Abstract
Purpose
The aim of this paper was to investigate H19 expression in gastric cancer (GC) and its effects on the biological behavior of gastric cancer cells (GCCs), and at exploring its potential mechanism.
Methods
H19 expression in the patients’ tissues and serum was detected, and the correlation of the expression with the patients’ pathological data and survival rate was analyzed. Overexpression or inhibitory vectors of H19, microRNA-138 (miR-138) and E2F2 were constructed and transfected into GCCs to observe their effects on the cells’ proliferation, invasion and apoptosis.
Results
H19 rose in GC and was higher in GC patients with a tumor size ≥5 cm, high stages (III+IV) and lymph node metastasis. High H19 expression was associated with the poorer survival rate of the patients, so serum H19 had a certain diagnostic value for GC. H19 knockdown could inhibit GCCs to proliferate and invade and induce their apoptosis. miR-138 can be used as the target gene of H19, and E2F2 can be negatively regulated by this miR, so miR-138 knockdown or E2F2 upregulation can weaken GCCs’ biological behavior changes that were caused by H19 knockdown.
Conclusion
H19 can be used as a biological indicator for diagnosing GC and predicting patients’ poor prognosis. Additionally, it promotes GCCs to proliferate and invade through miR-138/E2F2 axis.
Keywords: H19, miR-138, E2F2, gastric cancer
Introduction
As one of the most common cancers and the third leading cause of cancer-related death across the world,1 gastric cancer (GC) is currently treated based on the combination of surgical operation, radiotherapy and chemotherapy.2 Although progress has been made in its diagnosis and treatment, patients with the disease still have a poor prognosis due to the high metastasis and recurrence of GC.3 Therefore, it is urgent to know the potential molecular mechanism of GC development and progression and to find potential therapeutic targets for the disease.
There is more and more evidence that tumor patients experience many kinds of long-chain non-coding RNA (lncRNA) disorders, and that partial disordered lncRNAs are closely correlated with patients’ disease progression and prognosis.4,5 Over 200 nt long, lncRNAs are a series of RNA molecules without protein-coding ability, and they are involved in a variety of biological events via regulating chromatin and transcription as well as gene expression after transcription.6,7 Having found to be highly expressed in various tumors, lncRNA H19 (H19) is a member of lncRNAs and is considered as a tumor promoter. For instance, it plays a carcinogenic role in thyroid cancer through PI3K/AKT pathway,8 and promotes lung cancer cells to proliferate and invade by regulating markers for epithelial-mesenchymal transition.9 Additionally, it upregulates in GC and predicts the poor prognosis of patients, so downregulating its expression can inhibit gastric cancer cells (GCCs) to invade and migrate.10 However, the pathway H19 functions in GC are still unclear.
In this study, the expression and clinical value of H19 in GC was investigated. Its effects on the biological behavior of GCCs and its potential mechanism were also analyzed. The results may be helpful to further understand the pathogenesis of GC and to find potential therapeutic targets for it.
Materials and Methods
Sources of Clinical Samples
One hundred and twelve GC patients who underwent surgical treatment in the Department of Oncology of the First Affiliated Hospital of Nanchang University from April 2015 to January 2016 were enrolled. Their cancer tissues, normal adjacent tissues and blood were collected during the surgery. Further 112 healthy people undergoing physical examinations during the same period were recruited. Their blood was obtained and stored in a refrigerator (−70°C). Inclusion criteria for patients: Those aged >18 years; those confirmed with GC by pathological examinations; those who signed the informed consent form. Exclusion criteria for patients: Those with other tumors; those who had previously received treatment for cancers; those with incomplete clinical data. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University and strictly abided by the Declaration of Helsinki.
Cell Sources and Treatment
GCC lines (HGC-27, AGS, MKN-45) and normal gastric mucosal cells (GES-1) were purchased from ATCC cell bank, USA. The cells were placed in a RPMI-1640 medium (Gibco, USA) that contained 10% fetal bovine serum (PBS; Gibco, USA), and the medium was placed in an incubator (37°C, 5% CO2). Cell transfection: With pcDNA 3.1 plasmids considered as vectors, inhibitory or overexpression plasmids of H19, microRNA-138 (miR-138) and E2F2 as well as corresponding negative controls were established. Their transfection was performed using Lipofectamine™ 2000 kits (Invitrogen, USA). Six hours later, the transfected cells were transferred to a 10% PBS-containing medium for culture.
qRT-PCR
qRT-PCR was carried out to detect H19 and miR-138 expression, with TRIzol kits (Invitrogen, USA) for total RNA extraction and an ultraviolet spectrophotometer and agarose gel electrophoresis for detecting the purity, concentration and integrity of the RNA. Subsequently, qualified RNA (2 μg) was reversely transcribed into cDNA using reverse transcription kits (TaKaRa, Japan). Next, based on the instruction of SYBR_Premix ExTaq II kits (TaKaRa, Japan), PCR amplification was conducted. The system was 10 μL of SYBR Premix Ex Taq II (2X), 2 μL of cDNA, each 0.8 μL of upstream and downstream primers, and Sterile purified water that was added to supplement to 20 μL. Conditions for the amplification were pre-denaturation (95°C, 30 s), denaturation (95°C, 5 s), and annealing and extension (60°C, 30 s), for a total of 40 cycles. Data analysis was performed using 2−ΔΔct.11 GAPDH and U6 were internal reference genes of H19 and miR-138, respectively. The upstream and downstream primer sequences of H19 were 5ʹ-TGATGACGGGTGCAGGGGCTA-3ʹ and 5ʹ-TGATGTTCGCCCTGTCTGCACC-3ʹ. Those of GAPDH were 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ and 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ. Those of miR-138 were 5ʹ-CGAGAGCTGGTGTTGTGAAT-3ʹ and 5ʹ-GTGCAGGGTCCGAGGTAT-3ʹ. Those of U6 were 5ʹ-GTAGCGTCGTGAAGCGTTC-3ʹ and 5ʹ-GTGCAGGGTCCGAGGTAT-3ʹ.
Western Blotting (WB)
After culture, the cells were cleaved with RIPA lysis buffer (Thermo Fisher Scientific, USA), with BCA protein assay kits (Thermo Fisher Scientific, USA) for protein concentration determination. The loading quantity of sample (40 μg) was taken for 10% polyacrylamide gel electrophoresis (100 V) and PVDF membrane transfer. After sealed in 5% skimmed milk powder for 2 hours, the membrane was cleaned with TBST buffer solution for three times, and then added with E2F2 (1:1000) and β-catenin (1:1000) primary antibodies (Abcam, USA) for sealing all night at 4°C. After washed to remove the antibodies, it was added with goat anti-rabbit IgG antibody (1:1500; Sigma, USA), incubated at 37°C for 1 hour, and finally rinsed with PBS over 5 min for 3 times. After unnecessary liquid was removed, ECL (Millipore, USA) was used for luminescence and developing. The protein bands were scanned to analyze their gray values in Quantity One. Relative expression levels of the protein = the gray value of target protein band/the gray value of β-Actin protein band.
Dual-Luciferase Reporter Gene Assay (DLRGA)
H19-3ʹUTR wild type (Wt), H19-3ʹUTR mutant (Mut), E2F2-3ʹUTR Wt and E2F2-3ʹUTR Wt were, respectively, established, for respective co-transfection with miR-138-mimics or miR-NC into HEK293 cells. Luciferase activities were detected by DLRGA kits (Solarbio, Beijing).
Flow Cytometry for Cell Apoptosis
After digestion with 0.25% trypsin, the cells were added with 100 μL of buffer solution to prepare into a 1*106 cells/mL suspension. Next, the suspension was added with 10 μL of AnnexinV-FITC/PI (Yisheng Biotechnology Co., Ltd., Shanghai) in sequence, for 5-minute incubation at room temperature in dark. A flow cytometry (BD CantoII) was used for analysis, and the apoptotic rate was calculated.
CCK-8 for Cell Proliferation
CCK-8 assay kits (Beyotime Biotechnology, Shanghai, China) were used for detection. Cells were collected after 24-hour transfection, then adjusted to 4*106 cells/well, and finally inoculated on a 96-well plate. After 24-, 48-, 72-, and 96-hour culture, each well was, respectively, added with CCK-8 solution (10 μL) and DMEM (90 μL) for 2-hour culture at 37°C. Next, optical value (OD) values were measured at 490 nm using a microplate reader.
Transwell for Cell Invasion
The cells were adjusted to 1*106 cells/well and inoculated on the upper chamber of the 24-well plate, with 600 μL of the 10% FBS-containing medium added on the lower one. After the Transwell was cultured in an incubator for 24 hours, the matrix and the cells without penetrating the membrane surface in the upper chamber were wiped off. After the cells were cleaned with PBS (3 times), they were fixed with paraformaldehyde (20min), cleaned with double distilled water (3 times), and finally stained with 0.5% crystal violet after they were dried. The cell invasion was observed with a microscope.
Statistical Analysis
In this study, SPSS21.0 (IBM Corp, Armonk, NY, USA) was used to statistically analyze the collected data. GraphPad 7 was used to plot the required figures. The comparison of count data was analyzed by chi-square test or Fisher’s exact test, while that of measurement data between two groups was analyzed by independent samples t test. One-way analysis of variance (ANOVA) was used for the comparison between multiple groups, followed by Tukey HSD method for verification. Repeated measures ANOVA was used for the comparison between multiple time points, and then Bonferroni was used for test. Kaplan-Meier (K-M) survival curves were plotted to show patients’ survival, and Log rank test was used for analysis. A receiver operating characteristic (ROC) curve was plotted to assess the diagnostic value of serum H19 for GC. When P<0.05, the difference was statistically significant.
Results
H19 Expression in GC and Its Clinical Value
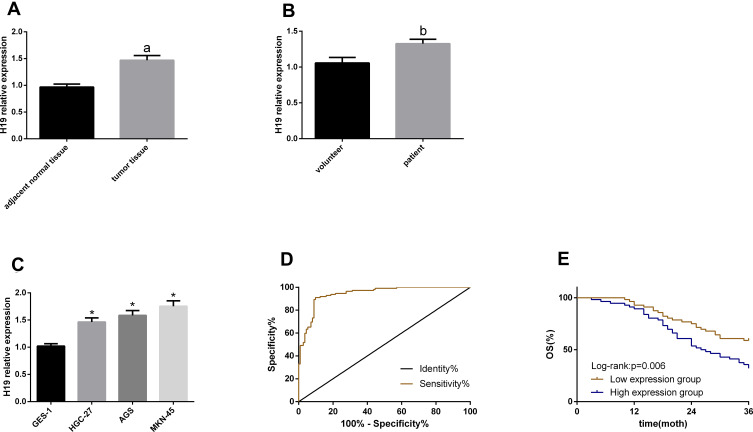
According to the qRT-PCR, H19 upregulated in GC serum, tissues and cells. Based on the analysis of the diagnostic value of serum H19 for GC, its sensitivity, specificity and area under the curve (AUC) were 90.07%, 90.18 and 0.943, respectively. With H19 expression in patient tissues as the median, the patients were divided into low and high expression groups, to analyze the correlation of H19 with patients’ clinical pathological data and survival rate. High H19 expression was related to a tumor size ≥5 cm, high stages (III+IV), lymph node metastasis and a poor survival rate. This suggests that this RNA is involved in GC pathogenesis and may act as an oncogene. See Table 1 and Figure 1.
Table 1.
Correlation of H19 with Clinical Pathological Data
| Groups | n | H19 | χ2 | P | ||
|---|---|---|---|---|---|---|
| Low Expression Group (n=56) | High Expression Group (n=56) | |||||
| Gender | 2.396 | 0.122 | ||||
| Male | 68 | 30 (53.57) | 38 (67.86) | |||
| Female | 44 | 16 (46.43) | 28 (432.14) | |||
| Age | 2.939 | 0.087 | ||||
| <60 years old | 49 | 29 (51.79) | 20 (35.71) | |||
| ≥60 years old | 63 | 27 (48.21) | 36 (64.29) | |||
| Tumor size | 1.820 | 0.177 | ||||
| ≥5cm | 45 | 19 (33.93) | 26 (46.43) | |||
| <5cm | 67 | 37 (66.07) | 30 (53.57) | |||
| TNM staging | 6.196 | 0.013 | ||||
| Stages I + II | 65 | 39 (69.64) | 26 (46.43) | |||
| Stages III+IV | 47 | 17 (30.34) | 30 (53.57) | |||
| Lymph node metastasis | 6.053 | 0.014 | ||||
| Yes | 53 | 20 (35.71) | 33 (58.93) | |||
| No | 59 | 36 (64.29) | 23 (41.07) | |||
| Degree of differentiation | 5.486 | 0.019 | ||||
| Lowly differentiated | 42 | 27 (48.21) | 15 (26.79) | |||
| Moderately + highly differentiated | 74 | 29 (51.79) | 41 (73.21) | |||
Figure 1.
H19 expression in GC and its clinical value. (A–C) H19 upregulated in GC serum, tissues and cells. (D) The ROC curve of serum H19 for diagnosing GC. (E) The 3-year overall survival in the high expression group was lower than that in the low expression group.
Notes:aIndicates P<0.05 when compared with normal adjacent tissues. bIndicates P<0.05 when compared with volunteer. *Indicates P<0.05 when compared with GES-1.
H19 Knockdown Can Inhibit GCCs to Proliferate and Invade
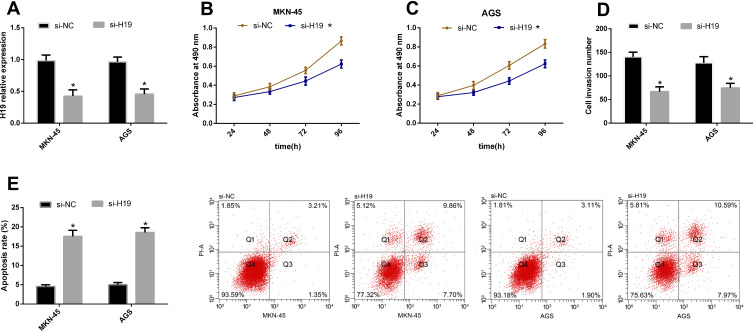
In this part, we explored the effects of H19 knockdown on the biological behavior of MKN-45 and AGS cells (H19 had the largest differential expression in the two cells). According to the CCK-8 assay, cell proliferation was inhibited in the si-H19 group compared with the si-NC group. According to the Transwell, the number of cells penetrating the membrane reduced in the si-H19 group compared with the si-NC group. According to the flow cytometry, the apoptotic rate rose in the si-H19 group compared with the si-NC group. See Figure 2.
Figure 2.
H19 knockdown can inhibit GCCs to proliferate and invade. (A) si-H19 transfection can inhibit H19 in GCCs. (B and C) Cell proliferation reduced after si-H19 transfection. (D) The number of cells penetrating the membrane sharply reduced after si-H19 transfection. (E) The apoptotic rate remarkably rose after si-H19 transfection.
Note: *Indicates P<0.05 when compared with the si-NC group.
H19 Can Negatively Regulate miR-138
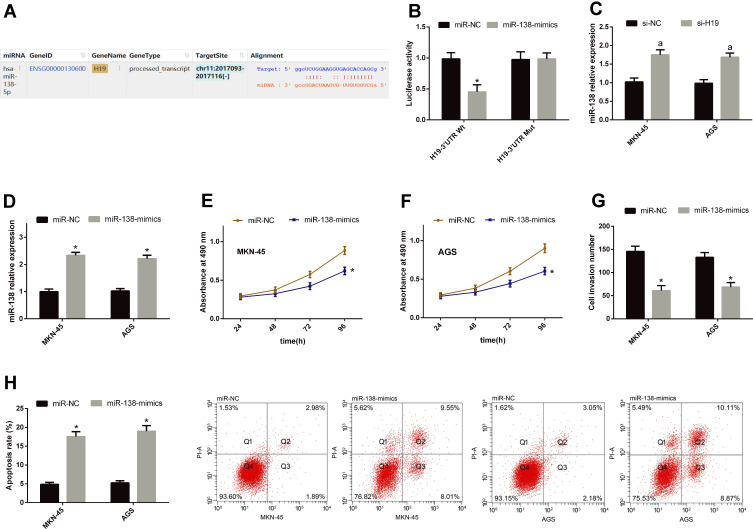
For understanding the potential mechanism of H19 in regulating the biological behavior of GCCs, we predicted its potential targeted miRs using starBase3.0 finding that miR-138 and H19 have targeted binding sites. For further verifying their relationship, we conducted DLRGA, and found that miR-138-mimics could inhibit H19-3ʹUTR Wt luciferase activity, with no remarkable effect on H19-3ʹUTR Mut luciferase activity. In addition, we tested the effects of si-H19transfection on miR-138 in GCCs. This miR rose in the cells after transfection. Subsequently, we explored the effects of miR-138 upregulation on the biological behavior of GCCs. Compared with the miR-NC group, miR-138 rose, cell proliferation and invasion reduced, and the apoptotic rate rose in the miR-138-mimics group. See Figure 3.
Figure 3.
H19 can negatively regulate miR-138. (A) miR-138 and H19 had targeted binding sites. (B) miR-138-mimics could inhibit H19-3ʹUTR Wt luciferase activity, with no remarkable effect on H19-3ʹUTR Mut luciferase activity. (C) miR-138 rose in GCCs after si-H19 transfection. (D) miR-138 rose in GCCs after miR-138-mimics transfection. (E and F) Cell proliferation reduced after miR-138-mimics transfection. (G) Cells penetrating the membrane were relatively few after miR-138-mimics transfection. (H) The apoptotic rate rose after miR-138-mimics transfection.
Notes: *Indicates P<0.05 when compared with the miR-NC group. aIndicates P<0.05 when compared with the si-NC group.
miR-138 Can Play an Anti-Cancer Role in GC Through E2F2
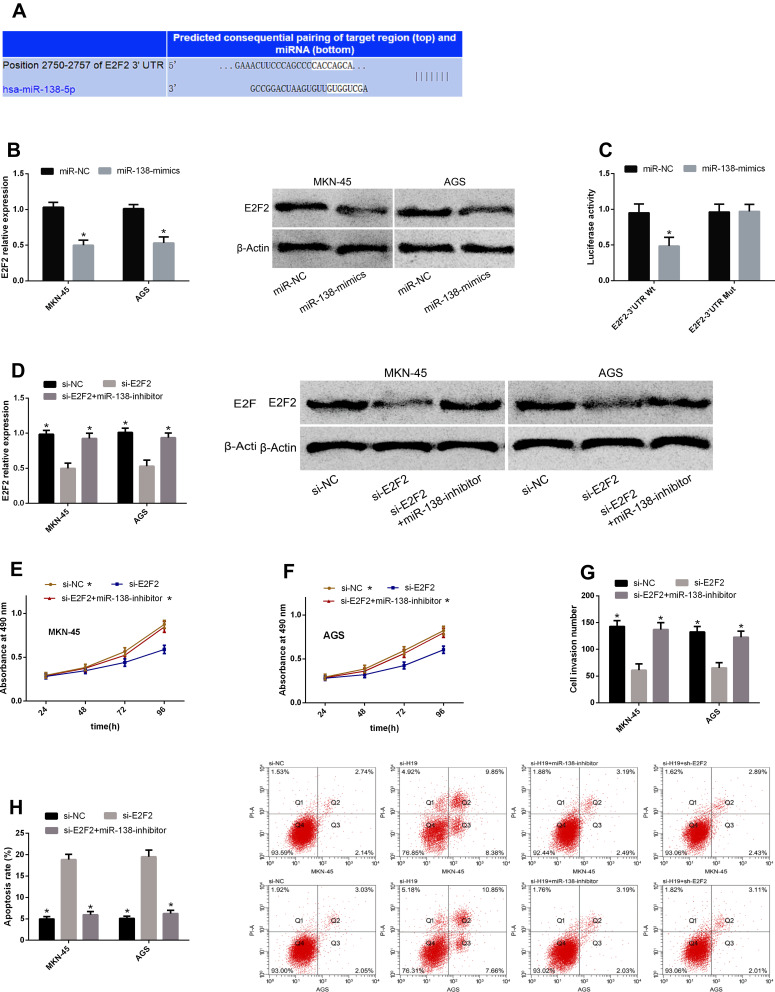
miRs can affect the basic biological behavior of cells by regulating target genes. For further exploring the mechanism of H19/miR-138 regulating GCCs’ biological behavior, we found that E2F2 and miR-138 had targeted binding sites through a prediction software for miR target genes. Therefore, we detected E2F2 changes in cells transfected with miR-NC and miR-138-mimics, and found that E2F2 reduced after miR-138-mimics transfection. According to the DLRGA, miR-138-mimics could inhibit E2F2-3ʹUTR Wt luciferase activity, with no remarkable effect on E2F2-3ʹUTR Mut luciferase activity. Next, we tested the effects of E2F2 knockdown on GCCs’ proliferation, invasion and apoptosis. Similar to the results of si-H19 and miR-138-mimics transfection, si-E2F2 transfection could also enhance the proliferation and invasion and reduce the apoptotic rate, and the co-transfection of si-E2F2 and miR-138-inhibitor could reverse the effects of si-E2F2 on the biological behavior. See Figure 4.
Figure 4.
miR-138 can play an anti-cancer role in GC through E2F2. (A) E2F2 and miR-138 had targeted binding sites. (B) E2F2 reduced after miR-138-mimics transfection. (C) miR-138-mimics could inhibit E2F2-3ʹUTR Wt luciferase activity, with no remarkable effect on E2F2-3ʹUTR Mut luciferase activity. (D) E2F2 rose after si-E2F2 transfection, which could be reversed after co-transfection with miR-138-inhibitor. (E and F) Cell proliferation enhanced after si-E2F2 transfection, which could be reduced after co-transfection with miR-138-inhibitor. (G) Cell invasion enhanced after si-E2F2 transfection, which could be reduced after co-transfection with miR-138-inhibitor. (H) The apoptotic rate rose after si-E2F2 transfection, which could be reduced after co-transfection with miR-138-inhibitor.
Note: *Indicates P<0.05 when compared with the si-E2F2 group.
H19 Promotes GCCs’ Proliferation and Invasion by Regulating miR-138/E2F2 Axis
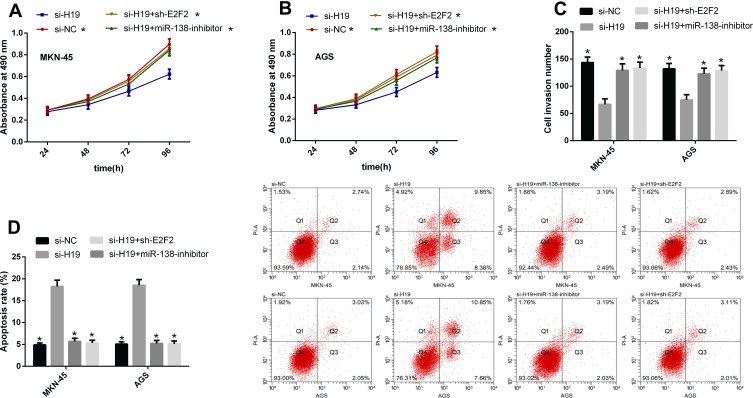
According to the above results, we know that the tumor-promoting effect of H19 may be related to the miR-138/E2F2 axis. For further understanding the correlations of the two, we co-transfected GCCs with si-H19 + miR-138-inhibitor and si-H19 + sh-E2F2, respectively, to observe whether such treatment could reverse or weaken the effects of H19 knockdown on the cells’ biological behavior. The results showed that both co-transfections could reduce changes in cell proliferation, invasion and apoptosis caused by si-H19 transfection. This shows that H19 can promote GCCs to proliferate and invade by regulating miR-138/E2F2 axis See Figure 5.
Figure 5.
H19 promotes GCCs’ biological behavior by regulating miR-138/E2F2 axis. (A and B) The co-transfection of both si-H19 + miR-138-inhibitor and si-H19 + sh-E2F2 could reduce changes in cell proliferation caused by si-H19 transfection. (C) The co-transfection of both si-H19 + miR-138-inhibitor and si-H19 + sh-E2F2 could reduce changes in cell invasion caused by si-H19 transfection. (D) The co-transfection of both si-H19 + miR-138-inhibitor and si-H19 + sh-E2F2 could reduce changes in cell apoptosis caused by si-H19 transfection.
Note: *Indicates P<0.05 when compared with the si-H19 group.
Discussion
At present, targeted therapy in tumor treatment has been recognized by medical personnel, so finding potential therapeutic targets for it is a hot research topic currently. With the continuous understanding of lncRNAs, it has been found that they have a great effect on tumor progression and are considered as potential targets for tumor therapy.12–14 As one of the earliest lncRNAs found, H19 (also an oncogene) is located on chromosome 11 and highly expressed in various tumors.8,9 In our study, we explored the role and mechanism of action of H19 in GC. H19 rose in tissues, serum and cells of GC patients, and this increased RNA was related to a tumor size ≥5 cm, high stages (III+IV), lymph node metastasis and a poor survival rate. Based on the analysis of the diagnostic value of serum H19 for GC, the AUC of H19 was 0.943. According to the in vivo experiments, H19 could promote GCCs to proliferate and invade and inhibit their apoptosis. These findings show that H19 has an extremely high research value, because it is not only a potential biomarker for GC diagnosis and prognosis, but also an important potential therapeutic target for the disease.
More and more studies have revealed that lncRNAs can act as a sponge for miRs, thus regulating levels of target genes downstream of miRs and regulating cell biology.15 For example, lncRNA SNGG1 increases CDK7 expression by competitively binding to miR-199a-3p, and then promotes the proliferation and cycle process of prostate cancer cells.16 LncRNA CCAT1 may promote retinoblastoma cells to proliferate, migrate and invade, and reduce their apoptosis via the negative regulation of miR-218-5p.17 For exploring the mechanism of action of H19 in GC and through the biological prediction software starBase3.0, we found targeted binding sites between miR-138 and H19. miR-138-mimics could inhibit H19-3ʹUTR Wt luciferase activity, with no remarkable effect on H19-3ʹUTR Mut luciferase activity. miR-138 rose in GCCs after si-H19 transfection. These results indicate that there is a certain regulatory effect between H19 and miR-138, so we believe that the effect of H19 on inhibiting GC progression may be related to this miR. As a kind of highly conservative endogenous short-chain non-coding RNAs, miRs can regulate approximately one third of human genes.18 Their abnormal expression in human body is considered as an important factor for the development and progression of various diseases.19–21 Previous studies have found that miR-138 as a member of miRs downregulates in various tumors including GC.22–24 In our study, GCCs’ proliferation and invasion enhanced but the apoptotic rate rose after miR-138 upregulation, showing that this miR is a tumor suppressor gene in GC, and that it is expected to become a potential therapeutic target for the disease.
It is well known that miRs are involved in biological processes by regulating downstream target genes. For further improving the mechanism of H19 and miR-138 on regulating GCCs’ biological behavior, we predicted the downstream target genes of this miR. There were binding sites between E2F2 and miR-138. E2F2 reduced in GCCs after miR-138 increased. miR-138-mimics could inhibit E2F2-3ʹUTR Wt luciferase activity, with no remarkable effect on E2F2-3ʹUTR Mut luciferase activity. These results confirm that there is interaction between E2F2 and miR-138. E2F2 is a member of E2F2 family, which is an important transcription factor that regulates cell cycle and apoptosis.25 In tumors, E2F2 and H19 are considered as cancer-promoting factors.26,27 In this article, after E2F2 knockdown, GCCs’ proliferation and invasion reduced but the apoptotic rate rose, and the co-transfection of si-E2F2 and miR-138-inhibitor could reverse effects of the knockdown on the biological behavior. This suggests that miR-138 and E2F2 exert a great function in GC progression, and that the two have a mutual regulatory effect. For confirming that H19 can promote GCCs to proliferate and invade through the miR-138/E2F2 axis, we knocked down miR-138 and upregulated E2F2 while transfecting si-H19. The results showed that after such treatment, the effects of si-H19 on the biological behavior were reversed.
Accordingly, H19 rises in GC and predicts the poor prognosis of patients, so H19 knockdown inhibits GCCs to proliferate and invade and induces their apoptosis, which is related to the miR-138/E2F2 axis. This study has certain limitations. Firstly, due to the short time of this study, more research samples cannot be collected and longer follow-ups cannot be conducted, which results in possibly limited clinical data. Secondly, other mechanisms of H19 are still unclear. We have only studied the miR-138/E2F2 axis, so whether H19 can promote GC progression through other pathways needs further research.
In summary, H19 that rises in GC can be used as a biological indicator for diagnosing the disease and predicting patients’ poor prognosis, because it promotes GCCs to proliferate and invade through miR-138/E2F2 axis.
Conclusion
In summary, H19 that rises in GC can be used as a biological indicator for diagnosing the disease and predicting patients’ poor prognosis, because it promotes GCCs to proliferate and invade through miR-138/E2F2 axis.
Funding Statement
The authors received no funding for this work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Liu K, Chen E. LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol Biol Lett. 2020;25(1):4. doi: 10.1186/s11658-020-0201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei J, Xu H, Wei W, et al. circHIPK3 promotes cell proliferation and migration of gastric cancer by sponging miR-107 and regulating BDNF expression. Onco Targets Ther. 2020;13:1613. doi: 10.2147/OTT.S226300 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Cui Y, Fan Y, Zhao G, et al. Novel lncRNA PSMG3-AS1 functions as a miR-143-3p sponge to increase the proliferation and migration of breast cancer cells. Oncol Rep. 2020;43(1):229–239. doi: 10.3892/or.2019.7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B-G, Lv Z, Ding H-X, et al. The association of lncRNA-HULC polymorphisms with hepatocellular cancer risk and prognosis. Gene. 2018;670:148–154. doi: 10.1016/j.gene.2018.05.096 [DOI] [PubMed] [Google Scholar]
- 6.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bian Z, Jin L, Zhang J, et al. LncRNA—UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Li Q, Jin X, et al. Long non-coding RNA H19 knockdown inhibits the cell viability and promotes apoptosis of thyroid cancer cells through regulating the PI3K/AKT pathway. Exp Ther Med. 2019;18(3):1863–1869. doi: 10.3892/etm.2019.7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao S, Yu C, Liu H, et al. Long non-coding RNA H19 promotes the proliferation and invasion of lung cancer cells and regulates the expression of E-cadherin, N-cadherin, and vimentin. Onco Targets Ther. 2019;12:4099. doi: 10.2147/OTT.S185156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JS, Wang YF, Zhang XQ, et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63(2):223–230. doi: 10.4149/207_150821N454 [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 12.Sun Z, Wu XY, Wu CL. The association between LncRNA HOTAIR and cancer lymph node metastasis and distant metastasis: a meta-analysis. Neoplasma. 2018;65(2):178–184. doi: 10.4149/neo_2018_170114N34 [DOI] [PubMed] [Google Scholar]
- 13.Cao Y, Luo X, Ding X, et al. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J Cell Biochem. 2018;119(8):6935–6942. doi: 10.1002/jcb.26894 [DOI] [PubMed] [Google Scholar]
- 14.Liu Z, Dai J, Shen H. Systematic analysis reveals long noncoding RNAs regulating neighboring transcription factors in human cancers. Biochim Biophys Acta Mol Basis Dis. 2018;1864(9):2785–92. [DOI] [PubMed] [Google Scholar]
- 15.Wieczorek E, Reszka E. mRNA, microRNA and lncRNA as novel bladder tumor markers. Clin Chim Acta. 2018;477:141–153. doi: 10.1016/j.cca.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–152. doi: 10.1016/j.bbrc.2017.03.169 [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Zhong J, Bian Z, et al. Long non-coding RNA CCAT1 promotes human retinoblastoma SO-RB50 and Y79 cells through negative regulation of miR-218-5p. Biomed Pharmacother. 2017;87:683–691. doi: 10.1016/j.biopha.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Liang X, Wang Y, et al. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep. 2016;6:27157. doi: 10.1038/srep27157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei C, Du F, Sun L, et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8(10):e3101. doi: 10.1038/cddis.2017.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pheiffer C, Dias S, Rheeder P, et al. Decreased expression of circulating miR-20a-5p in South African women with gestational diabetes mellitus. Mol Diagn Ther. 2018;22(3):345–352. doi: 10.1007/s40291-018-0325-0 [DOI] [PubMed] [Google Scholar]
- 21.Li SD, Yang J M, Xia Y, et al. Long noncoding RNA NEAT1 promotes proliferation and invasion via targeting miR-181a-5p in non-small cell lung cancer. Oncol Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang L, Li B, Zheng B, et al. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol Rep. 2017;38(2):1295–1302. doi: 10.3892/or.2017.5745 [DOI] [PubMed] [Google Scholar]
- 23.Ye Z, Fang B, Pan J, et al. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep. 2017;37(6):3244–3252. doi: 10.3892/or.2017.5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Zhao L-J, Tan Y-X, Ren H, Qi,Z-T. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33(5):1113–1120. doi: 10.1093/carcin/bgs113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indovina P, Pentimalli F, Casini N, et al. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget. 2015;6(20):17873–17890. doi: 10.18632/oncotarget.4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Yu JH, Lu ZH, et al. E2F2 induction in related to cell proliferation and poor prognosis in non-small cell lung carcinoma. Int J Clin Exp Pathol. 2015;8(9):10545–10554. [PMC free article] [PubMed] [Google Scholar]
- 27.Feliciano A, Garcia-Mayea Y, Jubierre L, et al. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8(10):e3141. doi: 10.1038/cddis.2017.544 [DOI] [PMC free article] [PubMed] [Google Scholar]