Abstract
Gibberellins (GAs) are a major class of plant hormones that regulates diverse developmental programs. Both acquiring abilities to synthesize GAs and evolving divergent GA receptors have been demonstrated to play critical roles in the evolution of land plants. In contrast, little is understood regarding the role of GA‐inactivating mechanisms in plant evolution. Here we report on the origin and evolution of GA methyltransferases (GAMTs), enzymes that deactivate GAs by converting bioactive GAs to inactive GA methylesters. Prior to this study, GAMT genes, which belong to the SABATH family, were known only from Arabidopsis. Through systematic searches for SABATH genes in the genomes of 260 sequenced land plants and phylogenetic analyses, we have identified a putative GAMT clade specific to seed plants. We have further demonstrated that both gymnosperm and angiosperm representatives of this clade encode active methyltransferases for GA methylation, indicating that they are functional orthologs of GAMT. In seven selected seed plants, GAMT genes were mainly expressed in flowers and/or seeds, indicating a conserved biological role in reproduction. GAMT genes are represented by a single copy in most species, if present, but multiple copies mainly produced by whole genome duplications have been retained in Brassicaceae. Surprisingly, more than 2/3 of the 248 flowering plants examined here lack GAMT genes, including all species of Poales (e.g., grasses), Fabales (legumes), and the large Superasterid clade of eudicots. With these observations, we discuss the significance of GAMT origination, functional conservation and diversification, and frequent loss during the evolution of flowering plants.
Keywords: gene loss, plant hormone, reproduction, seed plants
1. INTRODUCTION
Gibberellins (GAs) are a class of diterpenoid plant hormones that has played an important role in land plant evolution (Zi et al., 2014). Bryophytes, the basal lineage of land plants, do not contain GAs, but some of them have been demonstrated to use GA precursors to regulate development (Hirano et al., 2007; Yasumura et al., 2007). All vascular plants, including lycophytes, ferns, gymnosperms, and angiosperms, synthesize GAs as an essential plant hormone (MacMillan, 2001). In addition to their conserved roles in regulating some fundamental development programs such as stem elongation and leaf expansion (Sun, 2008), GAs have acquired lineage‐specific functions among vascular plants. In seed plants (gymnosperms and angiosperms), GAs promote seed germination (Urbanova & Leubner‐Metzger, 2016). In angiosperms, GAs regulate flowering (Blazquez et al., 1998). Such lineage/developmental program‐specific functions of GAs may have played an important role in the diversification of vascular plants and their adaptations. Thus, it is of fundamental interest to ask how GAs achieve such lineage/developmental program‐specific functions.
For biosynthesis of GAs, three types of genes are involved: terpene synthases, cytochrome P450 monooxygenases, and 2‐oxoglutarate‐dependent dioxygenases (Yamaguchi, 2008). The inability to synthesize GAs by the moss Physcomitrella patens has been partly attributed to the lack of one key P450 gene of the CYP88 family (Rensing et al., 2008). Therefore, evolving the complete set of the three types of genes is essential to enable GA biosynthesis in vascular plants. Recent studies have shown the importance of evolution of GA perception in defining specific functions of GAs. GID1, the receptor of GAs, evolved from carboxylesterase in ancestral vascular plants after the split from the bryophyte lineage (Ueguchi‐Tanaka et al., 2007; Yoshida et al., 2018). The lycophyte GID1s have been termed initial GID1s because of their inferior affinity toward bioactive GAs than those of GID1s in seed plants. The fern GID1s have been called adapted GID1s, which exhibit improved adjustments for binding different GAs. The seed plant GID1s have been diversified. For instance, nearly all eudicots contain two types of GID1, named A‐ and B‐type, with the latter type associated with organ‐specific functions (Griffiths et al., 2006; Yoshida et al., 2018). Besides biosynthesis and perception, inactivation of GAs also plays a role in regulating GA activities (Hedden & Phillips, 2000; Olszewski et al., 2002), for which multiple mechanisms are known to exist. These include 2β‐hydroxylation catalyzed by GA 2‐oxidases (Thomas et al., 1999), conjugation to form glucosyl esters and glucosides (Schneider et al., 1992), epoxidation catalyzed by a cytochrome P450 monooxygenase (Zhu et al., 2006) and methylation of the carboxyl group catalyzed by GA methyltransferase (GAMT) to form GA methylesters (Varbanova et al., 2007). Little is understood on the role of GA inactivation in plant evolution.
GAMT‐catalyzed deactivation of GAs is the most recently discovered mechanism of GA inactivation (Varbanova et al., 2007). The model plant Arabidopsis contain two GAMT genes designated AtGAMT1 and AtGAMT2. Both AtGAMT1 and AtGAMT2 showed the highest levels of expression during seed development (Varbanova et al., 2007). Using overexpression and knockout lines, the function of AtGAMTs in Arabidopsis was demonstrated to be deactivating bioactive GAs during seed development (Varbanova et al., 2007). Transgenic tobacco, petunia, and tomato plants overexpressing Arabidopsis GAMTs exhibit the phenotypes of GA deficit (Nir et al., 2014; Varbanova et al., 2007), supporting the role of GAMT in GA catabolism. GAMTs belong to the methyltransferase family called SABATH (D'Auria et al., 2003). Other known members of the SABATH family that methylate phytohormones include indole‐3‐acetic acid methyltransferase (IAMT) (Qin et al., 2005; Zhao et al., 2007), salicylic acid methyltransferase (SAMT) (Chen et al., 2003; Ross et al., 1999), and jasmonic acid methyltransferase (JAMT) (Seo et al., 2001). IAMT has been demonstrated to be ancient and conserved in seed plants (Zhao et al., 2008), while SAMT and JAMT appear to have arisen multiple times during the evolution of seed plants (Chaiprasongsuk et al., 2018). Despite discovery in Arabidopsis more than a decade ago (Varbanova et al., 2007), the origin, evolution, and function of GAMT genes in other plants is completely unknown. In this study, we use a comparative genomics approach to identify putative GAMT genes, and investigate their origin and evolution in the context of land plant evolution.
2. METHODS
2.1. Sequence retrieval and analysis
All protein models of the 260 sequenced plant genomes were downloaded from Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html), Brassica Database (http://brassicadb.org/brad/index.php), Citrus Genome Database (CGD, https://www.citrusgenomedb.org), Cucurbit Genomics Database (CuGenDB, http://cucurbitgenomics.org), Hardwood Genomics Project (HWG, https://www.hardwoodgenomics.org) or respective databases referred in literatures (Table S1). This dataset was searched for SABATH proteins by HMM search with E‐value of 1e‐5 against the HMM profile Methyltransf_7 (PF03492) (Finn et al., 2016). To identify and categorize GA2ox proteins, a method was applied based on two rounds of HMM searches (Johnson et al., 2010). An HMM‐based in‐house script was first used to identify proteins that contain both DIOX_N (PF14226) and 2OG‐FeII_Oxy (PF03171) conserved domains. Next, two HMM profiles, one for C19‐GA2ox (C19G) and the other for C20‐GA2ox (C20G), were made with specific conserved domains of GA2ox proteins from selected plant species (Table S4) as previously reported (Huang et al., 2015). Lastly, individual GA2ox proteins were separated into the C19‐GA2ox group and the C20‐GA2ox group by being subjected to HMM search against C19G and C20G HMM profiles with an E‐value of 1e‐5.
2.2. Phylogenetic reconstruction
All newly identified SABATH methyltransferases with a minimum length of 250 amino acids were used for phylogenetic reconstruction. Multiple protein sequence alignments were made with MAFFT version 7.369b under L‐INS‐I strategy (Katoh & Standley, 2013). The phylogenetic tree was generated by RAxML v8.2 using the LG + G+F model with 1,000 bootstraps (Stamatakis, 2014). The phylogenetic tree based on plant taxonomy was constructed using phyloT (https://phylot.biobyte.de). All phylogenetic trees were visualized by iTOL (Letunic & Bork, 2016).
2.3. Gene cloning, protein expression, and enzyme assays
Full‐length cDNAs for two GAMT genes from Ginkgo biloba, three GAMT genes from Brassica rapa and 11 SABATH genes from Brachypodium distichton were cloned from respective plant tissues by RT‐PCR with primers(Tabel S5) as previously described (Zhao et al., 2008). Putative full‐length cDNAs for all other GAMT or SABATH genes analyzed in this study were synthesized. All cDNAs were cloned into pET‐32a vector (MilliporeSigma) and confirmed by sequencing. Proteins were expressed in the Escherichia coli strain BL21 (DE3) (Stratagene) then tested for methyltransferase activities using radiochemical assays. Each assay was performed with a 50 μL volume containing 50 mM Tris‐HCl, pH 8.0, 1mM substrates, 3 μL 14C‐S‐adenosyl‐L‐methionine (SAM) (PerkinElmer), and 1 μL purified enzyme. After incubation at 30°C for 30 min, the assays were extracted with 150 μL ethyl acetate. The organic phase was counted in a scintillation counter (Beckman Coulter) to measure the relative methyltransferase activity.
2.4. Gene expression data retrieval
The gene expression data for Ginkgo biloba were retrieved from http://gigadb.org/dataset/100209 (Guan et al., 2016). The gene expression data for Camelina sativa and Vitis vinifera were analyzed through http://bar.utoronto.ca/ (Fucile et al., 2011). The gene expression data for Picea abies were retrieved from http://congenie.org (Sundell et al., 2015). The gene expression data for Phalaenopsis equestris were retrieved from http://orchidstra2.abrc.sinica.edu.tw (Chao et al., 2017). The gene expression data for Musa acuminata were retrieved from https://banana‐genome‐hub.southgreen.fr (Droc et al., 2013). The gene expression data for Citrus sinensis were retrieved from http://citrus.hzau.edu.cn (Wang et al., 2014). Read counts, fragments per kilobase million (FPKM) values, reads per kilobase million (RPKM) values or relative expression values were acquired via gene id search or blast search with putative GAMTs of that species in each database. Tissue specific expression data were later entered into tables, standardized to relative expression values by dividing highest expression value in each group and applied to drawing histograms in Excel, respectively. Standard deviations were marked if such information is available from that database.
3. RESULTS AND DISCUSSION
3.1. Comparative analysis of the SABATH family in 260 sequenced land plants and the identification of a putative GAMT clade
We compiled a total of 260 land plants with sequenced genomes, including 248 species of angiosperms, six species of gymnosperms, two species of ferns, one species of lycophyte and three species of bryophytes, from various public sources (Table S1). Then, the complete proteome for each of the 260 sequenced land plants was downloaded to a local server and the entire dataset was searched for SABATH proteins. A total of 6,458 SABATH proteins was identified with an average of 25 proteins per plant genome. The sizes of the SABATH family ranged from 1 (Apostasia shenzhenica and Pogostemon cablin) to 115 (Triticum aestivum). Next, the SABATH proteins were subject to phylogenetic analysis. SABATHs from seed plants were placed into five groups (I to V) (Figure 1). Group I contains SABATHs from all major lineages of land plants bryophytes, lycophytes, ferns, gymnosperms and angiosperms. Arabidopsis GAMT1 and GAMT2, the only two known GAMTs, belong to group I. Group II is specific to seed plants. It is noteworthy that all IAMTs that have been functionally characterized, including those from the angiosperms Arabidopsis, rice and poplar and the gymnosperm spruce, belong to group II. Group III is specific to gymnosperms. Group IV contains SABATHs from both gymnosperms and angiosperms. In contrast, group V is specific to angiosperms. Within group I, the SABATHs from angiosperms including the two Arabidopsis GAMTs and a subset of the SABATHs from gymnosperms form a clade with strong bootstrap support (100%) (Figure 1). This was defined as the putative GAMT clade. The GAMT clade was clustered with the SABATHs from bryophytes, lycophytes, and ferns with poor bootstrap support (53%).
FIGURE 1.
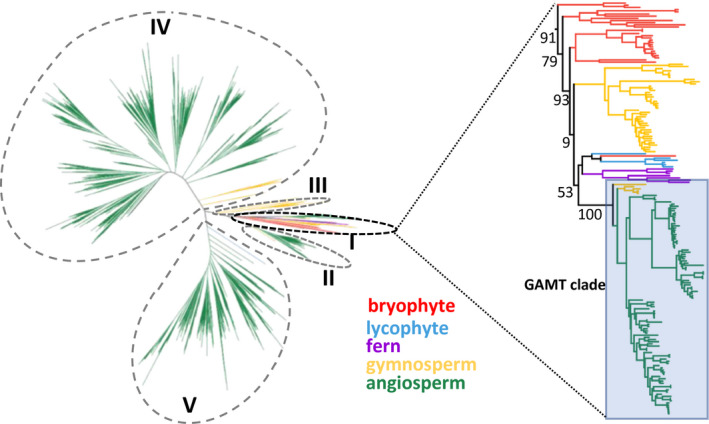
Phylogenetic analysis of SABATH proteins from 260 sequenced plants (Table S1). In this unrooted phylogenetic tree, the SABATHs were clustered into five groups I to V. Group I was enlarged to illustrate individual plant lineages with bootstrap values (percent out of 1,000 iterations) shown. The shaded clade indicates the putative GAMT clade
3.2. The catalytic activity of selected members in the GAMT clade
Within the putative GAMT clade, the phylogeny of the putative GAMTs (Figure 2a) is largely congruent to the species tree of seed plants established by APG IV (2016), implying that GAMT is conserved in seed plants. To determine whether any of the members in this putative GAMT clade besides the two Arabidopsis GAMTs encode enzymes with GAMT activity, we conducted biochemical analyses with representatives for methyltransferase activity via in vitro assays using gibberellin A1(GA1), gibberellin A3 (GA3), and gibberellin A4 (GA4) (Figure 2b), three of the most widely occurring bioactive GAs (MacMillan, 2001), as substrates. A total of 24 putative GAMTs from 20 species in the GAMT clade (Figure 2c) was selected for enzyme assays. A full‐length cDNA for each of the 24 GAMT genes was expressed in Escherichia coli and the recombinant protein tested for methyltransferase activity in in intro assays. Nineteen of the 24 proteins showed activity with GA4. Eight and eleven of the 19 active SABATHs also had catalytic activity with GA1 and GA3 as a substrate, respectively (Figure 2c). None of the 19 proteins with GAMT activity showed activity with IAA, JA, or SA as substrates, indicating that these GAMTs have strict substrate specificity towards GAs.
FIGURE 2.
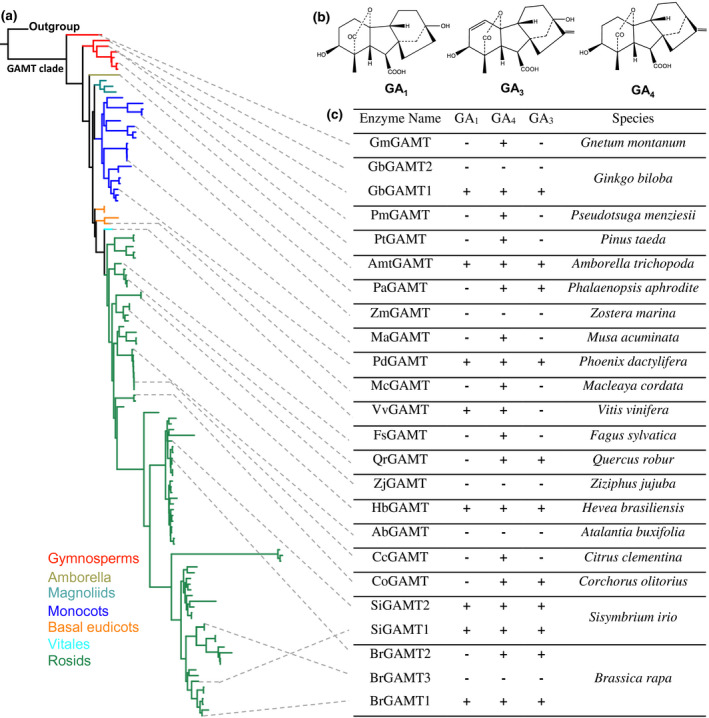
GAMT clade and biochemical activities. (a) Phylogeny of the GAMT clade with major lineages illustrated. (b) The chemical structures of gibberellin A1(GA1), gibberellin A3 (GA3), and gibberellin A4 (GA4). (c) Representative GAMTs and their activity towards to GA1, GA3, and GA4. “+” and “−” indicate “active” and “inactive,” respectively
3.3. Expressed patterns of GAMT genes in selected seed plants
To gain insight into the biological processes in which GAMT genes may be involved in seed plants we examined their expression patterns in seven species representing gymnosperms (Ginkgo biloba, Picea abies) and angiosperms, including monocots (Phalaenopsis equestris, Musa acuminate) and eudicots (Vitis vinifera, Citrus sinensis, and Camelina sativa) using public expression databases (Figure 3). In G. biloba, only one of two putative GAMTs showed activity with GAs and its bona fide GAMT gene expressed mainly in ovules (Figure 3a). The similar expression pattern was observed in another gymnosperm P. abies (Figure 3b) In P. equestris, GAMT was mainly expressed in the flower, especially in the labellum (Figure 3c). In M. acuminata, GAMT expression was observed in the fruit, with higher transcript levels detected during ripening (Figure 3d). In grapevine, its GAMT gene showed highest level of expression in senesced leaves. It also showed expression in young flowers, roots and pericarp (Figure 3e). In C. sinensis, GAMT was mainly expressed in flowers (Figure 3f). There are seven GAMT genes in the C. sativa genome; all copies were expressed mainly during reproductive growth, with five showing the highest level of expression in early or early‐mid stages of seed development; the other three gene copies showed the highest levels of expression in flowers (Figure 3g).
FIGURE 3.
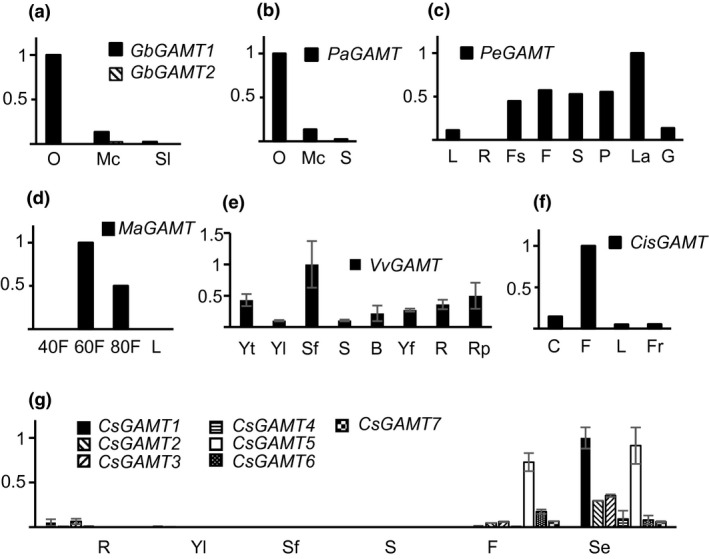
Expression patterns of GAMT genes in selected species based on their expression data in public sources. (a) GbGAMT1 and GbGAMT2 from Ginkgo biloba; (b) PaGAMT from Picea abies; (c) PeGAMT from Phalaenopsis equestris; (d) MaGAMT from Musa acuminata; (e) VvGAMT from Vitis vinifera; (f) CisGAMT from Citrus sinensis; (g) CsGAMT1 to CsGAMT7 from Camelina sativa. O, ovules; Mc, male cones; Sl, dtem and leaves; S, stem; L, leaf; R, root; Fs, floral stalk; F, flower; S, sepal; P, petal; La, labellum; G, gynostemium; 40F, 40‐day‐fruit; 60F, 60‐day‐fruit; 80F, 80‐day‐fruit; Yt, young tendril; Yl, young leaf; Sf, senescent leaf; B, bud; Yf, young flower; Rp, ripening pericarp; C, callus; Fr, fruit; Ro, rosette; Cl, cauline leaf; Se, seed. The highest level of expression in each species was arbitrarily set at 1.0. Standard deviations were marked with error bars in (e) and (g), not in other figures due to lack of such information
3.4. Retention after duplication of GAMT genes in Brassicaceae
Among the 69 flowering plants that contains GAMT genes, about a third (26 species) contains more than one copy of GAMT gene (Figure 4a). It is interesting to note most of the 26 species with two or more copies of GAMT belong to Brassicaceae. In fact, 18 of the 19 species of Brassicaceae, except Schrenkiella parvula, that were analyzed in this study contain two or more copies of GAMT genes (Figure 4b). Fourteen species, including Arabidopsis, contain two GAMT genes. In contrast, Brassica rapa, B. oleracea, B. napus, and Camelina sativa contain 4, 4, 9, and 7 GAMT genes, respectively. GAMTs of Brassicaceae forms two clades I and II (Figure 4b). Except S. parvula, all other 17 species contain GAMT in both clade I and clade II. This implies the duplication of GAMT in the common ancestor of Brassicaceae, most likely as an outcome of the whole genome duplication event that occurred in the common ancestor of Brassicaceae known as At‐α (Cardinal‐McTeague et al., 2016). This proposition is supported by the localization of AtGAMT1 (At4g26420) and AtGAMT2 (At5g56300) on two duplicated chromosomal segments. Within clade II, GAMTs from Brassica occurred in separate groups, which is likely due to a Brassica‐specific whole genome triplication event (Cheng et al., 2014). B. napus is a recent allopolyploid obtained by a cross between B. oleracea and B. rapa (Chalhoub et al., 2014). Consistent with this evolutionary history, each orthologus pair of GAMTs has one copy in B. oleracea, one copy in B. rapa and two copies in B. napus (Figure 4b). Notably, one of the two groups of Brassica GAMTs in clade II (Brassica 2) contains GAMTs from B. oleracea, B. rapa, and B. napus all as tandem repeats (Figure 4b), indicating that tandem duplication contributed to the expansion of the GAMT family in Brassica, although not to other Brassicaceae. Similarly, the three‐GAMT clusters of C. sativa within clade I and clade II were also likely an outcome of whole genome triplication (Kagale et al., 2014). Genome duplication is common in land plant evolution (Panchy et al., 2016; Qiao et al., 2019); it is one important mechanism leading to gene duplication and functional divergence. The doubling or further amplification of GAMT genes in Brassicaceae suggests that some of the Brassicaceae GAMT genes may have acquired specific specificity towards different GAs, as demonstrated for the two GAMTs in Arabidopsis (Varbanova et al., 2007) and the two active GAMTs in B. rapa (Figure 2c).
FIGURE 4.
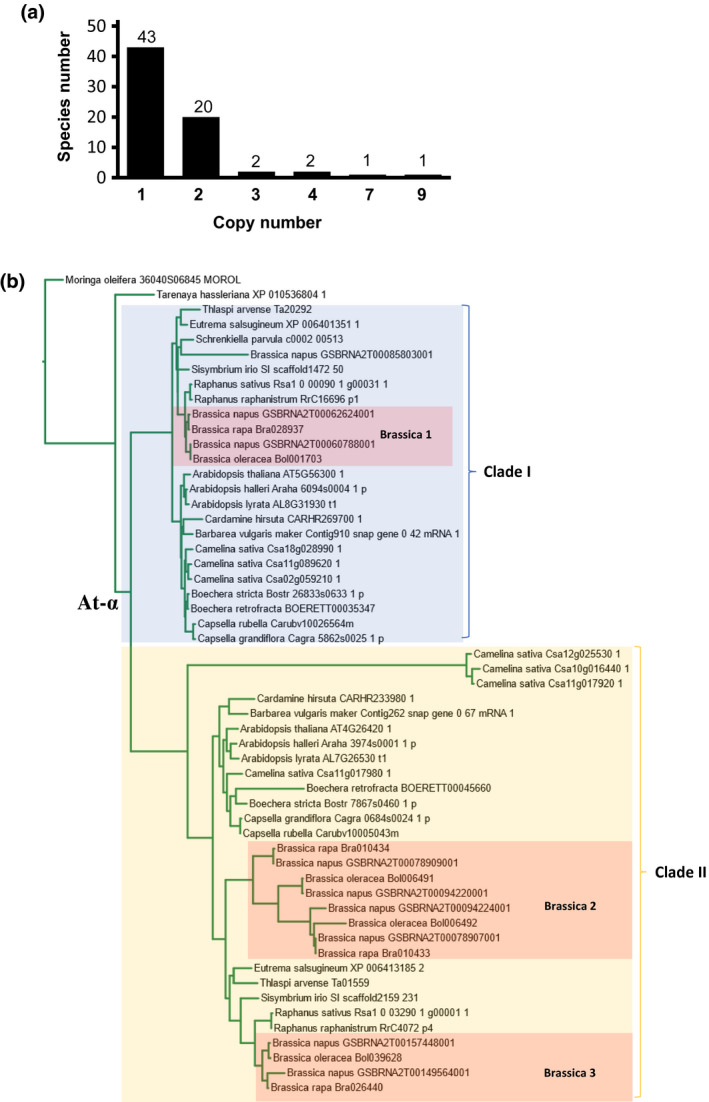
Copy number of GAMT and its duplication in Brassicaceae. (a) Distribution of copy numbers of GAMT genes among 69 GAMT‐containing flowering plants. (b) Phylogenetic tree of GAMTs in Brassicales. The two clades (clade I and clade II) in blue and in yellow, respectively, depict two clades that resulted from a whole gene duplication event occurred in the ancestor of Brassicaceae known as At‐α. The three smaller blocks (Brassica 1, Brassica 2, and Brassica 3) for Brassica GAMTs indicate a possible outcome of whole genome triplication event
3.5. The apparent ortholog of GAMT gene is absent in about 2/3 of the 248 flowering plants
When the GAMT genes in the GAMT clade were mapped to individual species, all six species of gymnosperms contain GAMT genes. In contrast, only 69 out of the 248 flowering plants contain GAMT genes (Figure 5), including some species‐rich lineages. No GAMT gene was identified in any of the 41 species in the order Poales. Among eudicots, GAMT appeared to be absent in all 22 species examined in Fabales and all 62 species included here from Superasterids. The absence of GAMT genes in certain land plant species could be due to incomplete coverage and/or poor quality of genome sequencing. Nonetheless, their absence in certain angiosperm lineages with a large number of species having sequenced genomes (e.g., Poales, Fabales, and Superasterids) can be concluded with confidence; these absences from entire clades imply multiple independent losses during angiosperm evolution.
FIGURE 5.
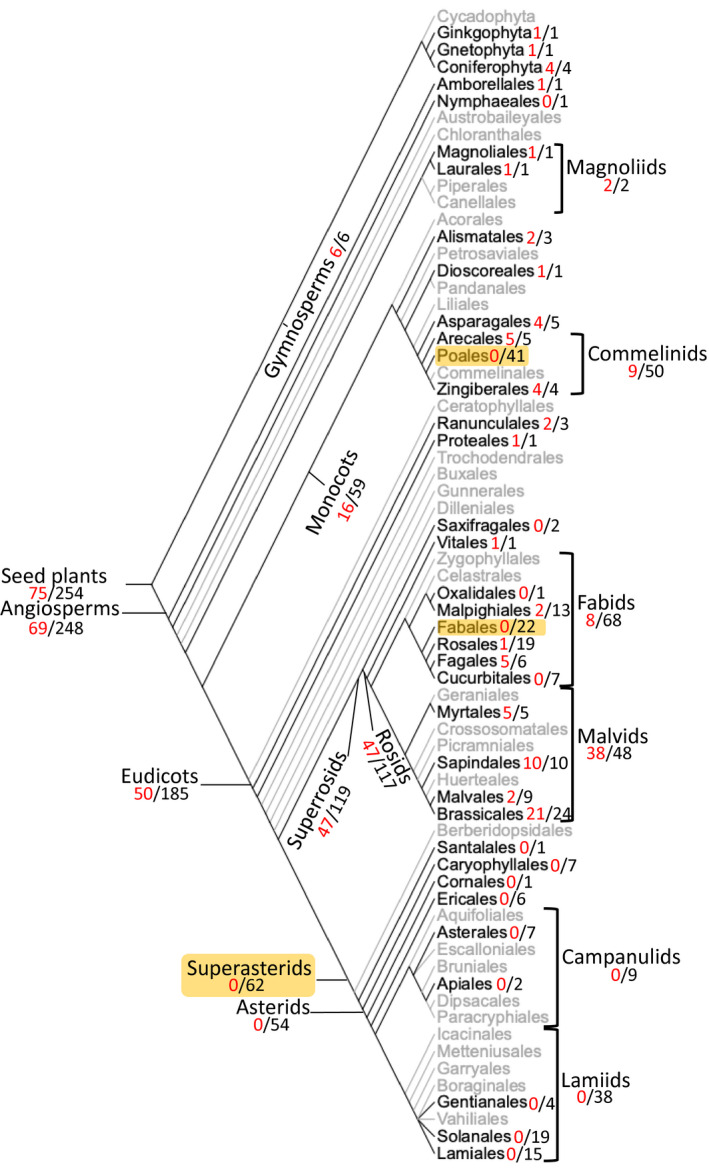
Presence/absence of GAMT genes in seed plants. The phylogeny was redrawn from APG IV (2016). The lineages in gray indicated that no species from those lineages was analyzed in this study. The two numbers (red and black) represent the number of species containing the GAMT gene and the total number of species from that specific lineage analyzed in this study. Three taxa with complete loss of GAMTs were shaded
As described earlier, there are several known mechanisms of GA inactivation, with GA 2β‐hydroxylation catalyzed by GA 2‐oxidases (GA2ox) considered the most important mechanism (Thomas et al., 1999). There are two types of GA2ox: C19‐GA2ox using C19 GAs as substrates and C20‐GA2ox using C20 GAs as substrates. Arabidopsis and rice contain five and seven C19‐GA2ox genes, and three and four C20‐GA2ox genes, respectively (Huang et al., 2015). For comparison, we also analyzed the occurrence of GA2ox genes in other flowering plants in our dataset. Putative GA2ox genes were identified in all the flowering plants analyzed except Pogostemon cablin and Zostera muelleri (Table S2). It remains to be determined whether the absence of GA2ox genes in P. cablin and Z. muelleri is a fact or due to the poor assembling and/or annotation of their respective genome. Consistent with the observation in Arabidopsis and rice, most plants contain more putative C19‐GA2ox genes than C20‐GA2ox genes (Table S2). The presence of GA2ox gene in 246 plants out of the 248 flowering plants analyzed indicate its ubiquitous occurrence, a sharp contrast to the sporadic distribution of GAMT gene among flowering plants.
Given the absence of GAMT orthologs in ~ 70% of the plant species analyzed in this study, we asked whether GAMT catalytic activity may have been maintained by SABATHs from the non‐GAMT clades. To test this possibility, we chose Brachypodium distachyon, a monocot in Poales, as a model species. The B. distachyon genome contains 12 SABATH genes with 10 of them being intact (Table S3). Full‐length cDNAs for all 10 intact SABATH genes were expressed in E. coli, and their respective recombinant proteins tested with GA3 and GA4. None of the 10 SABATHs proteins had methylating activity with GA3 or GA4, supporting the loss of GAMT activity in GAMT‐absent plants.
4. CONCLUSIONS AND IMPLICATIONS
By analyzing SABATH genes from a wide spectrum of land plants ranging from basal lineages (liverwort, moss), non‐seed vascular plants (lycophyte and ferns) to gymnosperms and angiosperms, we identified a GAMT clade (Figure 1) that arose early in the evolution of seed plants. In vitro enzyme assays and gene expression analysis led to two observations. We found that the catalytic activity of GAMTs for GA‐methylation (Figure 2c) and their biological function in reproduction (Figure 3) are generally conserved. The second observation is that functional divergence has also occurred, evidenced by different substrate specificity with GA3 and GA4 (Figure 2c) and by tissue‐specific expression of GAMT in certain species (e.g., in the senesced leaves of grapevine) (Figure 3). Such properties of GAMTs as a GA‐inactivating mechanism may have contributed to achieving lineage/developmental program‐specific functions of GAs. Equally important is the finding that GAMT gene is absent in approximately 2/3 of the flowering plants analyzed in this study (Figure 5). The direct consequence for the loss of GAMT gene is the lack of ability to inactivate GAs through methylation. While genetic innovations through gene duplication have been an engine for speciation, lineage‐specific losses of genes have also occurred frequently during eukaryote evolution (Aravind et al., 2000), including plants (Cannell et al., 2020; Gu et al., 2016). Loss‐of‐function may accompany key evolutionary transitions. For example, floral scent, which evolved early in flowering plants, has experienced repeated independent losses due to the transitions in pollinator types or modes of pollination (Raguso, 2016). It will be of great importance to determine the significance of the repeated loss of GAMT gene in the radiation of some of the largest lineages of flowering plants, including Poales, Fabales, and Superasterids (Figure 5). Finally, it is noteworthy that many major crops (e.g., cereal grasses and legumes) do not contain GAMT genes. While the lack of a GAMT gene may be advantageous, for certain agronomic traits (such as bushy phenotype), GAMT could be a useful new molecular tool for the genetic improvement of some of these GAMT‐lacking crops.
5. ACCESSION NUMBERS
The sequences for the biochemically characterized GAMT reported in this paper have been deposited in the GenBank database (accession numbers: MW149492 ‐ MW149515).
Supporting information
Table S1‐S5
ACKNOWLEDGMENTS
Chi Zhang was partly supported by a scholarship from the China Scholarship Council. Minta Chaiprasongsuk was supported by a Royal Thai Government Scholarship. The authors thank Dr. Eran Pichersky for stimulating discussions and his critical reading of the manuscript. The authors also thank the plant genome research community for the genome sequences of diverse plant species that enabled this study.
Zhang C, Chaiprasongsuk M, Chanderbali AS, Fu J, Soltis DE, Chen F. Origin and evolution of a gibberellin‐deactivating enzyme GAMT. Plant Direct. 2020;4:1–10. 10.1002/pld3.287
REFERENCES
- Aravind, L. , Watanabe, H. , Lipman, D. J. , & Koonin, E. V. (2000). Lineage‐specific loss and divergence of functionally linked genes in eukaryotes. Proceedings of the National Academy of Sciences of the United States of America, 97, 11319–11324. 10.1073/pnas.200346997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez, M. A. , Green, R. , Nilsson, O. , Sussman, M. R. , & Weigel, D. (1998). Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. The Plant Cell, 10, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell, N. , Emms, D. M. , Hetherington, A. J. , MacKay, J. , Kelly, S. , Dolan, L. , & Sweetlove, L. J. (2020). Multiple metabolic innovations and losses are associated with major transitions in land plant evolution. Current Biology, 30, 1783–1800. 10.1016/j.cub.2020.02.086 [DOI] [PubMed] [Google Scholar]
- Cardinal‐McTeague, W. M. , Sytsma, K. J. , & Hall, J. C. (2016). Biogeography and diversification of Brassicales: A 103 million year tale. Molecular Phylogenetics and Evolution, 99, 204–224. 10.1016/j.ympev.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Chaiprasongsuk, M. , Zhang, C. , Qian, P. , Chen, X. , Li, G. , Trigiano, R. N. , Guo, H. , & Chen, F. (2018). Biochemical characterization in Norway spruce (Picea abies) of SABATH methyltransferases that methylate phytohormones. Phytochemistry, 149, 146–154. 10.1016/j.phytochem.2018.02.010 [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I. A. , Tang, H. , Wang, X. , Chiquet, J. , Belcram, H. , Tong, C. , Samans, B. , Correa, M. , Da Silva, C. , Just, J. , Falentin, C. , Koh, C. S. , Le Clainche, I. , Bernard, M. , Bento, P. , Noel, B. , … Wincker, P. (2014). Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- Chao, Y. T. , Yen, S. H. , Yeh, J. H. , Chen, W. C. , & Shih, M. C. (2017). Orchidstra 2.0‐A transcriptomics resource for the Orchid family. Plant and Cell Physiology, 58, e9. 10.1093/pcp/pcw220 [DOI] [PubMed] [Google Scholar]
- Chen, F. , D'Auria, J. C. , Tholl, D. , Ross, J. R. , Gershenzon, J. , Noel, J. P. , & Pichersky, E. (2003). An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant Journal, 36, 577–588. [DOI] [PubMed] [Google Scholar]
- Cheng, F. , Wu, J. , & Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Horticulture Research, 1, 14024 10.1038/hortres.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria, J. C. , Chen, F. , & Pichersky, E. (2003). Chapter eleven The SABATH family of MTS in Arabidopsis Thaliana and other plant species In Romeo J. T. (Ed.), Recent Advances in Phytochemistry, (Vol. 37, pp. 253–283). Elsevier. [Google Scholar]
- Droc, G. , Larivière, D. , Guignon, V. , Yahiaoui, N. , This, D. , Garsmeur, O. , Dereeper, A. , Hamelin, C. , Argout, X. , Dufayard, J.‐F. , Lengelle, J. , Baurens, F.‐C. , Cenci, A. , Pitollat, B. , D’Hont, A. , Ruiz, M. , Rouard, M. , & Bocs, S. (2013). The Banana Genome Hub. Database, 2013, bat035. 10.1093/database/bat035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R. D. , Coggill, P. , Eberhardt, R. Y. , Eddy, S. R. , Mistry, J. , Mitchell, A. L. , Potter, S. C. , Punta, M. , Qureshi, M. , Sangrador‐Vegas, A. , Salazar, G. A. , Tate, J. , & Bateman, A. (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research, 44, D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucile, G. , Di Biase, D. , Nahal, H. , La, G. , Khodabandeh, S. , Chen, Y. , Easley, K. , Christendat, D. , Kelley, L. , & Provart, N. J. (2011). ePlant and the 3D data display initiative: Integrative systems biology on the world wide web. PLoS One, 6, e15237 10.1371/journal.pone.0015237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, J. , Murase, K. , Rieu, I. , Zentella, R. , Zhang, Z. L. , Powers, S. J. , Gong, F. , Phillips, A. L. , Hedden, P. , Sun, T. P. , & Thomas, S. G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis . The Plant Cell, 18, 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. Z. , Xing, S. L. , & He, C. Y. (2016). Genome‐wide analysis indicates lineage‐specific gene loss during papilionoideae evolution. Genome Biology and Evolution, 8, 635–648. 10.1093/gbe/evw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, R. , Zhao, Y. , Zhang, H. , Fan, G. , Liu, X. , Zhou, W. , Shi, C. , Wang, J. , Liu, W. , Liang, X. , Fu, Y. , Ma, K. , Zhao, L. , Zhang, F. , Lu, Z. , Lee, S. M. , Xu, X. , Wang, J. , Yang, H. , … Chen, W. (2016). Draft genome of the living fossil Ginkgo biloba . Gigascience, 5, 49 10.1186/s13742-016-0154-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden, P. , & Phillips, A. L. (2000). Gibberellin metabolism: New insights revealed by the genes. Trends in Plant Science, 5, 523–530. 10.1016/S1360-1385(00)01790-8 [DOI] [PubMed] [Google Scholar]
- Hirano, K. , Nakajima, M. , Asano, K. , Nishiyama, T. , Sakakibara, H. , Kojima, M. , Katoh, E. , Xiang, H. , Tanahashi, T. , Hasebe, M. , Banks, J. A. , Ashikari, M. , Kitano, H. , Ueguchi‐Tanaka, M. , & Matsuoka, M. (2007). The GID1‐mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens . The Plant Cell, 19, 3058–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Wang, X. , Ge, S. , & Rao, G. Y. (2015). Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evolutionary Biology, 15, 207 10.1186/s12862-015-0490-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L. S. , Eddy, S. R. , & Portugaly, E. (2010). Hidden Markov model speed heuristic and itrative HMM search procedure. BMC Bioinformatics, 11, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale, S. , Koh, C. , Nixon, J. , Bollina, V. , Clarke, W. E. , Tuteja, R. , Spillane, C. , Robinson, S. J. , Links, M. G. , Clarke, C. , Higgins, E. E. , Huebert, T. , Sharpe, A. G. , & Parkin, I. A. (2014). The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Communications, 5, 3706 10.1038/ncomms4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I. , & Bork, P. (2016). Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research, 44, W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan, J. (2001). Occurrence of gibberellins in vascular plants, fungi, and bacteria. Journal of Plant Growth Regulation, 20, 387–442. 10.1007/s003440010038 [DOI] [PubMed] [Google Scholar]
- Nir, I. , Moshelion, M. , & Weiss, D. (2014). The Arabidopsis GIBBERELLIN METHYL TRANSFERASE 1 suppresses gibberellin activity, reduces whole‐plant transpiration and promotes drought tolerance in transgenic tomato. Plant, Cell & Environment, 37, 113–123. [DOI] [PubMed] [Google Scholar]
- Olszewski, N. , Sun, T. P. , & Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. The Plant Cell, 14(Suppl), S61–S80. 10.1105/tpc.010476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy, N. , Lehti‐Shiu, M. , & Shiu, S.‐H. (2016). Evolution of gene duplication in plants. Plant Physiology, 171, 2294–2316. 10.1104/pp.16.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, X. , Li, Q. , Yin, H. , Qi, K. , Li, L. , Wang, R. , Zhang, S. , & Paterson, A. H. (2019). Gene duplication and evolution in recurring polyploidization‐diploidization cycles in plants. Genome Biology, 20, 38 10.1186/s13059-019-1650-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, G. , Gu, H. , Zhao, Y. , Ma, Z. , Shi, G. , Yang, Y. , Pichersky, E. , Chen, H. , Liu, M. , Chen, Z. , & Qu, L. J. (2005). An indole‐3‐acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. The Plant Cell, 17, 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguso, R. A. (2016). More lessons from linalool: Insights gained from a ubiquitous floral volatile. Current Opinion in Plant Biology, 32, 31–36. 10.1016/j.pbi.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Rensing, S. A. , Lang, D. , Zimmer, A. D. , Terry, A. , Salamov, A. , Shapiro, H. , Nishiyama, T. , Perroud, P. F. , Lindquist, E. A. , Kamisugi, Y. , Tanahashi, T. , Sakakibara, K. , Fujita, T. , Oishi, K. , Shin‐I, T. , Kuroki, Y. , Toyoda, A. , Suzuki, Y. , Hashimoto, S. , … Boore, J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science, 319, 64–69. 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Ross, J. R. , Nam, K. H. , D'Auria, J. C. , & Pichersky, E. (1999). S‐Adenosyl‐L‐methionine:Salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Archives of Biochemistry and Biophysics, 367, 9–16. 10.1006/abbi.1999.1255 [DOI] [PubMed] [Google Scholar]
- Schneider, G. , Jensen, E. , Spray, C. R. , & Phinney, B. O. (1992). Hydrolysis and reconjugation of gibberellin A20 glucosyl ester by seedlings of Zea mays L. Proceedings of the National Academy of Sciences of the United States of America, 89, 8045–8048. 10.1073/pnas.89.17.8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. S. , Song, J. T. , Cheong, J. J. , Lee, Y. H. , Lee, Y. W. , Hwang, I. , Lee, J. S. , & Choi, Y. D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate‐regulated plant responses. Proceedings of the National Academy of Sciences of the United States of America, 98, 4788–4793. 10.1073/pnas.081557298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.‐P. (2008). Gibberellin metabolism, perception and signaling pathways in Arabidopsis. The Arabidopsis Book, 6, e0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell, D. , Mannapperuma, C. , Netotea, S. , Delhomme, N. , Lin, Y. C. , Sjodin, A. , Van de Peer, Y. , Jansson, S. , Hvidsten, T. R. , & Street, N. R. (2015). The plant genome integrative explorer resource: PlantGenIE.org. The New Phytologist, 208, 1149–1156. [DOI] [PubMed] [Google Scholar]
- Thomas, S. G. , Phillips, A. L. , & Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2‐ oxidases, multifunctional enzymes involved in gibberellin deactivation. Proceedings of the National Academy of Sciences of the United States of America, 96, 4698–4703. 10.1073/pnas.96.8.4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi‐Tanaka, M. , Nakajima, M. , Motoyuki, A. , & Matsuoka, M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annual Review of Plant Biology, 58, 183–198. 10.1146/annurev.arplant.58.032806.103830 [DOI] [PubMed] [Google Scholar]
- Urbanova, T. , & Leubner‐Metzger, G. (2016). Gibberellins and seed germination. Annual Plant Reviews, 49, 253–284. [Google Scholar]
- Varbanova, M. , Yamaguchi, S. , Yang, Y. , McKelvey, K. , Hanada, A. , Borochov, R. , Yu, F. , Jikumaru, Y. , Ross, J. , Cortes, D. , Ma, C. J. , Noel, J. P. , Mander, L. , Shulaev, V. , Kamiya, Y. , Rodermel, S. , Weiss, D. , & Pichersky, E. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. The Plant Cell, 19, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Chen, D. , Lei, Y. , Chang, J. W. , Hao, B. H. , Xing, F. , Li, S. , Xu, Q. , Deng, X. X. , & Chen, L. L. (2014). Citrus sinensis annotation project (CAP): A comprehensive database for sweet orange genome. PLoS One, 9, e87723 10.1371/journal.pone.0087723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annual Review of Plant Biology, 59, 225–251. 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yasumura, Y. , Crumpton‐Taylor, M. , Fuentes, S. , & Harberd, N. P. (2007). Step‐by‐step acquisition of the gibberellin‐DELLA growth‐regulatory mechanism during land‐plant evolution. Current Biology, 17, 1225–1230. 10.1016/j.cub.2007.06.037 [DOI] [PubMed] [Google Scholar]
- Yoshida, H. , Tanimoto, E. , Hirai, T. , Miyanoiri, Y. , Mitani, R. , Kawamura, M. , Takeda, M. , Takehara, S. , Hirano, K. O. , Kainosho, M. , Akagi, T. , Matsuoka, M. , & Ueguchi‐Tanaka, M. (2018). Evolution and diversification of the plant gibberellin receptor GID1. Proceedings of the National Academy of Sciences, 115(33), E7844–E7853. 10.1073/pnas.1806040115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, N. , Ferrer, J. L. , Ross, J. , Guan, J. , Yang, Y. , Pichersky, E. , Noel, J. P. , & Chen, F. (2008). Structural, biochemical, and phylogenetic analyses suggest that indole‐3‐acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiology, 146, 455–467. 10.1104/pp.107.110049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, N. , Guan, J. , Lin, H. , & Chen, F. (2007). Molecular cloning and biochemical characterization of indole‐3‐acetic acid methyltransferase from poplar. Phytochemistry, 68, 1537–1544. 10.1016/j.phytochem.2007.03.041 [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Nomura, T. , Xu, Y. , Zhang, Y. , Peng, Y. , Mao, B. , Hanada, A. , Zhou, H. , Wang, R. , Li, P. , Zhu, X. , Mander, L. N. , Kamiya, Y. , Yamaguchi, S. , & He, Z. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. The Plant Cell, 18, 442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi, J. , Mafu, S. , & Peters, R. J. (2014). To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annual Review of Plant Biology, 65, 259–286. 10.1146/annurev-arplant-050213-035705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny Group . (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV Botanical Journal of the Linnean Society, 181, (1–20). 10.1111/boj.12385. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S5


