Abstract
FOXO3 is a prominent longevity gene. To date, no-one has examined whether longevity-associated FOXO3 genetic variants protect against mortality in all individuals, or only in those with aging-related diseases. We therefore tested longevity-associated FOXO3 single nucleotide polymorphisms in a haplotype block for association with mortality in 3,584 elderly American men of Japanese ancestry, 2,512 with and 1,072 without a cardiometabolic disease (CMD). At baseline (1991–1993), 1,010 CMD subjects had diabetes, 1,919 had hypertension, and 738 had coronary heart disease (CHD). Follow-up until Dec 31, 2019 found that in CMD-affected individuals, longevity-associated alleles of FOXO3 were associated with significantly longer lifespan: haplotype hazard ratio 0.81 (95% CI 0.72-0.91; diabetes 0.77, hypertension 0.82, CHD 0.83). Overall, men with a CMD had higher mortality than men without a CMD (P=6x10-7). However, those men with a CMD who had the FOXO3 longevity genotype had similar survival as men without a CMD. In men without a CMD there was no association of longevity-associated alleles of FOXO3 with lifespan. Our study provides novel insights into the basis for the long-established role of FOXO3 as a longevity gene. We suggest that the FOXO3 longevity genotype increases lifespan only in at-risk individuals by protection against cardiometabolic stress.
Keywords: longevity, genetics, mortality, FOXO3, resilience
INTRODUCTION
Longevity can be accompanied by either healthy aging (i.e., an absence of life-threatening diseases and chronic conditions), or unhealthy aging, in which elderly individuals live long lives with chronic conditions. Genetic factors make a 15–40% contribution to lifespan [1], and may be as high as 48% in the oldest-old [2]. At very old age (> 90 years) specific longevity genes may dominate over environmental influences in determination of eventual lifespan, with evidence that there is a stronger effect in males [3].
Our study focused on FOXO3, the gene encoding the forkhead/winged helix box-O3 (FoxO3) transcription factor. Minor alleles of single nucleotide polymorphisms (SNPs) located in FOXO3 have been found to be strongly associated with human longevity in multiple population subgroups worldwide [4–6]. This includes genome-wide association study data [7]. We found previously that a haplotype comprised of 14 FOXO3 SNPs is associated with increased likelihood of living close to 100 years in the Kuakini Honolulu Heart Program/ Kuakini and Honolulu-Asia Aging Study (KHHP/KHAAS) cohort [8]. FoxO3 can differentially induce gene expression programs depending on chromatin architecture by binding to specific enhancers, activating these by causing changes in histone acetylation, and subsequent recruitment of RNA polymerase II [9]. In response to cellular stress, FOXO3 may also function at the genomic level by facilitating long-range gene-gene interactions, changes in chromatin conformation, and interaction of topologically associated domains to regulate multiple neighboring genes involved in various processes that contribute to cell resilience, namely autophagy, stress response, energy/nutrient sensing, cell proliferation, apoptosis, and stem cell maintenance [8, 10]. The expression of at least some of these neighboring genes is increased following cellular stress (see supplement to ref [8]). Thus, FOXO3 is positioned at the center of an “aging hub.” The longevity alleles of FOXO3 enable enhanced FOXO3 expression, thereby activating other genes in its functional neighborhood. Physiologically, we now know that the major reason for the increase in lifespan conferred by protective alleles of FOXO3 is protection against death from coronary heart disease (CHD) [11], although in other cohorts such protection may extend to cancer and stroke [12, 13].
The present study addresses the hypothesis that the FOXO3 longevity-associated alleles increase cell and organism resilience by protecting against cellular stress caused by chronic conditions of aging. To test this hypothesis, we compared the effect of longevity-associated alleles with lifespan in subjects with and without type 2 diabetes, CHD and hypertension (collectively referred to as cardiometabolic diseases, CMDs).
RESULTS
Characteristics of subjects
Supplementary Table 1 shows age-adjusted baseline (1991–1993) characteristics of men in the study, adjusting for age, genotypes of the strongest SNP (rs2802292), and prevalence of medical conditions. By 31 December 2019, 3,548 out of 3,584 of the subjects had died during the 29 years of follow-up from exam 4. At exam 4 (baseline) among the 3,584 participants, there were 21% who had been diagnosed with CHD, 29% with diabetes, 54% with hypertension, and 14% with cancer. Seventy percent had at least one CMD, and 5.2% had all 3 components of CMD. Mean age at death was 88.6 ± 6.1 years for those with at least one of these CMDs, and 89.5 ± 6.0 years for those who did not have any CMD (p < 0.0001).
FOXO3 genotype and survival in CMD and non-CMD subjects.
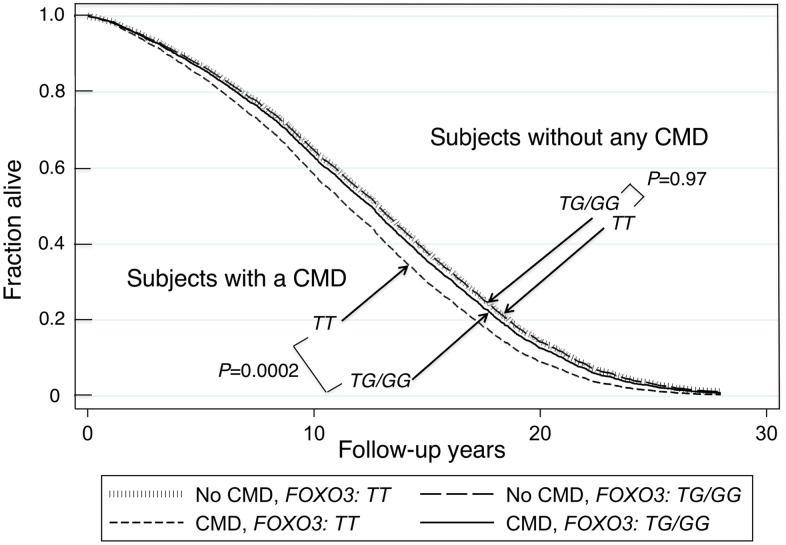
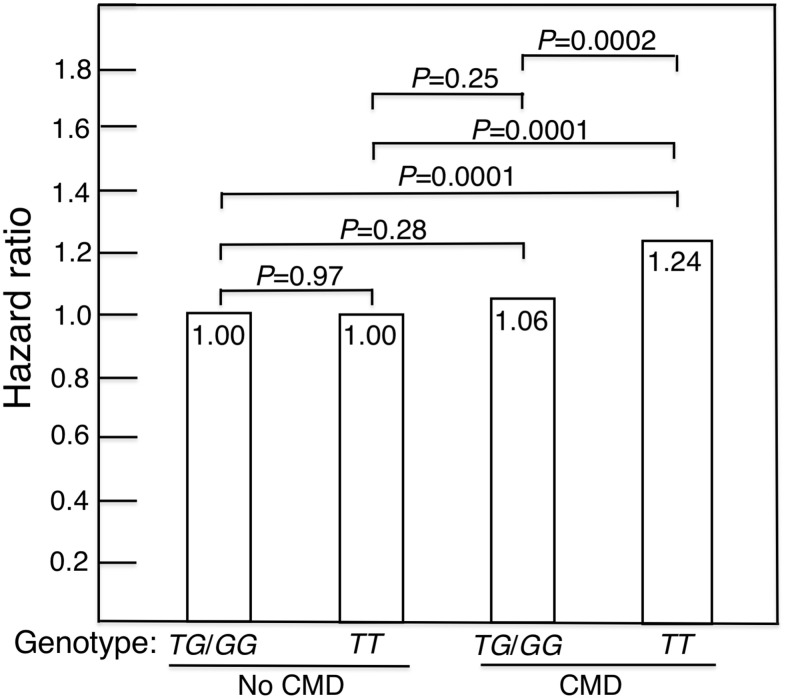
The 14 FOXO3 SNPs (shown in Supplementary Figures 1, 2) likely function as a cis-regulatory unit. Survival curves for men without a CMD and those with a CMD showed that the former lived longer (Kaplan-Meier Log-rank χ2 = 24.8, p = 6.4x10-7). Figure 1 shows survival curves for CMD subjects and subjects without a CMD according to whether they were carriers of the longevity-associated (G) allele of the strongest FOXO3 SNP, rs2802292, or were homozygous for the major (T) allele. These curves were determined using a Cox proportional hazard model adding an interaction term of FOXO3 with CMD. Only in CMD subjects was the G allele associated with greater lifespan than TT (p = 0.0002). Subjects without a CMD had the longest lifespans. Moreover, in contrast to CMD subjects, FOXO3 genotype was not associated with lifespan in subjects without a CMD. Figure 2 shows mortality risk as hazard ratio for CMD subjects and subjects without a CMD according to whether they were carriers of the longevity-associated (G) allele of the strongest FOXO3 SNP, rs2802292, or were homozygous for the major (T) allele. In men with CMD and the FOXO3 longevity genotype, mortality was reduced to that of men without CMD.
Figure 1.
Survival curves spanning the period from baseline (1991–1993) to Dec 31, 2019 for subjects with and without a CMD according to whether they were carriers of the longevity-associated G allele of SNP rs2802292. The survival probabilities were estimated from the Cox proportional hazard model (see Methods) h(t) = h(t0) * exp(β1*Age + β2*BMI + β3*Glucose + β4*CMD + β5*FOXO3_G + β6* (CMD*FOXO3_G)) by fixing age at 75 years, BMI at the mean, 23.5 kg/m2, and glucose at the mean, 113 mg/dL (where β6 is the effect of the interaction of CMD with FOXO3 genotype (G carriers vs. TT genotype) on mortality, giving P(β6) = 0.04). The P values for comparison of survival curves for the group without any CMD for FOXO3-G carriers vs. FOXO3-TT, and comparison of survival curves for the group with a CMD for FOXO3-G carriers vs. FOXO3-TT, were P=0.97 and P=0.0002, respectively. The P-values for comparison of survival curves for FOXO3-TT carriers or for FOXO3-G carriers for those with a CMD versus those without any CMD, were P=0.000039 and P=0.28 respectively.
Figure 2.
Mortality risk expressed as hazard ratio, adjusted for age, BMI and glucose, for CMD subjects and subjects without a CMD according to whether they were carriers of the longevity-associated (G) allele of the FOXO3 SNP rs2802292 (TG/GG) or were homozygous for the major (T) allele (i.e., were TT). In men with CMD and the FOXO3 longevity genotype, mortality risk was reduced to normal (i.e., did not differ significantly from that of men without CMD).
Table 1 shows age-adjusted baseline characteristics of men with CMD and men without a CMD. Analyses found no evidence of population stratification in the dataset (data not shown). In subjects with either diabetes, CHD, or hypertension (CMD) at baseline, G allele carriers had significantly lower CHD prevalence, but higher diabetes and hypertension (Table 1). In CMD subjects, after adjusting for co-variates, minor allele carriers of each of the 8 FOXO3 SNPs tested exhibited an association with longer lifespan (Table 2). In contrast, in individuals without any CMD component, there was no difference in lifespan associated with FOXO3 genotype. Table 3 shows the association with just CHD, just diabetes, and just hypertension for each of the 8 FOXO3 SNPs and the haplotype of each. Possession of the longevity-associated minor allele was associated with lower hazard ratio for mortality compared with major allele homozygotes in the CMD group when adjusted for age, and, in the full model, after adjustment for additional covariates that represent mortality risk factors.
Table 1. Age-adjusted baseline characteristics by CMD* and FOXO rs2802292 genotype.
| Variable | With a CMD | Without a CMD | ||||
| TT | GG/GT | P | TT | GG/GT | P | |
| n | 1331 | 1181 | 570 | 502 | ||
| Age (years) | 77.6 ± 4.5 | 78 ± 4.6 | 0.03 | 77.3 ± 4.4 | 78.1 ± 4.8 | 0.053 |
| BMI (kg/m²) | 23.7 ± 3.1 | 23.7 ± 3.0 | 0.62 | 22.8 ± 3 | 23 ± 3.0 | 0.37 |
| Fasting plasma glucose (mg/dL) | 116.8 ± 31.9 | 119.3 ± 35.2 | 0.060 | 101.3 ± 8.5 | 101.3 ± 8.6 | 1.00 |
| Smoking (pack-years) | 25.4 ± 33.4 | 28.5 ± 36.1 | 0.036 | 24.8 ± 33.4 | 23.9 ± 32.6 | 0.67 |
| Alcohol intake (oz/mo) | 19.6 ± 42.5 | 20.2 ± 44.3 | 0.76 | 16.2 ± 32.4 | 16.6 ± 33.4 | 0.85 |
| Physical activity index | 30.8 ± 4.8 | 30.8 ± 4.3 | 0.81 | 30.9 ± 4.3 | 31.2 ± 4.9 | 0.23 |
| Depressive symptoms (%) | 9.8 | 11.0 | 0.36 | 10.4 | 11.2 | 0.67 |
| Stroke (%) | 5.2 | 4.9 | 0.71 | 2.5 | 2.3 | 0.86 |
| Cancer (%) | 14.1 | 12.5 | 0.24 | 14.6 | 12.8 | 0.39 |
| CHD (%) | 31.7 | 26.7 | 0.01 | – | – | – |
| Diabetes (%) | 38.5 | 43.2 | 0.02 | – | – | – |
| Hypertension (%) | 75.9 | 77.0 | 0.50 | – | – | – |
Values shown are age-adjusted mean ± SD for indirect measures and proportion for direct measurements, except age.
*CMD: CHD or diabetes or hypertension
Table 2. Hazard ratios (HR) of FOXO3 minor allele carriers vs. major allele homozygotes with total mortality by cardiometabolic disease (CMD)* status.
| Model | FOXO3 SNP | With a CMD (n=2512) | Without a CMD (n=1072) | ||
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age adjusted | rs2802292 | 0.88 (0.81-0.95) | 0.0011 | 0.99 (0.88-1.12) | 0.93 |
| Age adjusted | rs1935952 | 0.90 (0.83-0.97) | 0.0070 | 0.98 (0.87-1.12) | 0.81 |
| Age adjusted | rs2253310 | 0.88 (0.81-0.95) | 0.0013 | 0.99 (0.88-1.12) | 0.92 |
| Age adjusted | rs2764264 | 0.90 (0.83-0.98) | 0.012 | 0.97 (0.86-1.09) | 0.61 |
| Age adjusted | rs2802288 | 0.88 (0.81-0.95) | 0.0012 | 0.99 (0.87-1.12) | 0.85 |
| Age adjusted | rs3800230 | 0.92 (0.84-1.00) | 0.053 | 0.97 (0.84-1.11) | 0.61 |
| Age adjusted | rs9398171 | 0.90 (0.83-0.98) | 0.012 | 0.98 (0.87-1.11) | 0.77 |
| Age adjusted | rs12212067 | 0.90 (0.81-1.00) | 0.040 | 0.98 (0.84-1.14) | 0.76 |
| Age adjusted | Haplotype | 0.86 (0.77-0.96) | 0.0069 | 0.99 (0.84-1.17) | 0.92 |
| Covariate adjusted† | rs2802292 | 0.82 (0.75-0.89) | 8.1E–06 | 1.03 (0.90-1.18) | 0.63 |
| Covariate adjusted | rs1935952 | 0.83 (0.76-0.91) | 4.2E–05 | 1.06 (0.92-1.21) | 0.44 |
| Covariate adjusted | rs2253310 | 0.82 (0.75-0.90) | 1.0E–05 | 1.04 (0.90-1.19) | 0.61 |
| Covariate adjusted | rs2764264 | 0.85 (0.78-0.93) | 0.0002 | 1.03 (0.90-1.18) | 0.65 |
| Covariate adjusted | rs2802288 | 0.82 (0.75-0.89) | 8.1E–06 | 1.03 (0.90-1.18) | 0.70 |
| Covariate adjusted | rs3800230 | 0.88 (0.80-0.97) | 0.010 | 1.03 (0.89-1.20) | 0.68 |
| Covariate adjusted | rs9398171 | 0.84 (0.77-0.92) | 0.0002 | 1.05 (0.92-1.21) | 0.46 |
| Covariate adjusted | rs12212067 | 0.88 (0.79-0.98) | 0.023 | 1.05 (0.88-1.25) | 0.59 |
| Covariate adjusted | Haplotype¥ | 0.81 (0.72-0.91) | 0.00031 | 1.07 (0.89-1.28) | 0.49 |
*CMD: At least one of CHD, diabetes and hypertension.
†Covariates in Cox model: age, BMI, glucose, smoking pack-year, alcohol consumption (ounces per month), physical activity index (PAI), depressive symptoms, cancer and stroke.
¥Haplotype was obtained for 1,740 subjects with a CMD, and 742 subjects without a CMD.
Haplotype comprised minor allele frequencies of SNPs rs2253310 (allele C), rs2802288 (allele A), rs2802292 (allele G), rs2764264 (allele C), rs9398171 (allele C), rs12212067 (G), and rs3800230 (allele G).
Table 3. Hazard ratios (HR) of FOXO3 minor allele carriers vs. major allele homozygotes with total mortality by diabetes, CHD, and hypertension.
| With a CMD | Without a CMD | |||||
| SNP | Cardiometabolic disease | Model* | HR (95% CI) ** | P | HR (95% CI) | P |
| rs2802292 | Diabetes | 1 | 0.84 (0.74-0.95) | 0.0064 | 0.94 (0.87-1.01) | 0.11 |
| (1010, 2525) *** | 2 | 0.78 (0.68-0.89) | 0.0003 | 0.93 (0.85-1.01) | 0.08 | |
| CHD | 1 | 0.84 (0.73-0.97) | 0.021 | 0.94 (0.88-1.02) | 0.13 | |
| (738, 2846) | 2 | 0.79 (0.67-0.93) | 0.0055 | 0.92 (0.85-1.00) | 0.04 | |
| Hypertension | 1 | 0.89 (0.81-0.97) | 0.0085 | 0.94 (0.85-1.04) | 0.24 | |
| (1919, 1665) | 2 | 0.83 (0.75-0.92) | 0.0003 | 0.94 (0.84-1.04) | 0.22 | |
| rs1935952 | Diabetes | 1 | 0.84 (0.74-0.96) | 0.0075 | 0.95 (0.88-1.03) | 0.25 |
| (1010, 2521) | 2 | 0.78 (0.68-0.90) | 0.0005 | 0.95 (0.87-1.03) | 0.22 | |
| CHD | 1 | 0.85 (0.73-0.98) | 0.030 | 0.95 (0.88-1.03) | 0.20 | |
| (738, 2842) | 2 | 0.81 (0.68-0.96) | 0.015 | 0.93 (0.85-1.01) | 0.08 | |
| Hypertension | 1 | 0.89 (0.82-0.98) | 0.017 | 0.95 (0.86-1.05) | 0.34 | |
| (1917, 1663) | 2 | 0.83 (0.75-0.92) | 0.0002 | 0.97 (0.87-1.08) | 0.60 | |
| rs2253310 | Diabetes | 1 | 0.84 (0.74-0.96) | 0.0076 | 0.94 (0.87-1.01) | 0.11 |
| (1010, 2523) | 2 | 0.78 (0.69-0.90) | 0.0004 | 0.93 (0.85-1.01) | 0.08 | |
| CHD | 1 | 0.84 (0.73-0.97) | 0.021 | 0.95 (0.88-1.02) | 0.14 | |
| (738, 2844) | 2 | 0.79 (0.67-0.93) | 0.0055 | 0.92 (0.85-1.00) | 0.04 | |
| Hypertension | 1 | 0.89 (0.81-0.97) | 0.0088 | 0.94 (0.86-1.04) | 0.25 | |
| (1918, 1664) | 2 | 0.83 (0.75-0.92) | 0.0003 | 0.94 (0.84-1.04) | 0.24 | |
| rs2764264 | Diabetes | 1 | 0.85 (0.75-0.96) | 0.0097 | 0.95 (0.88-1.03) | 0.20 |
| (1009, 2521) | 2 | 0.80 (0.70-0.92) | 0.0014 | 0.95 (0.87-1.04) | 0.25 | |
| CHD | 1 | 0.85 (0.73-0.99) | 0.034 | 0.95 (0.89-1.03) | 0.21 | |
| (736, 2843) | 2 | 0.81 (0.69-0.96) | 0.015 | 0.94 (0.86-1.02) | 0.13 | |
| Hypertension | 1 | 0.91 (0.83-1.00) | 0.041 | 0.94 (0.85-1.03) | 0.20 | |
| (1915, 1664) | 2 | 0.86 (0.78-0.95) | 0.0028 | 0.95 (0.85-1.06) | 0.38 | |
| rs2802288 | Diabetes | 1 | 0.84 (0.74-0.95) | 0.0062 | 0.93 (0.86-1.01) | 0.10 |
| (1009, 2516) | 2 | 0.78 (0.68-0.89) | 0.0002 | 0.92 (0.85-1.01) | 0.07 | |
| CHD | 1 | 0.84 (0.72-0.97) | 0.0182 | 0.94 (0.88-1.02) | 0.13 | |
| (736, 2838) | 2 | 0.79 (0.67-0.93) | 0.0047 | 0.92 (0.84-0.99) | 0.03 | |
| Hypertension | 1 | 0.89 (0.81-0.97) | 0.0084 | 0.94 (0.85-1.04) | 0.22 | |
| (1915, 1659) | 2 | 0.83 (0.75-0.92) | 0.0003 | 0.93 (0.84-1.04) | 0.19 | |
| rs3800230 | Diabetes | 1 | 0.89 (0.77-1.02) | 0.1038 | 0.96 (0.88-1.05) | 0.38 |
| (998, 2501) | 2 | 0.88 (0.76-1.02) | 0.095 | 0.93 (0.85-1.03) | 0.17 | |
| CHD | 1 | 0.96 (0.81-1.14) | 0.68 | 0.94 (0.86-1.02) | 0.12 | |
| (731, 2817) | 2 | 0.92 (0.76-1.11) | 0.3832 | 0.93 (0.85-1.01) | 0.10 | |
| Hypertension | 1 | 0.92 (0.83-1.02) | 0.099 | 0.94 (0.85-1.05) | 0.30 | |
| (1901, 1647) | 2 | 0.89 (0.79-0.99) | 0.033 | 0.95 (0.84-1.07) | 0.38 | |
| rs9398171 | Diabetes | 1 | 0.86 (0.76-0.97) | 0.015 | 0.95 (0.88-1.03) | 0.21 |
| (1002, 2508) | 2 | 0.81 (0.70-0.92) | 0.0017 | 0.95 (0.87-1.04) | 0.24 | |
| CHD | 1 | 0.85 (0.73-0.99) | 0.032 | 0.96 (0.89-1.03) | 0.26 | |
| (732, 2827) | 2 | 0.81 (0.68-0.96) | 0.013 | 0.94 (0.87-1.02) | 0.14 | |
| Hypertension | 1 | 0.91 (0.83-0.99) | 0.033 | 0.95 (0.86-1.05) | 0.30 | |
| (1905, 1654) | 2 | 0.85 (0.77-0.94) | 0.0019 | 0.96 (0.86-1.07) | 0.50 | |
| rs12212067 | Diabetes | 1 | 0.92 (0.78-1.08) | 0.31 | 0.95 (0.86-1.05) | 0.28 |
| (1005, 2502) | 2 | 0.88 (0.74-1.05) | 0.15 | 0.94 (0.84-1.05) | 0.27 | |
| CHD | 1 | 0.90 (0.74-1.10) | 0.30 | 0.94 (0.86-1.03) | 0.19 | |
| (732, 2824) | 2 | 0.90 (0.73-1.12) | 0.36 | 0.93 (0.84-1.03) | 0.17 | |
| Hypertension | 1 | 0.90 (0.80-1.00) | 0.056 | 0.96 (0.85-1.09) | 0.51 | |
| (1903, 1653) | 2 | 0.88 (0.78-0.99) | 0.037 | 0.96 (0.84-1.11) | 0.60 | |
| Haplotype**** | Diabetes | 1 | 0.83 (0.70-0.99) | 0.034 | 0.94 (0.85-1.05) | 0.26 |
| (661, 1793) | 2 | 0.77 (0.64-0.93) | 0.0071 | 0.93 (0.82-1.04) | 0.19 | |
| CHD | 1 | 0.85 (0.69-1.05) | 0.12 | 0.93 (0.84-1.03) | 0.15 | |
| (526, 1956) | 2 | 0.83 (0.66-1.04) | 0.10 | 0.90 (0.81-1.01) | 0.07 | |
| Hypertension | 1 | 0.87 (0.77-0.98) | 0.022 | 0.94 (0.81-1.09) | 0.43 | |
| (1331, 1151) | 2 | 0.82 (0.72-0.93) | 0.0003 | 0.93 (0.84-1.04) | 0.23 | |
* 1: Age-adjusted; 2: Covariate-adjusted, Covariates in Cox model: age, BMI, glucose, smoking (pack-year), alcohol intake (oz/mo), physical activity index, depression, cancer, and stroke.
** HR (95% CI) of FOXO3 minor allele carrier vs. major allele homozygotes on mortality estimated from the Cox regression model.
*** Number of subjects with a CMD, without a CMD.
**** Haplotype comprised minor allele frequencies of SNPs rs2253310 (C), rs2802288 (A), rs2802292 (G), rs2764264 (C), rs9398171 (C), rs12212067 (G), and rs3800230 (G).
DISCUSSION
Here we show for the first time that longevity-associated genetic variants of FOXO3 are associated with much lower mortality in elderly individuals who have one or more of the common aging-related conditions consisting of diabetes, CHD and hypertension. In contrast, no reduction in mortality was observed in men with a CMD who did not have the longevity genotype. In addition, surprisingly, in men with CMD and the FOXO3 longevity genotype, mortality was reduced to normal (i.e., to that of men without a CMD). We propose that longevity-associated FOXO3 variants confer resilience against the adverse medical conditions that comprise CMD.
FoxO3 serves as a core regulator of cellular homeostasis, stress response, and longevity through its ability to modulate a variety of stress responses during nutrient shortage, oxidative stress, hypoxia, heat shock, and DNA damage [14–17]. By reducing oxidative damage responsible for aging, FoxO3-mediated responses to stress are pivotal to health-span and lifespan [18]. Depending on the stress stimulus and subcellular context, once activated, FoxO3 can induce specific sets of nuclear genes, including cell cycle inhibitors, pro-apoptotic genes, scavengers of reactive oxygen species, autophagy effectors, gluconeogenic enzymes, and others [17]. On the other hand, under glucose restriction, FoxO3 translocates to mitochondria to stimulate transcription of oxidative phosphorylation genes, thus restoring cellular ATP levels [17]. FoxO3 target genes and the pathways that their gene products serve are diverse and sometimes antagonistic, meaning FoxO3 is an adaptable player in the dynamic homeostasis of normal and stressed cells [17].
In unstimulated lymphoblast cell lines, FOXO3 mRNA expression is significantly higher in rs2802292 GT vs. TT cells, and in response to H2O2-mediated stress, expression increases for each genotype (P < 0.0001), reaching a 3-fold higher level in GT than TT cells [8]. In this study fluorescence in situ hybridization showed that the nuclear position of FOXO3 changed with regards to its neighbours over a >7 Mb region following stress induction, bringing distant genes into proximity with FOXO3. These data indicated that the longevity haplotype is better “primed” for stress response than the common haplotype [8]. If there is no cellular stress, then FOXO3 appears to play only a small part in cell resilience.
The coronary artery and aortic arch are prone to atherosclerosis and thus CHD. FoxO3 is a central protective factor in safeguarding primate vascular homeostasis – it serves as a master regulator and key driver of aortic and coronary vascular endothelial aging [19]. FoxO3 is the top regulatory factor affecting primate vascular aging, having the highest number of target genes [19]. In various tissues, dysregulation of the developmental program is increasingly regarded as pivotal to cellular aging [20, 21]. A key player in this process is the developmental regulator FOXA2, which becomes upregulated in aging vessels. Crucially, in human aortic endothelial cells, FOXA2 binding to the FOXO3 promoter suppresses FOXO3 expression and compromises cell proliferation, the effect being reversed by siRNA-mediated knockdown of FOXA2 expression [19]. Interestingly the most 5’ SNP in our FOXO3 longevity haplotype was rs768023, located in the promoter region, and which we noted previously is the binding site for both FOXA2 and HDAC2 [8]. HDAC2 also deacetylates FoxO3 thus activating it [22]. For the rs768023 minor allele, binding of FOXA2 and HDAC is abolished, which would increase FOXO3 expression. The SNP immediately 3’ of rs768023 in the 14-SNP haplotype block, but not included in the present study, is rs1536057, the minor (A) allele of which abolishes NRF1 and E2F sites and creates a POU5F1 site. The inhibition of FOXO3 expression by E2F [23] would thereby be lost, so causing an elevation in FOXO3 expression. NRF1 and POU5FI are involved in cell proliferation and stem cell maintenance, while E2F is involved in DNA damage response.
The full complement of transcription factors that exhibit allele-dependent binding to SNPs in the 14-SNP FOXO3 haplotype block are shown in Supplementary Table 2. Of these, HDAC1, NRF1, POU5F1, TFCP2L1, MYOG, NKX3-1, MITF, MEF2A, MZF1, NR2F1, PRDM1, FOXP1, MZF1, and ONECUT1 are involved in cell proliferation/stem cell maintenance, whereas HNF3, FOXA2, HNF4A, and ONECUT1 (HNF6) are involved in diabetes/energy response, and E2F1 is involved in DNA damage repair.
We have shown previously that the G allele of FOXO3 SNP rs2802292 protects against risk of death from CHD [11]. G allele carriers had lower plasma TNF-α than non-carriers, suggesting an ability to reduce inflammation as a potential mediating factor for reduction of CHD mortality risk [24]. At the cell and molecular level, cardiovascular benefits of FoxO3 involve stimulation of stress resistance by up-regulation of mitochondrial superoxide dismutase, peroxisomal catalase, and peroxiredoxin expression [25], as well as the promotion of multiple beneficial vascular functions, and reversal of cellular aging through pro-proliferative effects that involve downregulation of CSRP1, which recruits sirtuin-1 to create a suppressive chromatin environment [26]. Longevity-associated genetic variants of FOXO3 are also associated with lower blood pressure and reduced hypertension prevalence [27]. The association of FOXO3 SNPs with self-rated health in individuals aged 75–87 is influenced solely by cardiovascular disease [28].
Various types of cellular stress induce the recruitment of the evolutionarily conserved transcription factor HSF1 to binding sites involved in controlling gene expression [29]. The fact that only in G allele carriers of FOXO3 SNP rs2802292 possess the HSF1 binding site [30] explains, at least in part, the association we found between this allele and resistance to mortality in CMD subjects, presumably mediated by HSF1-induced expression of FOXO3. Binding of HSF1 to its target site in rs2802292 leads to formation of a complex resulting in a promoter-enhancer interaction involving the upstream promoter region and the rs2802292 region of intron 2 [30]. Stress resistance and elevated FOXO3 transcription were seen following application of the potent oxidative stress inducer H2O2 in human dermal fibroblasts having the GG genotype, but not in those with the TT genotype, thus confirming the mechanism [30]. Unlike cells with the TT genotype, GG cells also exhibited higher expression of FoxO3 target genes SOD2, CAT, GADD45A, CCND1, RBL2, BCL2L11 and BCL6 in a HSF1-dependent manner [30]. Furthermore, GG cells displayed a significantly better DNA repair response. The FOXO3 SNP rs2802292 region also contains transcription factor response elements for SP1, GATA1 and ESR1 [30].
Our finding of 13% higher prevalence of diabetes at baseline in FOXO3 rs2800292 G-allele carriers is likely due to attrition of non-protective TT genotypes at earlier ages owing to mortality from cardiovascular disease (e.g., CHD, stroke) and other complications of diabetes, thereby enriching G-allele frequency in the cohort. In a twin study, the FOXO3 rs2800292 G allele was associated with more favourable insulin sensitivity and increased FOXO3 expression [31]. The protection against mortality afforded by this mechanism would explain our finding that diabetes alone was associated with longer lifespan in G-allele carriers compared with TT subjects. As we have reported previously, the FOXO3 longevity genotype involves a haplotype that includes at least 14 SNPs that work in concert [8].
We reported previously that the FOXO3 rs2800292 G allele was associated with reduced risk of hypertension in women, but not men, from a cohort of Japanese individuals living in Japan [32]. Our finding in the present study that G-allele carriers have lower mortality could, as shown here for individuals with diabetes, be due to a higher mortality rate for those with the TT genotype, in this instance from hypertension-related cardiovascular events, similar to what has been demonstrated by one of us for ACE [33].
In conclusion, we have found for the first time that remaining lifespan of elderly men with longevity-associated alleles of FOXO3 who have one or more chronic conditions of aging, specifically diabetes, and/or hypertension, and/or CHD (collectively, CMD), live as long as elderly men who lack any of these life-threatening conditions. Thus, favorable FOXO3 genotype can abrogate the increased risk for mortality in men with prevalent CMD. Since FOXO3 genotype has no effect on lifespan of men without a CMD, the well-established understanding of FOXO3 as being a longevity gene is because of an effect on elderly individuals who are at higher risk because they have a CMD.
MATERIALS AND METHODS
Study cohort
Participants were American men of Japanese ancestry living on the island of Oahu, Hawaii. They were recruited in 1965–1968 from World War II Selective Service records for the Kuakini Honolulu Heart Program (KHHP) [34], which continued from 1991 onwards as the Kuakini and Honolulu-Asia Aging Study (KHAAS) [4, 34–36]. The analysis was conducted as part of the Kuakini Hawaii Lifespan Study and the Kuakini Hawaii Healthspan Study, an embedded cohort study of healthy aging drawn from the original KHHP-KHAAS population. Subjects had parents who were both from a limited geographic area of Japan, mostly the western, central and southern regions [34, 37]. Subjects were recruited at the same time and place (Oahu), meaning there was no apparent reason why genetic background should be substantially different. The KHHP cohort is quite robust for phenotype-genotype associations, since the data collection was exceptionally accurate and involved cross validation utilizing an expert Morbidity and Mortality Committee. The Hawaii Japanese population is from Japan, with little outbreeding and, based on the authors’ unpublished data, exhibits a smaller degree of genetic diversity than the overall population of Japan.
All participants in the current study were interviewed at Examination 4 of the KHHP (1991–1993). Archived phenotypic data and blood samples from Examination 4 of the KHHP (1991–1993), which coincided with the commencement of the KHAAS, were used as the baseline examination for our study. The KHAAS was begun as an expansion of the KHHP for the study of neurodegenerative diseases, cognitive function, and other aging phenotypes in elderly persons. From 1991–1993, all of the survivors of the KHHP cohort, ranging in age from 71–93 years (mean age: 77.9 ± 4.7 years), were invited to the 4th examination. Response rate was 80% of survivors (including clinic, home, and nursing home visits; n = 3,741).
The study involved 3,584 of 3,741 men aged 71 to 93 (mean age 77.9 ± 4.7 SD years) for whom we had banked DNA so making them eligible for inclusion. Of the 3,584 men, 3,548 had died (mean age at death 89.0±6.2 SD years; range 72–108 years) and 36 were still alive (mean age 101.6 ± 1.9 SD years; range 100–108 years) at the end of the follow-up period, 31 December 2019.
The KHHP was a longitudinal observation study. Subjects from the KHHP/KHAAS population [4] had been followed with regular examinations and blood work until 2015, or death up to the end of 2019.
Information on the prevalence of CHD, stroke, and cancer was identified by the KHHP surveillance system (review of hospital records by an expert panel or matching to Tumor Registry for Cancer). Hypertension was defined as systolic/diastolic blood pressure ≥160/95 mmHg or on anti-hypertensive medication at baseline. Diabetes was defined by fasting serum glucose ≥126 mg/dL or 2-hour post-load glucose ≥200 mg/dL or taking insulin and/or oral hypoglycemic medications at baseline.
Procedures performed were in accord with institutional guidelines and were approved by the Institutional Review Board of Kuakini Medical Center. Written informed consent was obtained from all study participants or from family representatives, if participants could not provide consent.
Genotyping
Leucocyte DNA obtained from participants was used for genotyping. Recruitment, demographic characteristics, and DNA extraction were as described previously [4, 38]. In the HHP cohort, the FOXO3 longevity haplotype was defined by 14 SNPs (in order of location 5’ to 3’ with minor [longevity-associated] allele shown in brackets): rs768023 (G allele), rs1536057 (T), rs2253310 (C), rs2802288 (A), rs2802292 (G), rs2764264 (C), rs12202234 (G), rs17069665 (G), rs12213895 (A), rs12212067 (G), rs9398171 (C), rs73763159 (T), rs3800230 (G), and rs1935952 (G)) [8]. Longitudinal genotype data were obtained for 8 of these SNPs. All 14 affect transcription factor binding sites. Specifically, the minor allele of rs2802292 creates a HSF1 binding site in intron 2 of FOXO3 [30], rs1935952 disrupts an MZF1 binding site, rs2253310 minor allele creates a TFCP2L1 binding site, rs2764264 disrupts a NKX3 binding site, rs2802288 creates a MYF binding site, rs3800230 disrupts a FoxP1 binding site, rs9398171 creates NR2F1 and HNF4 site and abolishes a HNF6 site, and rs12212067 creates a MZF1 site [8]. The SNP rs768023 (not included in the present longitudinal study) abolishes both a FOXA2 and a HDAC2 recognition site [8].
Genotyping was performed by allelic discrimination assays using TaqMan® (Applied Biosystems, Inc.) and a Life Technologies QuantStudio 12K Flex OpenArray system. Although 88% of participants were born in Hawaii, there is a theoretical possibility of confounding of case vs. control status for allele frequencies due to geographic origin. Therefore, for certain analyses, cases and controls were stratified by parental prefecture of origin using conditional logistic regression models.
Statistical analyses
General linear models were used to compare age-adjusted indirect measurements between groups, and logistic models were used to compare the age-adjusted direct measurements. Cox proportional models were used to assess the association of FOXO3 minor-allele carriers with mortality stratified by disease status, such as by CHD, by diabetes, by hypertension, and by chronic conditions defined by presence of any of CHD, diabetes, or hypertension. The Cox proportional hazard assumption was tested for each Cox model. The effect of interaction of disease with FOXO genotype on mortality was tested in the Cox model. All statistical analyses were performed using the Statistical Analysis System version 9.4 [39]. Figures were generated using the STATA 12 Graphics [40].
Supplementary Material
ACKNOWLEDGMENTS
The authors thank all study participants and their families for their cooperation and the Hawaii State Department of Health for its help. The authors wish to acknowledge Dr. Alvin T. Onaka, Brian Horiuchi, and Caryn Tottori of the Hawaii State Department of Health for providing death certificate data on cause of death for the KHHP participants, Ms. Ayako Elliott and Ms. Eva Ardo for assistance with genotyping, and Ms. Hiromi Nakada and Ms. Ka-on Fong for monitoring the vital status of KHHP participants.
Footnotes
AUTHOR CONTRIBUTIONS: R.C., B.J.M., T.A.D., K.H.M., D.C.W., R.C.A., B.J.W. contributed to the study concept and design; R.C. carried out the statistical analyses; K.H.M. supervised recruitment and data collection; T.A.D. supervised the genotyping; B.J.M. drafted the manuscript; R.C., B.J.M., T.A.D., K.H.M., P.M.C.D., R.C.A., D.C.W. and B.J.W. provided critical input into data interpretation and manuscript preparation.
CONFLICTS OF INTEREST: These authors declare no conflicts of interest.
FUNDING: This work was supported by the Kuakini Medical Center, the US National Institutes of Health (contract N01-AG-4-2149, Grants 5 U01 AG019349-05, 5R01AG027060 [Kuakini Hawaii Lifespan Study], 5R01AG038707 [Kuakini Hawaii Healthspan Study]), 1P20GM125526-01A1 [Kuakini Center of Biomedical Research Excellence for Clinical and Translational Research on Aging], and contract N01-HC-05102 from the National Heart Lung and Blood Institute.
REFERENCES
- 1.Morris BJ, Willcox BJ, Donlon TA. Genetic and epigenetic regulation of human aging and longevity. Biochim Biophys Acta Mol Basis Dis. 2019; 1865:1718–44. 10.1016/j.bbadis.2018.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet. 2013; 132:1323–38. 10.1007/s00439-013-1342-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg NV, Rodríguez-Girondo M, de Craen AJ, Houwing-Duistermaat JJ, Beekman M, Slagboom PE. Longevity around the turn of the 20th century: life-long sustained survival advantage for parents of today’s nonagenarians. J Gerontol A Biol Sci Med Sci. 2018; 73:1295–302. 10.1093/gerona/gly049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008; 105:13987–92. 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, Yu L, Arnold AM, Aspelund T, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015; 70:110–18. 10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris BJ, Willcox DC, Donlon TA, Willcox BJ. FOXO3: a major gene for human longevity—a mini-review. Gerontology. 2015; 61:515–25. 10.1159/000375235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deelen J, Evans DS, Arking DE, Tesi N, Nygaard M, Liu X, Wojczynski MK, Biggs ML, van der Spek A, Atzmon G, Ware EB, Sarnowski C, Smith AV, et al. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat Commun. 2019; 10:3669. 10.1038/s41467-019-11558-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlon TA, Morris BJ, Chen R, Masaki KH, Allsopp RC, Willcox DC, Elliott A, Willcox BJ. FOXO3 longevity interactome on chromosome 6. Aging Cell. 2017; 16:1016–25. 10.1111/acel.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijkelenboom A, Mokry M, de Wit E, Smits LM, Polderman PE, van Triest MH, van Boxtel R, Schulze A, de Laat W, Cuppen E, Burgering BM. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol. 2013; 9:638. 10.1038/msb.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlon TA, Willcox BJ, Morris BJ. FOXO3 cell resilience gene neighborhood. Aging (Albany NY). 2017; 9:2467–68. 10.18632/aging.101349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willcox BJ, Tranah GJ, Chen R, Morris BJ, Masaki KH, He Q, Willcox DC, Allsopp RC, Moisyadi S, Poon LW, Rodriguez B, Newman AB, Harris TB, et al. The FoxO3 gene and cause-specific mortality. Aging Cell. 2016; 15:617–24. 10.1111/acel.12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, et al. , and Study of Osteoporotic Fractures. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009; 8:460–72. 10.1111/j.1474-9726.2009.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuningas M, Mägi R, Westendorp RG, Slagboom PE, Remm M, van Heemst D. Haplotypes in the human Foxo1a and Foxo3a genes; impact on disease and mortality at old age. Eur J Hum Genet. 2007; 15:294–301. 10.1038/sj.ejhg.5201766 [DOI] [PubMed] [Google Scholar]
- 14.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002; 419:316–21. 10.1038/nature01036 [DOI] [PubMed] [Google Scholar]
- 15.Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011; 286:7468–78. 10.1074/jbc.M110.179242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014; 39:159–69. 10.1016/j.tibs.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fasano C, Disciglio V, Bertora S, Lepore Signorile M, Simone C. FOXO3a from the nucleus to the mitochondria: a round trip in cellular stress response. Cells. 2019; 8:1110. 10.3390/cells8091110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurkar AU, Robinson AR, Cui Y, Li X, Allani SK, Webster A, Muravia M, Fallahi M, Weissbach H, Robbins PD, Wang Y, Kelley EE, Croix CM, et al. Dysregulation of DAF-16/FOXO3A-mediated stress responses accelerates oxidative DNA damage induced aging. Redox Biol. 2018; 18:191–99. 10.1016/j.redox.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Zhang S, Yan P, Ren J, Song M, Li J, Lei J, Pan H, Wang S, Ma X, Ma S, Li H, Sun F, et al. A single-cell transcriptomic landscape of primate arterial aging. Nat Commun. 2020; 11:2202. 10.1038/s41467-020-15997-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ermolaeva M, Neri F, Ori A, Rudolph KL. Cellular and epigenetic drivers of stem cell ageing. Nat Rev Mol Cell Biol. 2018; 19:594–610. 10.1038/s41580-018-0020-3 [DOI] [PubMed] [Google Scholar]
- 21.Whitton H, Singh LN, Patrick MA, Price AJ, Osorio FG, López-Otín C, Bochkis IM. Changes at the nuclear lamina alter binding of pioneer factor Foxa2 in aged liver. Aging Cell. 2018; 17:e12742. 10.1111/acel.12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beharry AW, Sandesara PB, Roberts BM, Ferreira LF, Senf SM, Judge AR. HDAC1 activates FoxO and is both sufficient and required for skeletal muscle atrophy. J Cell Sci. 2014; 127:1441–53. 10.1242/jcs.136390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q, Peng S, Tao L, Ruan H, Yang Y, Li TM, Adams U, Meng S, Bi X, Dong MQ, Yuan Z. E2F transcription factor 1 regulates cellular and organismal senescence by inhibiting forkhead box O transcription factors. J Biol Chem. 2014; 289:34205–13. 10.1074/jbc.M114.587170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willcox BJ, Morris BJ, Tranah GJ, Chen R, Masaki KH, He Q, Willcox DC, Allsopp RC, Moisyadi S, Gerschenson M, Davy PM, Poon LW, Rodriguez B, et al. Longevity-associated FOXO3 genotype and its impact on coronary artery disease mortality in Japanese, whites, and blacks: a prospective study of three American populations. J Gerontol A Biol Sci Med Sci. 2017; 72:724–28. 10.1093/gerona/glw196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiribau CB, Cheng L, Cucoranu IC, Yu YS, Clempus RE, Sorescu D. FOXO3A regulates peroxiredoxin III expression in human cardiac fibroblasts. J Biol Chem. 2008; 283:8211–17. 10.1074/jbc.M710610200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan P, Li Q, Wang L, Lu P, Suzuki K, Liu Z, Lei J, Li W, He X, Wang S, Ding J, Chan P, Zhang W, et al. FOXO3-engineered human ESC-derived vascular cells promote vascular protection and regeneration. Cell Stem Cell. 2019; 24:447–61.e8. 10.1016/j.stem.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 27.International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011; 478:103–9. 10.1038/nature10405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zettergren A, Kern S, Rydén L, Östling S, Blennow K, Zetterberg H, Falk H, Skoog I. Genetic variation in FOXO3 is associated with self-rated health in a population-based sample of older individuals. J Gerontol A Biol Sci Med Sci. 2018; 73:1453–58. 10.1093/gerona/gly021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vihervaara A, Sistonen L. HSF1 at a glance. J Cell Sci. 2014; 127:261–66. 10.1242/jcs.132605 [DOI] [PubMed] [Google Scholar]
- 30.Grossi V, Forte G, Sanese P, Peserico A, Tezil T, Lepore Signorile M, Fasano C, Lovaglio R, Bagnulo R, Loconte DC, Susca FC, Resta N, Simone C. The longevity SNP rs2802292 uncovered: HSF1 activates stress-dependent expression of FOXO3 through an intronic enhancer. Nucleic Acids Res. 2018; 46:5587–600. 10.1093/nar/gky331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banasik K, Ribel-Madsen R, Gjesing AP, Wegner L, Andersson A, Poulsen P, Borglykke A, Witte DR, Pedersen O, Hansen T, Vaag A. The FOXO3A rs2802292 G-allele associates with improved peripheral and hepatic insulin sensitivity and increased skeletal muscle-FOXO3A mRNA expression in twins. J Clin Endocrinol Metab. 2011; 96:E119–24. 10.1210/jc.2010-0881 [DOI] [PubMed] [Google Scholar]
- 32.Morris BJ, Chen R, Donlon TA, Evans DS, Tranah GJ, Parimi N, Ehret GB, Newton-Cheh C, Seto T, Willcox DC, Masaki KH, Kamide K, Ryuno H, et al. Association analysis of FOXO3 longevity variants with blood pressure and essential hypertension. Am J Hypertens. 2016; 29:1292–300. 10.1093/ajh/hpv171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris BJ, Zee RY, Schrader AP. Different frequencies of angiotensin-converting enzyme genotypes in older hypertensive individuals. J Clin Invest. 1994; 94:1085–89. 10.1172/JCI117423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worth RM, Kagan A. Ascertainment of men of Japanese ancestry in hawaii through world war II selective service registration. J Chronic Dis. 1970; 23:389–97. 10.1016/0021-9681(70)90022-6 [DOI] [PubMed] [Google Scholar]
- 35.Kagan A, Harris BR, Winkelstein W Jr, Johnson KG, Kato H, Syme SL, Rhoads GG, Gay ML, Nichaman MZ, Hamilton HB, Tillotson J. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California: demographic, physical, dietary and biochemical characteristics. J Chronic Dis. 1974; 27:345–64. 10.1016/0021-9681(74)90014-9 [DOI] [PubMed] [Google Scholar]
- 36.Yano K, Reed DM, McGee DL. Ten-year incidence of coronary heart disease in the Honolulu Heart Program. Relationship to biologic and lifestyle characteristics. Am J Epidemiol. 1984; 119:653–66. 10.1093/oxfordjournals.aje.a113787 [DOI] [PubMed] [Google Scholar]
- 37.Kagan A, Ed. (1996) The Honolulu Heart Program: An Epidemiological Study of Coronary Heart Disease and Stroke. Amsterdam, The Netherlands: Harwood Academic Publishers. [Google Scholar]
- 38.Donlon TA, Curb JD, He Q, Grove JS, Masaki KH, Rodriguez B, Elliott A, Willcox DC, Willcox BJ. FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci. 2012; 67:1132–39. 10.1093/gerona/gls067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Statistical Analysis System (SAS) version 9.4. SAS Institute, Cary, NC, USA. https://libguides.library.kent.edu/statconsulting/SAS
- 40.StataCorp LP. Stata Statistical Software: Release 12. College Station, TX. 2011. https://www.coursehero.com/file/16695740/stata/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.