Abstract
Multidrug resistance (MDR) and hypervirulence (hv) have been long considered distinct evolutionary traits for Klebsiella pneumoniae (Kp), a versatile human pathogen. The recent emergence of Kp strains combining these traits poses a serious global threat. In this article, we describe the phenotypic and genomic characteristics of an MDR hvKp isolate, MAR14-456, representative of a nosocomial outbreak in Moscow, Russia, that was recovered from a postoperative wound in a patient who later developed multiple abscesses, fatal sepsis, and septic shock. Broth microdilution testing revealed decreased susceptibility of MAR14-456 to carbapenems (MICs 0.5–2 mg/L) and a high-level resistance to most β-lactams, β-lactam-β-lactamase-inhibitor combinations, and non-β-lactam antibiotics, except ceftazidime-avibactam, amikacin, tigecycline, and colistin. Whole-genome sequencing using Illumina MiSeq and ONT MinION systems allowed to identify and completely assemble two conjugative resistance plasmids, a typical ‘European’ epidemic IncL/M plasmid that carries the gene of OXA-48 carbapenemase, and an IncFIIK plasmid that carries the gene of CTX-M-15 ESBL and other resistance genes. MLST profile, capsular, lipopolysaccharide, virulence genes encoded on chromosome and IncHI1B/FIB plasmid, and the presence of apparently functional type I-E* CRISPR-Cas system were all characteristic of hvKp ST23, serotype K1-O1v2. Phylogenetic analysis showed the closest relatedness of MAR14-456 to ST23 isolates from China. This report highlights the threat of multiple resistance acquisition by hvKp strain and its spread as a nosocomial pathogen.
Keywords: Klebsiella pneumoniae, OXA-48, hypervirulent ST23, multidrug resistance, whole-genome sequencing
1. Introduction
Klebsiella pneumoniae (Kp) is a versatile and increasingly important bacterial pathogen capable of causing various infections. The population of Kp is composed of genetically diverse strains that evolved into two distinct pathotypes: classical Kp (cKp) associated with nosocomial and opportunistic infections and less common hypervirulent Kp (hvKp) associated with more severe and aggressive infections, usually in healthy and community-residing individuals [1]. cKp is notorious for its remarkable ability to acquire antibiotic resistance determinants, of which the most important are the genes that encode extended-spectrum β-lactamases (ESBLs) and carbapenemases conferring resistance to newer-generation cephalosporins and carbapenems, respectively. This ability has earned Kp a place among the most difficult to treat ‘ESKAPE’ pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) and among the top-priority pathogens identified by WHO and CDC [2,3]. In contrast, hvKp strains are usually susceptible to most antibiotics [4]. They belong to a relatively small number of clonal groups and sequence types (STs) as defined by multilocus sequence typing (MLST), with ST23 being the most prevalent [5,6,7]. Initially, reports of infections caused by ST23, capsular type K1 Kp, manifesting as pyogenic liver abscess and often complicated with bacteremia and metastatic lesions to other organs, emerged across East Asia: Taiwan, China, Hong-Kong, Singapore, and South Korea, and more recently, from many other parts of the world, including Europe, Americas, South Asia, Middle East, Africa, and Australia [5,6,7].
Moreover, in the last few years, alarming reports have documented the acquisition of ESBL and carbapenemase genes by hvKp of ST23, thus, raising the concern of the emergence of a new “superbug” combining virulence and resistance traits [8,9,10,11,12,13]. The studies from UK and China described the isolation of ST23 isolates carrying the blaNDM-1 gene for metallo-β-lactamase [8,11] conferring resistance to all β-lactams except monobactams, while reports from Argentina and the United States revealed the presence of blaKPC-2 gene for class A carbapenemase gene in isolates of ST23 [9,10]. Another two reports from Russia described virulence characteristics and draft genome sequences of epidemiologically related ST23 isolates harboring blaOXA-48 gene for class D carbapenemase providing resistance to penicillins and carbapenems from an outbreak in a neurosurgical intensive care unit (ICU) in Moscow, Russia [12,13], which included at least six documented cases of infections in different patients.
The hvKp isolate MAR14-456 representing this outbreak was isolated on 26 February 2014 from wound biopsy of a 54-year old female patient after surgery for severe traumatic brain injury followed by prolonged (48 days) stay in the ICU with mechanical ventilation. The patient later developed multiple abscesses, fatal sepsis, and septic shock due to the same strain despite multiple surgical interventions and combination therapy with high-dose (8 g/day continuous infusion) meropenem, amikacin, and tigecycline. In this article, we describe the detailed phenotypic and genomic characteristics of the index isolate based on antimicrobial susceptibility testing by reference methods, hybrid whole-genome assembly, and phylogenetic comparison to other published sequence data.
2. Results
The results of antibiotic susceptibility testing of MAR14-456 are shown in Table 1. The isolate revealed a decreased susceptibility to carbapenems (MICs 0.5–2 mg/L) and high-level resistance to all penicillins, penicillin-β-lactamase-inhibitor combinations, cephalosporins, aztreonam, and non-β-lactam antibiotics, except ceftazidime-avibactam, amikacin, tigecycline, and colistin. A synergy between oxyimino-cephalosporins and clavulanic acid and positive results of carbapenem inactivation method (CIM test) were indicative of ESBL and carbapenemase production, respectively.
Table 1.
Antibiotic resistance phenotype of MAR14-456.
| Antibiotic/Combination | MIC, mg/L |
ECOFF- Based Category |
Clinical Susceptibility Category |
|---|---|---|---|
| Ampicillin | ≥256 | non-WT 1 | R |
| Amoxicillin-clavulanic acid | ≥256/4 | non-WT | R |
| Piperacillin-tazobactam | ≥256/4 | non-WT | R |
| Cefoxitin | 4 | WT | - |
| Cefotaxime | ≥256 | non-WT | R |
| Cefotaxime-clavulanic acid | 8/4 | - | - |
| Ceftazidime | 64 | non-WT | R |
| Ceftazidime-clavulanic acid | 2/4 | - | - |
| Ceftazidime-avibactam | 0.25/4 | - | S |
| Cefepime | 32 | non-WT | R |
| Cefepime-clavulanic acid | 4/4 | - | - |
| Aztreonam | 64 | non-WT | R |
| Ceftazidime-avibactam | 0.25/4 | WT | S |
| Imipenem | 1 | WT | S |
| Meropenem | 0.5 | non-WT | S |
| Ertapenem | 2 | non-WT | R |
| Tobramycin | 8 | non-WT | R |
| Gentamicin | 0.5 | WT | S |
| Netilmicin | 4 | non-WT | I |
| Amikacin | 4 | WT | S |
| Ciprofloxacin | 8 | non-WT | R |
| Doxycycline | 8 | non-WT | - |
| Tigecycline | 0.25 | WT | S |
| Chloramphenicol | 16 | - | R |
| Colistin | 0.125 | WT | S |
| Fosfomycin | 64 | non-WT | R |
| Trimethoprim-sulfamethoxazole | ≥256/4864 | non-WT | R |
1 WT, wild type; non-WT, non-wild type; S, susceptible; I, increased exposure susceptible; R, resistant; -, no breakpoints established by EUCAST.
The hybrid assembly of short- and long-read genome sequences of MAR14-456 included four contigs of 5,446,456 bp, 232,362 bp, 127,014 bp, and 63,589 bp, with the largest one corresponding to chromosome and the remaining three corresponding to IncHI1B/FIB, IncFIIK, and IncL/M plasmids, respectively. Additional analysis performed by remapping of short reads to hybrid assembly and polishing the resulting contigs allowed to reconstruct the circular structure of all three plasmids.
The results of the annotation of resistance determinants (Table 2) were fully consistent with the phenotype of resistance. BlaOXA-48 gene was identified on a composite Tn1999 transposon inserted in IncL/M plasmid showing complete sequence identity to the ‘European epidemic’ pOXA-48 plasmids (for example, GenBank acc. Nos. LR025105, MT989343, and CP039938). Other acquired resistance genes: qnrB1, tet(A), blaCTX-M-15, dfrA14, strAB, sul2, catB3::IS26, aac(6’)Ib-cr, blaTEM-1b and blaOXA-1 were found on an IncFIIK plasmid almost identical (98% coverage; 99.98% sequence identity) to pKDO1 (GenBank acc. No. JX424423).
Table 2.
Antibiotic resistance genotype of MAR14-456.
| Gene | Location | Function | Affected Antibiotics |
|---|---|---|---|
| bla SHV-11 | chromosome | Kp intrinsic penicillinase | Penicillins |
| oqxB2 | chromosome | Efflux pump | Quinolones |
| oqxA | chromosome | Efflux pump | Quinolones |
| fosA | chromosome | Fosfomycin thiol transferase | Fosfomycin |
| gyrA (WT) | chromosome | WT quinolone-sensitive catalytic subunit A of DNA gyrase | Quinolones |
| parC (WT) | chromosome | WT quinolone-sensitive catalytic subunit A of DNA topoisomerase IV | Quinolones |
| ompK36 (N49S, L59V, L191S, F207W, A217S, N218H, D224E, L228V, E232R, T254S) | chromosome | Outer membrane porin | Cephalosporins, carbapenems |
| ompK37 (I70M, I128M) | chromosome | Outer membrane porin | Cephalosporins, carbapenems |
| bla OXA-48 | IncL/M pl. | Carbapenemase | Penicillins ± inhibitors, carbapenems |
| qnrB1 | IncFIIK pl. | Quinolone resistance protein | Quinolones |
| tet(A) | IncFIIK pl. | Tetracycline efflux pump | Tetracyclines |
| bla CTX-M-15 | IncFIIK pl. | ESBL | Penicillins, cephalosporins, aztreonam |
| dfrA14 | IncFIIK pl. | Dihydrofolate reductase | Trimethoprim |
| strA | IncFIIK pl. | Aminoglycoside phosphotransferase | Streptomycin |
| strB | IncFIIK pl. | Aminoglycoside phosphotransferase | Streptomycin |
| bla TEM-1b | IncFIIK pl. | Penicillinase | Penicillins |
| sul2 | IncFIIK pl. | Sulfonamide resistant dihydropteroate synthase | Sulfonamides |
| catB3::IS26 | IncFIIK pl. | Chloramphenicol acetyltransferase | Chloramphenicol |
| aac(6’)Ib-cr | IncFIIK pl. | Aminoglycoside and ciprofloxacin acetyltransferase | Aminoglycosides, ciprofloxacin |
| bla OXA-1 | IncFIIK pl. | Penicillinase | Penicillins ± inhibitors |
pl.—plasmid.
The MLST profile, capsular, lipopolysaccharide, and chromosomal virulence gene clusters (for type III fimbriae, microcin, colibactin, and yersiniabactin) of MAR14-456 were characteristic of hvKp ST23, serotype K1-O1v2. Other virulence gene clusters for aerobactin, iron uptake, heavy-metal ion resistance, and rpmA/rmpA2 regulator of mucoid phenotype were identified on a typical IncHI1B/FIB plasmid nearly identical to pSGH10 (GenBank acc. No. CP025081.1) (Table 3).
Table 3.
Virulence and heavy metal resistance-associated genes of MAR14-456.
| Gene Cluster—Function | Location |
|---|---|
| Allantoinase cluster—Metabolism of allantoin: allA(1), allB(1), allC(2), allD(1), allR(1), allS(1), arcC(1), fdrA(1), gcl(2), glxK(1), glxR(1), hyi(1), ybbW(1), ybbY(1), ylbE(1), ylbF(1), KP1 1364(1), KP1 1371(1) |
chromosome |
| Type III fimbrial gene cluster—Mannose-resistant Klebsiella-like (type III) fimbria production and biofilm formation: mrkA(1), mrkB(1), mrkC(1), mrkD(1), mrkF(1), mrkH(1), mrkI(2), mrkJ(1) |
chromosome |
| Microcin E492 cluster—Bacteriocin production: mceA(1), mceB(1), mceC(1), mceD(2), mceE(1), mceG(2), mceH(1), mceI(2), mceJ(2) |
chromosome |
| Colibactin cluster—Toxin production: clbA(2), clbB(2), clbC(2), clbD(2), clbE(2), clbF(2), clbG(2), clbH(3), clbI(2), clbJ(5 ins.), clbK(3), clbL(2), clbM(2), clbN(2), clbO(2), clbP(2), clbQ(2), clbR(2) |
chromosome |
| Yersiniabactin cluster—Iron acquisition system: ybtA(2), ybtE(2), ybtP(2), ybtQ(2), ybtS(2), ybtU(2), ybtX(2), fyuA(2), irp1(6) |
chromosome |
| Iron uptake cluster kfuABC—Iron acquisition system: kfuA(1), kfuB(1), kfuC(1) |
chromosome |
| Aerobactin cluster—Iron acquisition system: iucA(1), iucB(1), iucC(1), iucD(1) |
IncHI1B/FIB plasmid |
| Iron uptake (salmochelin) cluster iroBCDN—Iron acquisition system: iroB(1), iroC(4), iroD(1), iroN(1) |
IncHI1B/FIB plasmid |
|
rmpA/rmpA2—Regulators of mucoid phenotype: rmpA(2), rmpA2(8 frameshift) |
IncHI1B/FIB plasmid |
|
pbrABCR cluster—Lead resistance: pbrA(4), pbrBC(1), pbrR(1) |
IncHI1B/FIB plasmid |
|
pcoABCDERS cluster—Copper resistance: pcoA(1), pcoB(1), pcoC(1), pcoD(1), pcoE(4), pcoR(1), pcoS(1) |
IncHI1B/FIB plasmid |
|
silCERS cluster—Silver resistance: silC(1), silE(1), silR(1), silS(1) |
IncHI1B/FIB plasmid |
|
terABCDEWXYZ cluster—Tellurite resistance: terA(2), terB(2), terC(2), terD(2), terE(2), terW(2), terX(2), terY(2), terZ(2) |
IncHI1B/FIB plasmid |
Allele variants of the genes are shown in parentheses according to BIGSdb-Kp database nomenclature (https://bigsdb.pasteur.fr).
In addition, a type I-E* CRISPR-Cas system was identified in MAR14-456 chromosome (start = 2,570,102; end = 2,578,395) that included intact and apparently functional cas3, csr1, cse2, cas6, cas7, cas5, cas1, and cas2 genes, and was flanked by two CRISPR arrays of 6 and 28 spacers, respectively. No genes of anti-CRISPR proteins were found.
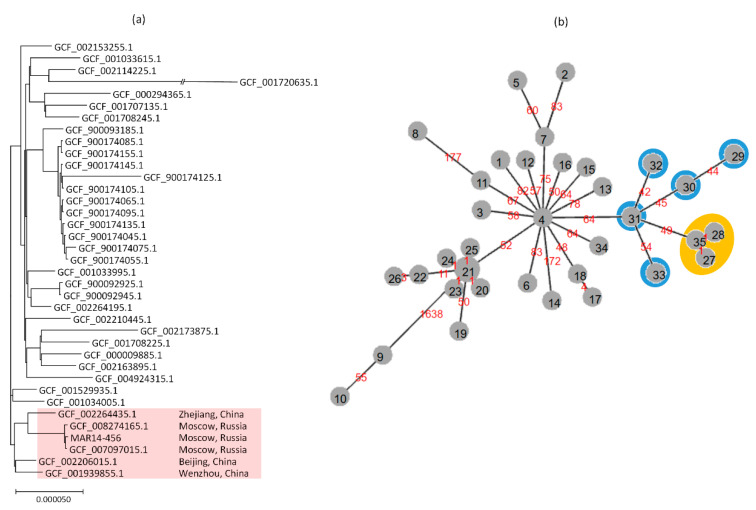
The cgSNP analysis confirmed the close relatedness of MAR14-456 and the ST23 isolates from the same hospital in Moscow whose draft genome sequences were reported recently [13]. The core genomes of MAR14-456 differed from those of the isolates KPB3188 (SCPM-O-B-7883, GenBank assembly acc. No. GCF_007097015.1) and KPB470 (SCPM-O-B-8739, GenBank assembly acc. No. GCF_008274165.1) at only eight SNPs each. The maximal likelihood tree for the cgSNP analysis is shown in Figure 1a. The Moscow isolates formed a distinct cluster and were most closely related to the Chinese ST23 isolates that lack blaOXA-48. There were from 106 to 151 core SNP differences between the MAR14-456 and the closest isolates from Zhejiang (GenBank assembly acc. No. GCF_002264435.1), Beijing (GenBank assembly acc. No. GCF_002206015.1), and Wenzhou (GenBank assembly acc. No. GCF_001939855.1).
Figure 1.
(a) Maximum likelihood tree for cgSNP analysis of ST23 K. pneumoniae isolates (b) Minimum-spanning tree (MST) of cgMLST allelic profiles of ST23 isolates. The node numbers on MST indicate distinct profiles; the branch labels indicate the number of differing alleles. The nodes outlined in orange correspond to Russian isolates (35: MAR14-456; 27: GCF_007097015.1; 28: GCF_008274165.1), the nodes outlined in blue correspond to Chinese isolates (29: GCF_001034005.1; 30: GCF_001529935.1; 31: GCF_001939855.1; 32: GCF_002206015.1; 33: GCF_002264435.1).
These findings were also supported by results of cgMLST analysis and minimum-spanning tree (MST) clustering. Out of the 1995 cgMLST loci that were present in all analyzed genomes of ST23 isolates, the Moscow isolates differed from each other at only one locus and diverged from ‘ancestral’ Chinese cluster at 49 to 70 loci (Figure 1b). cgMLST profiles of MAR14-456 and other related isolates are presented in Table S1.
3. Discussion
Our study details the phenotypic and genomic characteristics of an unusual multidrug-resistant (MDR) hvKp strain of ST23 that caused a nosocomial outbreak in Moscow, Russia. Most notably, the strain was found to carry the plasmids encoding CTX-M-15 ESBL and OXA-48 carbapenemase. The expression of the former resulted in high-level resistance to cephalosporins and aztreonam, while the expression of the latter conferred resistance to ertapenem and decreased susceptibility to meropenem and imipenem with the corresponding MICs exceeding the ECOFFs, but falling below the EUCAST clinical breakpoints. This phenotype was consistent with previous reports of low-level resistance conferred by OXA-48 [14]. Most worrying, however, is the fact that, despite in vitro “susceptibility”, the combination therapy with high-dose (8 g/day CI) meropenem, amikacin, and tigecycline, which was considered the most suitable treatment in the absence of ceftazidime-avibactam, has failed to eradicate the infection caused by OXA-48 producing hvKp. While the severe patient’s underlying condition and the strain’s high virulence could have also attributed to the negative treatment outcome, our report adds to the published evidence that the production of OXA-48 in K. pneumoniae is associated with a high risk of failure with carbapenems [15,16,17].
The combined use of short- and long-read genome sequencing allowed us to fully assemble the plasmid sequences of MAR14-456. The isolate harbored a large IncHI1B/FIB virulence plasmid, typical for ST23 [18], but also the two conjugative resistance plasmids: one, IncFIIK, carrying blaCTX-M-15 along with several other genes for penicillin, fluoroquinolone, aminoglycoside, tetracycline, trimethoprim, and sulfonamide resistance, and the other, IncL/M, carrying blaOXA-48. The latter plasmid was identical to other published pOXA-48 plasmids that were known to be the major vehicle for the dissemination of blaOXA-48 among different K. pneumoniae lineages in Europe and beyond [19]. Yet, to the best of our knowledge, the acquisition of pOXA-48 by ST23 hvKp was not described outside Russia.
Interestingly, an apparently functional type I-E* CRISPR-Cas system with two flanking CRISPR arrays of 6 and 28 spacers was identified in MAR14-456, and no genes for anti-CRISPR proteins were detected. The type I-E* CRISPR system was previously suggested to efficiently limit the acquisition of antibiotic resistance genes and plasmids in ST23 strains [20]. Nevertheless, our report, along with other recent findings, provide compelling evidence for the possibility of acquisition of common MDR plasmids by the ST23 strain despite the presence of the CRISPR-Cas system.
The cgSNP- and cgMLST-based phylogenetic analysis of MAR14-456 and related isolates from Moscow revealed their closest similarity to ST23 strains from China, all of which lack blaOXA-48 gene. We, therefore, propose that the most likely scenario of the emergence of MDR strain that caused an outbreak in Moscow involved an international transmission of ST23 hvKp followed by acquisition of pOXA-48-like plasmid.
In conclusion, our report highlights the threat of acquisition of multiple resistance by hvKp strain and its spread as a nosocomial pathogen.
4. Materials and Methods
4.1. Antimicrobial Susceptibility Testing (AST) and Phenotypic Detection of ESBL and Carbapenemase Production
Minimum inhibitory concentrations (MICs) of a wide range of antibiotics were determined using a reference broth microdilution method according to ISO 20776-1:2019 (https://www.iso.org/standard/70464.html). The AST to fosfomycin was performed by agar dilution method as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST). The results were interpreted according to EUCAST clinical breakpoints v.10.0 (http://www.eucast.org) and epidemiological cut-off (ECOFF) values (https://www.eucast.org/mic_distributions_and_ecoffs/). Escherichia coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as control strains. ESBL production was inferred from comparison of MICs of oxyimino-cephalosporins (cefotaxime, ceftazidime, and cefepime) alone and in combination with clavulanic acid (4 mg/L). Carbapenemase activity was detected by CIM test [21].
4.2. Whole Genome Sequencing (WGS)
The genomic DNA was isolated with a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) and used for the paired-end library preparation with a Nextera DNA Sample Prep Kit (Illumina, San Diego, CA, USA). Short-read WGS was performed on a MiSeq platform (Illumina, San Diego, CA, USA). Long-read sequencing was performed using a MinION sequencing system (Oxford Nanopore Technologies, Oxford, UK) according to the manufacturer’s protocols. Base calling of the raw MinION data was performed with Guppy Version 3.4.4 (Oxford Nanopore Technologies, Oxford, UK). Long and short sequencing reads were subjected to hybrid assembly using Unicycler version 0.4.8-beta [22]. Additional processing was performed by custom scripts, as described earlier [23].
4.3. Analysis of WGS Data
The antimicrobial resistance-associated genes and mutations were identified using AMRFinder [24], ResFinder [25] , PointFinder [26], and CARD databases [27]; MLST and virulence genes—using the Institute Pasteur BIGSdb software (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Capsular polysaccharide serotype was determined in silico using Kaptive [28]. CRISPR/Cas clusters were detected using CRISPRCasFinder [29].
Core-genome (cg) SNP phylogeny was constructed for MAR14-456 and published K. pneumoniae ST23 genomes using Prokka [30], Roary [31], and RAxML [32] software. cgMLST analysis was performed using MentaLiST [33], and the minimum spanning tree (MST) was constructed using PHYLOViZ 2.0 [34].
The complete genome sequence of MAR14-456 was submitted to GenBank under PRJNA667838, acc. no CP063277-CP063280.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/12/862/s1, Table S1. cgMLST profiles of MAR14-456 and other related K. pneumoniae isolates.
Author Contributions
I.A. isolated and performed initial characterization of MAR14-456; O.E. contributed clinical and epidemiological information; A.S., Y.Y., Y.M., and D.S. performed sequencing experiments, genome assembly and phylogenetic analysis; E.S. and M.E. performed reference susceptibility testing, annotation of antibiotic resistance and virulence genes and plasmids; V.A. and R.K. supervised the study; V.A. acquired funding; M.E., E.S., and A.S. wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Science and Higher Education of the Russian Federation within the framework of a grant in the form of a subsidy for the creation and development of the «World-class Genomic Research Center for Ensuring Biological Safety and Technological Independence under the Federal Scientific and Technical Program for the Development of Genetic Technologies», agreement No. 075-15-2019-1666.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boucher H.W., Talbot G.H., Bradley J.S., Edwards J.E., Gilbert D., Rice L.B., Scheld M., Spellberg B., Bartlett J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.WHO . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 3.CDC . Antibiotic Resistance Threats in the United States. CDC; Atlanta, GA, USA: 2019. [Google Scholar]
- 4.Bialek-Davenet S., Criscuolo A., Ailloud F., Passet V., Jones L., Delannoy-Vieillard A.S., Garin B., Le Hello S., Arlet G., Nicolas-Chanoine M.H., et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shon A.S., Bajwa R.P., Russo T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: A new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siu L.K., Yeh K.-M., Lin J.-C., Fung C.-P., Chang F.-Y. Klebsiella pneumoniae liver abscess: A new invasive syndrome. Lancet Infect. Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 7.Lam M.M.C., Wyres K.L., Duchene S., Wick R.R., Judd L.M., Gan Y.H., Hoh C.H., Archuleta S., Molton J.S., Kalimuddin S., et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roulston K.J., Bharucha T., Turton J.F., Hopkins K.L., Mack D.J.F. A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep. 2018;5:e005130. doi: 10.1099/jmmcr.0.005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson M., Stanton R.A., Ansari U., McAllister G., Chan M.Y., Sula E., Grass J.E., Duffy N., Anacker M.L., Witwer M.L., et al. Identification of a Carbapenemase-Producing Hypervirulent Klebsiella pneumoniae Isolate in the United States. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00519-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cejas D., Fernandez Canigia L., Rincon Cruz G., Elena A.X., Maldonado I., Gutkind G.O., Radice M.A. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J. Clin. Microbiol. 2014;52:3483–3485. doi: 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B.T., Su W.Q. Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. J. Med. Microbiol. 2019;68:866–873. doi: 10.1099/jmm.0.000996. [DOI] [PubMed] [Google Scholar]
- 12.Lev A.I., Astashkin E.I., Kislichkina A.A., Solovieva E.V., Kombarova T.I., Korobova O.V., Ershova O.N., Alexandrova I.A., Malikov V.E., Bogun A.G., et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog. Glob. Health. 2018;112:142–151. doi: 10.1080/20477724.2018.1460949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volozhantsev N.V., Kislichkina A.A., Mukhina T.N., Fursova N.K. Draft Genome Sequences of Clinical K1-Type Klebsiella pneumoniae Strains Isolated in Russia. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/MRA.01250-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore D.M., Nicolau D.P., Hopkins K.L., Meunier D. Carbapenem-Resistant Enterobacterales, Carbapenem Resistant Organisms, Carbapenemase-Producing Enterobacterales, and Carbapenemase-Producing Organisms: Terminology Past its “Sell-By Date” in an Era of New Antibiotics and Regional Carbapenemase Epidemiology. Clin. Infect. Dis. 2020;71:1776–1782. doi: 10.1093/cid/ciaa122. [DOI] [PubMed] [Google Scholar]
- 15.Cuzon G., Ouanich J., Gondret R., Naas T., Nordmann P. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob. Agents Chemother. 2011;55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro-San Francisco C., Mora-Rillo M., Romero-Gomez M.P., Moreno-Ramos F., Rico-Nieto A., Ruiz-Carrascoso G., Gomez-Gil R., Arribas-Lopez J.R., Mingorance J., Pano-Pardo J.R. Bacteraemia due to OXA-48-carbapenemase-producing Enterobacteriaceae: A major clinical challenge. Clin. Microbiol. Infect. 2013;19:E72–E79. doi: 10.1111/1469-0691.12091. [DOI] [PubMed] [Google Scholar]
- 17.Balkan I.I., Aygun G., Aydin S., Mutcali S.I., Kara Z., Kuskucu M., Midilli K., Semen V., Aras S., Yemisen M., et al. Blood stream infections due to OXA-48-like carbapenemase-producing Enterobacteriaceae: Treatment and survival. Int. J. Infect. Dis. 2014;26:51–56. doi: 10.1016/j.ijid.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Wu K.M., Li L.H., Yan J.J., Tsao N., Liao T.L., Tsai H.C., Fung C.P., Chen H.J., Liu Y.M., Wang J.T., et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J. Bacteriol. 2009;191:4492–4501. doi: 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David S., Cohen V., Reuter S., Sheppard A.E., Giani T., Parkhill J., European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. ESCMID Study Group for Epidemiological Markers (ESGEM) Rossolini G.M., Feil E.J., et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA. 2020;117:25043–25054. doi: 10.1073/pnas.2003407117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J., Lv L., Wang X., Xiu Z., Chen G. Comparative analysis of CRISPR-Cas systems in Klebsiella genomes. J. Basic Microbiol. 2017;57:325–336. doi: 10.1002/jobm.201600589. [DOI] [PubMed] [Google Scholar]
- 21.Van der Zwaluw K., de Haan A., Pluister G.N., Bootsma H.J., de Neeling A.J., Schouls L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE. 2015;10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shelenkov A., Mikhaylova Y., Yanushevich Y., Samoilov A., Petrova L., Fomina V., Gusarov V., Zamyatin M., Shagin D., Akimkin V. Molecular Typing, Characterization of Antimicrobial Resistance, Virulence Profiling and Analysis of Whole-Genome Sequence of Clinical Klebsiella pneumoniae Isolates. Antibiotics. 2020;9:261. doi: 10.3390/antibiotics9050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldgarden M., Brover V., Haft D.H., Prasad A.B., Slotta D.J., Tolstoy I., Tyson G.H., Zhao S., Hsu C.H., McDermott P.F., et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zankari E., Allesoe R., Joensen K.G., Cavaco L.M., Lund O., Aarestrup F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcock B.P., Raphenya A.R., Lau T.T.Y., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.V., Cheng A.A., Liu S., et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wick R.R., Heinz E., Holt K.E., Wyres K.L. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Neron B., Rocha E.P.C., Vergnaud G., Gautheret D., Pourcel C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018;46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 31.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 33.Feijao P., Yao H.T., Fornika D., Gardy J., Hsiao W., Chauve C., Chindelevitch L. MentaLiST—A fast MLST caller for large MLST schemes. Microb. Genom. 2018;4 doi: 10.1099/mgen.0.000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nascimento M., Sousa A., Ramirez M., Francisco A.P., Carrico J.A., Vaz C. PHYLOViZ 2.0: Providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.