecular diversity of this marker within SA (18–24 nucleotides, 1–4 indels) and SN (26–29 nucleotides, 4 indels) isolates was also similar among them. ITS1 for X. malaka sp. nov. (MT584088-MT584099) differed from the closest related species, X. subbaetense (MT026293–MT026295) by 132–136 different nucleotides and 28–29 indels (88% similarity), X. hispanum (GU725061) by 84–142 different nucleotides and 22–38 indels (87–90% similarity), and from X. adenohystherum (GU725063, MT584100–MT584102) by 133–139 different nucleotides and 40–45 indels (87–88% similarity).
For the 18S rRNA, two new identical sequences for X. malaka sp. nov. (MT584086–MT584087) were obtained in this study and both of them showed very high similarity values with other accessions from Xiphinema spp. deposited in GenBank, being 98–99% similar. From the closet related species they differed by 1–2 nucleotides and 0 indels from X. subbaetense (MT039135–MT039140), X. adenohystherum (GU725084) by two nucleotides different and 0 indels, and X. hispanum (GU725083) by one nucleotide different and 0 indels. Finally, thirteen new coxI sequences for X. malaka sp. nov. (MT580263–MT580274) were deposited in GenBank in this study. This gene showed an intraspecific variability with 3–48 different nucleotides and 0 indels (88–99% similarity). The molecular diversity of this marker within SA (0–2 nucleotides, 0 indels) and SN (0–9 nucleotides, 0 indels) isolates was similar among them. coxI for X. malaka sp. nov. (MT580263–MT580274) differed from the closest related species, X. subbaetense (MT040280–MT010300) by 59–66 different nucleotides and 0 indels (82% similarity), X. hispanum (KY816614, MT040301-MT040305) by 51–78 different nucleotides and no indels (78–81% similarity), and from X. adenohystherum (KY816588–KY816592) by 58–65 different nucleotides and no indels (82–85% similarity).
2.4. Phylogenetic Relationships
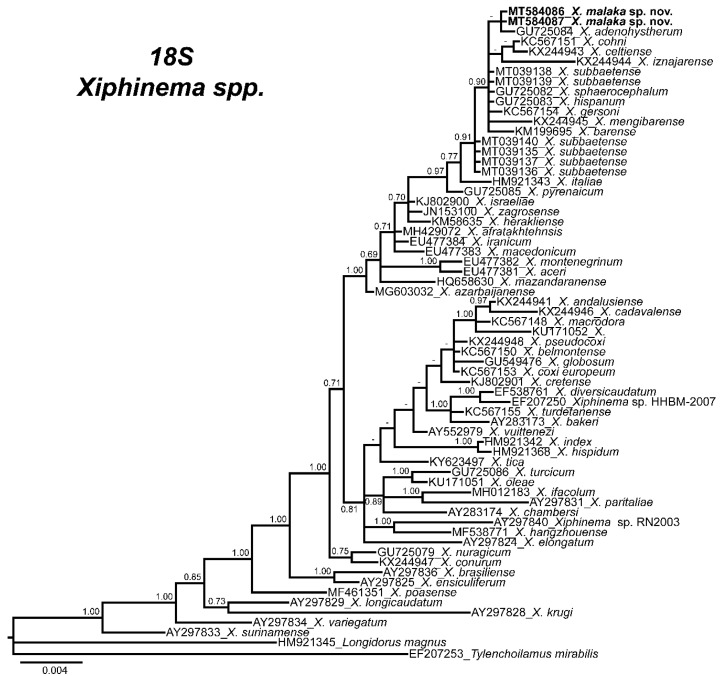
Phylogenetic relationships among Xiphinema species inferred from analyses of D2-D3 expansion domains of 28S rRNA, ITS1, the partial 18S rRNA and the partial coxI mtDNA gene sequences using BI are shown in Figure 3C, Figure 4, Figure 5 and Figure 6, respectively. The phylogenetic trees generated with the nuclear and mitochondrial markers included 136, 49, 65 and 95 sequences with 747, 1106, 1547 and 372 positions in length, respectively (Figure 3C, Figure 4, Figure 5 and Figure 6). The D2-D3 tree of Xiphinema spp. showed a well-supported clade (PP = 1.00), including 10 species from morphospecies Groups 5 and 6, seven of them belonging to morphospecies Group 5 and three to Group 6, all of them reported from the Iberian peninsula, and included X. malaka sp. nov. (MT584052–MT584085). All other clades followed the same pattern as previous studies. Xiphinema malaka sp. nov. was phylogenetically related with X. hispanum, X. celtiense and X. cohni in a moderately supported clade (PP = 0.88), but clearly separate from all of them (Figure 3C).
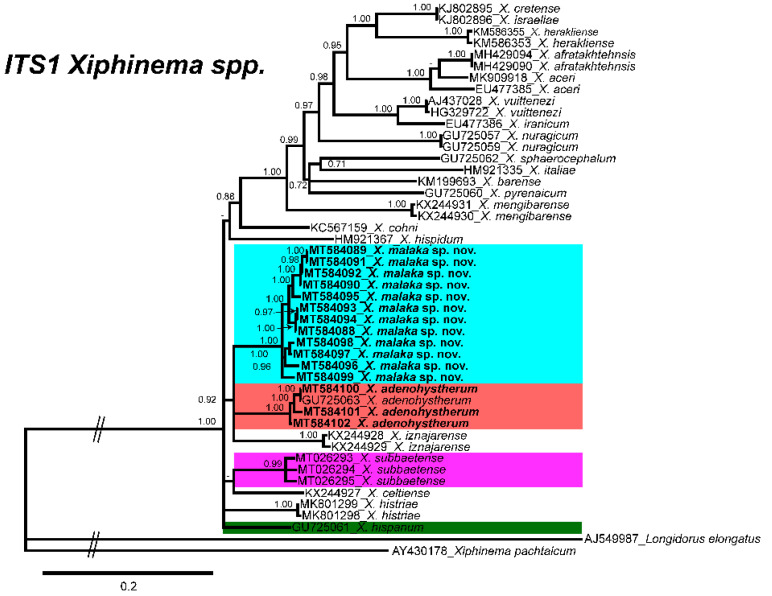
Figure 4.
Phylogenetic relationships of Xiphinema malaka sp. nov. within the genus Xiphinema. Bayesian 50% majority-rule consensus trees as inferred from ITS1 sequence alignments under transition model with a proportion of invariable sites and a rate of variation across sites (TIM2 + I + G). Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are in bold letters, and each colour is associated with each species of the complex.
Figure 5.
Phylogenetic relationships of Xiphinema malaka sp. nov. within the genus Xiphinema. Bayesian 50% majority-rule consensus trees as inferred from 18S sequence alignments under the GTR + I + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are in bold letters.
Figure 6.
Phylogenetic relationships of Xiphinema malaka sp. nov. within the genus Xiphinema. Bayesian 50% majority-rule consensus trees as inferred from cytochrome c oxidase subunit I (coxI) mtDNA gene sequence alignments under the GTR + I + G model. Posterior probabilities more than 70% are given for appropriate clades. Newly obtained sequences in this study are in bold letters, and each colour is associated with each species of the complex.
Difficulties were experienced with alignment of the ITS1 sequences due to scarce similarity. Thus, only related sequences were used for phylogeny. The 50% majority rule consensus ITS1 BI tree showed several clades low to moderately supported (Figure 4). Xiphinema malaka sp. nov. was phylogenetically related to X. adenohystherum and X. iznajarense in a moderately supported clade (PP = 0.92), but clearly separate from all of them (Figure 4).
The 50% majority rule consensus 18S rRNA gene BI tree showed several major clades (Figure 5). Phylogenetic inferences based on 18S suggest that X. malaka sp. nov. was related to other species of the X. hispanum-complex in a moderately supported clade (PP = 0.91), together with other species such as X. barense, X. celtiense, X. cohni, X. gersoni, X. iznajarense, X. mengibarense, and X. sphaerocephalum (Figure 5).
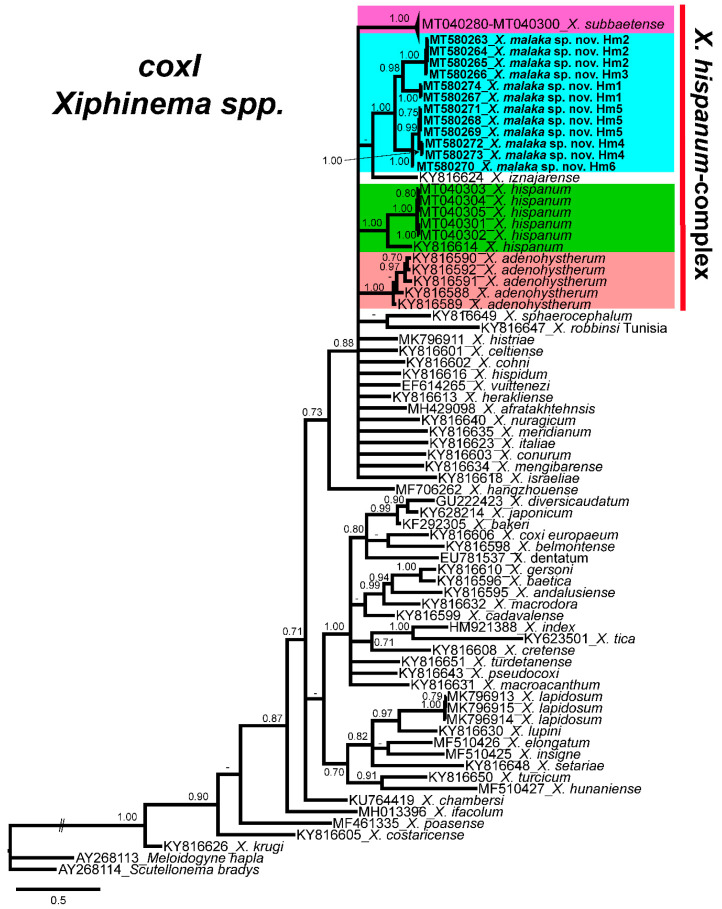
Finally, the phylogenetic relationships of Xiphinema species inferred from analysis of partial coxI gene sequences showed several clades that were not well defined (Figure 6). Xiphinema malaka sp. nov. (MT580263-MT580274) was phylogenetically related to X. hispanum-complex species in a low supported clade (PP = 0.65), but clearly separate from all of them (Figure 6).
2.5. Morphology and Morphometry of Xiphinema malaka sp. nov
Xiphinema malaka sp. nov.
http://zoobank.org/urn:lsid:zoobank.org:act: BDBF964D-71E8-4E4F-B61C-50C5A7C51083
(Figure 7, Figure 8, Figure 9 and Figure 10, Table 3 and Table 4)
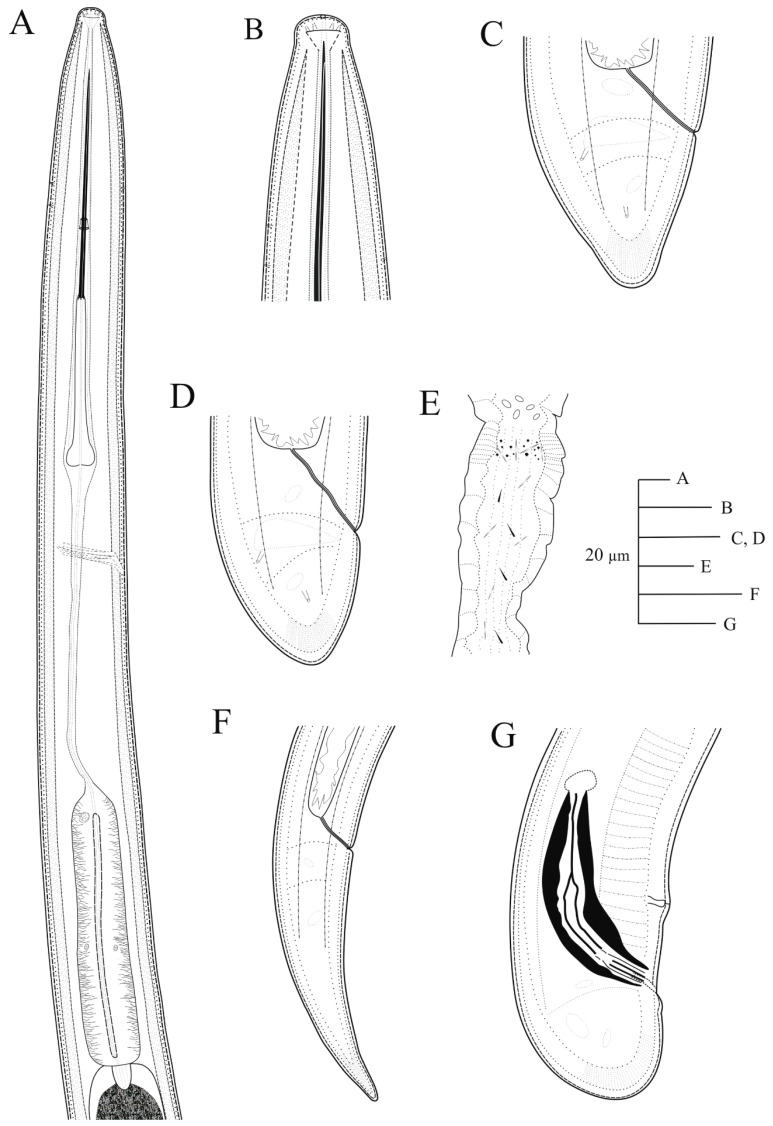
Figure 7.
Line drawings of holotype for Xiphinema malaka sp. nov. (A), pharyngeal region; (B), detail of lip region; (C,D), female tails; (E), detail of uterine pseudo Z-differentiation.; (F), tail of first-stage juvenile (J1); (G), male tail.
Figure 8.
Light photomicrographs of Xiphinema malaka sp. nov. females holotype and paratypes: (A), anterior region holotype; (B,C) anterior regions paratypes; (D), detail of odontophore and guiding ring in holotype; (E), vulval region; (F–H), detail of female genital track showing Z-differentiation in holotype; (I), tail region of holotype; (J–N), tail region in paratypes; (O), detail of first-stage anterior region; (P–S), tail region of 1st, 2nd, 3rd and 4th stage juveniles. Abbreviations: a = anus; af = amphidial fovea; cb = crystalloid bodies; fl = odontophore flanges; gr = guiding ring; odp = odontophore; odt = odontostyle; psZ = pseudo-Z organ; rodt = replacement odontostyle; sp = spine; v = vulva. Scale bars: 20 μm.
Figure 9.
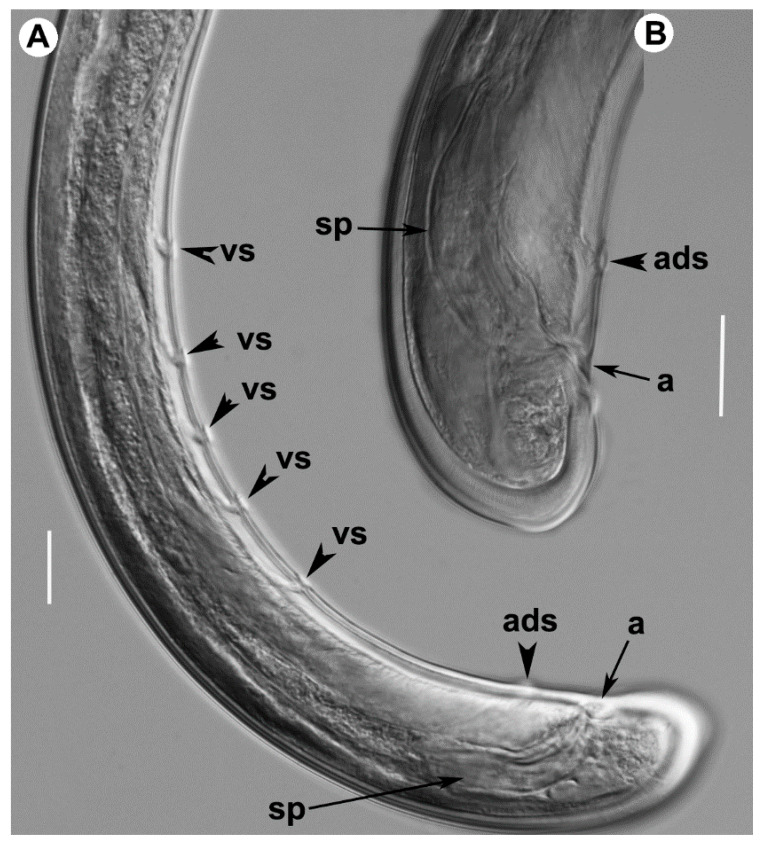
Light photomicrographs of Xiphinema malaka sp. nov. male: (A), posterior region; (B), detail of tail showing spicules. Abbreviations: a = anus; ads = adanal supplements; sp = spicules; vs = ventromedian supplements. Scale bars: 20 μm.
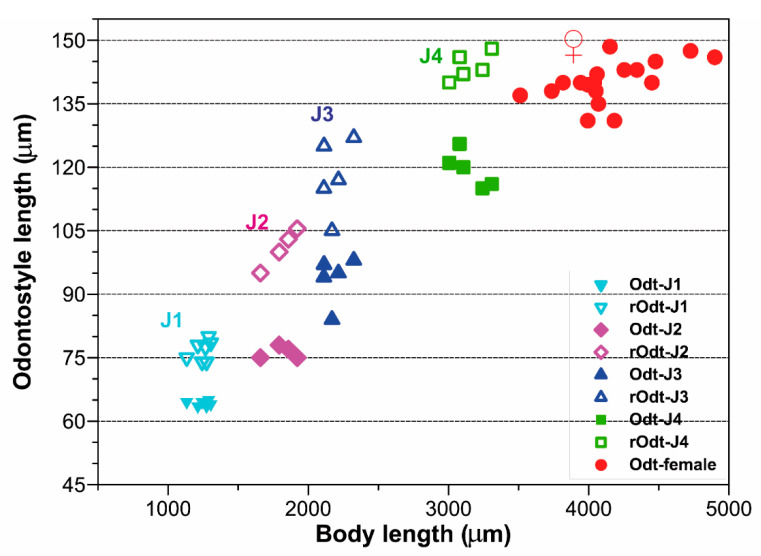
Figure 10.
Relationship of body length to length of functional and replacement odontostyle (Ost and rOst, respectively) length in all developmental stages from first-stage juveniles (J1) to mature females of Xiphinema malaka sp. nov.
2.5.1. Material Examined
Holotype. Adult female was found in the rhizosphere of maritime pine (Pinus pinaster Aiton) at 1312 m a.s.l. from Canillas de Albaida, Málaga province, Spain (GPS: 36°52′21.81″ N; 3°55′41.00″ W) collected by A. Archidona-Yuste on 12 December 2019; mounted in pure glycerin and deposited in the nematode collection at Institute for Sustainable Agriculture (IAS) of Spanish National Research Council (CSIC), Córdoba, Spain (Slide number X-SA3-02).
Paratypes. Female and juvenile paratypes were collected from the same soil sample as the holotype (Table 3); mounted in pure glycerin and deposited in the Institute for Sustainable Agriculture (IAS) of the Spanish National Research Council (CSIC), Córdoba, Spain (Slide numbers X-SA3-03–X-SA3-08); one female at Istituto per la Protezione delle Piante (IPP) of Consiglio Nazionale delle Ricerche (C.N.R.), Sezione di Bari, Bari, Italy (X-SA3-011); one female at the USDA Nematode Collection (T-7474p).
Additional material examined. Additional nematode isolates were studied and characterized from the rhizosphere of maritime pine, black pine, cork oak and yellow broom at several localities at Málaga and Almería provinces (Table 4). Morphometric measurements were taken for 62 individuals, 40 females, one male and 21 juveniles from J1 to J4 from several localities in Málaga province, Table 3 and Table 4. Unfortunately, the scarce nematode isolate detected in the isolte of Tabernas (Almería) did not allow us to take measurements of adult females.
Type locality. Canillas de Albaida, Málaga province, Spain (GPS: 36°52′21.81″ N; 3°55′41.00″ W); 1254 m above sea level (a.s.l.) collected by A. Archidona-Yuste on 12 December 2019.
Etymology. The species epithet refers to the Phoenician word Malaka, the name of the province of Málaga where the species was found in several localities.
2.5.2. Diagnosis. Xiphinema malaka sp. nov.
Belongs to morphospecies Group 5 from the Xiphinema nonamericanum-group species [18]. It is characterized by a moderate long body (3.5–4.9 mm), assuming a J-shaped when heat-relaxed; lip region hemispherical, separate from the body contour by a depression, 14.0–15.0 μm wide; a relatively long odontostyle 131.0–148.5 μm; vulva located at 47.1–53.8% of body length; female reproductive system didelphic-amphidelphic having both branches about equally developed, pseudo Z-differentiation containing numerous small granular bodies, uterus tripartite with small crystalloid bodies and spines in low number and presence of prominent wrinkles in the uterine wall that may be confused with spiniform structures; female tail short convex-conoid on both sides, and bearing 3 caudal pores, ending in a rounded and broad terminus with a very small bulge at the end in some specimens; c’ ratio (0.9–1.0); male rare one individual out of 75 females. Four developmental juvenile stages were identified, the 1st-stage juvenile with tail elongate-conoid with characteristic subdigitate rounded terminus (c’ ratio 3.2–3.8). According to the polytomous key of Loof & Luc [18] and matrix codes sorted by Archidona-Yuste et al. [19], codes for the new species are (codes in parentheses are exceptions): A4-B23-C6-D6-E65-F4(5)-G3-H2-I3-J6-K2-L1. The DNA sequences of D2-D3 expansion domains of 28S, ITS1 rRNA, 18S rRNA, and partial coxI were deposited in GenBank under the accession numbers MT584052-MT584085, MT584088-MT584099, MT584086-MT584087 and MT580263-MT580274, respectively.
2.5.3. Description
Female. Body cylindrical, slightly tapering towards anterior end in a J-shape when heat relaxed. Cuticle with fine transverse striae visible in tail region, 3.2 ± 0.3 (3.0–3.5) μm thick at mid body but thicker just posterior to anus. Lateral chord 13.2 ± 2.5 (11.5–16.0) μm wide, occupying ca. 25% of corresponding body diam. Lip region hemispherical, slightly offset from body contour by a depression, 14.6 ± 0.4 (14.0–15.0) μm wide and 6.5 ± 0.6 (6.0–7.5) μm high. Odontostyle moderately long, 1.7–1.9 times longer than odontophore, the latter with well-developed flanges (13.0–15.5 μm wide). Guiding ring and guiding sheath variable in length depending on degree of protraction/retraction of stylet. Pharynx composed by a slender narrow flexible part 335–582 μm long, and a posterior muscular, cylindrical and expanded part with three nuclei. Terminal pharyngeal bulb variable in length, 112.0–149.0 μm long and 24.0–31.0 μm wide. Glandularium 110.0–129.0 μm long. Dorsal gland nucleus (DN) located at beginning of basal bulb (8.5–14.3%), ventrosublateral gland nuclei (SVN) situated ca halfway along bulb (52.3–67.9%) (position of gland nuclei calculated as described by Loof & Coomans [27]). Cardia conoid-rounded and variable in length, 12.0–15.0 μm long. Intestine simple, prerectum variable in size 471–516 μm long. Rectum 32.0–40.0 μm long ending in anus as a small rounded slit. Reproductive system didelphic-amphidelphic with two equally developed branches. Each branch composed of a short ovary 47–78 μm long, a reflexed oviduct 93–104 μm long with well-developed pars dilatata oviductus, a sphincter, a well-developed pars dilatata uteri, and a 254–286 μm long uterus with pseudo-Z differentiation containing numerous small granular bodies with small crystalloid bodies (6.0–12.5 μm long) and spines in low number, and presence of prominent wrinkles in the uterine wall that may be confused with spiniform structures (Figure 7 and Figure 8); a 35.5–47.0 μm long vagina perpendicular to body axis (having 28–32% corresponding body diam. ingrowth), ovejector well-developed 36.5–50.0 μm wide, pars distalis vaginae 16.8 ± 2.4 (13.0–19.5) μm long, and pars proximalis vaginae 24.3 ± 2.4 (20.5–27.5) μm long and 26.3 ± 1.0 (21.5–29.5) μm wide, and vulva as a transverse slit. Tail short, convex-conoid on both sides, and bearing three caudal pores, ending in a rounded and broad terminus, with a very small bulge at the end in some specimens (Figure 8).
Male. Extremely rare, only one male individual out of 75 female specimens was found in one sample near the type locality. Morphologically similar to female except for genital system and secondary sexual features. Male genital tract diorchic with testes containing multiple rows of different stages of spermatogonia. Tail short, convex-conoid with a broadly rounded terminus and thickened outer cuticular layer. Adanal supplements paired, preceded anteriorly by a row of five irregularly spaced ventromedians supplements. Spicules paired, dorylaimoid, moderately long and slightly curved ventrally, approximately 2.5 times longer than tail length; lateral guiding pieces more or less straight or with curved proximal end.
Juveniles. Four developmental juvenile stages were detected and distinguished by relative body length, odontostyle and replacement odontostyle length. The 1st-stage juveniles were characterized by the replacement odontostyle inserted into odontophore base (Figure 8). In all other stages, the replacement odontostyle was posterior to the flanges of odontophore in its resting position. The correlation between body length, replacement and functional odontostyle of the type population is given in Figure 10. Lip region in all juvenile stages looks similar to that in females. Other morphological characters similar to female, except for their size and immature sexual characteristics (developing genital primordium 16.0–87.0 μm long). The first-stage juvenile was characterized by a tail elongate-conoid with characteristic subdigitate rounded terminus (c’ ratio 3.2–3.8). Tail of other developmental stages becoming progressively shorter and wider after each moult (Figure 8).
2.5.4. Remarks
According to the polytomous key by Loof & Luc [18] and matrix codes sorted by Archidona-Yuste et al. [6]: A (type of female genital apparatus), C (tail shape), D (c’ ratio), E (vulva position), F (body length), and G (total stylet length) (in this order of main features), X. malaka sp. nov. is closely related to X. subbaetense Cai, Archidona-Yuste, Cantalapiedra-Navarrete, Palomares Rius & Castillo [11], X. hispanum Lamberti, Castillo, Gomez-Barcina & Agostinelli [22], X. adenohystherum Lamberti, Castillo, Gomez-Barcina & Agostinelli [22], X. cohni Lamberti, Castillo, Gomez-Barcina & Agostinelli [22], and X. sphaerocephalum Lamberti, Castillo, Gomez-Barcina & Agostinelli [22].
Xiphinema malaka sp. nov. is morphometrically almost undistinguishable from X. subbaetense and X. hispanum, from the former can only be differentiated in females by a higher a ratio (65.6–99.8 vs. 49.0–70.0), a shorter odontophore (75.0–88.0 vs. 82.0–96.5 μm), narrower lip region (14.0–15.5 vs. 15.5–18.5 μm), higher c’ ratio in J1 (3.2–3.8 vs. 2.6–3.1, 2.7–3.1, respectively), and presence of male (very rare vs. absent) [11,20]. Morphologically can be differentiated from X. subbaetense and X. hispanum in pseudo-Z differentiation containing numerous small granular bodies vs. 4–5 granular bodies. It can be differentiated from X. adenohystherum by slightly shorter odontostyle (131.0–149.0 vs. 143.0–152.0 μm), longer tail (26.0–47.0 vs. 29.0–35.0 μm), and slightly higher a ratio (65.6–96.5 vs. 65.2–73.3). It can be differentiated from X. sphaerocephalum by its shorter odontostyle (131.0–149.0 vs. 143.5–168.0 μm), and shorter oral aperture-guiding ring distance (96.0–135.0 vs. 126.0–162.0 μm). Finally, X. malaka sp. nov. can be differentiated from X. cohni by its shorter odontostyle (131.0–149.0 vs. 149–174 μm), shorter oral aperture-guiding ring distance (96.0–135.0 vs. 137.0–161.0 μm), and higher c ratio (97.3–178.6 vs. 82.6–115.2). Nevertheless, it can be clearly separated by specific 28S rRNA, ITS1 rRNA and coxI sequences.
3. Discussion
The primary objective of this study was to decipher the cryptic diversity of the X. hispanum-complex by applying an integrative taxonomical approaches on several new unidentified Xiphinema isolates from Málaga and Almería provinces (southern Spain), appearing morphologically and morphometrically indistinguishable from this species complex. Multivariate morphometric analyses proved to be useful tools for species delimitation within the genera Longidorus and Xiphinema [11,15,19,28]. These data support that X. hispanum-complex species comprise a model example of morphostatic speciation (genetic modifications not reflected in morphology and morphometry) [23,24], since independent approaches based on molecular analyses using ribosomal and mitochondrial sequences (haploweb and haplonet) revealed high levels of genetic diversity within these species complexes which clearly separated X. malaka sp. nov. from all other X. hispanum-complex species. These results, as well as those from previous studies, may suggest that X. hispanum-complex species comprises a Xiphinema endemic lineage, with members morphologically and morphometrically very similar, that have diversified in the Iberian peninsula, since no other records on these species have been reported outside this area [20,22,29].
Phylogenetic analyses based on three rDNA molecular markers (D2–D3 expansion domains of 28S rRNA gene, ITS1 region and the partial 18S rRNA) resulted in a general consensus of species phylogenetic positions for the majority of them, and were generally congruent with those given by previous phylogenetic analysis [6,11,19,30,31,32,33].
The results of this research support our hypothesis that biodiversity of Longidoridae in southern Spain is still not fully clarified and needs additional sampling efforts given the significant gaps in soil nematode biodiversity regarding the large number of undescribed species [34,35] and the hypothesis suggesting the Iberian Peninsula as a possible center of speciation for some groups of the family Longidoridae [6,15,36]. The recognition of this extraordinary cryptic diversity has a direct bearing on estimates of global nematode biodiversity and concepts of nematode biogeography. Regional endemicity in plant-parasitic nematodes has seldom been recognized and cosmopolitan distributions in nematodes, like other microscopic organisms, are reportedly common [37,38].
In summary, the present study confirmed the extraordinary cryptic diversity of X. hispanum-complex species in Andalusia and comprises a paradigmatic example of morphostatic speciation of dagger nematodes in southern Spain, which can be a potential explanation of the unusual high biodiversity within Longidoridae, considering Andalusia as a hot spot of biodiversity. However, additional similar intensive taxonomic studies are needed in other areas which can confirm this statement.
4. Materials and Methods
4.1. Nematode Isolates and Morphological Studies
No specific permits were required for the indicated fieldwork studies. The soil samples were obtained in public areas, forests and other natural areas and did not involve any endangered species or those protected in Spain, nor were the sites protected in any way.
A total of 62 individuals including 41 adults and 21 juvenile specimens from several localities in Málaga and Almería provinces (southern Spain) were used for morphological analyses (Table 1, Figure 1). Nematodes were surveyed during spring season in 2019 in natural ecosystems in Andalusia, southern Spain (Table 1). Soil samples were collected for nematode analysis with a shovel randomly selecting four to five cores at each point, and considering the upper 5–50 cm depth of soil that was close to the active plant root at each sampling spot. Nematodes were extracted from a 500-cm3 sub-sample of soil by centrifugal flotation [39] and a modification of Cobb’s decanting and sieving [40] methods. For morphometric studies, Xiphinema specimens were killed and fixed by a hot solution of 4% formalin + 1% glycerol, then processed in pure glycerin [41] as modified by De Grisse [42].
Specimens for light microscopy were killed by hot fixative using a solution of 4% formaldehyde +1% propionic acid and embedded in pure glycerine using Seinhorst’s [41] method. The morphometric study of each nematode isolate included morphology-based diagnostic features in Xiphinema (i.e., de Man body ratios), lip region width, amphid shape, oral aperture-guiding-ring, odontostyle and odontophore length and female tail shape [7]. For line drawings of the new species, light micrographs were imported to CorelDraw ver. X7 and redrawn. The light micrographs and measurements of each nematode isolate, including important diagnostic characteristics (i.e., de Man indices, body length, odontostyle length, lip region, tail shape, amphid shape and oral aperture-guiding ring; [7]) were performed using a Leica DM6 (Wetzlar, Germany) compound microscope with a Leica DFC7000 T digital camera. For the line drawings of the new species, CorelDraw software version X7 (Corel Corporation, London, UK) was used to redraw according to the selected light micrographs.
4.2. DNA Extraction, Polymerase Chain Reaction (PCR) and Sequencing
For molecular analyses, in order to ensure the selected nematodes for extracting DNA were from the same species, two live nematodes from each sample were temporary mounted in a drop of 1M NaCl containing glass beads (to avoid nematode crushing/damaging specimens) to ensure specimens conformed to the unidentified isolates of Xiphinema. Thus, 34 individuals collected from several sampling points in Andalusia were molecularly analyzed (Table 1). All necessary morphological and morphometric data, by taking pictures and measurements using the above camera-equipped microscope, were recorded. This was followed by DNA extraction from a single specimen and polymerase chain reaction (PCR) cycle conditions as previously described [6,15]. Several sets of primers were used for PCR. A partial region of the 28S rRNA gene including the expansion domains D2 and D3 (D2-D3) was amplified by using the primers D2A (5′-ACAAGTACCGTGAGGGAAAGTTG-3′) and D3B (5′-TCGGAAGGAACCAGCTACTA-3′) [43]. The Internal Transcribed Spacer region 1 (ITS1) separating the 18S rRNA gene from the 5.8S rRNA gene was amplified using forward primer 18S (5′-TTGATTACGTCCCTGCCCTTT-3′) [44] and reverse primer rDNA1 5.8S (5′-ACGAGCCGAGTGATCCACCG-3′) [45]. A partial sequence of the 18S rRNA gene (18S) was amplified as previously described [46] using primers 988F (5′-CTCAAAGATTAAGCCATGC-3′), 1912R (5′-TTTACGGTCAGAACTAGGG-3′), 1813F (5′-CTGCGTGAGAGGTGAAAT-3′), and 2426R (5′-GCTACCTTGTTACGACTTTT -3′. Finally, the portion of the cytochrome c oxidase subunit I gene (coxI) was amplified using the primers COIF (5′-GATTTTTTGGKCATCCWGARG-3′) and COIR (5′-CWACATAATAAGTATCATG-3′) [47]. The newly obtained sequences were deposited in the GenBank database under accession numbers indicated in Table 1 and on the phylogenetic trees.
PCR cycle conditions were one cycle of 94 °C for two min, followed by 35 cycles of 94 °C for 30 s, annealing temperature of 55 °C for 45 s, 72 °C for three min, and finally one cycle of 72 °C for 10 min. PCR products were purified after amplification using ExoSAP-IT (Affimetrix, USB products, High Wycombe, UK), quantified using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and used for direct sequencing in both directions using the primers noted above. The resulting products were purified and run on a DNA multicapillary sequencer (Model 3130XL genetic analyser; Applied Biosystems, Foster City, CA, USA), using the BigDye Terminator Sequencing Kit v.3.1 (Applied Biosystems, Foster City, CA, USA), at the Stab Vida sequencing facilities (Caparica, Portugal). The newly obtained sequences were submitted to the GenBank database under accession numbers indicated in Table 1 and on the phylogenetic trees.
4.3. Species Delimitation via Multivariate Morphometric Analysis and Haplotype Networks Construction
The nine new Xiphinema isolates detected in this study were included in the X. hispanum-complex species group given the close relationships morphologically with X. hispanum as outlined above. An iterative analysis of morphometric and molecular data using two independent strategies of species delimitation was utilized to asses described and undescribed specimens and to determine species boundaries within this species complex.
Species delineation using morphometry was conducted with principal component analysis (PCA) in order to estimate the degree of association among species within the X. hispanum-complex [48]. PCA was based upon the following morphological characters: L (body length), the ratios a, c, c’, d, d’, V, odontostyle and odontophore length, lip region width and hyaline region length (Table 2) [6,7,13]. Prior to the statistical analysis, diagnostic characters were tested for collinearity [49]. We used the collinearity test based on the values of the variance inflation factor (VIF) method that iteratively excludes numeric covariates showing VIF values > 10 as suggested by Montgomery and Peck [50]. PCA was performed by a decomposition of the data matrix amongst isolates using the principal function implemented in the package psych [51]. Orthogonal varimax raw rotation was used to estimate the factor loadings. Only factors with sum of squares (SS) loadings > 1 were extracted. Finally, a minimum spanning tree (MST) based on the Euclidean distance was superimposed on the scatter plot of the X. malaka sp. nov.-specimens complex against the PCA axes. MST was performed using the ComputeMST function implemented in the package emstreeR [52]. All statistical analyses were performed using the R v. 3.5.1 freeware [53].
In order to detect distinct phylogenetic groups possibly representing separate species, haplotype networks (briefly, haplonet) were constructed to each of the two separate datasets, i.e., the D2-D3 and coxI. Alignments were converted to the NEXUS format using DnaSP V.6 [54]; TCS networks [55] were applied in the program PopART V.1.7 [56]. Illustrations of networks were prepared using the program Adobe illustrator to add connecting curves between the haplotypes found co-occurring in heterozygous individuals [57].
4.4. Phylogenetic Analysis
Sequenced genetic markers in the present study (after discarding primer sequences and ambiguously aligned regions), and several Xiphinema spp. sequences obtained of GenBank, were used for phylogenetic reconstruction (Table 1). Outgroup taxa for each dataset were selected based on previous published studies [6,11,30,45,58]. Multiple sequence alignments of the newly obtained and published sequences were made using the FFT-NS-2 algorithm of MAFFT v. 7.450 [59]. Sequence alignments were visualized using BioEdit [60] and edited by Gblocks ver. 0.91b [61] in the Castresana Laboratory server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) using options for a less stringent selection (minimum number of sequences for a conserved or a flanking position: 50% of the number of sequences + 1; maximum number of contiguous no conserved positions: 8; minimum length of a block: 5; allowed gap positions: with half).
Phylogenetic analyses of the sequence data sets were based on Bayesian inference (BI) using MRBAYES 3.2.7a [62]. The best-fit model of DNA evolution was calculated with the Akaike information criterion (AIC) of JMODELTEST v. 2.1.7 [63]. The best-fit model, the base frequency, the proportion of invariable sites and the gamma distribution shape parameters and substitution rates in the AIC were then used in phylogenetic analyses. BI analyses were performed under a general time reversible, with a proportion of invariable sites and a rate of variation across sites (GTR + I + G) model for D2-D3, the partial 18S rRNA, and the partial coxI gene, and under a transition model with a proportion of invariable sites and a rate of variation across sites (TIM2 +I + G). These BI analyses were run separately per dataset with four chains for 2 × 106 generations. The Markov chains were sampled at intervals of 100 generations. Two runs were conducted for each analysis. After discarding burn-in samples of 30% and evaluating convergence, the remaining samples were retained for more in-depth analyses. The topologies were used to generate a 50% majority-rule consensus tree. Posterior probabilities (PP) were given on appropriate clades. Trees from all analyses were visualized using FigTree software version v.1.42 [64].
Acknowledgments
We would like to thanks J. Martin Barbarroja and G. León Ropero (IAS-CSIC) for their excellent technical assistance in surveys and management of soil samples, and further anonymous reviewers and editors for their effort in reviewing the manuscript and helping improve this study. The first author is a recipient of Humboldt Research Fellowship for Postdoctoral Researchers at Helmholtz Centre for Environmental Research-UFZ, Leipzig, Germany. The second author acknowledges the China Scholarship Council (CSC) for financial support. The sixth author acknowledges Spanish Ministry of Economy and Competitiveness for the “Ramon y Cajal” Fellowship RYC-2017-22228.
Author Contributions
Conceptualization, A.A.-Y., R.C., C.C.-N., J.A.C., A.R., B.V., G.L., J.E.P.-R. and P.C., methodology, A.A.-Y., R.C., C.C.-N., J.A.C., A.R., G.L., J.E.P.-R. and P.C., software, A.A.Y., R.C., J.E.P.-R. and P.C., analysis, A.A.Y., R.C., J.E.P.-R. and P.C., resources, J.A.C., A.R., G.L. and P.C., writing, A.A.-Y., R.C., C.C.-N., J.A.C., A.R., G.L., J.E.P.-R. and P.C. All authors contributed to the final discussion data, and read and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Spanish Ministry of Science, Innovation and Universities, grant number RTI2018-095345-B-C21, LITHOFOR (“Modulating role of LITHOlogy in the response of Mediterranean FORest ecosystems to climate change: growth, edaphological processes and future predictions”) and the Humboldt Research Fellowship for Postdoctoral Researchers awarded for the first author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Decraemer W., Hunt D. Structure and classification. In: Perry R.N., Moens M., editors. Plant Nematology. CABI; Wallingford, UK: 2006. pp. 3–32. [Google Scholar]
- 2.Wyss U. Root parasitic nematodes: An overview. In: Fenoll C., Grundler F.M.W., Ohl S.A., editors. Cellular and Molecular Aspects of Plant-Nematode Interactions. 10. Kluwer Academic Publisher; Dordrecht, The Netherlands: 1997. pp. 5–22. [Google Scholar]
- 3.Singh S.K., Hodda M., Ash G.J. Plant-parasitic nematodes of potential phytosanitary importance, their main hosts and reported yield losses. Bull. OEPP. 2013;43:334–374. doi: 10.1111/epp.12050. [DOI] [Google Scholar]
- 4.Davies L.J., Elling A.A. Resistance genes against plant-parasitic nematodes: A durable control strategy? Nematology. 2015;17:249–263. doi: 10.1163/15685411-00002877. [DOI] [Google Scholar]
- 5.Coomans A., Huys R., Heyns J., Luc M. Character Analysis, Phylogeny, and Biogeography of the Genus Xiphinema Cobb, 1973 (Nematoda, Longidoridae) Volume 287 Annales du Musée Royal de l’Afrique Centrale (Zoologie); Tervuren, Belgique: 2001. [Google Scholar]
- 6.Archidona-Yuste A., Navas-Cortés J.A., Cantalapiedra-Navarrete C., Palomares-Rius J.E., Castillo P. Remarkable diversity and prevalence of dagger nematodes of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) in olives revealed by integrative approaches. PLoS ONE. 2016;11:e0165412. doi: 10.1371/journal.pone.0165412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jairajpuri M.S., Ahmad W. Dorylaimida: Free-Living, Predaceous and Plant-Parasitic Nematodes. IBH Publishing Co; New Delhi, India: 1992. [Google Scholar]
- 8.Siddiqi M.R. Tylenchida. Parasites of Plants and Insects. CABI Bioscience; Egham, UK: 2000. [DOI] [Google Scholar]
- 9.Blaxter M.L., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., et al. A molecular evolutionary framework for the Phylum Nematoda. Nature. 1998;392:71–75. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 10.Decraemer W., Robbins R.T. The who, what and where of Longidoridae and Trichodoridae. J. Nematol. 2007;39:295–297. [PMC free article] [PubMed] [Google Scholar]
- 11.Cai R., Archidona-Yuste A., Cantalapiedra-Navarrete C., Palomares-Rius J.E., Castillo P. New evidence of cryptic speciation in the family Longidoridae (Nematoda: Dorylaimida) J. Zool. Syst. Evol. Res. 2020;58 doi: 10.1111/jzs.12393. [DOI] [Google Scholar]
- 12.Gutiérrez-Gutiérrez C., Cantalapiedra-Navarrete C., Decraemer W., Vovlas N., Prior T., Palomares-Rius J.E., Castillo P. Phylogeny, diversity, and species delimitation in some species of the Xiphinema americanum-group complex (Nematoda: Longidoridae), as inferred from nuclear and mitochondrial DNA sequences and morphology. Eur. J. Plant Pathol. 2012;134:561–597. doi: 10.1007/s10658-012-0039-9. [DOI] [Google Scholar]
- 13.Archidona-Yuste A., Navas-Cortés J.A., Cantalapiedra-Navarrete C., Palomares-Rius J.E., Castillo P. Unravelling the biodiversity and molecular phylogeny of needle nematodes of the genus Longidorus (Nematoda: Longidoridae) in olive and a description of six new species. PLoS ONE. 2016;11:e0147689. doi: 10.1371/journal.pone.0147689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czekanski-Moir J., Rundell R. The ecology of nonecological speciation and nonadaptive radiations. Trends Ecol. Evol. 2019;34:400–415. doi: 10.1016/j.tree.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Archidona-Yuste A., Navas-Cortés J.A., Cantalapiedra-Navarrete C., Palomares-Rius J.E., Castillo P. Cryptic diversity and species delimitation in the Xiphinema americanum-group complex (Nematoda: Longidoridae) as inferred from morphometrics and molecular markers. Zool. J. Linn. Soc. 2016;176:231–265. doi: 10.1111/zoj.12316. [DOI] [Google Scholar]
- 16.Taylor C.A., Brown D.J.F. Nematode Vectors of Plant Viruses. CAB International; Wallingford, UK: 1997. [Google Scholar]
- 17.Lamberti F., Molinari S., Moens M., Brown D.J.F. The Xiphinema americanum group. I. Putative species, their geographical occurrence and distribution, and regional polytomous identification keys for the group. Russ. J. Nematol. 2000;8:65–84. [Google Scholar]
- 18.Loof P.A.A., Luc M. A revised polytomous key for the identification of species of the genus Xiphinema Cobb, 1913 (Nematoda: Longidoridae) with exclusion of the X. americanum-group. Syst. Parasitol. 1990;16:35–66. doi: 10.1007/BF00009600. [DOI] [Google Scholar]
- 19.Archidona-Yuste A., Navas-Cortés J.A., Cantalapiedra-Navarrete C., Palomares-Rius J.E., Castillo P. Molecular phylogenetic analysis and comparative morphology resolve two new species of olive-tree soil related dagger nematodes of the genus Xiphinema (Dorylaimida: Longidoridae) from Spain. Invertebr. Syst. 2016;30:547–565. doi: 10.1071/IS16002. [DOI] [Google Scholar]
- 20.Gutiérrez-Gutiérrez C., Palomares-Rius J.E., Cantalapiedra-Navarrete C., Landa B.B., Esmenjaud D., Castillo P. Molecular analysis and comparative morphology to resolve a complex of cryptic Xiphinema species. Zool. Scr. 2010;39:483–498. doi: 10.1111/j.1463-6409.2010.00437.x. [DOI] [Google Scholar]
- 21.Lazarova S., Oliveira C.M.G., Prior T., Peneva V., Kumari S. An integrative approach to the study of Xiphinema brevicolle Lordello and Da Costa 1961, supports its limited distribution worldwide (Nematoda: Longidoridae) Eur. J. Plant Pathol. 2019;153:441–464. doi: 10.1007/s10658-018-1571-z. [DOI] [Google Scholar]
- 22.Lamberti F., Castillo P., Gomez-Barcina A., Agostinelli A. Descriptions of six new species of Xiphinema (Nematoda, Dorylaimida) from the Mediterranean region. Nematol. Mediterr. 1992;20:125–139. [Google Scholar]
- 23.Davis G.M. Evolution of prosobranch snails transmitting Asian Schistosoma: Coevolution with Schistosoma: A review. Prog. Clin. Parasitol. 1992;3:145–204. doi: 10.1007/978-1-4612-2732-8_6. [DOI] [PubMed] [Google Scholar]
- 24.Gittenberger E. What about non-adaptive radiation? Biol. J. Linn. Soc. 1991;3:263–272. doi: 10.1111/j.1095-8312.1991.tb00598.x. [DOI] [Google Scholar]
- 25.Cai R., Prior T., Lawson B., Cantalapiedra-Navarrete C., Palomares-Rius JECastillo P., Archidona-Yuste A. An integrative taxonomic study of the needle nematode complex Longidorus goodeyi Hooper, 1961 (Nematoda: Longidoridae) with description of a new species. Eur. J. Plant Pathol. 2020;157:59–81. doi: 10.1007/s10658-020-02055-0. [DOI] [Google Scholar]
- 26.Barsi L., Fanelli E., De Luca F. A new record of Xiphinema dentatum Sturhan, 1978 and description of X. paradentatum sp. n. (Nematoda: Dorylaimida) from Serbia. Nematology. 2017;19:925–949. doi: 10.1163/15685411-00003098. [DOI] [Google Scholar]
- 27.Loof P.A.A., Coomans A. On the development and location of the oesophageal gland nuclei in the Dorylaimina. Nematologica. 1968;14:596. doi: 10.1163/187529268X00345. [DOI] [Google Scholar]
- 28.Archidona-Yuste A., Cantalapiedra-Navarrete C., Castillo P., Palomares-Rius J.E. Molecular phylogenetic analysis and comparative morphology reveals the diversity and distribution of needle nematodes of the genus Longidorus (Dorylaimida: Longidoridae) from Spain. Contrib. Zool. 2019;88:1–41. doi: 10.1163/18759866-20191345. [DOI] [Google Scholar]
- 29.Roca F., Bravo M.A. The occurrence of Xiphinema sphaerocephalum Lamberti et al. and X. hispanum Lamberti et al. (Nematoda: Longidoridae) in Portugal with descriptions of X. lanceolatum sp. n. and X. lapidosum sp. n. Fundam. Appl. Nematol. 1993;16:455–465. [Google Scholar]
- 30.Gutiérrez-Gutiérrez C., Cantalapiedra-Navarrete C., Montes-Borrego M., Palomares-Rius J.E., Castillo P. Molecular phylogeny of the nematode genus Longidorus (Nematoda: Longidoridae) with description of three new species. Zool. J. Linn. Soc. 2013;167:473–500. doi: 10.1111/zoj.12019. [DOI] [Google Scholar]
- 31.Tzortzakakis E.A., Archidona-Yuste A., Cantalapiedra-Navarrete C., Nasiou E., Lazanaki M.S., Kabourakis E.M., Palomares-Rius J.E., Castillo P. Integrative diagnosis and molecular phylogeny of dagger and needle nematodes of olives and grapevines in the island of Crete, Greece, with description of Xiphinema cretense n. sp. (Nematoda, Longidoridae) Eur. J. Plant Pathol. 2014;140:563–590. doi: 10.1007/s10658-014-0488-4. [DOI] [Google Scholar]
- 32.Palomares-Rius J.E., Cantalapiedra-Navarrete C., Archidona-Yuste A., Subbotin S.A., Castillo P. The utility of mtDNA and rDNA for barcoding and phylogeny of plant-parasitic nematodes from Longidoridae (Nematoda, Enoplea) Sci. Rep. 2017;7:10905. doi: 10.1038/s41598-017-11085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fouladvand Z.M., Pourjam E., Castillo P., Pedram M. Genetic diversity, and description of a new dagger nematode, Xiphinema afratakhtehnsis sp. nov., (Dorylaimida: Longidoridae) in natural forests of southeastern Gorgan, northern Iran. PLoS ONE. 2019;14:e0214147. doi: 10.1371/journal.pone.0214147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron E.K., Martins I.S., Lavelle P., Mathieu J., Tedersoo L., Gottschall F., Eisenhauer N. Global gaps in soil biodiversity data. Nat. Ecol. Evol. 2018;2:1042–1043. doi: 10.1038/s41559-018-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decaëns T. Macroecological patterns in soil communities. Global Ecol. Biogeogr. 2010;19:287–302. doi: 10.1111/j.1466-8238.2009.00517.x. [DOI] [Google Scholar]
- 36.Coomans A. Phylogeny of the Longidoridae. Russ. J. Nematol. 1996;4:51–60. [Google Scholar]
- 37.Finlay B.J. Global dispersal of free-living microbial eukaryotic species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 38.Olson M., Harris T., Higgins R., Mullin P., Powers K., Olson S., Powers T.O. Species delimitation and description of Mesocriconema nebraskense n. sp. (Nematoda: Criconematidae), a morphologically cryptic, parthenogenetic species from North American Grasslands. J. Nematol. 2017;49:42–66. doi: 10.21307/jofnem-2017-045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coolen W.A. Methods for extraction of Meloidogyne spp. and other nematodes from roots and soil. In: Lamberti F.F., Taylor C.E., editors. Root-knot Nematodes (Meloidogyne species). Systematics, Biology, and Control. Academic Press; London, UK: 1979. pp. 317–329. [Google Scholar]
- 40.Flegg J.J.M. Extraction of Xiphinema and Longidorus species from soil by a modification of Cobb’s decanting and sieving technique. Ann. Appl. Biol. 1967;60:429–437. doi: 10.1111/j.1744-7348.1967.tb04497.x. [DOI] [Google Scholar]
- 41.Seinhorst J.W. Killing nematodes for taxonomic study with hot f.a. 4:1. Nematologica. 1966;12:178. doi: 10.1163/187529266X00239. [DOI] [Google Scholar]
- 42.De Grisse A.T. Redescription ou modification de quelques techniques utilisées dans l’étude des nematodes phytoparasitaires. Meded. Rijksfak. Landbouwwet. Gent. 1969;34:351–369. [Google Scholar]
- 43.De Ley P., Felix M.A., Frisse L.A., Nadler S., Sternberg P., Thomas W. Molecular and morphological characterisation of two reproductively isolated species with mirror-image anatomy (Nematoda: Cephalobidae) Nematology. 1999;1:591–612. doi: 10.1163/156854199508559. [DOI] [Google Scholar]
- 44.Vrain T.C., Wakarchuk D.A., Levesque A.C., Hamilton R.I. Intraspecific rDNA Restriction Fragment Length Polymorphism in the Xiphinema americanum group. Fund. Appl. Nematol. 1992;15:563–573. [Google Scholar]
- 45.Cherry T., Szalanski A.L., Todd T.C., Powers T.O. The internal transcribed spacer region of Belonolaimus (Nemata: Belonolaimidae) J. Nematol. 1997;29:23–29. [PMC free article] [PubMed] [Google Scholar]
- 46.Holterman M., Van Der Wurff A., Van Den Elsen S., Van Megen H., Bongers T., Holovachov O., Bakker J., Helder J. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Phylogenet. Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 47.Lazarova S.S., Malloch G., Oliveira C.M.G., Hübschen J., Neilson R. Ribosomal and mitochondrial DNA analyses of Xiphinema americanum-group populations. J. Nematol. 2006;38:404–410. [PMC free article] [PubMed] [Google Scholar]
- 48.Legendre P., Legendre L. Numerical Ecology. Elsevier; Oxford, UK: 2012. 1006 [Google Scholar]
- 49.Zuur A.F., Ieno E.N., Elphick C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010;1:3–14. doi: 10.1111/j.2041-210X.2009.00001.x. [DOI] [Google Scholar]
- 50.Montgomery D.C., Peck E.A. Introduction to Linear Regression Analysis. Wiley; New York, NY, USA: 1992. 688p [Google Scholar]
- 51.Revelle W. psych: Procedures for Personality and Psychological Research. North-western University; Evanston, IL, USA: 2020. [(accessed on 14 May 2020)]. Delay Discounting of Different Outcomes 21. Available online: https://CRAN.R-project.org/package=psychVersion=1.9.12.31. [Google Scholar]
- 52.Quadros A. emstreeR: Tools for Fast Computing and Plotting Euclidean Minimum Spanning Trees. R Package Version 2.2.0. [(accessed on 14 May 2020)];2019 Available online: https://CRAN.R-project.org/package=emstreeR.
- 53.R Core Team . R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. [Google Scholar]
- 54.Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 55.Clement M., Snell Q., Walker P., Posada D., Crandall K. TCS: Estimating Gene Genealogies; Proceedings of the 6th International Parallel and Distributed Processing Symposium; Fort Lauderdale, FL, USA. 15–19 April 2002; Fort Lauderdale, FL, USA: IEEE Computer Society; 2002. p. 184. [Google Scholar]
- 56.Leigh J.W., Bryant D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 57.Flot J.F., Couloux A., Tillier S. Haplowebs as a graphical tool for delimiting species: A revival of Doyle’s “field for recombination” approach and its application to the coral genus Pocillopora in Clipperton. BMC Evol. Biol. 2010;10:372. doi: 10.1186/1471-2148-10-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y., Subbotin S.A., Rubtsova T.V., Lamberti F., Brown D.J.F., Moens M. A molecular phylogenetic approach to Longidoridae (Nematoda: Dorylaimida) Nematology. 2005;7:111–124. doi: 10.1163/1568541054192108. [DOI] [Google Scholar]
- 59.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 61.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 62.Ronquist F., Huelsenbeck J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 63.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rambaut A. FigTree v1.4.2, A Graphical Viewer of Phylogenetic Trees. [(accessed on 15 September 2020)];2014 Available online: http://tree.bio.ed.ac.uk/software/figtree/