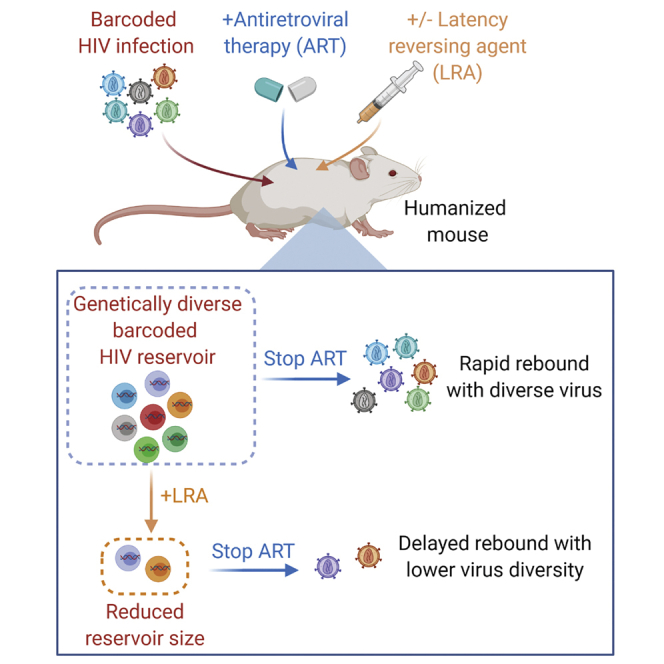
Summary
HIV latency prevents cure of infection with antiretroviral therapy (ART) alone. One strategy for eliminating latently infected cells involves the induction of viral protein expression via latency-reversing agents (LRAs), allowing killing of host cells by viral cytopathic effects or immune effector mechanisms. Here, we combine a barcoded HIV approach and a humanized mouse model to study the effects of a designed, synthetic protein kinase C modulating LRA on HIV rebound. We show that administration of this compound during ART results in a delay in rebound once ART is stopped. Furthermore, the rebounding virus appears composed of a smaller number of unique barcoded viruses than occurs in control-treated animals, suggesting that some reservoir cells that would have contributed virus to the rebound process are eliminated by LRA administration. These data support the use of barcoded virus to study rebound and suggest that LRAs may be useful in HIV cure efforts.
Keywords: HIV, latency, LRA, reservoir, cure, protein kinase C, antiretroviral therapy, ART, humanized mice
Graphical Abstract
Highlights
A genetically barcoded HIV swarm has been constructed and characterized
This swarm forms a latent reservoir in antiretroviral (ART)-treated humanized mice
Administering an HIV latency-reversing agent (LRA) during ART delays rebound
LRA administration during ART also reduces barcode diversity of rebounding virus
Marsden et al. construct a barcoded HIV swarm to study HIV latency formation and rebound in humanized mice. They find that the administration of a PKC modulating HIV latency-reversing agent delays rebound and reduces the genetic diversity of rebounding virus when antiretroviral therapy is stopped, suggesting the reduction of the reservoir.
Introduction
In 2018, an estimated 38 million people worldwide were living with HIV, and 32 million people had already died of AIDS-related illnesses since the start of the epidemic.1 Antiretroviral therapy (ART) can inhibit HIV replication and prevent disease progression, but ART does not cure the infection.2,3 The most well-defined reservoir of HIV that persists during ART is within latently infected CD4+ T cells.4,5 These very-long-lived cells harbor intact, integrated HIV genomes that express little or no viral RNA and no viral proteins (reviewed by Marsden and Zack6), making them effectively invisible to the immune system. The central relevance of latently infected cells and other persistent reservoirs lies in their ability to maintain infectious HIV during ART and then supply virus that can emerge and replicate to high levels or “rebound” if ART is stopped.
Animal models of HIV infection provide a system for testing strategies to eliminate latent reservoirs and delay or prevent the rebound process. Bone marrow-liver-thymus (BLT) mice are one of the most advanced small animal models of the human immune system available (reviewed by Marsden and Zack7). These mice are constructed by implanting fetal liver and thymus tissue under the kidney capsule of an immunodeficient (typically Nod-SCID-common gamma chain knockout) mouse, followed by irradiation and then transfusion of hematopoietic stem cells.8 This results in the generation of multilineage immune reconstitution with human cells including CD4+ and CD8+ T cells, natural killer (NK) cells, monocytes, macrophages, and dendritic cells.8,9 We and others have shown that the BLT mouse model can be efficiently infected with HIV and forms authentic post-integration latency in resting CD4+ T cells.9, 10, 11, 12, 13 Viral loads can be suppressed using clinically relevant ART, and if ART is stopped, then viral replication quickly rebounds, as occurs with infected patients.
One area in which humanized mice have been particularly useful is the exploration of “kick and kill” strategies to deplete latent HIV,14 where non-expressing HIV proviruses are induced to express viral proteins with a latency-reversing agent (LRA) allowing the host cell to be killed by viral cytopathic effects, immune effector mechanisms, or other approaches targeting productively infected cells. Various LRAs that function through several different intracellular signaling pathways have been tested in different in vitro or in vivo models15, 16, 17, 18, 19, 20, 21, 22 and one of the most effective pathways for reversing HIV latency has proven to be via PKC (protein kinase C) modulation.23 We have previously demonstrated that the latency-reversing capabilities of naturally occurring PKC modulators, including bryostatin 1 and prostratin, can be improved by designed synthetic analogs of these compounds.19,24,25 Importantly, certain modifications to bryostatin 1 improved in vivo tolerability and bioactivity in mice, and one particularly potent bryostatin analog (SUW133) is capable of inducing latent HIV expression in vivo in BLT mice leading some of the previously latently infected cells to die 1 day after compound administration during ART.12 These results show that a strong “kick” in the context of an in vivo infection is sufficient to induce the death of some latently infected cells. However, it is unknown whether in vivo induction of HIV expression by highly efficient LRAs would result in biologically meaningful consequences, the most important of which is delaying or eliminating HIV rebound.
We reasoned that if we possessed an accurate means of tracking virus replication and reservoir formation, we would be able to more readily quantify the effects of LRAs (and potential addition of separate “kill” approaches) on the latent reservoir. Advances in molecular biology, nucleotide sequencing, and bioinformatic approaches have allowed the incorporation and analysis of short, unique nucleotide sequences to tag individual molecules and study their behaviors within a mixed population. Several prior studies have leveraged the use of this “genetic barcoding” to study the expression of a non-replicating HIV-based vector in vitro26 or simian immunodeficiency virus replication and rebound in vivo.27 Here, we have developed a barcoded HIV system to track individual viral lineages as they form latent reservoirs in BLT mice treated with ART, and then rebound if ART is stopped. Using this system, we show that a bryostatin analog PKC modulator can meaningfully affect the HIV reservoir present during ART, leading to a reduction in barcode diversity, indicating a depletion of the reservoir itself. This impact on the reservoir resulted in a corresponding delay or prevention of viral rebound.
Results and Discussion
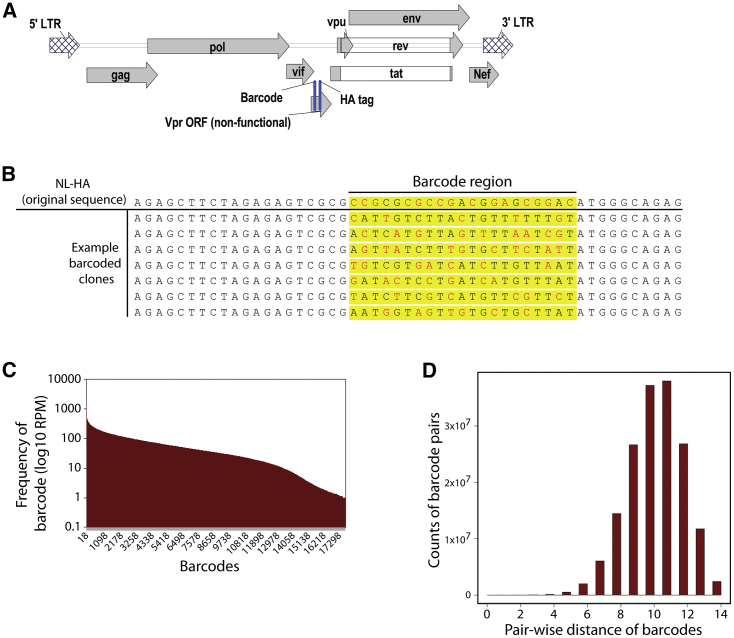
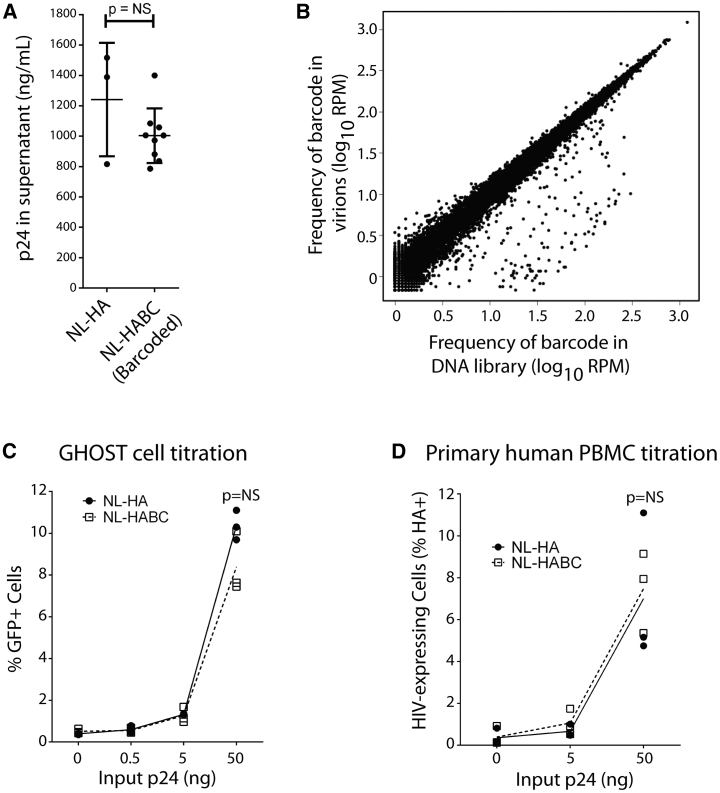
We reasoned that the ability to detect the clonal origin of HIV virions during rebound would be important for assessing viral reservoirs. Consequently, we constructed a swarm of genetically barcoded HIV viruses that differ only in the presence of a short, non-expressed, phenotypically neutral sequence. We used the NL-hemagglutinin (HA) backbone plasmid as template for modification. This is a near-full-length, replication-competent, pathogenic HIV strain derived from NL4-3, which has been modified to express an HA epitope in place of Vpr,28 and has proven useful in prior in vivo latency studies.11,12 This virus therefore affords the benefit of encoding a selectable marker for the identification of productively infected cells and enriching/depleting latently infected cells in HIV latency studies. However, it is not intended to study the effects of Vpr in this process. A 21-nt barcode sequence was inserted in a non-expressed region of this vector immediately upstream of the HA tag and outside any known cis elements or splice sites (Figure 1A). Each 3rd nucleotide of this barcode was constrained to a thymidine to avoid random strings of consecutive GC bases, providing an overall theoretical complexity of 414 unique sequences (Figure 1B). The resultant collection of plasmids was expanded in bacteria, with >20,000 bacterial colonies harvested, and then subjected to Illumina HiSeq deep sequencing, which revealed a barcode complexity of ∼18,000 unique sequences present at relatively similar frequencies (Figure 1C). The average number of differences between any 2 barcode sequences was 11 bp (Figure 1D), meaning that the original barcodes could easily be distinguished from one another in downstream virologic studies. The barcoded plasmid was transfected into 293FT cells to generate virions and yielded similar amounts of cell-free virus to the parental plasmid, as quantified by ELISA specific for HIV p24 (Figure 2A). Collected virions were treated with DNase, viral RNA was extracted, and then the barcode region was amplified and deep sequenced. As expected, the virions retained a similar barcode virus complexity and frequency to that present in the original plasmid preparation. The close correlation between barcode frequencies in the DNA library versus in virions emphasizes the neutrality of these barcodes for viral replication (Spearman correlation ρ = 0.99; Figure 2B).
Figure 1.
Generation of Barcoded HIV Plasmid Library
(A) Location of barcode upstream of the HA tag in the genome of HIV strain NL-HA.
(B) Example of individual 21 nucleotide “barcode” sequences in which each 3rd nucleotide is constrained to a thymine.
(C) Complexity of barcode region as quantified by HiSeq sequencing, showing ~18,000 barcodes with relatively equal distribution.
(D) Average pairwise distance between barcodes is ~11 bp.
Figure 2.
In Vitro Analysis of Barcoded HIV Swarm
(A) Transfection with NLHABC plasmid produces similar HIV virion concentrations to transfection with the parental NL-HA plasmid as quantified by HIV p24 protein levels. n = 3 transfections (biological replicates) for NL-HA and 9 transfections (biological replicates) for NLHABC. Error bars, mean ± 1 SD.
(B) Comparison between barcode occurrence in original plasmid DNA and resultant virion-associated viral RNA after HIV production in 293FT cells (Spearman correlation ρ= 0.99).
(C) Infection of GHOST cells for 2 days shows similar infectivity between NL-HA and NLHABC in a cell line model (n = 3 independent experiments).
(D) Infection of co-stimulated primary human PBMCs (n = 3 biological replicate infections per condition using cells from different human donors) for 3 days shows similar in vitro replication rate between NL-HA and NLHABC.
Statistical comparisons in (A), (C), and (D) performed with unpaired, 2-sided t test.
See also Figure S1.
We then validated that the newly constructed barcoded HIV swarm was infectious during in vitro assays. The infection of GHOST CXCR4+CCR5+ cells with parental NL-HA virus and the barcoded NLHABC yielded similar frequencies of infected (GFP+) cells during parallel 2-day infections (Figures 2C and S1A). Primary human peripheral blood mononuclear cells (PBMCs) were also infected with different concentrations of either NL-HA or NL-HABC (Figures 2D and S1B), which resulted in statistically indistinguishable frequencies of HIV-expressing (HA+) cells. Virus produced by primary PBMCs after 3 days of infection was then harvested and the barcode region amplified by RT-PCR, subjected to deep sequencing, and compared with the diversity of the input viral RNA. There was good correlation between barcode frequencies before and after PBMC infection (Spearman correlation ρ= 0.74; Figures S1C and S1D), indicating as expected that individual barcodes are not strongly selected for or against during replication in primary cells.
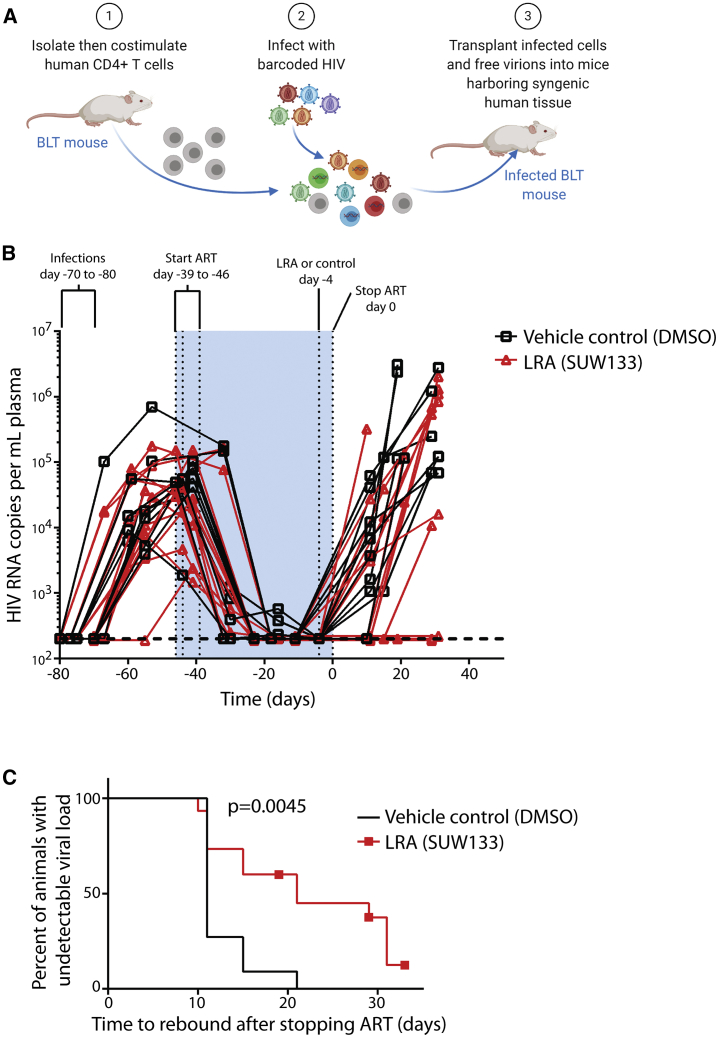
Humanized BLT mice were then constructed and infected with barcoded HIV. Our goal was to establish infections with a large number of barcoded HIV variants, and since primary infection with HIV is well known to be associated with a profound genetic bottleneck of virus populations, we devised a cell-associated virus infection strategy in which CD4+ T cells are isolated from some animals in a series, co-stimulated, infected with HIV using standard procedures, suspended in free virus, and then transplanted into other mice from the same series harboring syngeneic human tissue (Figure 3A). This approach led to efficient infection of the animals (Figures 3B and S2A). Three mice were sacrificed at 4 weeks post-infection and were found to harbor between 18 and 42 unique original barcode sequences in spleen and between 28 and 49 barcode sequences in plasma (Figures S2B and S2C). For other animals, after 4–5 weeks of infection, we initiated ART (Figure 3B), which was administered in animal feed for a further 6–7 weeks to suppress viral loads. Mice were then either treated with an intraperitoneal injection of vehicle (DMSO) control or a synthetic, designed, bryostatin analog (SUW133), which we have previously demonstrated can induce the expression of latent HIV in BLT mice and cause some of the newly HIV-expressing cells to die.12 There were no significant differences in humanization (percentage of human CD45+ cells) between control and LRA-treated animal groups either before or after LRA administration (p > 0.05), and the levels of humanization did not significantly change after LRA or vehicle control administration in either group compared with the pre-administration time point (Figure S2D; Table S1). We allowed 4 days for potential reactivation and death of latently infected cells to occur via viral cytopathic effects or immune effector mechanisms, and then stopped ART and monitored viral rebound over the next several weeks (Figures 3B, S3A, and S3B). Notably, 4 of the 15 animals treated with SUW133 did not rebound during this time period, and overall rebound in this group was delayed (p = 0.0045, log-rank [Mantel-Cox] test) when compared with vehicle control-treated animals (Figure 3C). To further explore the effect of LRA on viral reservoirs, splenocytes from several control and treated animals were subjected to ex vivo co-stimulation with anti-CD3+ anti-CD28 antibodies and then co-cultured with CEM cells to facilitate viral outgrowth. HIV p24 concentrations were measured in the supernatants following 14–15 days of culture. Notably, replication-competent HIV was recovered from all of the animals tested, including non-rebounding mouse #26 (Figure S4). Therefore, while the LRA treatment in this animal was sufficient to delay rebound, it does not eliminate all viral reservoirs capable of producing infectious virus. Hence, it is likely that if experiments were extended longer, rebound would eventually be observed.
Figure 3.
In Vivo Barcode Virus Infection and Effects of LRA on Rebound
(A) Schematic representation of infection procedure intended to overcome potential transmission bottlenecks and increase barcode diversity during initial infection of humanized mice.
(B) Viral loads of mice at each phase of infection. Primary infections lasted 4–5 weeks, followed by 6–7 weeks of antiretroviral therapy (ART). SUW133 (LRA) (n = 15 mice) or control injections (n = 11 mice) were administered 4 days before stopping ART, to allow latently infected cells sufficient time to die after reactivation. See Figure S3 for individual viral load plots for each animal.
(C) Delay in viral rebound induced by SUW133 versus DMSO control-treated animals compared using a log-rank (Mantel-Cox) test.
See also Table S1 and Figures S2–S5.
We also conducted additional evaluations to determine whether peak viral loads or area under the curve (AUC: viral load over time) before LRA administration differed between control and treatment groups or whether there was a significant relationship between these parameters and time to rebound. There was no overall significant differences in peak viral load or AUC between groups (Figures S5A and S5B), and these parameters did not correlate with time to rebound (Figures S5C–S5F). However, two of the three animals with the lowest peak viral loads and AUC were non-rebounders after LRA administration. This is consistent with the concept that lower levels of pre-ART viral replication result in a smaller rebound-competent reservoir that is easier to reduce, resulting in delayed rebound, as may have occurred when the “Mississippi baby” received potent ART very soon after birth and displayed significantly delayed rebound once ART was stopped.29
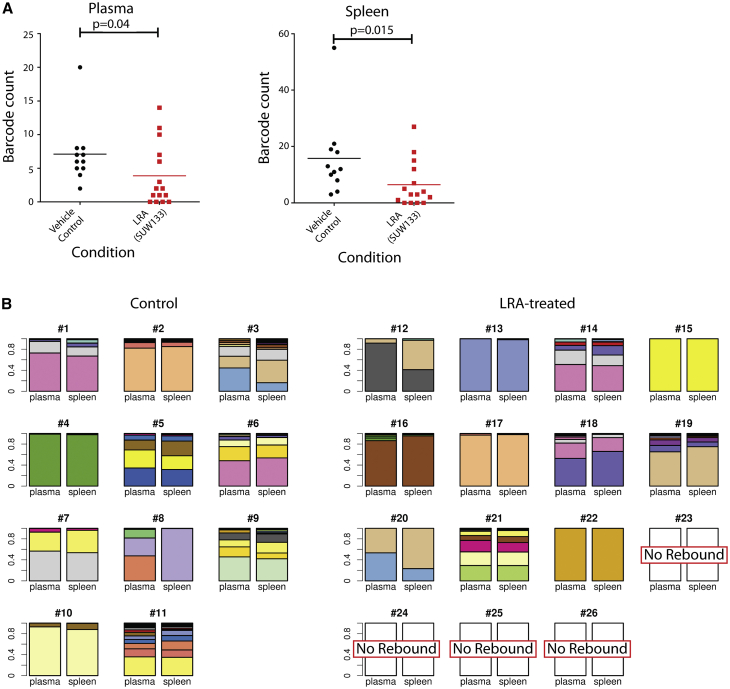
To determine whether the number of barcoded HIV variants that seeded the rebound viremia was also reduced, as may be expected if some latently infected cells were killed by the LRA treatment, we used an error-free deep sequencing strategy30 to accurately quantify the number of rebounding viral clones in plasma and spleen. Consistent with this hypothesis, in both compartments a smaller number of rebounding barcoded virus clones was present in the SUW133-treated group of animals than in controls (Figure 4A). Interestingly, if we eliminate the 4 non-rebounding mice from these calculations, then we see only a trend toward decline in rebounding clones. This could be influenced by the reduced sample size in the analysis or could suggest that complex factors regulate the number of reactivating cells. The use of barcoded virus also allowed tracking and comparison of the specific HIV variants that populated the different compartments during the rebound process. In general, following rebound, the major barcoded HIV variants present in the spleen also predominated in the plasma and vice versa (Figure 4B). The barcode diversities were significantly less in the SUW133-treated group than those in the DMSO-treated group (p = 0.028 for plasma, p = 0.022 for spleen, 2-sided Mann-Whitney U test on Shannon entropy). This type of approach may therefore be useful in future LRA studies, in which the animals are maintained on ART during the stimulation process, to determine the tissue origin of plasma virus induced by LRA stimulation in the absence of subsequent HIV replication.
Figure 4.
HIV Barcode Diversity Present in Plasma and Spleen at Necropsy
(A) Plasma and spleen barcode diversity quantified by deep sequencing of viral RNA. Statistical comparisons between groups performed using a 2-tailed Mann-Whitney test. If the 4 non-rebounding mice are excluded from analysis, then comparisons yield a trend lower in the LRA-treated group (p > 0.05).
(B) Stacked barplots showing the relationship between plasma and spleen virus. For each animal, the relative frequencies of major barcoded variants identified in the plasma and spleen are shown. Each plasma-spleen tissue pair shares a unique color scheme, with individual barcodes represented by 1 color.
These data both demonstrate that the administration of a single LRA can eliminate a subset of latently infected cells and delay HIV rebound in this in vivo model system and validate the barcode HIV approach as a method for tracking HIV latency reversal and rebound in vivo. Combination approaches with synergistic LRAs or augmented HIV-specific killing approaches including NK cells, cytotoxic T lymphocytes, or antibodies may well produce further improvement in these results.31 These proof-of-concept studies provide a convenient system for head-to-head comparisons of “kick and kill” approaches and show that a strong in vivo “kick” can have measurable and biologically relevant effects on the subsequent HIV rebound, thus supporting the continued investigation of “kick and kill” approaches to HIV cure efforts.
Limitations of Study
The virus used in this study encodes a reporter gene and barcode in place of Vpr. While we have found that the replacement of Vpr with this reporter does not prevent the establishment of or reactivation from latency in humanized mice,9,12,32 this particular virus is not suitable for capturing the effects of Vpr on latency. In addition, HIV latency reversal and reservoir cell killing were demonstrated using the LRA SUW133 in BLT mice in a prior study,12 but those experiments were not repeated here (doing so may provide additional information on reservoir depletion during ART). Finally, rebound barcode diversity was significantly lower in mice that received SUW133 when all of the mice were analyzed. However, if non-rebounders are excluded, then the difference in diversity becomes a trend. None of these nuances affect the main findings that the administration of an LRA can delay HIV rebound in this model.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA-Biotin, High Affinity (3F10) | Millipore Sigma | Cat#12158167001 RRID:AB_390915 |
| Streptavidin, R-Phycoerythrin Conjugate (SAPE) - 1 mg/mL | Fisher Scientific | Cat#S866A |
| CD45 Monoclonal Antibody (HI30), eFluor 450 | eBioscience | Cat#48-0459-42 RRID:AB_2016677 |
| Pacific Blue anti-human CD45 Antibody | BioLegend | Cat#304029, RRID:AB_2174123 |
| Anti-human CD3 - PURE HU CD3 (OKT3) | Fisher Scientific | Cat#50-105-5018 |
| Anti-human CD28 - IVR HU CD28 (CD28.2) | Fisher Scientific | Cat#50-105-4571 |
| Goat Anti-Mouse antibody - IgG (H+L) Goat anti-Mouse | Fisher Scientific | Cat#PI-31160 RRID:AB_228297 |
| Human-CD4 MicroBeads | Miltenyi Biotec | Cat#130-045-101 |
| Bacterial and Virus Strains | ||
| NL-HA | Laboratory of Otto Yang28 | N/A |
| Barcoded NL-HA | This paper | N/A |
| MegaX DH10B T1R cells | Thermo Fisher | Cat# C640003 |
| Biological Samples | ||
| Human De-Identified Peripheral Blood Mononuclear Cells | UCLA Virology Core | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SUW133 | Laboratory of Paul Wender25 | N/A |
| Raltegravir | Merck | N/A |
| Tenofovir disoproxil fumarate (TDF) | Gilead Sciences | N/A |
| Emtricitibine (FTC) | Gilead Sciences | N/A |
| Critical Commercial Assays | ||
| Perkin Elmer Alliance HIV-1 p24 kits | Perkin Elmer | Cat#NEK050 |
| Deposited Data | ||
| Raw sequencing reads deposited on SRA (Short Read Archive) database | This study | Accession number: PRJNA562389 |
| Experimental Models: Cell Lines | ||
| 293FT cells | Invitrogen | Cat#R70007 RRID:CVCL_6911 |
| GHOST (3) CXCR4+CCR5+ | NIH AIDS Reagent Program | Cat#3942 |
| CEM CD4+ Cells | NIH AIDS Reagent Program | Cat#117 |
| Experimental Models: Organisms/Strains | ||
| Mouse: NSG: PrkdcscidIl2rgtm1wjl | The Jackson Laboratory | Stock#005557 RRID:IMSR_JAX:005557 |
| Oligonucleotides | ||
| See Table S2 for List of Oligonucleotide sequences | N/A | N/A |
| Software and Algorithms | ||
| Codes | This study | https://github.com/Tian-hao/BarcodeHIV |
| FlowJo (v7) | FlowJo, LLC | N/A |
| Prism 6 | GraphPad | N/A |
| Excel | Microsoft | N/A |
| Other | ||
| Phusion High-Fidelity DNA Polymerase | ThermoFisher | Cat#F530L |
| NEBNext Ultra II DNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7645 |
| RNeasy minikit | QIAGEN | Cat#74104 |
| QIAamp Viral RNA Mini Kit | QIAGEN | Cat# 52904 |
| One-Step RT-PCR kit | Bioland Scientific | Cat#RT01-01 |
| SuperScript IV First-Strand Synthesis System | ThermoFisher | Cat#18091050 |
| RNA clean and concentrator kit | Zymo Research | Cat#R1017 |
| DNA clean and concentrator kit | Zymo Research | Cat#D4033 |
Resource Availability
Lead Contact
Additional information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Jerome Zack (jzack@ucla.edu).
Materials Availability
The barcoded HIV plasmid library used in this study will be made available by the lead author upon request.
Data and Code Availability
All codes are available on https://github.com/Tian-hao/BarcodeHIV.
All raw sequencing reads were deposited on SRA (Short Read Archive) database under the accession number (PRJNA562389).
The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.
Experimental Model and Subject Details
Cell Lines
293FT cells were obtained from Invitrogen. These cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen) containing 10% fetal bovine serum (FBS: Omega Scientific), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (pen/strep: Invitrogen). Cells were cultured at 37°C in 5% CO2.
GHOST (3) CXCR4+CCR5+ cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Dr. Vineet N. KewalRamani and Dr. Dan R. Littman (Cat# 3942). These were cultured in DMEM containing 10% FBS, 500 μg/mL G418 (GIBCO Cat#10131-27) 100 μg/mL hygromycin (Sigma Cat#H3274-100MG), pen/strep, and 1 μg/mL puromycin (Sigma Cat#P8833-10MG). Cells were cultured at 37°C in 5% CO2.
Cell lines were obtained directly from NIH AIDS Reagent repository or Invitrogen. Their phenotype and characteristics were as expected therefore they were not subjected to further authentication procedures.
Primary Cells
De-identified peripheral blood mononuclear cells were provided by the UCLA AIDS Institute Virology Core Laboratory following IRB approval. Cells were costimulated for 3 days by culturing in RF10 medium (consisting of RPMI 1640 medium supplemented with 10% FBS and pen/strep) containing 20 U/mL of interleukin-2 (IL-2) in the presence of plate-bound anti-CD3 (1 μg/mL, Fisher Scientific Cat#50-105-5018) (adhered via pre-treatment of plates with Goat Anti-Mouse antibody [Fisher Scientific Cat#PI-31160]) and soluble anti-CD28 (100 ng/mL) using previously described procedures.9 Cells were cultured at 37°C in 5% CO2.
Humanized Mice
Animal experiments conformed to all local and national guidelines and regulations (including the Public Health Service Policy on Humane Care and Use of Laboratory Animals, the Guide for the Care and Use of Laboratory Animals, and the AVMA Guidelines for the Euthanasia of Animals), and procedures were approved by the UCLA Animal Research Committee (Approval number ARC # 1996–058). Euthanasia was performed by anesthetizing animals with isofluorane followed by cervical dislocation.
Humanized bone marrow liver thymus (BLT) mice8 were constructed by the UCLA humanized mouse core using techniques described previously.9,12,33,34 In brief, NOD.Cg-PrkdcscidIl2rgtm1wjl (Nod-SCID-common gamma chain knockout [NSG]) mice were irradiated with 270 rads and then transplanted under the kidney capsule with pieces of fetal thymus and liver tissue. Mice were then infused intravenously by retro-orbital injection with 5x105 human fetal liver-derived CD34+ cells isolated by immunomagnetic separation as previously described.12,35 At 8 weeks post-transplantation and approximately every 2 weeks thereafter mice were evaluated for reconstitution with human cells. Mice were bled as previously described9,12 and peripheral blood mononuclear cells analyzed by flow cytometry. To reduce the effects of potential donor-to-donor variability, results are derived from series of mice constructed with tissue from 4 different human donors. Only animals with undetectable plasma viral loads at the end of ART treatment and no graft-versus-host disease were included in analysis. Mouse groups were randomized to ensure similar pre-ART viral loads between groups and eliminate the possibility of variations in pre-ART viral load affecting post-ART results. This was pre-defined because these experimental factors would confound study of HIV latency and rebound. Both male and female mice were included in this study to avoid systematic biases associated with sex as a biological variable. Mice were purchased from The Jackson Laboratory and bred at UCLA. Mice were maintained in HEPA-filtered mouse racks in groups of up to 5 animals per cage. Mice were 3 months of age at time of initial transplant.
Method Details
Construction of HIV-1 barcode library
We utilized the HIV-1 construct NL43-HA12,28 as the backbone, which has a hemagglutinin (HA) tag inserted in place of the vpr gene, and expresses the HA epitope on the infected cell surface with a murine heat stable antigen. The barcode was inserted into a non-translated region before the HA tag start codon. This region does not contain known cis element or splicing sites, thus should be neutral for viral replication. The barcode is designed to be 21 base pairs in length, with 14 random nucleotides that contain a ‘T’ at each 3rd position to avoid unwanted start or stop codons. This randomization strategy allows a total complexity of 268,435,456 (414) unique barcodes. The barcode region was amplified by 2-step PCR with the following primers:
1st PCR:
Primer 1 (Table S2) Forward:
agagcttctagagagtcgcgNNTNNTNNTNNTNNTNNTNNTATGggcagagcgatggtg
Primer 2 Reverse:
cgcctattctgctatgtcgacaccc
Amplicon: 336bp
Primer 3 2nd PCR
Forward:
GAA GCA GAG CTA GAA CTG GCA GA
Reverse:
PCR product of first PCR
Amplicon: 2,513 bp
The randomized nucleotides were inserted into the forward primer of the 1st PCR. Then the amplified region was digested using AgeI and EcoRI and ligated back to NL43-HA vector and transformed into MegaX DH10B T1R cells (ThermoFisher Cat# C640003).
We anticipated that 10,000 barcodes would be more than sufficient for our in vivo experiments, thus we harvested plasmid DNA from > 20,000 bacteria colonies post transformation, each representing a uniquely barcoded virus, resulting in a very complex barcoded HIV-1 plasmid library. Plasmids from collected bacteria were midi-prepped as the input plasmid library. Classic Sanger sequencing across the barcode region were performed to examine the diversity at expected nucleotide positions.
Sequencing and analysis of in vitro barcode library
For in vitro samples, the barcode region was amplified by PCR. The PCR was performed using Phusion High-Fidelity DNA Polymerase (ThermoFisher Cat#F530L) and thermal cycling conditions of 98°C 30 sec, “98°C 10 sec, 59°C 20 sec, 72°C 30 sec” for 27 cycles, 72°C 5 min with the following primers:
Primer 4 1st PCR
Forward:
ggactggttttatagacatcactatg
Primer 5 Reverse:
cgcctattctgctatgtcgacaccc
2nd PCR
Primer 6 Forward:
ccccagaagaccaagggc
Primer 7 Reverse:
ctgcgtgggtaggagcag
For 1st round amplification of in vitro RNA samples by RT-PCR the One-Step RT-PCR kit (Bioland Sci Cat#RT01-01) was used with the same primers listed above. Thermal cycling conditions for the first-round PCR were 42°C 30 min, 94°C 3 min, “94°C 30 sec, 54°C 20 sec, 72°C 30 sec” for 27 cycles, then 72°C for 5 min. For the second-round PCR we utilized Phusion High-Fidelity DNA Polymerase (ThermoFisher Cat#F530L) and thermal cycling of 98°C 30 sec, “98°C 10 sec, 59°C 20 sec, 72°C 30 sec” for 35 cycles, then 72°C for 5 min.
The amplified fragment was ligated with the sequencing adaptor, which had six-nucleotide multiplexing ID to distinguish among different samples (NEBNext Ultra II DNA Library Prep Kit for Illumina Cat# E7645). Deep sequencing was performed with Illumina HiSeq PE150. Raw sequencing reads were de-multiplexed using the six-nucleotide ID. Sequencing error within the barcode region was corrected by filtering out low quality reads (quality score < 30) and un-matched base-pairs between forward and reverse reads. Barcodes with read depth < 10 were filtered.
PCR amplification of in vivo samples
Viral RNA was extracted from plasma samples using a QIAamp viral RNA minikit and tissue RNA was extracted from lysed splenic cells using an RNeasy minikit (QIAGEN). Prior to extraction, plasma samples were spiked with 250 ng of carrier RNA. Splenic RNA was treated with DNase during extraction (QIAGEN) while plasma RNA was treated with DNase post-extraction (Invitrogen). 8 μL of plasma RNA or 500 ng of splenic RNA was used as template for cDNA synthesis. cDNA was synthesized using a SuperScript IV first-strand synthesis kit (ThermoFisher Cat#18091050). The cDNA primer was Primer 8: 5′- GCCTTGCCAGCACGCTCACAG NNNNNNNNNNNNTAGGAGCAGTGCCAGAAGC-3′. We used a PrimerID strategy to accurately quantify RNA copy numbers.30 The cDNA primer was synthesized by Integrated DNA Technologies with machine mixing of random nucleotides and standard desalting for purification. Each cDNA reaction mixture contained 2 μM cDNA primer, 10 mM deoxynucleoside triphosphates (dNTPs), 100 mM dithiothreitol (DTT), 40 U of RNase inhibitor and 200 U of Superscript IV in a total volume of 20 μL. The primers, dNTPs, and templates were mixed and heated at 65°C for 5 min then cooled on ice for 1 min. The reaction mixtures were incubated at 52.5°C for 10 min and then at 80°C for 10 min to inactivate the enzymes. To remove the RNA template, 1 μL of RNase H (Invitrogen) was added to each reaction mixture, followed by incubation at 37°C for 20 min. To remove unused cDNA primer, samples were purified using an RNA clean and concentrator kit (Zymo Research Cat#R1017). cDNA was eluted in RNase/DNase-free water and the entire sample was used for PCR amplification.
cDNA was amplified using a hemi-nested PCR. We used Phusion DNA polymerase (ThermoFisher Cat#F530L) for both rounds of PCR. The first-round PCR forward primer was Primer 9: 5′- CTGACAGAGGACAGGTGGAACAAGC-3′ and the reverse primer was Primer 10: 5′- GCCTTGCCAGCACGCTCACAG-3′. Each first-round PCR reaction mixture contained 10 mM deoxynucleoside triphosphates (dNTPs), 0.4 μM forward primer, 0.4 μM reverse primer and 1U of Phusion in a total volume of 50 μL. The first-round PCR cycling protocol had an initial denaturation at 98°C for 30 s, 30 cycles of 98°C for 10 s, 69.4°C for 20 s, and 72°C for 7 s and a final extension at 72°C for 5 min. 2 μL of first-round PCR product was used as input for second-round PCR. The second-round forward primer was Primer 11: 5′-AAGGGCCACAGAGGGAGC −3′, and the reverse primer was Primer 12: 5′-GCCTTGCCAGCACGCTCACAG −3′. Each second-round PCR reaction mixture contained 10 mM deoxynucleoside triphosphates (dNTPs), 0.4 μM forward primer, 0.4 μM reverse primer and 1U of Phusion in a total volume of 100 μL. The second-round PCR cycling protocol had an initial denaturation at 98°C for 30 s, 35 cycles of 98°C for 10 s, 66.6°C for 20 s, and 72°C for 6 s and a final extension at 72°C for 5 min. PCR products were purified using a DNA clean and concentrator kit (Zymo Research Cat#D4033). Purified DNA was eluted in RNase/DNase-free water and 500 ng in 25 μL was sent for deep sequencing.
PCR product was phosphorylated and A-tailed by Ultra II End Repair Module (NEB). A sequencing adaptor with 6-nucleotide multiplexing tag was ligated to PCR product to distinguish different samples. Deep sequencing was performed on HiSeq3000 platform (Illumina) with PE150 settings.
Data analysis of in vivo sequences
Barcode sequences and PrimerID sequences were extracted by mapping flanking sequences. Barcodes and PrimerIDs with ambiguous base, or base quality score lower than 30 were filtered out. Barcodes with inconsistent forward and reverse reads were also filtered out. We calculated the read depth of PrimerIDs and filtered out low occurrence PrimerIDs according to the bimodal distribution of their read depth36. For each PrimerID, the most dominantly linked barcode was counted. We used CD-hit to cluster barcodes with less than 3-nucleotides difference.37 Mice sacrificed at week 4 post-infection (Figure S2C) were analyzed without incorporation of PrimerID.
Production of barcoded virions
For transfection, 107 293FT cells were seeded into 150 cm2 tissue culture flask and the next day were transfected with 30 μg of barcoded HIV plasmid using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. One day after transfection the media was replaced with 20 mL of Opti-MEM media (GIBCO) containing 10% FBS. Virus-containing supernatant was harvested at day 2 post-transfection and passed through a 0.45 μm filter. Aliquots of virus were frozen at −80°C and thawed immediately prior to use.
In vitro infections
During infection of costimulated PBMC, 5x105 cells were exposed to virus in a screw-capped 1.5 mL tube containing 250 μL of RF10 medium and 10 μg/mL of Polybrene. This was incubated on a rocking platform for 2 h at 37°C. After infection, cells were washed and resuspended in 1 mL of RF10 medium containing 20 U/mL of IL-2, and viral growth was quantified by analyzing supernatant samples using an HIV p24 enzyme-linked immunosorbent assay kit (Perkin Elmer Alliance Cat#NEK050).
GHOST (3) CXCR4+CCR5+ were seeded into 24-well tissue culture plates at 5x104 cells/well prior to infection. The next day media was aspirated and 250 μl of virus suspended in fresh media containing 10 μg/mL of Polybrene was added to cells. Plates were incubated for 2h at 37°C and then the media was replaced with 1 mL of fresh media without polybrene. Cells were incubated for a further 2 days and then harvested by exposure to 0.05% tripsin (GIBCO Cat#5400-54) in PBS for 5 min, and then removed by agitation with media. After removal from plate cells were pelleted and resuspended in 2% paraformaldehyde then analyzed for GFP expression by flow cytometry as described below.
Infection and analysis of BLT mice
Ex vivo infection of CD4+ T cells was performed by sacrificing 2-4 animals from a humanized BLT mouse series to isolate human cells. Mice were anesthetized with isoflurane and bled by cardiac puncture then euthanized and spleens removed. Spleen tissues was disaggregated and filtered through a 40 μm filter to produce a single-cell suspension. Splenocytes and peripheral blood mononuclear cells from all animals were then pooled and subjected to positive immunomagnetic selection using human-CD4 MicroBeads (Miltenyi Biotec Cat#130-045-101) following the manufacturer’s instructions. Cells were costimulated for 3 days as described above and then infected with barcoded HIV (182-560 ng of p24). Infections were conducted by spinoculating 2 million costimulated CD4+ T cells per well of a 24-well plate in 400 μL of virus containing 10 μg/mL of polybrene. Plates were centrifuged at 1200xg for 2 hours at 25°C.
After infection an aliquot of cells was removed for overnight culture in RF10 media to quantify infection efficacy by flow cytometry specific for the HA reporter gene, and the remaining cells were resuspended in 500 μl of barcoded HIV virions (ranging from 280-700ng of p24 in different experiments) and then injected intraperitoneally into mice from the same series (i.e., syngeneic mice generated with cells from the same human donor tissue).
HIV RNA copy numbers in plasma at each bleed were quantified using RT-PCR performed by the UCLA AIDS Institute virology core as previously described.9 After 4-5 weeks of infection, mice were treated with ART in animal feed consisting of emtricitibine (FTC) at 0.5 mg/g of feed, tenofovir disoproxil fumarate (TDF) at 0.75 mg/gram of feed, and raltegravir at 1 mg/g of feed for a further 6-7 weeks. PKC modulator for injection (2 μg/mouse) was initially suspended in 0.6 μL of DMSO and then this was added to 500 μL of phosphate buffered saline before administration to mice by intraperitoneal injection. Control animals were injected with the same concentration of DMSO in PBS without compound.
At the time of necropsy the mice were anesthetized with isoflurane and then exsanguinated by intracardiac bleed using a 1 mL syringe and 25G ½ inch needle rinsed with 0.5M EDTA to prevent clotting, and then transferred into a 1.5 mL screw-capped tube containing 2 μL of 0.5M EDTA. Animals were then euthanized and spleens removed. Blood was centrifuged at 1300 xg and the upper plasma layer was collected, aliquoted and stored at −80°C. The central 150 μL layer containing white blood cells was transferred into a new 1.5 mL screw-capped tube and then 1 mL of Ammonium Chloride Solution lysis buffer (StemCell Technologies, Inc.) was added to each tube. Tubes were then briefly vortexed and incubated for 5 min at room temperature, before pelleting by centrifugation at 1300 xg. Splenocytes were obtained by passing disaggregated tissue through a 40 μm filter. Blood and spleen cells were then washed with RF10 media and either used for flow cytometry as described below, frozen as cell pellets, or suspended in RLT buffer (QIAGEN) for RNA storage and then frozen at −80°C.
Ex vivo stimulations
During ex vivo stimulations for virus outgrowth, splenocytes comprising approximately 60% of spleen tissue were costimulated in RF10 medium containing 20 U/mL of interleukin-2 (IL-2) in the presence of plate-bound anti-CD3 (1 μg/mL) and soluble anti-CD28 (100 ng/mL) and then 106 CEM cells were added to the cultures to facilitate replication of any released virus. At day 3 post-stimulation cells were washed and stimulatory antibodies removed. Cultures were split every 3 to 4 days and p24 levels in the supernatant were quantified using an HIV p24 enzyme-linked immunosorbent assay kit (Perkin Elmer Alliance Cat:NEK050) at 14- or 15-days post-stimulation.
Flow cytometry
During cell staining 105 cells were suspended in a 50 μL volume of 1:1 dilution of phosphate buffered saline (PBS):Human AB serum (Sigma). For 2-step staining to detect cell surface HA expression, cells were first stained with Anti-HA-Biotin, High Affinity antibody (Sigma Cat#12158167001) at 4°C for 20 min and then after washing were stained with the secondary Streptavidin, R-PE Conjugate (FisherSci Cat#S866A) before resuspension in 2% paraformaldehyde in PBS. PBMC from humanized mice were evaluated for the presence of human immune cells by staining with either anti-human CD45 e450 (eBioscience Cat#48-0459-42) or Pacific Blue anti-human CD45 Antibody (HI30) (Biolegend Cat#304029). Stained samples were stored at 4°C, then samples were run using a Cytomics FC 500 flow cytometer (Beckman Coulter). Data was analyzed using FlowJo (v7) software.
SUW133 compound
PKC modulator SUW133 was synthesized as previously described.25
Quantification and Statistical Analysis
Statistical analyses were performed using Prism software (GraphPad) or Microsoft Excel. Statistical details of experiments and significance can be found in the figure legends.
Acknowledgments
This work was supported by grants from the National Institutes of Health (P01 AI131294 to J.A.Z., CA31845 to P.A.W., AI124763 to M.D.M., AI124743 to J.A.Z. and P.A.W.), the National Center for Advancing Translational Sciences UCLA CTSI Grant (UL1TR001881 and UL1TR000124), and the UCLA Center for AIDS Research (AI28697). The UCLA AIDS Institute and Charity Treks also provided support. ART drugs for humanized mouse studies were kindly provided by Merck (raltegravir) and Gilead Sciences (TDF and FTC). The illustrations were created with https://biorender.com/. Funding agencies did not play a specific role in the conception, design, data collection, analysis, decision to publish, or preparation of this manuscript.
Author Contributions
M.D.M., T.-h.Z., Y.D., M.D., X.W., M.S.A.S., J.T.K., A. Shimizu, and A. Schrier conducted the experiments and acquired the data. M.D.M., T.-h.Z., Y.D., M.D., X.W., M.S.A.S., J.T.K., R.S., P.A.W., and J.A.Z. participated in LRA analog selection and supply and data interpretation and analysis. All of the authors contributed text, edited the manuscript, and approved the final version.
Declaration of Interests
Stanford University has filed patent applications on SUW133 and related technology, which has been licensed by Neurotrope BioScience for the treatment of neurological disorders and by BryoLogx for use in HIV/AIDS eradication and cancer immunotherapy. P.A.W. is an adviser to both companies and a cofounder of the latter. J.A.Z. is on the scientific advisory board for BryoLogx and is a co-founder of CDR3 Therapeutics.
Published: December 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100162.
Contributor Information
Paul A. Wender, Email: wenderp@stanford.edu.
Ren Sun, Email: rsun@mednet.ucla.edu.
Jerome A. Zack, Email: jzack@ucla.edu.
Supplemental Information
References
- 1.UNAIDS Global HIV & AIDS statistics — 2020 fact sheet. https://www.unaids.org/en/resources/fact-sheet
- 2.Chun T.W., Davey R.T., Jr., Ostrowski M., Shawn Justement J., Engel D., Mullins J.I., Fauci A.S. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 2000;6:757–761. doi: 10.1038/77481. [DOI] [PubMed] [Google Scholar]
- 3.Davey R.T., Jr., Bhat N., Yoder C., Chun T.W., Metcalf J.A., Dewar R., Natarajan V., Lempicki R.A., Adelsberger J.W., Miller K.D. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun T.W., Stuyver L., Mizell S.B., Ehler L.A., Mican J.A., Baseler M., Lloyd A.L., Nowak M.A., Fauci A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D., Hermankova M., Pierson T., Carruth L.M., Buck C., Chaisson R.E., Quinn T.C., Chadwick K., Margolick J., Brookmeyer R. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Marsden M.D., Zack J.A. HIV/AIDS eradication. Bioorg. Med. Chem. Lett. 2013;23:4003–4010. doi: 10.1016/j.bmcl.2013.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsden M.D., Zack J.A. Humanized Mouse Models for Human Immunodeficiency Virus Infection. Annu. Rev. Virol. 2017;4:393–412. doi: 10.1146/annurev-virology-101416-041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melkus M.W., Estes J.D., Padgett-Thomas A., Gatlin J., Denton P.W., Othieno F.A., Wege A.K., Haase A.T., Garcia J.V. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 2006;12:1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 9.Marsden M.D., Kovochich M., Suree N., Shimizu S., Mehta R., Cortado R., Bristol G., An D.S., Zack J.A. HIV latency in the humanized BLT mouse. J. Virol. 2012;86:339–347. doi: 10.1128/JVI.06366-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denton P.W., Olesen R., Choudhary S.K., Archin N.M., Wahl A., Swanson M.D., Chateau M., Nochi T., Krisko J.F., Spagnuolo R.A. Generation of HIV latency in humanized BLT mice. J. Virol. 2012;86:630–634. doi: 10.1128/JVI.06120-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llewellyn G.N., Seclén E., Wietgrefe S., Liu S., Chateau M., Pei H., Perkey K., Marsden M.D., Hinkley S.J., Paschon D.E. Humanized mouse model of HIV-1 latency with enrichment of latent virus in PD-1+ and TIGIT+ CD4 T cells. J. Virol. 2019;93:e02086-e18. doi: 10.1128/JVI.02086-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsden M.D., Loy B.A., Wu X., Ramirez C.M., Schrier A.J., Murray D., Shimizu A., Ryckbosch S.M., Near K.E., Chun T.W. In vivo activation of latent HIV with a synthetic bryostatin analog effects both latent cell “kick” and “kill” in strategy for virus eradication. PLOS Pathog. 2017;13:e1006575. doi: 10.1371/journal.ppat.1006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai P., Wu G., Baker C.E., Thayer W.O., Spagnuolo R.A., Sanchez R., Barrett S., Howell B., Margolis D., Hazuda D.J. In vivo analysis of the effect of panobinostat on cell-associated HIV RNA and DNA levels and latent HIV infection. Retrovirology. 2016;13:36. doi: 10.1186/s12977-016-0268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsden M.D. Benefits and limitations of humanized mice in HIV persistence studies. Retrovirology. 2020;17:7. doi: 10.1186/s12977-020-00516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun T.W., Engel D., Mizell S.B., Hallahan C.W., Fischette M., Park S., Davey R.T., Jr., Dybul M., Kovacs J.A., Metcalf J.A. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 1999;5:651–655. doi: 10.1038/9498. [DOI] [PubMed] [Google Scholar]
- 16.Archin N.M., Liberty A.L., Kashuba A.D., Choudhary S.K., Kuruc J.D., Crooks A.M., Parker D.C., Anderson E.M., Kearney M.F., Strain M.C. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J., Gaiha G.D., John S.P., Pertel T., Chin C.R., Gao G., Qu H., Walker B.D., Elledge S.J., Brass A.L. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2:807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkosky J., Culnan D.M., Roman J., Dornadula G., Schnell M., Boyd M.R., Pomerantz R.J. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood. 2001;98:3006–3015. doi: 10.1182/blood.v98.10.3006. [DOI] [PubMed] [Google Scholar]
- 19.Marsden M.D., Wu X., Navab S.M., Loy B.A., Schrier A.J., DeChristopher B.A., Shimizu A.J., Hardman C.T., Ho S., Ramirez C.M. Characterization of designed, synthetically accessible bryostatin analog HIV latency reversing agents. Virology. 2018;520:83–93. doi: 10.1016/j.virol.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang G., Mendes E.A., Kaiser P., Wong D.P., Tang Y., Cai I., Fenton A., Melcher G.P., Hildreth J.E., Thompson G.R. Synergistic Reactivation of Latent HIV Expression by Ingenol-3-Angelate, PEP005, Targeted NF-kB Signaling in Combination with JQ1 Induced p-TEFb Activation. PLOS Pathog. 2015;11:e1005066. doi: 10.1371/journal.ppat.1005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spivak A.M., Nell R.A., Petersen M., Martins L., Sebahar P., Looper R.E., Planelles V. Synthetic Ingenols Maximize Protein Kinase C-Induced HIV-1 Latency Reversal. Antimicrob. Agents Chemother. 2018;62:e01361-e18. doi: 10.1128/AAC.01361-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brooks D.G., Hamer D.H., Arlen P.A., Gao L., Bristol G., Kitchen C.M., Berger E.A., Zack J.A. Molecular characterization, reactivation, and depletion of latent HIV. Immunity. 2003;19:413–423. doi: 10.1016/s1074-7613(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 23.Laird G.M., Bullen C.K., Rosenbloom D.I., Martin A.R., Hill A.L., Durand C.M., Siliciano J.D., Siliciano R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015;125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beans E.J., Fournogerakis D., Gauntlett C., Heumann L.V., Kramer R., Marsden M.D., Murray D., Chun T.W., Zack J.A., Wender P.A. Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc. Natl. Acad. Sci. USA. 2013;110:11698–11703. doi: 10.1073/pnas.1302634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeChristopher B.A., Loy B.A., Marsden M.D., Schrier A.J., Zack J.A., Wender P.A. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat. Chem. 2012;4:705–710. doi: 10.1038/nchem.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H.C., Martinez J.P., Zorita E., Meyerhans A., Filion G.J. Position effects influence HIV latency reversal. Nat. Struct. Mol. Biol. 2017;24:47–54. doi: 10.1038/nsmb.3328. [DOI] [PubMed] [Google Scholar]
- 27.Fennessey C.M., Pinkevych M., Immonen T.T., Reynaldi A., Venturi V., Nadella P., Reid C., Newman L., Lipkey L., Oswald K. Genetically-barcoded SIV facilitates enumeration of rebound variants and estimation of reactivation rates in nonhuman primates following interruption of suppressive antiretroviral therapy. PLOS Pathog. 2017;13:e1006359. doi: 10.1371/journal.ppat.1006359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali A., Yang O.O. A novel small reporter gene and HIV-1 fitness assay. J. Virol. Methods. 2006;133:41–47. doi: 10.1016/j.jviromet.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Persaud D., Gay H., Ziemniak C., Chen Y.H., Piatak M., Jr., Chun T.W., Strain M., Richman D., Luzuriaga K. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N. Engl. J. Med. 2013;369:1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou S., Jones C., Mieczkowski P., Swanstrom R. Primer ID Validates Template Sampling Depth and Greatly Reduces the Error Rate of Next-Generation Sequencing of HIV-1 Genomic RNA Populations. J. Virol. 2015;89:8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsden M.D., Zack J.A. HIV cure strategies: a complex approach for a complicated viral reservoir? Future Virol. 2019;14:5–8. [Google Scholar]
- 32.Pache L., Marsden M.D., Teriete P., Portillo A.J., Heimann D., Kim J.T., Soliman M.S.A., Dimapasoc M., Carmona C., Celeridad M. Pharmacological Activation of Non-canonical NF-kappa B Signaling Activates Latent HIV-1 Reservoirs In Vivo. Cell Rep. Med. 2020;1:100037. doi: 10.1016/j.xcrm.2020.100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bristol G.C., Gao L.Y., Zack J.A. Preparation and maintenance of SCID-hu mice for HIV research. Methods. 1997;12:343–347. doi: 10.1006/meth.1997.0488. [DOI] [PubMed] [Google Scholar]
- 34.McCune J.M., Namikawa R., Kaneshima H., Shultz L.D., Lieberman M., Weissman I.L. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–1639. doi: 10.1126/science.241.4873.1632. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu S., Ringpis G.E., Marsden M.D., Cortado R.V., Wilhalme H.M., Elashoff D., Zack J.A., Chen I.S., An D.S. RNAi-Mediated CCR5 Knockdown Provides HIV-1 Resistance to Memory T Cells in Humanized BLT Mice. Mol. Ther. Nucleic Acids. 2015;4:e227. doi: 10.1038/mtna.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert D., Di Giallonardo F., Topfer A., Singer J., Schmutz S., Gunthard H.F., Beerenwinkel N., Metzner K.J. A Comprehensive Analysis of Primer IDs to Study Heterogeneous HIV-1 Populations. J Mol Biol. 2016;428:238–250. doi: 10.1016/j.jmb.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All codes are available on https://github.com/Tian-hao/BarcodeHIV.
All raw sequencing reads were deposited on SRA (Short Read Archive) database under the accession number (PRJNA562389).
The authors declare that all other data supporting the findings of this study are available within the paper and its supplementary information files.