Abstract
Adult skeletal muscle robustly regenerates throughout an organism’s life, but as the muscle ages, its ability to repair diminishes and eventually fails1,2. Previous work suggests that the regenerativepotentialofmusclestemcells(satellitecells)isnottriggeredin the old muscle because of a decline in Notch activation, and that it can be rejuvenated by forced local activation of Notch3. Here we report that, in addition to the loss of Notch activation, old muscle produces excessive transforming growth factor (TGF)-β (but not myostatin), which induces unusually high levels of TGF-β pSmad3 in resident satellite cells and interferes with their regenerative capacity. Importantly, endogenous Notch and pSmad3 antagonize each other in the control of satellite-cell proliferation, such that activation of Notch blocks the TGF-β-dependent upregulation of the cyclin-dependent kinase (CDK) inhibitors p15, p16, p21 and p27, whereas inhibition of Notch induces them. Furthermore, in muscle stem cells, Notch activity determines the binding of pSmad3 to the promoters of these negative regulators of cell-cycle progression. Attenuation of TGF-β/pSmad3 in old, injured muscle restores regeneration to satellite cells in vivo. Thus a balance between endogenous pSmad3 and active Notch controls the regenerative competence of muscle stem cells, and deregulation of this balance in the old muscle microniche interferes with regeneration.
Satellite cells are muscle stem cells capable of lifelong maintenance and repair of myofibres, or differentiated muscle cells4–6. The decline in muscle tissue regeneration with age is largely due to a decreased activation of Notch pathway, which is required for satellite cells to break quiescence and prevents premature differentiation into myotubes by antagonizing Wnt pathway3–9. Forced activation of Notch-1, by antibody specific to its external domain (Notch-1 cleavage and activation), rejuvenates muscle repair; whereas inhibiting Notch in young muscle interferes with regeneration3, suggesting that Notch pathway is an essential and age-specific molecular determinant of adult myogenesis.
It is widely believed that the microenvironment controls resident stem-cell behaviour10, and our recent work established that aged muscle fibres inhibit the regenerative responses of muscle stem cells11. Here we examine the biochemical changes occurring in the aged microniche of muscle stem cells, for example differentiated myofibres, focusing on the age-specific interplay between active Notch and TGF-β/pSmad3 and on the ability of this signal integration to control levels of CDK inhibitors in satellite cells.
Binding of activated TGF-β proteins to their receptors induces the phosphorylation and activation of the Smad transcription factors, which form heteromers(Smad4asa common component)that translocate into the nucleus12–15. Different ligands, for example TGF-β1, - β2, - β3 and myostatin, are capable of activating the same Smad2, 3 proteins12,16,17. Increased TGF-β signalling has been implicated in the inhibition of cell-cycle progression (both generally and in myogenic lineage), by activating CDK inhibitors and inactivating cMyc14,18–22. Importantly, recent genetic-targeting experiments suggest that during ageing the necessity to impose cell-cycle checkpoints becomes antagonistic to the regenerative responses of adult stem cells23–26.
This report establishes the following causalities: (1) old myofibres inhibit their own repair by shifting balance from active Notch to over-pronounced pSmad3 in resident muscle stem cells, which upregulates p15, p16, p21 and p27 and thwarts satellite-cell regenerative capacity; (2) active Notch can override this block of satellite-cell responses by removal of pSmad3 from CDK inhibitor promoters.
Growth factors, including TGF-β family members, are typically localized and activated in a tissue’s extracellular matrix (ECM), which in skeletal muscle is the main component of the basement membrane surrounding myofibres and their associated satellite cells27.
As shown in Fig. 1, there is dramatic and constant upregulation of functional TGF-β, but not the muscle-specific family member myostatin28, in the aged, injured and resting muscle compared with young. As expected, when present at high levels, TGF-β co-localizes with the laminin1 basement membrane (the immediate microniche of muscle stem cells) (Fig. 1a)29. Isotype-matched antibody controls for these and other experiments were negative (Supplementary Fig. 1a). These data were confirmed in western blot analysis, which also established that with age both differentiated myofibres and satellite cells, located in resting and injured muscle, upregulate levels of TGF-β (inactive precursor plus bioactive protein) and pSmad3 (Fig. 1b–e and Supplementary Fig. 2). In contrast, levels of myostatin and follistatin are not changed with age (Fig. 1a–c). In agreement with previously published work3, the levels of active Notch were reciprocal to those of pSmad3 and TGF-β, which is high in young and low in old satellite cells (Fig. 1b and Supplementary Fig. 1b). The purity of satellite cells in these preparations is greater than 95% (Supplementary Fig. 3)3. High levels of nuclear pSmad3 were also detected in satellite cells residing in the aged muscle in vivo (Supplementary Fig. 4), and excessive TGF-β was detected in old muscle at days 1 and 3 after injury (not shown).
Figure 1 |. TGF-β/pSmad3, but not myostatin, increases in old skeletal muscle.
a, Immunodetection of TGF-β, myostatin or follistatin (green) and laminin (red) in 10-μm skeletal muscle cryosections. Hoechst labels nuclei (blue). Scale bar, 50 μm. b, Western blot on myofibres and satellite cells; quantified in c.d, Immunoprecipitation with anti-Smad3 antibody, followed by western blot with anti-phosphorylated Smad3 antibody; quantified in e. f, After overnight transwell co-culture with young or old myofibres, young satellite cells were analysed by western blotting; data are means ±s.d., n = 3. *P ≤ 0.05 compared with young.
Our recent report demonstrated that proliferation and myogenesis of even young satellite cells are inhibited by the aged myofibres11. Interestingly, factors secreted by aged myofibres rapidly upregulated TGF-β production by young satellite cells (Fig. 1f) in transwell co-cultures (impermeable to cell migration). This provides a molecular explanation for the pre-mature ‘ageing’ of young progenitor cells exposed to aged tissue.
These data establish that although both Notch and pSmad3 can be robustly activated in skeletal muscle, because their ligands are expressed by myofibres and satellite cells, with age the balance is shifted from active Notch to active TGF-β/pSmad3 (Fig. 1)3.
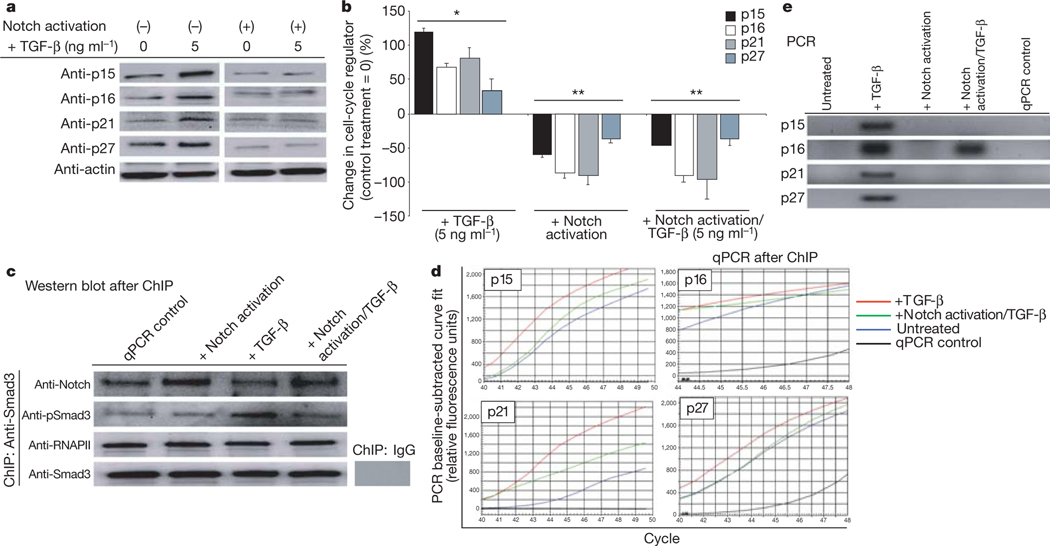
During the first days after injury, satellite cells need to break quiescence and proliferate. However, there is an age-specific elevation of pSmad3 and diminished Notch activation, either one of which is sufficient to inhibit cell-cycle progression17,22. We hypothesized that excessive levels of TGF-β/pSmad might upregulate the levels of CDK inhibitors in muscle stem cells, whereas activation of Notch might antagonize this process. After muscle injury, satellite cells were derived from young muscle3 and cultured with TGF-β1, with or without simultaneous forced activation of Notch. Compared with untreated cells, exogenously added TGF-β1 caused a prompt upregulation of p15, p16, p21 and p27 in satellite cells (Fig. 2a, quantified in Fig. 2b). When endogenous Notch was experimentally activated, simultaneously with TGF-β1 treatment, the inducing effects on p15, p16, p21 and p27 levels were significantly attenuated (Fig. 2a, b and Supplementary Fig. 5c) at a range of TGF-β1 concentrations (Supplementary Fig. 5). Manipulation of TGF-β/pSmad and active Notch balance not only controls CDK inhibitor levels, but also regulates proliferation of satellite cells in their endogenous microniches (Supplementary Fig. 6).
Figure 2 |. Notch removes pSmad3 from the 5’ regulatory regions of CDK inhibitors.
a, Satellite cells treated with TGF-β1, with or without Notch activation, were analysed by western blotting for p15, p16, p21 and p27; quantified in b. *P ≤ 0.05 compared with untreated control (0); **P ≤ 0.05 compared with TGF-β. Smad3 co-precipitated proteins (c) were resolved by western blot; genomic DNA (d) was analysed by RT–qPCR, using primers specific for 5’ regions of CDK inhibitors; data are means ±s.d., n = 3. e, qPCR reactions after 44 cycles revealed fragments of expected molecular weight on agarose gel.
To analyse this antagonistic interaction with higher precision, we examined whether active Notch and pSmad3 physically interact on promoters of the p15, p16, p21 and p27 genes. A pSmad3-specific chromatin immunoprecipitation assay (ChIP) was performed on satellite cells treated with TGF-β only, activation of Notch only, TGF-β and activation of Notch together or untreated.
As shown in Fig. 2c, both active Notch and RNA polymerase II are detected in a complex with pSmad3, suggesting that active Notch and pSmad3 physically interact on gene regulatory regions. Consistent with the idea of functional balance, forced activation of Notch yielded more endogenous Notch and treatment with TGF-β yielded more pSmad3 in these complexes (Fig. 2c).
DNA co-precipitated with pSmad3 was analysed by quantitative polymerase chain reaction (Q-PCR), using primers specific for 5’ promoter regulatory regions of p15, p16, p21 and p27 (Fig. 2d, e). These data provided a further understanding of the molecular mechanism of active Notch and TGF-β/pSmad3 antagonism. Namely, forced activation of Notch dramatically reduced pSmad3 presence on the promoter regions of p15, p16, p21 and p27, even in the presence of TGF-β treatment (Fig. 2d, e).
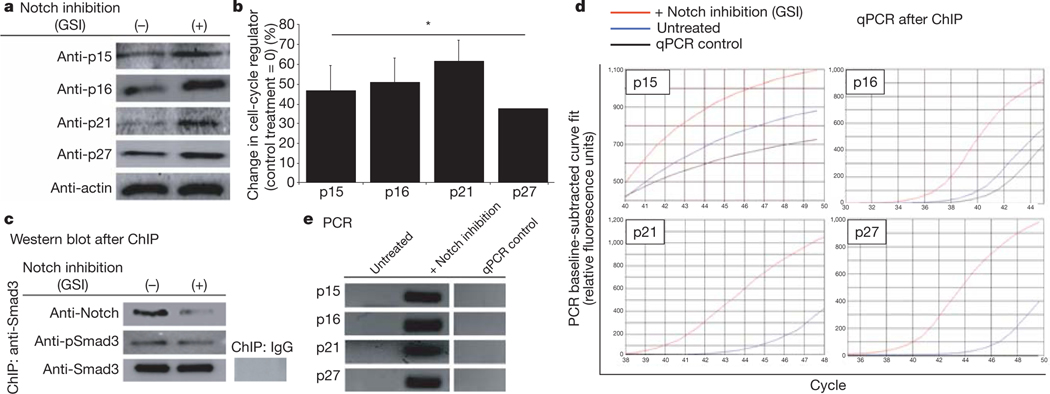
Interestingly, in the absence of Notch activation (by γ-secretase inhibitor (GSI)), young/low levels of TGF-β are sufficient to induce p15, p16, p21 and p27 proteins (Fig. 3a, b); and pSmad3 presence on the promoters of these genes increases in young muscle stem cells (Fig. 3d, e). Expectedly, less active Notch was detected in Notch–pSmad3complexes after treatment with GSI, whereas pSmad3 levels were not affected (Fig. 3c). These results were confirmed with several sets of PCR primers spanning about 1kilobase of the studied regulatory regions; and corroborated by negative controls, including the lack of amplification of a Smad3 non-enriched genome region (Supplementary Fig. 7).
Figure 3 |. Inhibition of endogenous Notch upregulates CDK inhibitors.
a, Compared with untreated cells (−), Notch inhibition by GSI caused prompt upregulation of p15, p16, p21 and p27; western blot assays quantified in b; *P ≤ 0.05 compared with untreated control. In ChIP assay, Smad3 co-precipitated proteins were resolved by western blot (c), genomic DNA (d) was analysed by RT–qPCR, using primers to p15, p16, p21 and p27 5’ regulatory regions; data are means ± s.d., n = 3. e, qPCR reactions after 44 cycles revealed fragments of expected molecular weight on agarose gel.
These data show that in muscle stem cells, active Notch attenuates age-specific induction of multiple CDK inhibitors by reducing the presence of pSmad3 on their promoter regions; and that either diminished Notch activation or increased TGF-β/pSmad3 levels are sufficient for upregulating these negative regulators of cell-cycle progression (Figs 2 and 3, and Supplementary Figs 6 and 7). Interestingly, p16 is sensitive to a slight increase in pSmad3, as inhibition of Notch robustly enhanced pSmad3 binding, whereas Notch activation did not significantly reduce TGF-βimposedpSmad3binding to the p16 promoter (Figs 2, 3 and Supplementary Fig. 7).
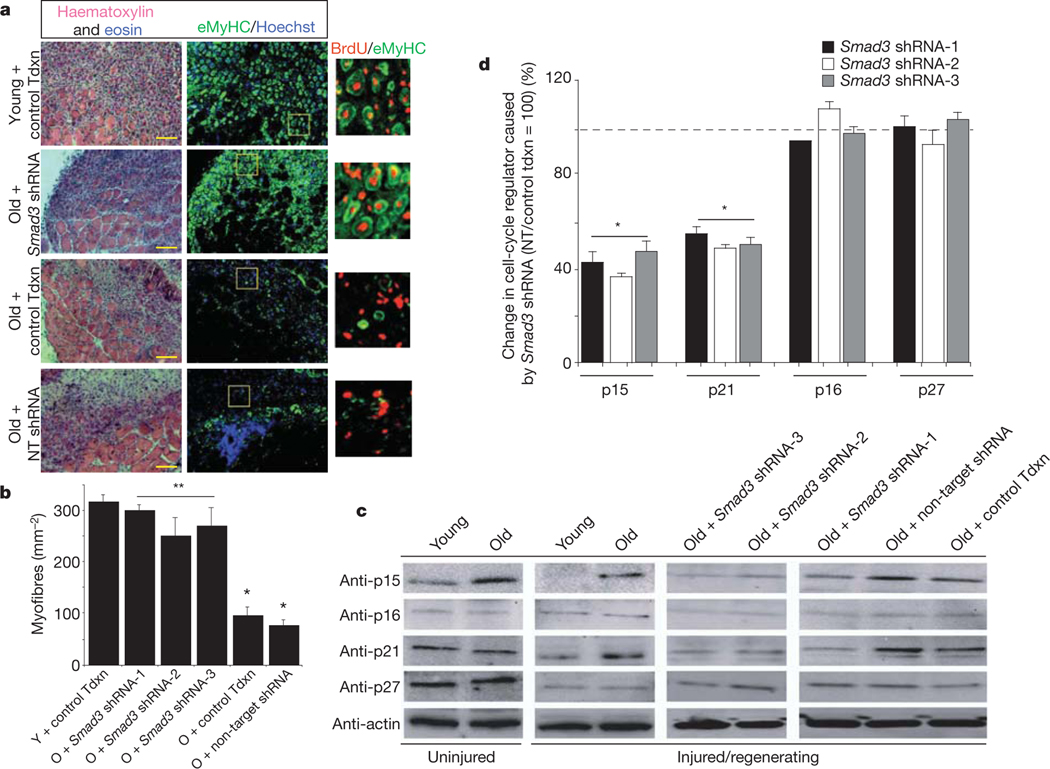
To confirm these findings and address their physiological significance in vivo, we examined whether aged muscle stem cells would effectively repair old tissue when pSmad3 is locally attenuated by lentivirally delivered short hairpin RNA (shRNA) to Smad3.
Young and old muscles were infected with the following lentiviral particles in vivo: three different Smad3-targeted shRNAs, non-target shRNA to control for non-specific RNA interference or transduction control; and followed by muscle injury (Fig. 4 and Supplementary Figs 8–10). As expected, at 5 days after injury, old control muscle contains fewer regenerated myofibres than young, based on haema-toxylin and eosin histology and immunodetection of eMyHC de novo myofibres with centrally located bromodeoxyuridine (BrdU+) nuclei that are the fusion product of myoblasts generated by satellite cells (Fig. 4)3. In contrast to control viral transduction and non-target shRNA control, the acute in vivo expression of each one of the Smad3-targeted shRNAs enhanced and rejuvenated regeneration of old muscle (Fig. 4a, b and Supplementary Fig. 10a). Furthermore, in old muscle satellite cells there was pronounced increase in p15 (at all times) and p21 (upon muscle injury); and importantly, p15 and p21 were attenuated in vivo to their ‘youthful’ levels by each tested Smad3-targeted shRNA, but not by non-target shRNA (Fig. 4c, d). The levels of p16 and of p21, p27 were not increased in quiescent satellite cells endogenous to old resting muscle, consistent with rapid activation of their myogenic responses in young environments (Fig. 4c, d)3,11. A slight increase in p16 and p27 levels was detected 24 h after injury, suggesting a potential physiological importance for early muscle stem-cell responses (Supplementary Fig. 9). Thus, elevated TGF-β/pSmad3 assures the age-dependent upregulation of at least one CDK inhibitor at all times, while specific time points are expectedly different for individual CDK inhibitors known to be under control of many molecular cues. These data strongly suggest that Smad3-specific (rather than non-target effects30) rescued the satellite-cell regenerative responses in old niches, as three different Smad3-targeted shRNAs similarly enhanced myogenesis of old muscle and diminished the levels of p15 and p21, but not of p16 and p27 in the aged satellite cells (Fig. 4a–d and Supplementary Fig. 10). Smad3 targeting of these shRNAs is confirmed by the expected downregulation of messenger RNA levels of Smad3 and nuclear levels of pSmad3 protein, as well as of Smad3, Smad6, Smad7, TGF-β and myostatin in satellite cells in vivo14,15 (Supplementary Fig. 10b–e). Notably, pan-neutralizing antibody against TGF-β rejuvenated repair of old muscle, and recombinant TGF-β resulted in scarring of young muscle (Supplementary Fig. 11), both of which are consistent with the effects of Smad3 targeting by three different shRNAs.
Figure 4 |. Smad3 shRNA rescues responses of satellite cells in old niche in vivo.
Lentiviral-infected muscle (control transduction (Tdxn), Smad3 shRNAs, non-target shRNA) was analysed 5 days after injury. a, Haematoxylin and eosin 10-μm cryosections of Smad3 shRNA-1; quantified in b; n = 15, *, **P ≤ 0.05; individual Smad3 shRNA-2, −3 data in Supplementary Fig. 10a; immunodetection of eMyHC myofibres with BrdU+ nuclei. Scale bar, 100μm; O, old; Y, young. c, Isolated satellite cells analysed by western blot for p15, p16, p21 and p27; quantified in d; *P ≤ 0.05, compared with non-target shRNA, control tdxn; data are means ± s.d., n = 3.
In contrast, inhibition of myostatin by follistatin resulted in multitudes of very small nascent myofibres, and proliferating Myf-5+ myogenic cells that persisted within the injured area (Supplementary Fig. 11). Such a defect in myogenic differentiation was also evident when both TGF-β and myostatin were neutralized; strongly suggesting that myostatin, which remains constant with age (Fig. 1), is required for productive differentiation of myogenic cells into myofibres. Distinct regenerative outcomes, where neutralization of TGF-β but not myostatin restores productive repair to old muscle, confirms that TGF-β is the main age-specific local inhibitor in skeletal muscle niche.
Comprehensively, this work establishes that productive muscle repair is determined by an antagonistic balance between the levels of TGF-β/pSmad3 (low in young and high in old) and the activation of Notch (high in young and low in old)3,which regulates the levels of four distinct CDK inhibitors in resident stem cells. An age-specific shift in either pathway would suffice for the upregulation of CDK inhibitors and diminished proliferation of muscle stem cells. Deregulation of both Notch and TGF-β/pSmad3 thus assures the lack of satellite-cell activation in the context of aged microniche.
Our data indicate that TGF-β does not directly inhibit the expression of Notch ligand Delta, or the levels of active Notch in satellite cells (Supplementary Fig. 12). Likewise, activation of Notch does not directly diminish the TGF-β/pSmad3 levels (Figs 2 and 3). Hence, the age-specific changes in reciprocal activation of Notch and TGF-β/ pSmad3 seem to initiate independently of each other, and future work will identify molecular triggers causing such imbalance. Additional studies will also answer whether excessive TGF-β found in old muscle ECM reflects only local secretion or also results from higher levels of TGF-β in the aged circulation, and whether local and systemic TGF-β are connected and provide feedbacks for each other. Importantly, this work has established that one factor in the ageing of skeletal muscle—and perhaps other organ niches—seems to be a selfimposed inhibition of regenerative potential.
METHODS SUMMARY
Young (~2 month) and old (~24 month) C57 BL/6 mice were from Jackson Laboratories and the National Instituteon Ageing. Skeletal muscle was cardiotoxin injured3. Co-injection with anti-TGF-β neutralizing antibody, TGF-β, follistatin or IgG was performed in some experiments. Muscle was isolated 1–5 days after injury and prepared for cryosectioning/histology, western blotting or in vitro culturing of myofibre explants and satellite cells.
Immunocytochemistry/histological analyses were performed as described3,11. Samples were analysed with a Zeiss Axio Imager A1, imaged with an Axiocam MRc camera and AxioVision software. Western blots were analysed with BioRad Gel Doc/Chemi Doc Imaging System and Quantity One software.
Transwell co-cultures (1μm) were performed with isolated satellite cells and young/old myofibres. Conditioned supernatants were analysed for secreted bioactive TGF-β1 levels in enzyme-linked immunosorbent assay (ELISA)-based cytokine antibody arrays (Raybiotech). CDK inhibitors were induced in vitro by culturing isolated satellite cells with TGF-β1 or myostatin. Notch was activated by immobilized Delta or EDTA exposure before seeding.
Lentiviral transduction was performed with distinct Smad3 shRNAs: accession number NM_016769.2. (shRNA-2): CCGGCCTTACCACTATCAGAGA GTACTCGAGTACTCTCTGATAGTGGTAAGGTTTTTG, (shRNA-3)-CCGGCTGTCCAATGTCAACCGGAATCTCGAGATTCCGGTTGACATTGGACAGTTTTTG. Smad3 shRNA cocktail (shRNA-1, see Methods) yielded results similar to shRNAs 2, 3. Control transduction with non-target shRNA or GFP transduction lentiviral particles was also performed.
BrdU was injected intraperitoneally, 3 days after injury. Tissues were analysed 5 days after injury for regenerative responses and transduction levels by histological analysis, western blotting and PCR with reverse transcription (RT–PCR) (Bio-Rad iQ5).
In ChIP assays (Upstate), satellite cells underwent treatments for 24h, followed by DNA shearing and immunoprecipitations. Proteins co-precipitated with pSmad3 were analysed by western blot. pSmad3 co-precipitated DNA was analysed using primers to the gene regulatory regions of p15, p16, p21 and p27. Real-time qPCR samples were analysed using a Bio-Rad iQ5 real-time PCR detection system, with iQ5 optical system software. Standard PCR samples were analysed with a Bio-Rad iQ5 thermal cycler.
METHODS
Animal strains.
Young (2–3 month) and old (22–24 month) C57 BL/6 male mice were obtained from pathogen-free breeding colonies at Jackson Laboratories and the National Institute on Ageing, respectively. Animals were housed at the Northwest Animal Facility, University of California, Berkeley. Animal procedures were conducted in accordance with the Administrative Panel on Laboratory Animal Care at University of California, Berkeley.
Muscle injury.
Tibialis anterior and gastrocnemius muscles of mice were injected with 5 ng cardiotoxin (CTX-1) (Sigma), suspended in 1 X PBS at four or five sites in each muscle (10 μl per site), using a 28-gauge needle. In some experiments anti-TGF-β neutralizing antibody, TGF-β or control goat IgG (all at 500 ng ml−1 final concentration) were co-injected with CTX-1, following the same protocol as described above for CTX alone. Uninjured or injured/regenerating muscle tissue was isolated 1–5 days after injury. Whole muscle was prepared for cryosectioning and histological analysis, western blotting or for use in culturing of isolated satellite cells in vitro.
Muscle isolation and satellite-cell culture.
Myofibre explants and satellite cells were generated from C57 BL/6 mice as described previously3,11. Briefly, whole muscle underwent enzymatic digestion at 37 °C in DMEM (Invitrogen)/1% penicillin–streptomycin (Invitrogen)/125U ml−1 Collagenase Type IIA (Sigma) solution. Bulk myofibres with associated satellite cells (located beneath the basement membrane and above the plasma membrane) were purified away from muscle interstitial cells, tendons, etc. by multiple rounds of trituration, sedimentation and washing. Satellite cells were isolated from purified myofibres by subsequent enzymatic digestion with Collagenase Type IIA and Dispase (Sigma), followed by sedimentation, washing and fine-mesh straining procedures, as in refs 3 and 11. Purified satellite cells and satellite cell-stripped myofibres were used in subsequent experiments, as described below. The approximate 95% purity of satellite cells was routinely confirmed by generation of proliferating fusion-competent myoblasts after 24h in growth medium (Ham’s F-10 (Mediatech), 20% FBS (Mediatech), 5ng ml−1 FGF (Chemicon) and 1% penicillin–streptomycin); and myotube formation after 48h in DMEM, 12% horse serum. The efficiency of satellite-cell depletion from myofibres was routinely confirmed by the absence of such myogenic potential. Satellite cells were cultured on ECM:PBS-coated plates (1:500; BD Biosciences).
Transwell co-cultures of myofibres and satellite cells.
Transwell co-cultures (1.0μm, Corning) were used for culturing isolated satellite cells with young or old myofibres. Satellite cells were seeded onto ECM-coated plates in OPTI-MEM (Invitrogen), +5% FBS. Transwell inserts, containing isolated myofibre explants from young or old muscle, were placed over satellite cells and cultured for 72–96 h before lysing for western blot analysis (see below). Supernatants conditioned for 24 h, from both young and old control myofibre explants, were used to analyse secreted bioactive TGF-β1 levels (immunodetection of active protein) in ELISA-based cytokine antibody arrays (RayBiotech).
Western blot analysis.
Myofibre and satellite-cell lysates were prepared in lysis buffer (50mM Tris, 150mM NaCl, 1% NP40, 0.25% sodium deoxycholate and 1 mM EDTA, pH 7.4). Protease inhibitor cocktail (Sigma) and 1mM PMSF were added before use. Phosphatase activity was inhibited by 1 mM sodium fluoride and 1 mM sodium orthovanadate for pSmad immunodetection. Approximately 30 μg protein extract were run on pre-cast SDS PAGE gels (Biorad). Primary antibodies were diluted in 5% non-fat milk/1 X PBST, and nitrocellulose membranes were incubated with antibody mixtures overnight at 4 °C. HRP-conjugated secondary antibodies (Santa Cruz Biotech) were diluted 1:1,000 in 1 X PBST/1% BSA and incubated for 1h at room temperature. Blots were developed using Western Lightning ECL reagent (Perkin Elmer), and analysed with Bio-Rad Gel Doc/Chemi Doc Imaging System and Quantity One software. Results of multiple assays were quantified by digitizing the data and normalizing pixel density of examined protein by actin-specific pixel density.
Immunocytochemistry and histological analysis.
Muscle tissue was treated in a 25% sucrose/PBS solution, frozen in OCT compound (Tissue Tek), cryosectioned at 10 μm (Thermo Shandon Cryotome E) and immunostained as previously described3,11. Immunostaining or haematoxylin and eosin staining were performed on cryosections. For indirect immunoflourescence, sections were permeabilized in (PBS, +1% FBS, +0.25% Triton X-100), incubated with primary antibodies for 1h at room temperature in PBS, +1%FBS, and then with fluorophore-conjugated, species-specific secondary antibodies for 1 h at room temperature (1:500 in PBS, +1%FBS). pSmad3- and BrdU-specific immunostaining required additional nuclear permeabilization and DNA-chromatin denaturation with 4N HCL. Nuclei were visualized by Hoechst staining for all immunostains. Biologically active TGF-β and myostatin proteins (immunodetectible ligands cleaved from the latent complex)14,29 were also examined. Samples were analysed at room temperature with a Zeiss Axio Imager A1, and imaged with an Axiocam MRc camera and AxioVision software.
In vitro induction of cell-cycle regulators by TGF-β and myostatin.
Activatedby-injury satellite cells were isolated from injured muscle. Cells were seeded onto ECM-coated, six-well plates at a uniform density of 1.2 X 105 cells per well, and cultured for 24 h in OPTI-MEM +1% mouse sera, containing various levels of TGF-β1 or myostatin. For Notch activation, ECM-coated plates were coated with 2 μg ml−1 Delta-4 (DLL4) overnight at 37°C, before seeding satellite cells. Alternatively, Notch activation was induced by exposing satellite cells to 5mM EDTA for 15 min at 37°C before seeding. Cells lysates were prepared and used for western blotting analysis (described above).
shRNA delivery by lentiviral transduction.
Young and old tibialis anterior and gastrocnemius muscles were infected, in vivo, with control non-target shRNA (Sigma SHC002V), GFP (Sigma SHC003V) or pLKO.1-puro (Sigma SHC001V) control transduction lentiviral particles (at least 106 transducing units per millilitre, as determined by p24 antigen ELISA titre). Mouse Smad3 shRNA-producing lentiviral particles (also obtained from Sigma) were used for in vivo transduction experiments (target-set generated from accession number NM_016769.2: (1) CCGGCCCATGTTTCTGCATGGATTTCTCGAGAAATCC ATGCAGAAACATGGGTTTTTG; (2) CCGGCCTTACCACTATCAGAGAGTA CTCGAGTACTCTCTGATAGTGGTAAGGTTTTTG; (3) CCGGCTGTCCAA TGTCAACCGGAATCTCGAGATTCCGGTTGACATTGGACAGTTTTTG; (4) CCGGGCACACAATAACTTGGACCTACTCGAGTAGGTCCAAGTTATTGTGTGCTTTTTG; (5) CCGGCATCCGTATGAGCTTCGTCAACTCGAGTTGAC GAAGCTCATACGGATGTTTTTG). shRNAs were used in a Smad3 shRNA cocktail (shRNAs 1–5), designated Smad3 shRNA-1, or individually (shRNA(2) designated Smad3 shRNA-2 and shRNA(3) designated Smad3 shRNA-3). Skeletal muscle was infected by intramuscular injection of lentiviral particles (about 50,000TU) with a 28-gauge needle on multiple consecutive days, before or coincident with CTX-1 injury. To examine cell proliferation, 50 μl of 10 μM BrdU was injected intraperitoneally at 3 days after injury. Tissues were analysed for regenerative responses and transduction levels at 5 days after injury by cryo-sectioning of whole tissues, or western blotting of satellite-cell lysates (described above). Transcript levels were analysed using SuperScript RT–PCR kit (Invitrogen) for amplification of Smad3 (F = CTGGCTACCTGAGTGAAGATGGAGA, R = AAAGACCTCCCCTCCGATGTAGTAG) and GAPDH (F = TGAGGCCGGTGCTGAGTATGTCGTG, R = TCCTTGGAGGCCATGTAGGCCAT). Amplification products (25–40 cycles on BioRad iQ5) were examined and confirmed for predicted molecular masses on EtBr-stained 2% agarose gels.
ChIP assays and RT–qPCR/PCR.
Isolated satellite cells were treated with TGF-β1 only, activation of Notch only, TGF-β1/Notch together, Notch inhibition (GSI) or untreated (as described above). After culture for 24 h, satellite cells were fixed with 1% PFA and ChIP assay was performed according to manufacturer’s guidelines (Upstate). Fragments of about 500 base pairs were produced by shearing DNA with attached proteins (confirmed by EtBr-stained gels), and precipitated with antibodies to DNA-bound protein. Proteins that co-precipitated with pSmad3 were analysed by western blot, using indicated antibodies. DNA that co-precipitated with pSmad3 was analysed using primers specific for the 5’ gene regulatory regions of p15, p16, p21 and p27 (p15 F = TCACCGAAGCTACTGGGTCT, R = GTTCAGGGCGTTGGGATCT; p16 F = GTCACACGACTGGGCGATT, R = GTTGCCCATCATCATCACCT and F = GATGACTTCACCCCGTCACT, R = AACACCCCTGAAAACACTGC/GTCCCTCCTTCCTTCCTCTG; p21 F = CCGCGGTGTCAGAGTCTA, R = CATGAGCGCATCGCAATC; p27 F = AGCCTACGCTCCGACTGTT, R = AGTTCTGCGACTGCACACAG and F = CTAGCCACCGAAGCTCCTAA, R = AGT TCTGCGACTGCACACAG and F = CTGGCTCTGCTCCATTTGAC, R = GGCTCCCGTTAGACACTCTC). GAPDH primers (see above) were used as control for Smad3 non-enriched genomic regions. Primers were designed with Oligo Perfect Designer (Invitrogen). For RT–qPCR, samples were analysed using a Bio-RadiQ5 real-time PCR detection system, with iQ5opticalsystem software. For PCR, samples were amplified with Platinum Taq DNA Polymerase (Invitrogen) and analysed on a Bio-Rad iQ5 system. After 40–55 cycles of amplification, fragments produced from each primer set were examined and confirmed for their predicted molecular masses on EtBr-stained 2% agarose gels. Fifty-five cycles of amplification were used for negative control PCR reactions.
Reagents.
Antibodies to BrdU (ab6326), activated Notch1 (ab8925) and CHiP grade Smad3 (ab287379) were purchased from Abcam. Antibody to developmental eMyHC (clone RNMy2/9D2) was acquired from Vector Laboratories. Antibodies to desmin (clone DE-U-10, Cat#D8281), laminin (L9393), actin (A5060) and follistatin (F2177) were acquired from Sigma. Bioactivity-neutralizing antibodies against TGF-β1/2/3 were obtained from R&D Systems (MAB1835) and Santa Cruz Biotechnologies (sc7892). Antibodies to myostatin (sc-34781), follistatin (sc-30194), TGF-β1 (sc146), phosphorylated-smad3 (sc11769), smad6 (sc13048), smad7 (sc11392), p15 (sc613), p16 (sc1207 and sc1661), p21 (sc756), p27 (sc776), RNAP II (sc899), Myf5 (sc31946) and goat/rabbit IgG were acquired from Santa Cruz Biotechnologies. Smad3 antibody (06–920) was obtained from Upstate. Fluorophore-conjugated secondary antibodies (Alexa Fluor) were supplied by Invitrogen. HRP-conjugated secondary antibodies were purchased from Santa Cruz Biotechnologies. BrdU labelling reagent was obtained from Sigma. Recombinant human TGF-β1 (RD 240B), recombinant mouse DLL4 (RD 1389) and recombinant mouse myostatin (788-G8-010) were obtained from R&D Systems. TGF-β RI Kinase Inhibitor (50 nM) (#616451) and gammaSecretase Inhibitor X (50nM) (#565771) were purchased from Calbiochem. Two3SYBR-Green RT–PCR reaction mixture was purchased from Bio-Rad, and SuperScript One-Step RT–PCR (#10928) from Invitrogen.
Statistical analysis.
Quantified data are expressed as mean ±s.d. Significance testing was performed using one-way analysis of variance, with an alpha level of 0.01–0.05, to compare data from different experimental groups. A minimum of three replicates were performed for each described experimental condition.
Supplementary Material
Acknowledgements
We thank R. Derynck and M. Conboy for discussions. This work was supported by National Institutes of Health (NIH) R01 (AG027252), NIH R21 (AG27892) and Ellison’s Medical Foundation grants to I.M.C., and a Pre-doctoral Training Fellowship from the California Institute for Regenerative Medicine training grant to M.E.C.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Grounds MD Age-associated changes in the response of skeletal muscle cells to exercise and regeneration. Ann. NY Acad. Sci 854, 78–91 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Renault V, Thornell LE, Eriksson PO, Butler-Browne G & Mouly V. Regenerative potential of human skeletal muscle during aging. Aging Cell 1, 132–139 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Conboy IM, Conboy MJ, Smythe GM & Rando TA Notch-mediated restoration of regenerative potential to aged muscle. Science 302, 1575–1577 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Wagers AJ & Conboy IM Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell 122, 659–667 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Collins CA & Partridge TA Self-renewal of the adult skeletal muscle satellite cell. Cell Cycle 4, 1338–1341 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Morgan JE et al. Myogenic cell proliferation and generation of a reversible tumorigenic phenotype are triggered by preirradiation of the recipient site. J. Cell Biol 157, 693–702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz E & Lipton BH Skeletal muscle satellite cells: changes in proliferation potential as a function of age. Mech. Ageing Dev 20, 377–383 (1982). [DOI] [PubMed] [Google Scholar]
- 8.Brack AS et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317, 807–810 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Conboy IM et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Li L & Xie T. Stem cell niche: structure and function. Annu. Rev. Cell Dev. Biol 21, 605–631 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Carlson ME & Conboy IM Loss of stem cell regenerative capacity within aged niches. Aging Cell, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massague J. TGF-βeta signal transduction. Annu. Rev. Biochem 67, 753–791 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Massague J & Chen YG Controlling TGF-βeta signaling. Genes Dev. 14, 627–644 (2000). [PubMed] [Google Scholar]
- 14.Derynck R & Zhang YE Smad-dependent and Smad-independent pathways in TGF-βeta family signalling. Nature 425, 577–584 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Feng XH & Derynck R. Specificity and versatility in TGF-βeta signaling through Smads. Annu. Rev. Cell Dev. Biol 21, 659–693 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Natarajan E et al. A keratinocyte hypermotility/growth-arrest response involving laminin 5 and p16INK4A activated in wound healing and senescence. Am. J. Pathol 168, 1821–1837 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T, Sawada R, Fujiwara Y, Seyama Y & Tsuchiya T. FGF-2 suppresses cellular senescence of human mesenchymal stem cells by down-regulation of TGF-βeta2. Biochem. Biophys. Res. Commun 359, 108–114 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Untergasser G et al. Profiling molecular targets of TGF-βeta1 in prostate fibroblast-to-myofibroblast transdifferentiation. Mech. Ageing Dev 126, 59–69 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Olson NE, Kozlowski J & Reidy MA Proliferation of intimal smooth muscle cells. Attenuation of basic fibroblast growth factor 2-stimulated proliferation is associated with increased expression of cell cycle inhibitors. J. Biol. Chem 275, 11270–11277 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Joulia D et al. Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp. Cell Res 286, 263–275 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Lin J et al. P27 knockout mice: reduced myostatin in muscle and altered adipogenesis. Biochem. Biophys. Res. Commun 300, 938–942 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Rao P & Kadesch T. The intracellular form of notch blocks transforming growth factor beta-mediated growth arrest in Mv1Lu epithelial cells. Mol. Cell. Biol 23, 6694–6701 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janzen V et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Molofsky AV et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443, 448–452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamurthy J et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443, 453–457 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Stepanova L & Sorrentino BP A limited role for p16Ink4a and p19Arf in the lossof hematopoietic stem cells during proliferative stress. Blood 106, 827–832 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husmann I, Soulet L, Gautron J, Martelly I & Barritault D. Growth factors in skeletal muscle regeneration. Cytokine Growth Factor Rev. 7, 249–258 (1996). [DOI] [PubMed] [Google Scholar]
- 28.McPherron AC, Lawler AM & Lee SJ Regulation of skeletal muscle mass inmice by a new TGF-βeta superfamily member. Nature 387, 83–90 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Barcellos-Hoff MH Latency and activation in the control of TGF-βeta. J. Mammary Gland Biol. Neoplasia 1, 353–363 (1996). [PubMed] [Google Scholar]
- 30.Svoboda P. Off-targeting and other non-specific effects of RNAi experiments inmammalian cells. Curr. Opin. Mol. Ther 3, 248–257 (2007). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.