Abstract
The sanitary emergency generated by the pandemic COVID-19, instigates the search for scientific strategies to mitigate the damage caused by the disease to different sectors of society. The disease caused by the coronavirus, SARS-CoV-2, reached 216 countries/territories, where about 20 million people were reported with the infection. Of these, more than 740,000 died. In view of the situation, strategies involving the development of new antiviral molecules are extremely important. The present work evaluated, through molecular docking assays, the interactions of 4′-acetamidechalcones with enzymatic and structural targets of SARS-CoV-2 and with the host’s ACE2, which is recognized by the virus, facilitating its entry into cells. Therefore, it was observed that, regarding the interactions of chalcones with Main protease (Mpro), the chalcone N-(4′[(2E)-3-(4-flurophenyl)-1-(phenyl)prop-2-en-1-one]) acetamide (PAAPF) has the potential for coupling in the same region as the natural inhibitor FJC through strong hydrogen bonding. The formation of two strong hydrogen bonds between N-(4[(2E)-3-(phenyl)-1-(phenyl)-prop-2-en-1-one]) acetamide (PAAB) and the NSP16-NSP10 heterodimer methyltransferase was also noted. N-(4[(2E)-3-(4-methoxyphenyl)-1-(phenyl)prop-2-en-1-one]) acetamide (PAAPM) and N-(4-[(2E)-3-(4-ethoxyphenyl)-1-(phenyl)prop-2-en-1-one]) acetamide (PAAPE) chalcones showed at least one strong intensity interaction of the SPIKE protein. N-(4[(2E)-3-(4-dimetilaminophenyl)-1-(phenyl)-prop-2-en-1-one]) acetamide (PAAPA) chalcone had a better affinity with ACE2, with strong hydrogen interactions. Together, our results suggest that 4′-acetamidechalcones inhibit the interaction of the virus with host cells through binding to ACE2 or SPIKE protein, probably generating a steric impediment. In addition, chalcones have an affinity for important enzymes in post-translational processes, interfering with viral replication.
Keywords: Chalcone, Molecular docking, SPIKE, ACE2
Graphical abstract
1. Introduction
The health problem caused by the COVID-19 pandemic can be measured by the numbers of cases and deaths confirmed by the disease globally. In Brazil, community transmission is observed, with the collapse of the Health System in some regions. As it is an infection triggered by a new coronavirus (SARS-CoV-2), the pathophysiology of COVID-19 is little known and there is no specific treatment for the disease [1,2].
The search for new candidates for antiviral drugs has made great progress in recent years with the discovery of molecular targets, the development of organic synthesis and the discovery of new bioactives substances. A big number of techniques have been used in the search for new antiviral drugs. Despite the great progress, the arsenal of antiviral drugs is still small [1]. In this sense, strategies involving the development and validation of new antiviral molecules have been considered.
Chalcones, known as α, β-unsaturated ketones (1,3-diaryl-2-propene-1-one) are a class of naturally occurring compounds belonging to the flavonoid family. They can be obtained from natural sources or by synthesis, and are widely distributed in fruits, vegetables, and tea [3]. The double connection together with carbonyl group are possibly responsible for diverse biological activities such as antibacterial, antioxidant, anti-inflammatory and antiparasitic [4].
Antiviral properties of chalcones have been recorded in studies with plant viruses and human rhinoviruses [5]. Antiviral studies [6] with chalcones containing hydroxy and methoxy groups, confirm that the activity is dependent on the nature of the group and its positions in the aromatic rings. Santos [7] reports in a recent study the evaluation of the antiviral activity of hydroxychalcones and synthetic curcuminoids against infection caused by HPV in vitro.
Therefore, in this work, for the first time acetamide chalcones will be study theoretically by the Molecular Docking to characterize the inhibition power of the chalcones with the enzyme Mpro, methyltransferase, the SPIKE, and ACE2 proteins by the interaction energy and the distance of the compounds and the target protein’s amino acids.
2. Material and methods
2.1. Chalcones
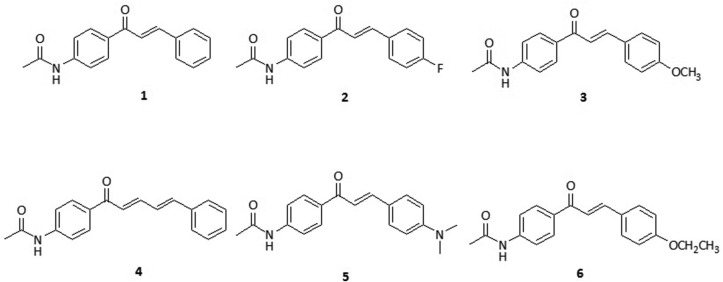
Using the methodological principle of synthesis the Claisen-Schmith reaction (in basic medium) [8], chalcones were synthesized from benzaldehydes and 4-aminoacetophenone, both at a concentration of 2 mmol. The reagents were added in a volumetric flask (25 mL), to which 5 mL of ethanolic NaOH solution (50%) were added. After adding the ethanol solution, the mixtures were kept under stirring for 48 h (at room temperature). TLC (n-hexane: ethyl acetate, 2: 1) was used to monitor the progress of the reaction. After 48 h, the reaction mixture was neutralized with diluted HCl (10%) and ice water added. The products were obtained using the filtration technique under reduced pressure, washing with cold water and recrystallization in ethanol. To obtain the 4′-acetamidecalcones (1–6) (Fig. 1 ), the acetylation reaction of the 4′-aminocalcones (2 mmol) with acetic anhydride (2 mmol) in buffered medium (5 mL) at pH = 5.0 with AcOH/AcONa was used [9].
Fig. 1.
Structural representation of the chalcones (1) PAAB (2) PAAPF (3) PAAPM (4) PAACN (5) PAAPA (6) PAAPE.
From the data available in the database, Protein Data Bank (https://www.rcsb.org/), the structures of the following target proteins were obtained: Main protease COVID-19 (Mpro), NSP16-NSP10 SARS-CoV −2, SPIKE, and ACE2. Then, the proteins were prepared for analysis by removing all residues and adding polar hydrogens, producing favorable protonation states for molecular docking. The Main protease COVID-19 (Mpro) was identified in the repository as “The crystal structure of COVID-19 main protease in complex with an inhibitor N3” (PDB ID: 6LU7). The structure of this enzyme is deposited in the Protein Data Bank with a resolution of 2.16 Å, determined from X-ray diffraction R-Value Free: 0.235, R-Value Work: 0.202, R-Value Observed: 0.204), classified as viral protein, Bat SARS-like coronavirus organism and Escherichia coli BL21 (DE3) expression system [10].
The enzyme structure of the methyltransferase complex, the NSP16-NSP10 SARS-CoV-2 heterodimer, was identified in the repository as “1.98 Å Resolution Crystal Structure of NSP16-NSP10 Heterodimer from SARS-CoV-2 in Complex with Sinefungin” (PDB ID: 6WKQ). The structure was deposited in the Protein Data Bank with a resolution of 1.98 Å, determined from X-ray diffraction (R-Value Free: 0.180, R-Value Work: 0.162), classified as viral protein, severe acute respiratory syndrome coronavirus 2, and expression system Escherichia coli BL21 (DE3), Escherichia coli BL21 [11].
The structures of the SPIKE protein and the ACE2 enzyme were identified in the repository as “Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2” (PDB ID: 6MOJ). The structures were deposited in the Protein Data Bank with a resolution of 2.45 Å, determined from X-ray diffraction (R-Value Free: 0.227, R-Value Work: 0.192, R-Value Observed: 0.194), classified as viral protein/hydrolase, Homo sapiens organism, severe acute respiratory syndrome coronavirus 2, and Trichoplusia ni expression system [12].
2.2. Molecular docking
The interaction simulations between the selected inhibitors and proteins were performed using AutoDock Vina code (version 1.1.2), using 3-way multithreading, Lamarkian Genetic Algorithm [13]. The docking parameters: grid box sizes, centers, spacing and exhaustiveness to the proteins are given in Table S1 (Supplementary material). All grid boxes were configured to fit all the protein in the simulation for seeking the greater amplitude in the selection of molecular positions. As a standard procedure, one hundred (100) independent simulations were performed for all the target proteins, and it was obtained ten (10) positions each. As selection criteria, the simulations that showed positions with free binding energy (ΔG) below −6.0 kcal mol−1 [14] and RMSD (Root Mean Square Deviation) values less than two (2.0) [15] were analyzed.
For results analysis, image plotting, and generation of bi and tri-country maps, the Discovery Studio Visualizer [16] and UCSF Chimera [17] codes were used. For statistical analysis, the Morpheus® online server (https://software.broadinstitute.org/morpheus/) was used, in which heat maps were generated to identify the ligand-residue interaction and similarity profiles by the Pearson statistical test [18].
Based on observations of the interactions of the molecules with the enzyme, the hydrogen bonds were plotted and classified according to previous studies that group interactions with distances between 2.5 and 3.1 Å as strong, from 3.1 to 3.55 Å as average and >3.55 Å as weak [19].
3. Results
3.1. Interaction between the 4′-acetamidechalcones molecules and the Main protease (Mpro)
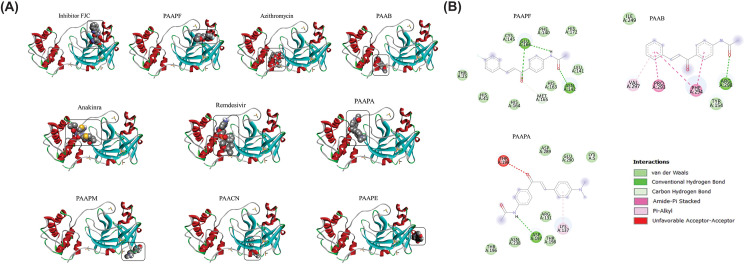
The positions shown in Fig. 2 A were obtained from the simulations of interactions of 4′-acetamidechalcones with Mpro by molecular docking. After comparative analyzes, it was noted that the PAAPF interacted with the enzyme at a site like that of the FJC inhibitor. Likewise, PAAB interacted at the same site as the antibiotic Azithromycin. Also, the reference drugs Anakinra and Remdesivir interacted in common sites, the same as for PAAPA. Additionally, PAAPM, PAACN, and PAAPE interacted with distinct sites from each other and different from any other reference inhibitor.
Fig. 2.
Theoretical calculations of the interaction between the FJC inhibitor, the reference drugs, and the chalcone derivatives with the enzyme Mpro (A). The bi-dimensional map of the hydrogen bonds and the hydrophobic interaction of the chalcone derivatives with the enzyme Mpro (B).
To analyze the intensity and affinity of the interactions obtained in the molecular docking simulations, the values of the interaction energy and RMSD were collected, plotted, and compared (Table S2, Supplementary material). Interactions with energy < −6.0 kcal mol−1 and RMSD <2.0 were considered satisfactory. Therefore, based on these criteria, only the chalcone PAACN showed an affinity of −6.1 kcal mol−1 with the enzyme Mpro, although this molecule does not have a binding site in common with the inhibitors, especially the natural inhibitor FJC. Concerning the reference inhibitor, azithromycin did not show good affinity when presenting energy of −5.8 kcal mol−1.
To examine and describe the intrinsic characteristics of the interactions of 4′-acetamidechalcones molecules with the enzyme, the hydrogen bonds, and hydrophobic interactions were highlighted and compared with the reference ligands (Fig. 2B). Since PAAPM, PAACN, and PAAPE did not show similarities with the reference ligands, those molecules were not considered for analysis. Table S3 (Supplementary material) shows the interaction distances between chalcones and the amino acid residues of the enzyme Mpro. Thus, detailed information about the interactions of the molecules with the enzyme was obtained. Initially, for the PAAPF chalcone, which has the potential for coupling in the same region as the FJC inhibitor, it has two significant interactions with the GLU166 residue, one of which is a strong hydrogen bond. The PAAB chalcone presented only interaction of hydrogen with the residue ARG298, which the Azithromycin did not interact; moreover, the Azithromycin presents most hydrophobic interactions, all with a distance higher than 3.5 Å. The PAAPA chalcone interacted with the enzyme through two hydrogen bonds, highlighting the ASP197 residue in common with Anakinra and Remdesivir; it is important to notice that, among the reference drugs, Remdesivir interacted with the enzyme through five hydrogen bonds, mostly of moderate to strong intensity.
3.2. Interaction with the methyltransferase heterodimer NSP16-NSP10 SARS-CoV-2
The interactions of chalcones with the NSP16-NSP10 are illustrated in Fig. 3 A. Among the studied structures, PAACN and PAAPE did not show significant interaction with the enzyme. Besides, the other chalcones interacted in similar places. Table S4 (Supplementary material) lists the energies of the interactions and the respective RMSD; thus, it was possible to observe affinity values < −6.0 kcal mol−1 for the four chalcones, obtaining values of up to −8.2 kcal mol−1 for PAAPF. All RMSD values were less than 2.0 Å, suggesting a satisfactory interaction between the molecules and the enzyme.
Fig. 3.
Simulated interaction between the chalcones derivatives and the enzyme NSP16-NSP10 SARS-CoV-2 heterodimer methyltransferase (A). The bi-dimensional map of the hydrogen bonds and the hydrophobic interaction between the chalcones derivatives and the enzyme NSP16-NSP10 SARS-CoV-2 heterodimer methyltransferase (B).
The specification of the interactions of chalcones with the enzyme is illustrated in the bi-dimensional maps contained in Fig. 3B. The distances of the interactions are specified in Table S4 (Supplementary material). Therefore, it is possible to understand the formation of two strong hydrogen bonds between PAAB and the methyltransferase. The other chalcones also had two hydrogen bonds each, however, with an intermediate to weak intensity.
3.3. Interactions with the SPIKE protein of the SARS-CoV-2 and with the ACE2
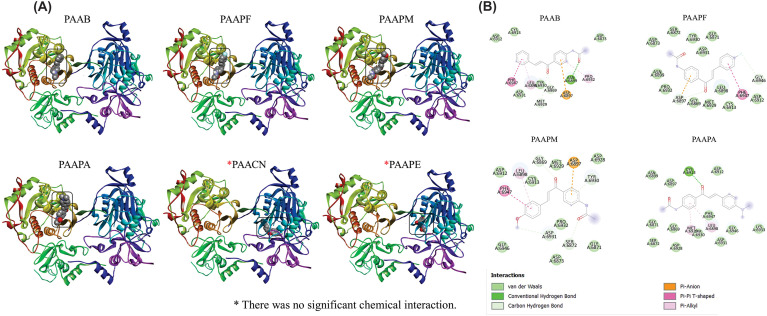
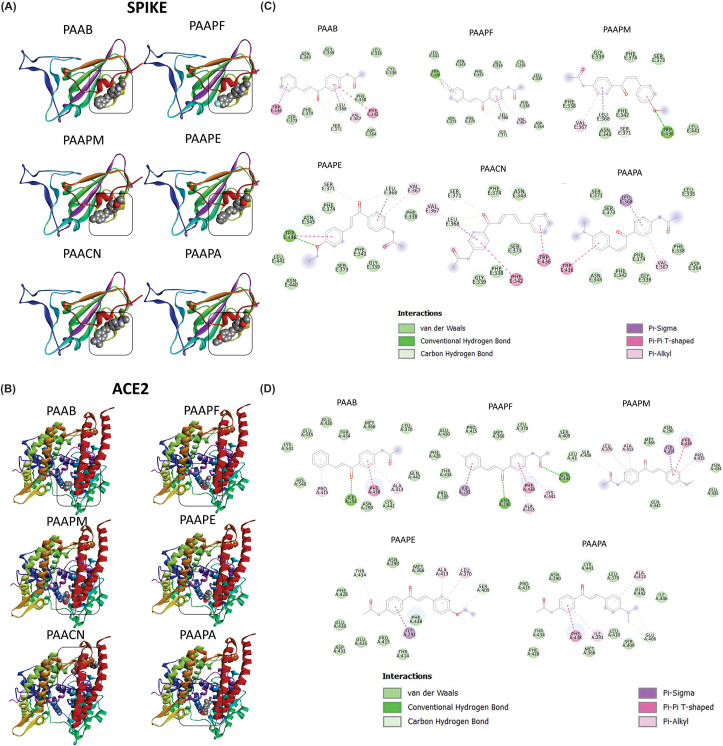
To study the potential of the chalcone acetamide derivatives to inhibit the interaction of the virus that causes COVID-19 with the target cells, a simulation of the interaction of the chalcones with the heterodimeric proteins of the virus, the Spike proteins, was carried out. The illustrations of the coupling simulations are contained in Fig. 4 A. It was possible to observe that all chalcones had connections with similar regions of the protein. According to data of interaction energy and RMSD contained in Table S2 (Supplementary material), it is observed that all chalcones had satisfactory affinity < −6.0 kcal/mol. In particular, the PAAPA and PAACN derivatives showed the best affinity values (−7.0 and −6.9, respectively), demonstrating the link of greater stability between chalcones.
Fig. 4.
Calculated interaction positions of the studied chalcones with the SPIKE (A) protein and the ACE2 protein of the human host (B). The bi-dimensional map of the hydrogen bonds and hydrophobic interactions between the chalcones and the SPIKE protein of the SARS-CoV-2 (C) and between the ACE2 protein of the human host (D).
The interactions with the amino acid residues of Spike protein heterodimer and the chalcones molecules were represented in bi-dimensional maps, corroborating with the information collected in the molecular docking simulations (Fig. 4C). Besides, the specific interactions of the chalcones derivatives with the Spike protein are shown in Table S5 (Supplementary material). It is possible to observe that the chalcone derivatives showed a pattern of interaction with the protein, as with the SER371 residue, which the derivatives interacted through the hydrogen of moderate intensity. Furthermore, the PAAPM and PAAPE chalcones showed, at least, one interaction of strong intensity, and the chalcone PAAPA did not show hydrogen bonds with the Spike protein.
Additionally, it is known that the interaction of the SPIKE protein with ACE2 is necessary for the virus to enter the host cells, hence the simulations of the interactions of the chalcone derivatives with this enzyme were performed, as shown in Fig. 4B. It is possible to observe that the chalcones interacted in similar sites, except for PAACN, which may have interacted at a non-specific site. Those data are reinforced by the affinity energy of the connections and RMSD, which the PAACN presented higher values, indicating an interaction of lower stability. PAAB and PAAPA had lower affinity values, with PAAPA having the best-suggested interaction, with an affinity of −8.0 kcal/mol associated with a low RMSD value (1355 Å).
Those results are reinforced when characterizing the interactions of the chalcones derivatives with the ACE2 protein, as illustrated in the bi-dimensional maps (Fig. 4D) and the interaction distances are shown in Table S5 (Supplementary material). The chalcones PAAPA and PAAPF stood out for presenting two interactions of strong hydrogen bonds. Also, the chalcones derivatives had common binding sites, such as the ILE291 residue, in which the PAAPA even had a hydrogen bond.
4. Discussion
Overall, the present work evaluated, through molecular docking assays, the interactions of chalcone acetamide derivatives with enzymatic and structural targets of SARS-CoV-2 and with the host’s ACE2, which is recognized by the virus, facilitating its entry into cells. Therefore, it was observed that, regarding the interactions of chalcones with Main protease (Mpro), the PAAPF derivative has the potential for coupling in the same region as the natural inhibitor FJC through strong hydrogen bonding. The formation of two strong hydrogen bonds between PAAB and the NSP16-NSP10 heterodimer methyltransferase was also noted. PAAPM and PAAPE showed at least one strong intensity interaction of the SPIKE protein. PAAPA had a better affinity with ACE2, with strong hydrogen interactions.
These results are relevant, as several chalcones have been described as having antiviral activity. A work performed by Park et al. [20] showed that chalcones isolated from Angelica Keiskei inhibit the chymotrypsin protease (3CL (pro)) and a papain protease (PL (pro)) in SARS-CoV. Proteases are important for post-translational modifications of structural proteins in the viral particle. Therefore, the inhibition of proteases interferes with the viral replication process [20]. Literature data suggest the inhibition of viral proteases by flavonoids and related compounds. For example, flavonoids such herbacetin, isobavachalcone, quercetin 3-β-d-glucoside and helicrysetin have been described as potent inhibitors of the Middle Eastern respiratory syndrome-coronavirus protease (MERS-CoV 3CLpro), indicating that flavonol and chalcones are favorite structures for binding to the catalytic site, suggesting that modifications in the more hydrophobic molecules or with carbohydrates attached to their main structures have a good inhibitory effect [21].
In fact, synthetic flavonoids and chalcones are described for potential antiviral properties. In a previous study, substituted chalcones, showed inhibition of viral translation in cells infected with hepatitis C virus (HCV) by the ablation of ribosomal protein phosphorylation 6 (rps6) [22]. Additionally, several reports in the literature demonstrate that derivatives of chalcones present antiviral activity better than the reference drugs in experimental models, including synergistic effects with these. For example, a previous study showed that thienyl-chalcone derivatives showed moderate to excellent antiviral activity, with higher in vitro potency against human cytomegalovirus compared to the standard drug Ganciclovir [23]. These data corroborate the findings of the present study, in which the chalcone derivatives interacted with the virus protease at sites and with similar affinities to the clinically used drugs and the theoretical inhibitor FJC.
Binding and inhibiting the enzymatic activity of proteases and methyltransferases can lead to a disruption in the viral capsid construction process, interrupting the flow of transmission. In this sense, the importance of in silico screening of phytochemical compounds is ratified, allowing a preliminary and rational analysis of a high number of molecules. For example, an investigation of phytochemicals as antiviral agents against dengue has shown that secondary phenolic plant metabolites such as alkaloids, terpenoids, chalcones, flavonoids, coumarins, and quinones have the potential to bind to targets such as protease (NS2B-NS3pro), helicase (NS3 helicase), methyltransferase (MTase) and dengue virus envelope protein [24].
Therefore, it is evident that it is feasible to study the interaction of drugs and antiviral molecules with structural proteins of these viruses, in addition to enzyme targets. Flavone derivatives, such as luteolin and semisynthetic derivatives of gallic acid have been described as potential binders to the SARS-CoV surface SPIKE protein and, therefore, may interfere with the entry of the virus into its host cells. This connection has been shown to happen with great avidity, with surface proteins linked to natural compounds being detected by frontal affinity chromatography coupled with mass spectrometry [25].
The inhibition of the interaction of virus structural proteins with receptors in host cells is a line of research that has been highlighted. It is described that when binding the SPIKE protein to the angiotensin-converting enzyme 2 (ACE2) in the lung and intestinal cells, the virus enters the cells, releasing proteins such as the high-mobility group box 1 protein (HMGB1), allowing the occurrence of sepsis. Phenolic compounds have been described as steric inhibitors of the interaction of the viral structure with ACE2 and/or the release of HMGB1, reducing infectivity [26].
Together, these results suggest that chalcone derivatives inhibit the interaction of the virus with host cells through binding to ACE2 or SPIKE protein, probably generating a steric impediment. In addition, chalcones have an affinity for important enzymes in post-translational processes, interfering with viral replication.
5. Conclusion
The 4′-acetamidechalcones presented inhibitory potential over the SARS-CoV-2 proteins, detaching the PAAPF that has the potential to couple to Mpro (same region as the natural inhibitor FJC through strong hydrogen bonds), PAAB that can bind to NSP16– NSP10 methyltransferase, PAAPM and PAAPE that can interact with the protein Spike and PAAPA that demonstrated a strong affinity with ACE2, these results being a strong indication of the pharmacological potential of 4′-acetamidechalcones on important enzymes in post-translational processes, interfering in the viral replication of SARS-CoV-2, being an indication for the feasibility of in vitro tests. Emphasizing that the present work represents a fundamental step in the process of developing a pharmacological tool for COVID-19 based on chalcones.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank PROGRAMA INOVA FIOCRUZ - CE/FUNCAP, CAPES and CNPq for financial support and scholarship. Hélcio S. dos Santos also acknowledges financial support from the FUNCAP-BPI (BP4-0172-00075.01.00/20).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2020.12.074.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J., Coronaviridae Study V. Group of the International Committee on Taxonomy of, the species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui A.A., Rahman A., Shaharyar, Mishra R. Synthesis and anticonvulsant activity of some substituted 3,5-diphenyl-2-pyrazoline-1-carboxamide derivatives. Chem. Sci. J. 2010;1 doi: 10.4172/2150-3494.1000006. [DOI] [Google Scholar]
- 4.da Silva P.T., daCunhaXavier J., Freitas T.S., Oliveira M.M., Coutinho H.D.M., Leal A.L.A.B., Barreto H.M., Bandeira P.N., Nogueira C.E.S., SenaJr D.M., Almeida-Neto F.W.Q., Marinho E.S., Santos H.S., Teixeira A.M.R. Synthesis, spectroscopic characterization and antibacterial evaluation by chalcones derived of acetophenone isolated from Croton anisodontus Müll.Arg. J. Mol. Struct. 2021;1226 doi: 10.1016/j.molstruc.2020.129403. [DOI] [Google Scholar]
- 5.Nowakowska Z. A review of anti-infective and anti-inflammatory chalcones. Eur. J. Med. Chem. 2007;42:125–137. doi: 10.1016/j.ejmech.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Onyilagha J.C., Malhotra B., Elder M., French C.J., Towers G.H.N. Comparative studies of inhibitory activities of chalcones on tomato ringspot virus (ToRSV) J. Indian Dent. Assoc. 1997;19:133–137. doi: 10.1080/07060669709500541. [DOI] [Google Scholar]
- 7.Santos I.A. Universidade Federal de Uberlândia-Instituto de Ciências Biomédicas; 2018. Atividade antiviral de compostos naturais brasileiros e hidroxichalconas e curcuminoides sintétiticos. [Google Scholar]
- 8.Bhat B.A., Dhar K.L., Puri S.C., Saxena A.K., Shanmugavel M., Qazi G.N. Synthesis and biological evaluation of chalcones and their derived pyrazoles as potential cytotoxic agents. Bioorg. Med. Chem. Lett. 2005;15:3177–3180. doi: 10.1016/j.bmcl.2005.03.121. [DOI] [PubMed] [Google Scholar]
- 9.Bandeira P.N., Lemos T.L.G., Santos H.S., de Carvalho M.C.S., Pinheiro D.P., de Moraes M.O., Pessoa C., Barros-Nepomuceno F.W.A., Rodrigues T.H.S., Ribeiro P.R.V., Magalhaes H.S., Teixeira A.M.R. Synthesis, structural characterization, and cytotoxic evaluation of chalcone derivatives. Med. Chem. Res. 2019;28:2037–2049. doi: 10.1007/s00044-019-02434-1. [DOI] [Google Scholar]
- 10.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 11.Rosas-Lemus M., Minasov G., Shuvalova L., Inniss N.L., Kiryukhina O., Wiersum G., Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Jaroszewski L., Godzik A., Joachimiak A., Satchell K.J.F. The Crystal Structure of Nsp10-Nsp16 Heterodimer from SARS-CoV-2 in Complex with S-Adenosylmethionine. bioRxiv. 2020 doi: 10.1101/2020.04.17.047498. 2020.2004.2017. [DOI] [Google Scholar]
- 12.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 13.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shityakov S., Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinform Chem. 2014;7:23–36. doi: 10.2147/aabc.s63749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yusuf D., Davis A.M., Kleywegt G.J., Schmitt S. An alternative method for the evaluation of docking performance: RSR vs RMSD. J. Chem. Inf. Model. 2008;48:1411–1422. doi: 10.1021/ci800084x. [DOI] [PubMed] [Google Scholar]
- 16.Biovia D.S., Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E., Darden T., York D., Pedersen L.G., Bussi G., Donadio D., Parrinello M., Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G., Parrinello M., Rahman A., Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C., Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E., Berendsen H.J.C., Postma J.P.M., Van Gunsteren W.F., Dinola A., Haak J.R., Hockney R.W., Goel S.P., Eastwood J.W., Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Dassault systèmes BIOVIA, discovery Studio visualizer, v.17.2, san diego: dassault systèmes. J. Chem. Phys. 2000;10 doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 17.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Ruffinatti F.A., Genova T., Mussano F., Munaron L. MORPHEUS: an automated tool for unbiased and reproducible cell morphometry. J. Cell. Physiol. 2020;235:10110–10115. doi: 10.1002/jcp.29768. [DOI] [PubMed] [Google Scholar]
- 19.Imberty A., Hardman K.D., Carver J.P., Perez S. Molecular modelling of protein-carbohydrate interactions. Docking of monosaccharides in the binding site of concanavalin A. Glycobiology. 1991;1:631–642. doi: 10.1093/glycob/1.6.631. [DOI] [PubMed] [Google Scholar]
- 20.Park J.-Y., Ko J.-A., Kim D.W., Kim Y.M., Kwon H.-J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzym. Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 21.Jo S., Kim H., Kim S., Shin D.H., Kim M.-S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mateeva N., Eyunni S.V.K., Redda K.K., Ononuju U., Hansberry T.D., Aikens C., Nag A. Functional evaluation of synthetic flavonoids and chalcones for potential antiviral and anticancer properties. Bioorg. Med. Chem. Lett. 2017;27:2350–2356. doi: 10.1016/j.bmcl.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patil V., Patil S.A., Patil R., Bugarin A., Beaman K., Patil S.A. Exploration of (hetero)aryl derived thienylchalcones for antiviral and anticancer activities. Med. Chem. 2019;15:150–161. doi: 10.2174/1573406414666180524074648. [DOI] [PubMed] [Google Scholar]
- 24.Powers C.N., Setzer W.N. An in-silico investigation of phytochemicals as antiviral agents against dengue fever. Comb. Chem. High Throughput Screen. 2016;19:516–536. doi: 10.2174/1386207319666160506123715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., Zhang H., Luo H., Zhu L., Jiang P., Chen L., Shen Y., Luo M., Zuo G., Hu J., Duan D., Nie Y., Shi X., Wang W., Han Y., Li T., Liu Y., Ding M., Deng H., Xu X. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78:11334. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyganowska-Swiatkowska M., Nohawica M., Grocholewicz K., Nowak G. Influence of herbal medicines on HMGB1 release, SARS-CoV-2 viral attachment, acute respiratory failure, and sepsis. A literature review. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.