Summary
Mode of delivery strongly influences the early infant gut microbiome. Children born by cesarean section (C-section) lack Bacteroides species until 6–18 months of age. One hypothesis is that these differences stem from lack of exposure to the maternal vaginal microbiome. Here, we re-evaluate this hypothesis by comparing the microbial profiles of 75 infants born vaginally or by planned versus emergent C-section. Multiple children born by C-section have a high abundance of Bacteroides in their first few days of life, but at 2 weeks, both C-section groups lack Bacteroides (primarily according to 16S sequencing), despite their difference in exposure to the birth canal. Finally, a comparison of microbial strain profiles between infants and maternal vaginal or rectal samples finds evidence for mother-to-child transmission of rectal rather than vaginal strains. These results suggest differences in colonization stability as an important factor in infant gut microbiome composition rather than birth canal exposure.
Keywords: infant gut microbiota, caesarean delivery, Bacteroides, delivery mode, transmission of maternal strains
Graphical Abstract
Highlights
Week 1 gut microbiota does not differ between infants born vaginally versus C-section
Week 2 gut microbiota of C-section infants lacks Bacteroides
Microbiota of infants born by C-section after labor resembles scheduled C-section
Bacterial strains in infants match maternal rectal rather than vaginal strains
Mitchell et al. compare early-life infant gut microbiota by delivery mode, suggesting early colonization by Bacteroides regardless of delivery mode, but loss of Bacteroides by 2 weeks in C-section-delivered infants, whether or not exposed to the vagina in labor. Infant strains matched maternal rectal rather vaginal strains.
Introduction
Infants begin life with a relatively simple gut microbial community and acquire a more complex, adult-like microbiome over the first 3 years of life.1, 2, 3, 4 While multiple factors influence composition and dynamics during the first year of life, delivery mode is one of the most significant.5,6 Infants born by cesarean section (C-section, CS) have significantly different early gut microbial features compared to vaginally delivered infants, including lower diversity and richness2,7 and lower prevalence of colonization with Bacteroides species.1,5, 6, 7, 8, 9, 10, 11, 12 This CS microbial signature is detectable to at least 6 months of age, despite subsequent effects of breastfeeding on the infant microbiome.1,4,5
Pioneering studies demonstrated the difference in early microbiome development between delivery modes, but did not identify the underlying causes of this difference.13 It is widely believed that the infant gut is seeded with microbes from the mother during labor, particularly from the maternal vaginal microbiome. The data supporting these hypotheses, however, are limited. Prior studies have relied on a single infant stool sample collected around birth,1,2,8,14, 15, 16 examined a small number of 20–30 mother-infant dyads,7,8,13,17,18 confounded vaginal microbiome exposure by combining pre- and post-labor CS groups (infants born by post-labor CS are exposed to the maternal vaginal microbiome, but eventually extracted by C-section),2,4,6,8,15 or excluded post-labor CS altogether.2,4,6,8,15, 16, 17
Infants born by post-labor CS provide a unique opportunity to study the impact of traveling through the birth canal on bacterial transmission from mother to infant. Comparing the microbiomes of infants born by pre- and post-labor CSs will more definitively address the importance of exposure to vaginal microbes independent of mode of exit. Furthermore, studying mother-to-infant microbial transmission requires a comparison of the microbes found in both mothers and infants at the strain-level using deep metagenomic sequencing, as we (and others) have recently reported.18, 19, 20, 21
Here, we examine the sources and timing of delivery mode-associated microbial signatures. We established a birth cohort to collect multiple newborn and maternal microbiome samples from the first 2 weeks of life. Using 16S and metagenomic sequencing, we compared the gut microbiome of newborns across delivery modes and found that (1) the vaginal delivery microbial signature is not simply the result of exposure to birth canal microbes, as children born by post-labor CS had a microbial signature resembling those born by pre-labor CS; (2) the lack of Bacteroides species in the guts of infants delivered by both pre- and post-labor CSs only appears in the second week of life, as many children had detectable Bacteroides in their first week of life that later disappeared; and (3) mother-to-child bacterial transmission events occur mostly in vaginally delivered children, and the maternal source is rectal rather than vaginal.
Results
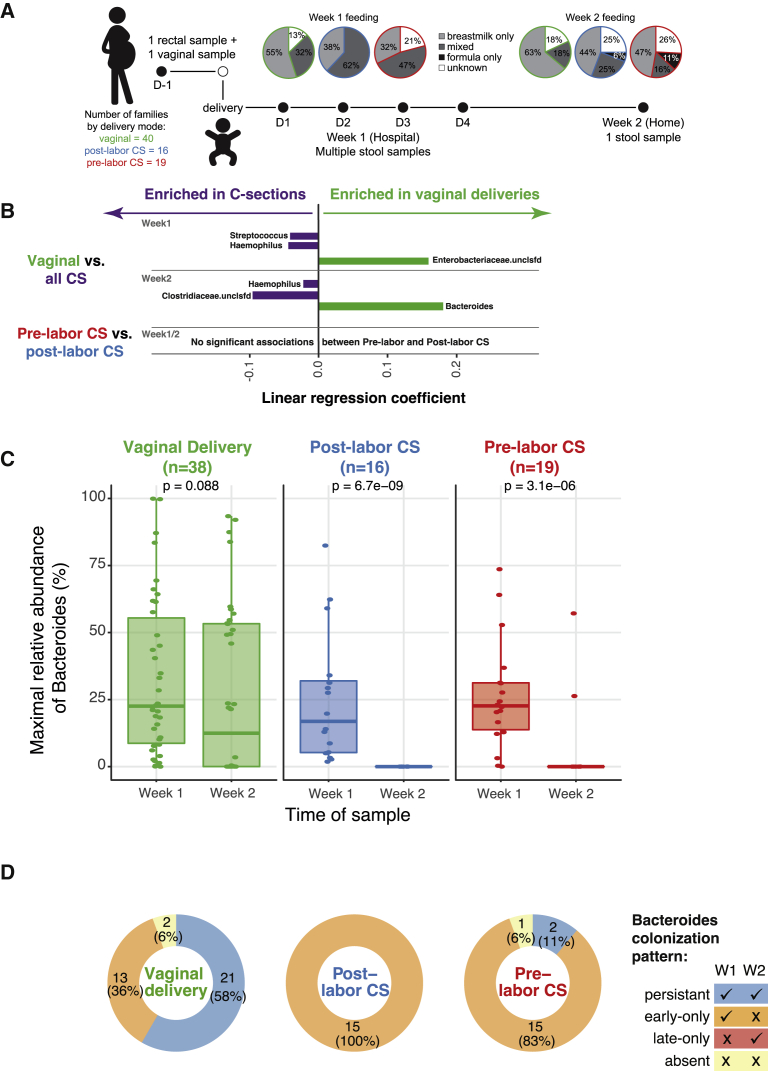
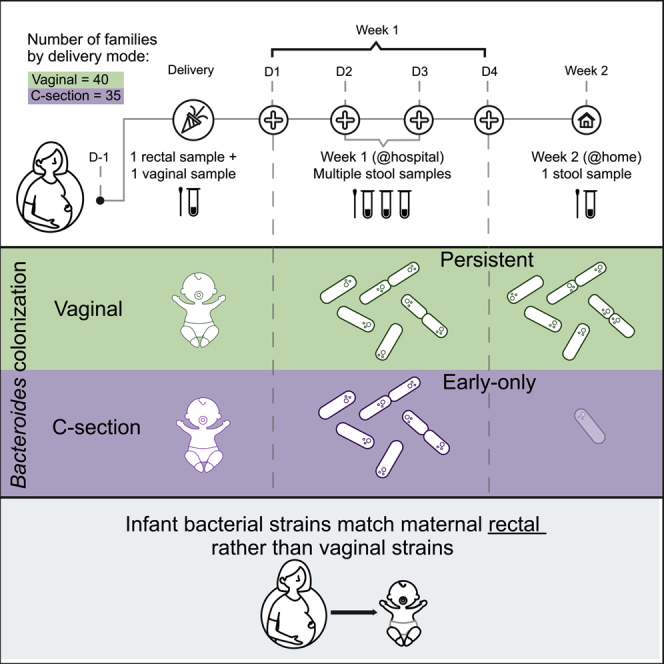
To study microbiome acquisition immediately following birth, we collected maternal and newborn samples from 75 families admitted to the Massachusetts General Hospital Labor and Delivery unit. We enrolled 40 children born vaginally, 19 children born by scheduled, pre-labor CS, and 16 children born by non-elective CS following labor. We collected maternal rectal and vaginal samples <24 h before delivery to evaluate potential maternal microbial sources at the time of delivery. In addition, we collected daily stool samples from the newborns while they were at the hospital (days 0–4 of life, 2–4 samples per infant) and 1 stool sample collected at home at 2 weeks of age (median 17 days of life) (see Figure 1A; Tables 1 and S2 for characteristics of the cohort). Neonatal stool samples often have low bacterial biomass, a high proportion of human DNA, and varying sample volumes, which motivated our collection of multiple samples from the first week of life to obtain a more complete picture of initial microbial exposure and colonization. We performed 16S rRNA gene sequencing on infant samples and metagenomic sequencing on both maternal and infant samples.
Figure 1.
Cohort Description and Key Differential Taxa across Delivery Modes
(A) Cohort information about delivery mode, feeding practices, and sampling. Women presenting to Labor and Delivery at term (≥37 weeks) with singleton gestation were enrolled and had a rectal and vaginal sample collected before delivery. A stool sample was collected from the infants’ diaper daily while in the hospital, and parents sent a single stool sample from home at 2 weeks of life. Feeding practices were abstracted from inpatient charts for week 1 and obtained from parent questionnaires for week 2.
(B) Impact of delivery mode on early life microbial composition. Multivariate linear regression was used to identify taxa that were enriched in vaginally delivered versus C-section-delivered (CS) infants and pre- versus post-labor CS (using the results of 16S rRNA sequencing). Analyses were adjusted for feeding practices in the week of interest. A positive coefficient represents a taxon more abundant in vaginally delivered infants. Only associations with absolute coefficients ≥0.015, and FDR corrected q value < 0.25 are included here.
(C) Loss of Bacteroides colonization in CS-delivered infants. Maximal abundance of Bacteroides in week 1 (i.e., the sample with the highest relative abundance) versus week 2 samples by 16S sequencing was compared within delivery mode using the t test.
(D) Differences in patterns of Bacteroides colonization by delivery mode. The Bacteroides colonization phenotype was assigned based on detection at ≥0.1% in week 1 samples only (early-only), week 2 only (late-only), both week 1 and week 2 (persistent), or neither (absent). Results are presented for the 67 infants with samples available from both time points.
Table 1.
Clinical Data Summary for the 75 Dyads in the Cohort
| Vaginal n = 40 | Post-labor n = 16 | Pre-labor n = 19 | pa | |
|---|---|---|---|---|
| Age, y | 33 ± 5 | 31 ± 5 | 38 ± 4 | <0.001 |
| White race (%) | 31 (78) | 11 (69) | 18 (95) | 0.19 |
| BMI | 28 ± 7 | 28 ± 5 | 28 ± 6 | 0.98 |
| Gestational diabetes (%) | 0 (0) | 1 (6) | 1 (6) | 0.3 |
| Hypertension (%) | 7 (18) | 2 (13) | 1 (5) | 0.43 |
| Maternal asthma (%) | 7 (18) | 3 (19) | 2 (11) | 0.75 |
| Choriamnionitis (%) | 7 (18) | 3 (19) | 0 (0) | 0.14 |
| Maternal antibiotics (%) | 18 (46) | 16 (100) | 19 (100) | <0.001 |
| Meconium-stained amniotic fluid (%) | 12 (31) | 6 (38) | 1 (5) | 0.06 |
| Birth weight, g | 3,505 ± 413 | 3,331 ± 421 | 3,687 ± 528 | 0.07 |
| Infant antibiotics (%) | 6 (16) | 4 (25) | 0 (0) | 0.09 |
| Any formula in hospital (%) | 10 (26) | 9 (56) | 9 (47) | 0.05 |
| Breastfeeding (hospital discharge) (%) | 27 (69) | 7 (44) | 10 (53) | 0.06 |
| Exclusive breastfeeding, 2 weeks (%) | 25 (63) | 7 (44) | 9 (47) | 0.63 |
| Duration of 2nd stage (median, IQR) | 0.7 (0.3, 2) | 4.2 (3, 4.2) | – | 0.002 |
| Duration of ruptured membranes (median, IQR) | 6 (2, 16) | 8 (1, 13) | – | 0.8 |
BMI, body mass index.
χ2, ANOVA, or Kruskal-Wallis.
To determine how delivery mode affects acquisition and persistence of bacterial taxa, we stratified samples into week 1 (multiple time points per infant) and week 2 (days 13–29, 1 sample per infant). Week 1 samples were collected between days 1 and 4 of life, with a median of 3 samples from both vaginally and CS-delivered infants. Due to gut microbiome variability in the first few days of life,19 we calculated the maximum abundance for each taxon across the multiple week 1 samples from each infant to consider all of the species that were observed in the infant gut in the first week of life. We focused our analyses on the 16S rRNA gene sequencing data due to the increased sensitivity of this method compared to metagenomics in low-microbial biomass samples.22,23 We compared genus-level gut microbiome profiles across delivery modes using multivariate linear regression.24 Because breastfeeding rates and exposure to formula differed by delivery mode (Figure 1A), we adjusted for infant feeding in regression analyses comparing delivery groups. Principal-component analysis did not reveal a significant difference using beta diversity across delivery modes (data not shown).
Surprisingly, the well-established absence of Bacteroides species in the gut microbiome of most infants born by CS was not evident in the first week of life (Figures 1B and 1C). We found that 33/35 (94%) infants born by pre- or post-labor CS had detectable levels of gut Bacteroides species (≥0.1%) during their first week of life. Consistent with the literature, we found that at week 2, infants born via CS (both pre- and post-labor) were much less likely to be colonized by Bacteroides relative to infants born vaginally (Figures 1B and 1C). The same analysis using metagenomic data identified Bacteroides in fewer samples from CS infants, but demonstrated a similar loss of colonization in the week 2 samples, although this did not reach statistical significance (Figure S1A). Notably, we found no significant differences in taxa between the microbiomes of infants born by pre- and post-labor CSs, suggesting that vaginal exposure may not be a driving force in Bacteroides colonization (Figure 1B).
To facilitate additional analyses, we grouped infants who had both week 1 and week 2 samples according to the pattern of Bacteroides colonization in weeks 1 and 2. We defined Bacteroides colonization as "persistent" if it was detected in weeks 1 and 2 samples, "early-only" if it was detected in week 1 but not week 2, and "absent" if it was not detected in either week 1 or 2. A small number of children (3) had the absent colonization pattern, and no infants in our study were only colonized at week 2. The vast majority of children born by CS (91%) have an early-only colonization pattern of Bacteroides, whereas most vaginally delivered children (58%) have the persistent colonization pattern (Figure 1D). To be clear, persistent by our definition refers only to week 2 colonization and does not imply that Bacteroides will persist into early life.
We confirmed this Bacteroides signature (presence in week 1 followed by loss at week 2 in CS-delivered infants) by re-examining published datasets. We analyzed the raw data from 2 published cohorts that used 16S sequencing,12,14 and 3 that used shotgun metagenomic sequencing.11,18,19 We observed the same pattern of early Bacteroides colonization and later disappearance in some children delivered by CS (Figure S1B; Table S3); however this difference between weeks 1 and 2 had not been identified or highlighted in the original articles. The early presence of Bacteroides species in CS-delivered infants suggests that their later absence cannot be attributed to lack of exposure.13,25 We also used data from a study by Ferretti et al.,18 which included several samples from the first week of life, as well as our own data to examine the impact of using maximal abundance from all week 1 samples compared to using data from a single, randomly selected sample. We found that in both cohorts, using maximal abundance was more likely to identify week 1 Bacteroides colonization (Figure S1B), suggesting that the additional samples allowed for the increased sensitivity.
We next wondered whether maternal factors other than delivery mode may explain the differential Bacteroides colonization patterns. However, we saw no difference across delivery modes in maternal race, body mass index (BMI), or prevalence of medical comorbidities, which suggests that these characteristics were not significant confounders (Table 1). Of note, the median maternal BMI in all three delivery mode groups was in the overweight range. Age was higher in women who underwent scheduled CS, and meconium-stained amniotic fluid was more common in women who labored (whether delivering vaginally or via CS). Finally, all women undergoing CS receive antibiotics before incision, and we wanted to confirm that the differential colonization pattern across delivery modes was not due to the antibiotic treatment. Some of the women who delivered vaginally were also given antibiotics—for example, if they tested positive for group B Streptococcus (GBS). To test the hypothesis that antibiotic treatment was associated with the loss of Bacteroides in week 2, we compared the relative abundance of Bacteroides in vaginally delivered children at week 2, but found no effect of maternal antibiotics during labor on the Bacteroides colonization pattern (Figure S1C). While maternal antibiotic regimens varied within each delivery group (Table S2), the primary difference between delivery modes is that most women who had a CS received cefazolin, while most women with a vaginal delivery received ampicillin (when treated with antibiotics). These two antibiotics are from related classes (cephalosporins and penicillins) and target similar microbial classes. Thus, while there may be subtle differences in the types of antibiotics used between delivery groups, this is likely not a significant contributor to differences in infant Bacteroides colonization.
We then explored what may cause the loss of Bacteroides in week 2. We compared infant characteristics across the Bacteroides colonization patterns. While delivery mode was significantly associated with Bacteroides colonization, infant characteristics such as exposure to formula in the hospital, breastfeeding at week 2, and infant receipt of antibiotics were not (Table S1A). Exposure to formula in the first week of life was higher after CS delivery (51% versus 26%, p = 0.02; Table 1), so analyses were controlled for this variable. As previous analyses have suggested that exposure to the vaginal microbiome is the source of differences in gut microbiota between vaginal and C-section-delivered infants, we asked whether longer exposure increased the chances of persistent Bacteroides colonization. We compared the duration of vaginal exposure in infants who experienced labor (vaginal or post-labor CS deliveries) using two variables: the time between membrane rupture and delivery and the length of the second stage of labor (time between full dilation and birth). The length of the second stage was in fact shorter in infants with persistent Bacteroides colonization (Tables S1A and S1B), suggesting that the persistence of Bacteroides colonization is not a result of vaginal exposure.
We next wondered whether maternal factors influenced the infant Bacteroides colonization patterns. We examined the relationship between maternal microbial communities and the infant Bacteroides colonization phenotype (using linear regression; see Method Details). We found no statistically significant difference in maternal rectal colonization between infant Bacteroides colonization phenotypes. Although vaginal exposure is proposed as a potential source for infant gut colonization, it cannot account for the Bacteroides phenotype, as only a single maternal vaginal sample had detectable (≥0.1%) Bacteroides colonization. Some authors have suggested that the differences seen in the infant gut microbiome due to delivery mode point to underlying differences in women who have CSs compared to those who deliver vaginally.26 When we compared the relative abundance of individual species in maternal rectal samples across delivery modes, we found a higher abundance of B. ovatus (q = 0.226), B. dorei (q = 0.234), and B. uniformis (q = 0.236) in mothers who had a vaginal delivery versus those who had a CS delivery, although the numbers were small.
Finally, we hypothesized that the later loss of Bacteroides may be due to competition within the infant gut. To address this question, we compared week 1 samples from all of the CS infants to those born vaginally to identify additional taxa that may be associated with the delivery mode and predict the loss of Bacteroides in week 2. Interestingly, we found a higher abundance of the genera Streptococcus and Haemophilus in CS-born children in week 1 samples, before the disappearance of Bacteroides (Figures 1B, S2A, and S2B). Across all samples and all time points, the relative abundances of Bacteroides and either Streptococcus or Haemophilus were inversely correlated (Figures S2C and S2D). When we compare the week 1 samples of infants with persistent Bacteroides colonization versus early-only, we found a higher relative abundance of Bacteroides (coefficient = 0.09, q = 0.006) and a lower relative abundance of Streptococcus (coefficient = −0.03, q = 0.23) in infants who have persistent Bacteroides colonization. The relative abundance of Haemophilus was not significantly different between the persistent and early-only groups (coefficient 0.02, q = 0.72). These results could suggest competition between Streptococcus and Bacteroides in early life as one factor contributing to the loss of Bacteroides in the second week of life.
We next asked, for children in whom we observe Bacteroides in week 1 samples, what is the source of these strains? Specifically, are the infant strains shared with their mothers? It is possible that the source of the Bacteroides strains plays a role in its persistence in an infant’s gut. The data above suggest that Bacteroides colonization is not dependent on whether an infant was exposed to the birth canal, as we saw no differences between our pre- and post-labor CS groups. Therefore, for analysis of the source of infant colonization, we considered all CSs together as a single group. Because exiting the birth canal brings the infant in proximity to the maternal rectum, we hypothesized that during a vaginal delivery, the infant is exposed to bacterial strains from the maternal gut microbiota. To obtain species-level resolution and be able to ask strain-level questions, we used metagenomic sequencing.
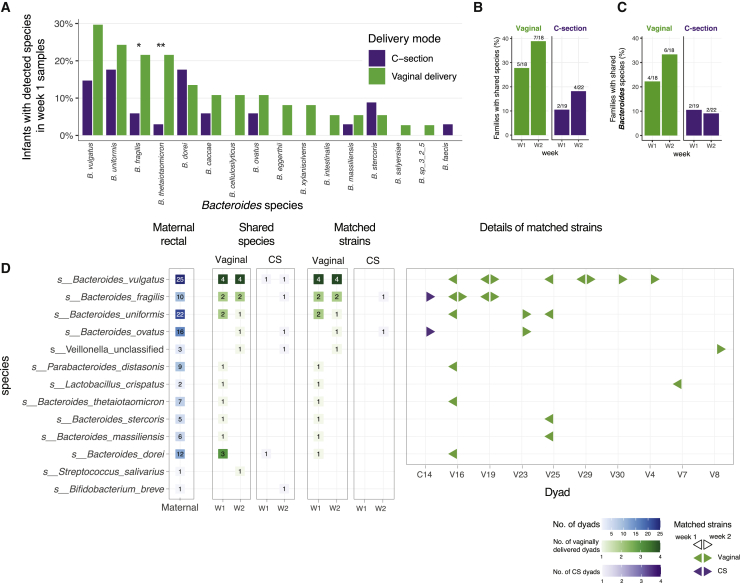
We first looked for species-level taxonomic differences between vaginally and CS-delivered infants. Among infants with Bacteroides detected in the first week of life, B. fragilis and B. thetaiotamicron were more common in vaginally delivered infants (Figure 2A). As noted previously, neither of these species were found more frequently in maternal samples from either delivery mode. To compare strains between mothers and infants using metagenomic data, we used strict criteria for sequencing depth (see Method Details). Among the dyads in which both maternal and infant samples met these criteria, 5/18 (28%) vaginally delivered infants shared at least one species of any genus with their mothers in week 1 samples, compared to 2/19 (11%) CS-delivered infants (Figure 2B). This difference persisted in week 2 samples: 7/18 (39%) versus 4/22 (18%). Most of the shared species were Bacteroides, meaning that vaginally delivered infants were more likely to share a Bacteroides species with their mothers (Figure 2C).
Figure 2.
Shared Species and Strain between Maternal Rectal and Infant Stool Samples Using Metagenomic Sequencing
(A) Comparison of Bacteroides species prevalence across delivery modes, among infants with detectable Bacteroides in week 1 samples (≥0.1%, n = 67).
(B) Mother-infant dyads with shared species between maternal rectal swabs and infant stool samples in either week 1 or week 2 samples (≥0.1%, n = 18 vaginal delivery, n = 22 CS delivery).
(C) Mother-infant dyads with shared Bacteroides species.
(D) Detailed view of shared species and matched strains in mother-infant dyads by delivery mode and week of sampling. A total of 10 dyads (9 vaginal, 1 CS) demonstrated evidence of shared strains between maternal rectal and infant stool samples.
We next turned to identify matched strains between mother and infant species that may suggest a possible transmission event, using single-nucleotide variants extracted from the metagenomic reads (see Method Details). As the infant gut and the adult gut harbor different microbial communities, we are limited in this analysis to species that are shared within at least one dyad with sufficient coverage (Figure 2D). Among the 7 dyads with shared species in week 1, the 5 with matching strains were all vaginally delivered children, while the two CS infants with shared species did not have the same strain as their mothers (Figure 2D). In week 2, among the 11 dyads who shared a species with their mother, 7/7 (100%) vaginally delivered infants had a matched strain, while only 1 of 4 (25%) CS infants with shared species also had shared strains (Figure 2D).
Finally, it has been suggested that vaginally delivered infants acquire Lactobacillus species from their mothers as they pass through the birth canal.13 Despite 75% of mothers having >80% relative abundance of Lactobacillus in vaginal samples (Figure S3A), we did not observe higher abundance of Lactobacillus species in the gut microbiome of children born vaginally. Using 16S rRNA sequencing, 23 infants had Lactobacillus detected (≥0.1% relative abundance) in week 1 stool samples, of which 16 (70%) were vaginally delivered. However, using shotgun sequencing (necessary for strain-matching analysis), only 2 infants had Lactobacillus species detected in week 1 samples (≥0.1%), both vaginally delivered. In both cases, maternal samples had a high proportion (>90%) of the same Lactobacillus species. If we extend this analysis to additional vaginal microbes, among 85 species found at ≥0.1% in any maternal vaginal samples, there were only 5 dyads (2 vaginal deliveries, 3 CS after labor) in which an infant had detectable colonization by a maternal vaginal species (Figure S3B), suggesting that the transmission of maternal vaginal species to the infant is not exclusive to vaginal deliveries and is infrequent or in low abundance.
Discussion
To summarize, we sequenced microbiomes from 75 newborns and their mothers close to the time of delivery to determine the impact of delivery mode on the origin of the infant gut microbiome.
Our results confirm the association between CS and lack of durable Bacteroides colonization, but challenge the dogma that this difference is due to lack of vaginal exposure. Identifying the canonical CS microbial signature in infants delivered by post-labor CS suggests that the major factor driving the association between birth mode and infant microbiota is not entering the birth canal, but rather exiting through it. The similarity between infants delivered by CS before or after labor is consistent with findings in the Baby Biome Study from the United Kingdom,11 which sampled infants once in the first week of life. We did not find evidence of vaginal microbiota transmission from mothers to their vaginally delivered infants, and very few infants had detectable levels of Lactobacillus (the most common vaginal microbiome member) in their gut community, also consistent with Baby Biome Study findings.11 The strongest strain-level evidence for bacterial transmission at birth is found in vaginally delivered children but stems from the maternal rectal source. Our results suggest that the establishment of the initial infant gut microbial community requires more than topical exposure to maternal vaginal microbes.
Multiple infant samples collected during the first week of life enabled us to question the timing of the canonical CS microbial signature, namely the depletion of Bacteroides species in the first 6–18 months. To our surprise, we found that a majority of infants born by pre- or post-labor CS had detectable levels of gut Bacteroides species during their first week of life, which disappeared in their second week of life. These results raise interesting questions: first, why do these Bacteroides disappear? Second, from where are they acquired? In 8 infants, we were able to identify matched strains in the maternal rectal swabs that appear to be the source of Bacteroides species; more often than not, however, standard metagenomic sequencing was insufficient to identify the source strain from mother vaginal or rectal samples. Likely, more targeted, species-specific methods would be necessary to bridge this gap.
Our data suggest that the origin of infant microbial differences may lie in both the source of the microbes and the infant environment that they are colonizing. The disappearance of Bacteroides species in the second week of life from infants born via CS suggests either the lack of a supporting factor, the introduction of an antagonist, or differences in the fitness of colonizing strains. A well-characterized cohort of vaginally delivered infants suggested that infant “seeding” likely originated from several maternal sites and that the ability of those microbes to persist may be related to the relative fitness of a given isolate for the gut environment.18 In our cohort, infants born by CS were less likely to have B. fragilis or B. thetaiotaomicron compared to vaginally delivered infants, two species of significant importance to gut health in adults. This may reflect differences in the source of colonization or colonization with less fit species after CS delivery. Alternatively, the experience of being delivered by CS may alter the infant gut environment in a way that makes it less receptive to colonization, although we could not assess this in our cohort. Competition with co-occurring taxa may also play a role in defining the community of microbes that persists beyond initial exposure. Our data support Streptococcus and Haemophilus as potential antagonists to Bacteroides persistence, as their abundance in week 1 samples was negatively associated with establishing persistent Bacteroides colonization. The striking negative correlation between Bacteroides and Streptococcus could be due to Streptococcus species filling a niche that was vacated or to direct competition between the two genera. In several body sites, Streptococcus species are known to compete with other members of the microbial community. For example, species in the S. mitis group can produce hydrogen peroxide, which inhibits the growth of other oral microbes,27,28 and S. oralis can inhibit pathogen growth and biofilm formation.29 Either of these mechanisms could account for the negative correlation between Streptococcus and Bacteroides. Haemophilus is more commonly thought of as an oropharyngeal colonizer, but has been found to be increased in CS-delivered infants1 and inversely correlated with Bacteroides abundance in other cohorts.30
Feeding practices may also be a significant influence on community composition. In the Canadian Healthy Infant Longitudinal Development (CHILD) cohort, even a single exposure to formula during the delivery hospitalization was associated with a lower prevalence of Bifidobacteriaceae and higher prevalence of Enterobacteriaceae at 3 months.31 Most infants in our cohort were primarily breastfed: 97% at hospital discharge, 91% at 2 weeks. However, upon a review of charts, we found a much higher rate of formula exposure during the delivery hospital stay in CS-delivered infants. The formula exposure was often a single feeding, or a few feedings, until a mother felt comfortable breastfeeding. Formula feeding may be one of several factors that contribute to a difference in the gut microbiota of infants born via CS. Few other studies have evaluated this type of limited, single-instance formula exposure, which can occur even when mothers plan to exclusively breastfeed. We adjusted for feeding practices in our analyses, but larger cohorts are needed to further study these confounding influences.
As infants grow, feeding practices play an increasing role in determining the composition of the infant gut microbiota;2,31 however, delivery mode has a stronger influence on the composition of the microbial community in the days immediately after birth. Understanding the earliest microbial communities and their determinants is an important component of designing microbiome-related interventions to improve infant health. The collection of multiple stool samples from early life, as well as maternal vaginal and gut microbiome, breast milk samples, and other possible sites of exposure such as the skin of family members may be necessary to provide a complete picture of how early colonization is established. Part of the challenge in early-life samples is the low biomass of the stool samples. Fortunately, constant improvement in the methods for profiling low-biomass samples23,32 will make it increasingly feasible to study the strain-level infant gut colonization at the very early stages of life.
Our results demonstrate that maternal seeding of the infant gut microbiome can occur, but is not a straightforward transfer of the maternal vaginal microbial community. We also show that the impact of CS delivery on the infant gut community is a delayed effect; this suggests that the influence of surgical delivery is not solely a difference in the source bacteria but also in colonization efficiency. This would suggest that the strategy of “vaginal seeding” or transferring maternal vaginal secretions to CS-delivered infants may not be effective. Our findings demonstrate the complexity of establishing a microbial community and our lack of understanding of how obstetric care influences infants.
Limitations of Study
Due to the within- and between-individual variability in composition of the gut microbiota, our sample size may be too small to identify the more subtle differences between groups. Both 16S and shotgun sequencing may lack the sensitivity to detect very-low-abundance Bacteroides colonization, which could lead to misclassification bias by assigning infants as “never colonized” when they may have had transient colonization. In addition, early infant stool samples have low microbial biomass, which leads to variability in read depth and quality of sequencing. The inclusion of more participants and the use of newer sequencing and analysis pipelines in future research would reduce these limitations.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Infant stool samples | This study | N/A |
| Maternal vaginal and rectal swabs | This study | N/A |
| Critical Commercial Assays | ||
| PowerSoil DNA Isolation Kit | QIAGEN Inc | Cat. 47014 |
| Chemagic MSM I | Perkin Elmer | CMG-717 |
| Deposited Data | ||
| Raw sequencing data | This project | NCBI Sequence Read Archive as BioProject PRJNA591079. |
| Software and Algorithms | ||
| KneadData v0.5.1 | Huttenhower Lab | https://github.com/biobakery/KneadData |
| MetaPhlAn v2.0 | Truong et al.35 | https://github.com/biobakery/MetaPhlAn |
| decontam v1.6.0 | Davis et al.36 | R package |
| MaAsLin2 | Mallick et al. | https://github.com/biobakery/Maaslin2 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Moran Yassour (Moranya@mail.huji.ac.il).
Materials Availability
This study did not generate any new reagents.
Data and Code Availability
The accession number for the sequences reported in this paper is BioProject PRJNA657818. Full metadata is available in Table S2. There is no custom code or unique algorithms that are central to this manuscript.
Experimental Model and Subject Details
Women with singleton gestation, presenting for delivery at ≥ 37 weeks of gestation to the Massachusetts General Hospital Labor & Delivery unit in Boston, MA were enrolled in this prospective cohort study. Participants were excluded if they had known HIV infection, fetal congenital anomaly, gestational age < 37 weeks, maternal age < 18, planned to give up the infant for adoption, or were a gestational carrier. If an enrolled dyad had subsequent admission of the infant to the NICU or special care nursery at any time after delivery, that dyad was excluded.
Human patient research in the OriGiN cohort was reviewed and approved by the Partners Human Research Committee (ref. 2015P000460/PHS). Each mother signed two informed consent forms (one for her and one for her child) prior to participation.
Method Details
Sample collection
At enrollment, mothers had vaginal and rectal swabs collected. Vaginal swabs were collected by a provider without a speculum, by parting the labia and inserting a foam swab 3-4 cm, then rotating 3-4 times. Rectal swabs were collected by a provider, who placed a swab 1-2cm into the anus and rotated it twice. Swabs were placed at 4C within 30 min after collection and then at −80C within 18 hours and stored until processed.
Infant stool was collected by parents from the diaper once daily during the delivery hospitalization (2-5 days), into tubes containing pure ethanol as a preservative. Study staff collected infant stool samples from parents each day. A sample collection kit was sent to parents, who collected another infant stool sample between day 13-29 of life (also into ethanol) and returned it at room temperature within 24 hours of collection, via a delivery service. Participants answered a short questionnaire about infant health status and mode of feeding, which was returned with the 2 week stool sample. All stool (home or hospital collected) was stored at room temperature in the ethanol preservative until placed in a −80C freezer within 2 days of collection. Participant and infant medical records were abstracted to obtain information about mode of delivery, receipt of antibiotics, reported infant feeding, duration of labor stages, obstetric complications, and health history.
DNA Extraction and sequencing
Vaginal and rectal swabs were eluted in 400uL of saline, vortexed for 1 minute and then spun at 10,000 xg for 10 minutes. The cell pellet underwent DNA extraction using the MoBio Power Soil kit (QIAGEN Inc, Waltham MA). Stool samples underwent DNA extraction using chemical and mechanical lysis with magnetic bead-based purification via the Chemagic MSM I with the Chemagic DNA Blood Kit-96 from Perkin Elmer. Prior to extraction on the MSM-I, TE buffer, lysozyme, proteinase K, and RLT buffer with beta-mercaptoethanol were added to each stool sample. DNA samples were quantified using a fluorescence-based PicoGreen assay.
Sequence processing and analysis
A total of 427 samples (308 infant stool, 63 vaginal, 56 maternal rectal) from 85 families underwent sequencing. Whole-genome shotgun (WGS) sequencing for all infant and maternal samples was performed on the Illumina NextSeq platform with 100 bp paired-end reads. Reads pairs with fewer than 60 observed bases on either read were excluded from downstream analysis. Read were trimmed based on the following criteria: (1) Removing leading low quality or N bases (below quality 3); (2) Removing trailing low quality or N bases (below quality 3); (3) Scanning the read with a 4-base wide sliding window, cutting when the average quality per base drops below 15. Samples with fewer than 500,000 raw reads were excluded. Read pairs that were attributed to human genomic DNA were removed using KneadData Tool, v0.5.1 using the hg19 human reference genome. To taxonomically profile the bacterial DNA sequences observed in the samples, MetaPhlAn 2.0 was used to align reads to the MetaPhlAn 2.0 unique marker database. Samples that were fully unclassified by metaphlan were excluded. Samples were also excluded if there were 100 or fewer reads mapping to the species-specific marker genes of a given taxa that was reported at > 50% abundance. For inclusion in downstream analysis, samples coming from a given dyad had to pass the quality measures above and meet at least one of the following dyad criteria: a) two or more child samples were available in week 1, b) One or more child samples for week 1 were available and one child sample at week 2, or c) one or more child samples and one or more mother sample available. A subset of 75 families met one or more of these criteria for inclusion. There were 71 infant stool samples from this subset that had sufficient DNA and underwent 16S rRNA sequencing of the V3-V4 region. 16S rRNA gene sequencing was performed as previously described.4 Taxonomy was assigned using version 1.8.0 of Qiime37 (with the closed_reference_otu function) and the Greengenes reference database of OTUs.38 Additionally, the R package decontam36 was used to identify contaminating taxa in week 1 taxonomic profiles. Taxa whose relative abundance was found to be significantly correlated to the DNA concentration of samples across the cohort were removed and taxonomic profiles consequently renormalized. The number of reads for both 16S and WGS methods was not different across delivery modes.
Mother-child strain transmission analysis
We tabulated the species that were observed most frequently in both mother and child samples of the same dyad (Figure 2D). To examine families for possible shared strains within these species, we applied an approach we recently developed to examine sub-species variation between samples.19 Briefly, coverage overlap events were identified in mother-child pairs where metagenomic reads mapped to the same segment of species-specific marker genes of various species. In these overlap regions, sites of single nucleotide variation (SNV) were tabulated across the set of marker genes for a given species. Samples with fewer than 1000 reads mapping to species-specific marker genes were not considered for this analysis. For analyses of week 1 samples, the sample with the most reads was used to assess strain matching. We report here a matched strain in a dyad, if the infant had a dominant or secondary maternal strain.
Quantification and Statistical Analysis
A Bacteroides colonization phenotype of early-only (week 1 only), persistent (both week 1 and 2) or absent was assigned using 16S rRNA sequencing results, using a detection threshold of 0.1% relative abundance. Associations between maternal and delivery characteristics and this phenotype were examined using chi-square or ANOVA. Linear regression models were run using MaAsLin2, to identify statistically significant associations between microbial features and delivery mode or Bacteroides phenotype. Analyses were adjusted for infant feeding,24 and a q-value threshold of 0.25 was considered significant. Additional comparisons of Bacteroides maximal abundance between week 1 and week 2 within delivery mode were made using a t test.
All figures were generated with the R ggplot2 package (version 3.1.0). Boxplots were generated with geom_boxplot default parameters, such that the box represents 25-75 percentile, and the whiskers from the box to the largest/smallest value no further than 1.5 ∗ IQR from the box (where IQR is the inter-quartile range, or distance between the first and third quartiles).
Acknowledgments
We thank T. Poon, L. Besse, S. Steelman, and C. Nusbaum (Broad Institute) for help in sequence production and sample and data management; V. Subramanian and T. Arthur for experimental methods support and helpful discussions; T. Reimels for valuable figure and text support; the nurses, midwives, and clinicians at the Labor and Delivery Department at Massachusetts General Hospital for their enthusiastic collaboration during enrollment; and the families that agreed to enroll and donate samples. C.M. was supported by the Vincent Memorial Research Funds; E.S.L. was supported by National Human Genome Research Institute grant 2U54HG003067-10; R.J.X. was supported by funding from JDRF, CCFA, and NIH grants R01 DK092405 and P30 DK043351; and M.Y. was supported by NIH grant DK113224-01, Israel Science Foundation grant 2660/18, an Azrieli Foundation Faculty Fellowship, and a BroadIgnite early career grant from the Broad Institute of MIT and Harvard.
Author Contributions
C.M.M. and M.Y. designed the study. C.M.M., A. Bryant, S.P., P.H., M.C., K.S., and M.Y. enrolled participants and conducted the study. A. Bergerat, A.C., and H.V. performed laboratory analyses. C.M.M., C.H., E.S.L., H.V., R.J.X., and M.Y. supervised the study. C.M.M., C.M., L.H., H.V., R.J.X., E.S.L., and M.Y. designed and conducted the analyses of data. All of the authors contributed to writing the manuscript and signed off on the final format.
Declaration of Interests
C.M.M. receives grant funding from Merck and has served as a consultant for Scynexis. C.H. is a member of the Seres Therapeutics, Empress Therapeutics, and ZOE Nutrition scientific advisory boards. E.S.L. serves on the board of directors for Codiak BioSciences and on the scientific advisory board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several non-profit organizations, including serving on the board of directors of the Innocence Project, Count Me In, and the Biden Cancer Initiative, and the board of trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees. R.J.X. is a consultant to Nestlé.
Published: December 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100156.
Supporting Citations
The following references appear in the supplemental information: Stockholm et al.33 and Hill et al.34
Supplemental Information
References
- 1.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Bokulich N.A., Chung J., Battaglia T., Henderson N., Jay M., Li H., D Lieber A., Wu F., Perez-Perez G.I., Chen Y. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yassour M., Vatanen T., Siljander H., Hämäläinen A.-M., Härkönen T., Ryhänen S.J., Franzosa E.A., Vlamakis H., Huttenhower C., Gevers D., DIABIMMUNE Study Group Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016;8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azad M.B., Konya T., Persaud R.R., Guttman D.S., Chari R.S., Field C.J., Sears M.R., Mandhane P.J., Turvey S.E., Subbarao P., CHILD Study Investigators Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016;123:983–993. doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 6.Madan J.C., Hoen A.G., Lundgren S.N., Farzan S.F., Cottingham K.L., Morrison H.G., Sogin M.L., Li H., Moore J.H., Karagas M.R. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr. 2016;170:212–219. doi: 10.1001/jamapediatrics.2015.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakobsson H.E., Abrahamsson T.R., Jenmalm M.C., Harris K., Quince C., Jernberg C., Björkstén B., Engstrand L., Andersson A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 8.Brumbaugh D.E., Arruda J., Robbins K., Ir D., Santorico S.A., Robertson C.E., Frank D.N. Mode of Delivery Determines Neonatal Pharyngeal Bacterial Composition and Early Intestinal Colonization. J. Pediatr. Gastroenterol. Nutr. 2016;63:320–328. doi: 10.1097/MPG.0000000000001124. [DOI] [PubMed] [Google Scholar]
- 9.Sordillo J.E., Zhou Y., McGeachie M.J., Ziniti J., Lange N., Laranjo N., Savage J.R., Carey V., O’Connor G., Sandel M. Factors influencing the infant gut microbiome at age 3-6 months: findings from the ethnically diverse Vitamin D Antenatal Asthma Reduction Trial (VDAART) J. Allergy Clin. Immunol. 2017;139:482–491.e14. doi: 10.1016/j.jaci.2016.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., Ross M.C., Lloyd R.E., Doddapaneni H., Metcalf G.A. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y., Forster S.C., Tsaliki E., Vervier K., Strang A., Simpson N., Kumar N., Stares M.D., Rodger A., Brocklehurst P. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyman M., van Houten M.A., van Baarle D., Bosch A.A.T.M., Man W.H., Chu M.L.J.N., Arp K., Watson R.L., Sanders E.A.M., Fuentes S., Bogaert D. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019;10:4997. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu D.M., Ma J., Prince A.L., Antony K.M., Seferovic M.D., Aagaard K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dogra S., Sakwinska O., Soh S.-E., Ngom-Bru C., Brück W.M., Berger B., Brüssow H., Lee Y.S., Yap F., Chong Y.S., GUSTO Study Group Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio. 2015;6:e02419-14. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller N.T., Shin H., Pizoni A., Werlang I.C., Matte U., Goldani M.Z., Goldani H.A., Dominguez-Bello M.G. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci. Rep. 2016;6:23133. doi: 10.1038/srep23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y.-C., Guo H., Chen J., Sun G., Ren R.-R., Guo M.Z., Peng L.H., Yang Y.S. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci. Rep. 2018;8:3255. doi: 10.1038/s41598-018-21657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yassour M., Jason E., Hogstrom L.J., Arthur T.D., Tripathi S., Siljander H., Selvenius J., Oikarinen S., Hyöty H., Virtanen S.M. Strain-Level Analysis of Mother-to-Child Bacterial Transmission during the First Few Months of Life. Cell Host Microbe. 2018;24:146–154.e4. doi: 10.1016/j.chom.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nayfach S., Rodriguez-Mueller B., Garud N., Pollard K.S. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korpela K., Costea P., Coelho L.P., Kandels-Lewis S., Willemsen G., Boomsma D.I., Segata N., Bork P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira-Marques J., Hout A., Ferreira R.M., Weber M., Pinto-Ribeiro I., van Doorn L.J., Knetsch C.W., Figueiredo C. Impact of Host DNA and Sequencing Depth on the Taxonomic Resolution of Whole Metagenome Sequencing for Microbiome Analysis. Front. Microbiol. 2019;10:1277. doi: 10.3389/fmicb.2019.01277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillmann B., Al-Ghalith G.A., Shields-Cutler R.R., Zhu Q., Gohl D.M., Beckman K.B., Knight R., Knights D. Evaluating the Information Content of Shallow Shotgun Metagenomics. mSystems. 2018;3 doi: 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan X.C., Tickle T.L., Sokol H., Gevers D., Devaney K.L., Ward D.V., Reyes J.A., Shah S.A., LeLeiko N., Snapper S.B. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Bello M.G., De Jesus-Laboy K.M., Shen N., Cox L.M., Amir A., Gonzalez A., Bokulich N.A., Song S.J., Hoashi M., Rivera-Vinas J.I. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stinson L.F., Payne M.S., Keelan J.A. A Critical Review of the Bacterial Baptism Hypothesis and the Impact of Cesarean Delivery on the Infant Microbiome. Front. Med. (Lausanne) 2018;5:135. doi: 10.3389/fmed.2018.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okahashi N., Nakata M., Sumitomo T., Terao Y., Kawabata S. Hydrogen peroxide produced by oral Streptococci induces macrophage cell death. PLOS ONE. 2013;8:e62563. doi: 10.1371/journal.pone.0062563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakubovics N.S., Gill S.R., Vickerman M.M., Kolenbrander P.E. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol. Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiedler T., Riani C., Koczan D., Standar K., Kreikemeyer B., Podbielski A. Protective mechanisms of respiratory tract Streptococci against Streptococcus pyogenes biofilm formation and epithelial cell infection. Appl. Environ. Microbiol. 2013;79:1265–1276. doi: 10.1128/AEM.03350-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong X., Xu W., Janton S., Henderson W.A., Matson A., McGrath J.M., Maas K., Graf J. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PLOS ONE. 2016;11:e0152751. doi: 10.1371/journal.pone.0152751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forbes J.D., Azad M.B., Vehling L., Tun H.M., Konya T.B., Guttman D.S., Field C.J., Lefebvre D., Sears M.R., Becker A.B., Canadian Healthy Infant Longitudinal Development (CHILD) Study Investigators Association of Exposure to Formula in the Hospital and Subsequent Infant Feeding Practices With Gut Microbiota and Risk of Overweight in the First Year of Life. JAMA Pediatr. 2018;172:e181161. doi: 10.1001/jamapediatrics.2018.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caruso V., Song X., Asquith M., Karstens L. Performance of Microbiome Sequence Inference Methods in Environments with Varying Biomass. mSystems. 2019;4 doi: 10.1128/mSystems.00163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokholm J., Blaser M.J., Thorsen J., Rasmussen M.A., Waage J., Vinding R.K., Schoos A.M., Kunøe A., Fink N.R., Chawes B.L. Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 2018;9:141. doi: 10.1038/s41467-017-02573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill C.J., Lynch D.B., Murphy K., Ulaszewska M., Jeffery I.B., O’Shea C.A., Watkins C., Dempsey E., Mattivi F., Tuohy K. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017;5:4. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truong D.T., Franzosa E.A., Tickle T.L., Scholz M., Weingart G., Pasolli E., Tett A., Huttenhower C., Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 36.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequences reported in this paper is BioProject PRJNA657818. Full metadata is available in Table S2. There is no custom code or unique algorithms that are central to this manuscript.