Summary
Current organoid technologies require intensive manual manipulation and lack uniformity in organoid size and cell composition. We present here an automated organoid platform that generates uniform organoid precursors in high-throughput. This is achieved by templating from monodisperse Matrigel droplets and sequentially delivering them into wells using a synchronized microfluidic droplet printer. Each droplet encapsulates a certain number of cells (e.g., 1,500 cells), which statistically represent the heterogeneous cell population in a tumor section. The system produces >400-μm organoids within 1 week with both inter-organoid homogeneity and inter-patient heterogeneity. This enables automated organoid printing to obtain one organoid per well. The organoids recapitulate 97% gene mutations in the parental tumor and reflect the patient-to-patient variation in drug response and sensitivity, from which we obtained more than 80% accuracy among the 21 patients investigated. This organoid platform is anticipated to fulfill the personalized medicine goal of 1-week high-throughput screening for cancer patients.
Keywords: microfluidics, droplet, printing, organoid, tumor
Graphical Abstract
Highlights
An automated platform produces and patterns uniform organoids in high throughput
The production of a >400-μm large organoid is shortened to 1 week
Organoids possess both inter-organoid homogeneity and inter-patient heterogeneity
Organoids recapitulate 97% gene mutations in tumors and predict drug responses
Jiang et al. present an automated system that comprises organoid fabrication, manipulation, and drug evaluation functions in sequence, aiming to fulfill the personalized medicine goal of 1-week high-throughput screening for cancer patients.
Introduction
Despite substantial progress achieved with the advents of new therapeutics, the treatment of metastatic solid tumors faces major challenges. The failure is mainly attributed to the intrinsic tumor heterogeneity, caused by the accumulation of gene mutations that result in diversified cell phenotypes.1, 2, 3 To predict the response of a patient tumor to a certain drug, in vitro tumor models reflecting the drug sensitivity and resistance with high efficacy and accuracy shall be established.
Two-dimensional (2D) culture of cancer cells reflects poorly of tumor properties due to loss of native cell microenvironment.4 Multicellular spheroids and cell clusters are lack of the organotypic cell constructs, or their growth is limited in size.5,6 Patient-derived xenograft models recapitulate the genotype and phenotype of patient tumors, but the model establishment is costly and time-consuming and has low success rates and limited scalability.7,8
Organoid is an in vitro three-dimensional (3D) cell-culture technology that captures and stably passes down the genomic and phenotypic profiles of human healthy organs and tumors, by growing from single or multiple cells embedded in an appropriate 3D matrix, such as Matrigel or basement membrane matrix.9, 10, 11 Organoids are scalable, easy to culture, and prospective to evaluate patient tumor sensitivity to anticancer drugs.4,12,13 Recapitulation of personalized immune responses to immuno-checkpoint drugs targeting PD-1/PD-L1 has also been technically proven in organoids.14,15
However, the current organoid technologies have limitations. Manual manipulation of unpatterned cell-suspension volumes introduces significant batch-to-batch and organoid-to-organoid variability.16,17 Though the exact causes are unclear, inconsistent cellular complexity among organoids and batch experiments is a contributing factor. Growing organoids from manually patterned cell-laden Matrigel volumes reduces the organoid-to-organoid variability, but it remains labor intensive and batch-to-batch variant.18 More importantly, current culturing protocol requires 4–6 weeks to obtain large organoids, which exerts timeliness stress on organoid-based therapy screenings.4
To capture patient tumor heterogeneity and meanwhile shorten the model establishment duration, an organoid should be grown from a collection of cells, e.g., 1,000–2,000 cells, that are statistically representative of the heterogeneous cell population of a parental tumor tissue, and patterned in defined Matrigel volume. It enables the simultaneous achievement of inter-organoid homogeneity and inter-patient heterogeneity. Additionally, to reduce labor cost and batch-to-batch variability, high-throughput and automated organoid distribution become necessary.
Microfluidics has been utilized widely to fabricate reproducible spherical cell-laden structures supported by engineerable scaffolds, such as alginate and gelatin.19 Matrigel, though proved supreme in supporting cell growth, has yet to be manipulated in microfluidics toward spherical structures or in printing for automated distribution. Here, we report an automated organoid platform that manipulates Matrigel spheres and fulfills the aforementioned requirements. The organoids are validated by displaying the analogous gene-expression profiles and histological characteristics as the healthy and cancerous organs of cell derivation, as well as patient-dependent variance in drug responses.
Results
The Automated Organoid Platform
Substantial inter-organoid variability20,21 has remained a major challenge of 3D-organoid technology since its early establishment from a single intestinal stem cell in 2009.22 To reduce its variability and labor cost, we developed an organoid platform that embraced the power of high throughputs, automation, and uniformity in organoid fabrication and manipulation, simultaneously.
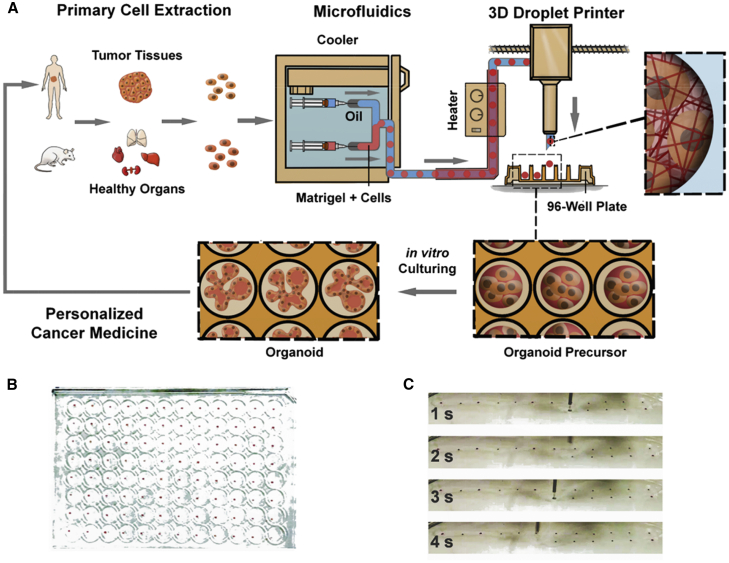
The organoid platform consisted of two modules in synchronization, a microfluidics-based module (M1) for organoid production and a 3D droplet printing module (M2) for automated organoid distribution. M1 was a customized droplet-based microfluidics system for Matrigel manipulation, assembled in a low-temperature controller (cooler), which maintained the Matrigel solution at 4°C to prevent its gelation. Two pieces of polytetrafluoroethylene (PTFE) injection tubing for Matrigel and volatile cell-compatible oil (HFE7000, ∼34°C b.p., 3M) were connected to a third tubing via a 3-way polydimethylsiloxane (PDMS) connector, where cell-laden growth-factor-reduced Matrigel solution was formulated into monodisperse droplets. Outside the cooler in M1, the tubing, 10 m long and rolled up (except for the ends), was incubated in a water tank warmed at 37°C, to accelerate the gelation of Matrigel droplets. Matrigel droplets were equally spaced by oil plugs, underwent a sol-gel transition in the same tubing, and transformed into gel spheres before approaching the tubing outlet (Figure 1A; Figure S1).
Figure 1.
The Automated Organoid Platform
(A) Sketch of the organoid platform. The platform contains an organoid fabrication module (M1) and an organoid printing module (M2). M1 is a customized droplet-based microfluidics system, where monodisperse cell-laden Matrigel droplets are generated and function as the structural templates for organoid precursors in the PTFE tubing. The tubing outlet is connected to the M2 module that is a modified 3D droplet printer and distributes individual organoid precursors into precision patterns.
(B) Display of precision distribution of identical organoid precursors, i.e., cell-laden Matrigel spheres, on the cover of a 96-well plate. Each well cover contains one single sphere and positioned in the center.
(C) Snapshot of the printing process, when the tubing outlet is placed on top of a well cover and controlled by the 3D droplet printer. The flow rates of the injection pumps were set at 30 μL/min for the oil phase and 20 μL/min for the droplet phase for formulation. The printer translation speeds were set as XY: 500 mm/s, Z: 20 mm/s, standing time 2 s for each well, at the oil flow rate of 30 μL/min for printing.
See also Figures S1.
The strategy of adopting a single piece of PTFE tubing to conduct the Matrigel droplet formation, incubation, transport, and positioning minimized the mechanical interference and flow disturbance on the Matrigel droplets and gelled spheres. The droplets took the entire cross-section of the tubing, which prevented the droplet collision and eliminated the dependence on surfactants to stabilize the droplets. The oil plug spacing sequenced the droplets, enabling them to be sequentially printed.
M2 was a droplet printer23 but customized to pattern Matrigel spheres with high spatiotemporal resolution. The XY motion of the printing head was stepwise and synchronized with the Matrigel droplet formation frequency. The droplet printer guided the locomotion of the tubing outlet to pattern individual gelled cell-laden Matrigel spheres, i.e., organoids precursors, into discrete culturing chambers, e.g., 96 micro-plate wells. The highly volatile oil, HFE7000, rapidly evaporated when its temperature was elevated approaching the outlet. No oil residual was observed at a few minutes after accomplishing a plate printing. The organoid precursors were detached from the tubing, patterned onto the substrate and developed into organoids during culture (Figure 1A).
Under the translocation guidance of the M2 module, the organoid precursors were printed into precision arrays on the cover plate with replica patterns as the 96-well plate for better visualization. The organoids were positioned to the micro-plate well centers, with each well containing one single organoid (Figure 1B; Videos S1, S2, and S3). The current success rate reached above 95%, i.e., up to 4 missing placements in a 96-well plate. The error was largely attributed to the lack of sufficient low temperature control on Matrigel loaded in the infusion syringe, as the cooling setup was a fridge and not customized for this task.
The play was accelerated by 6 times.
The play was accelerated by 6 times.
Figure 1C shows the printing head and the sequential placement of individual organoids during the organoid array production. The Matrigel spheres containing cells were consistent in appearance after being printed. High-throughput capability refers to the fact that the templating cell-laden Matrigel organoid precursors are generated at 10°–102 Hz; the amount of organoid precursors generated in one single experiment depends on the volume of parental tissues and Matrigel consumption. In our study, we generated 100–1,000 organoid precursors in less than 10 min and sequentially patterned individual organoids in approximately one organoid per second by machine.
Inter-organoid Homogeneity
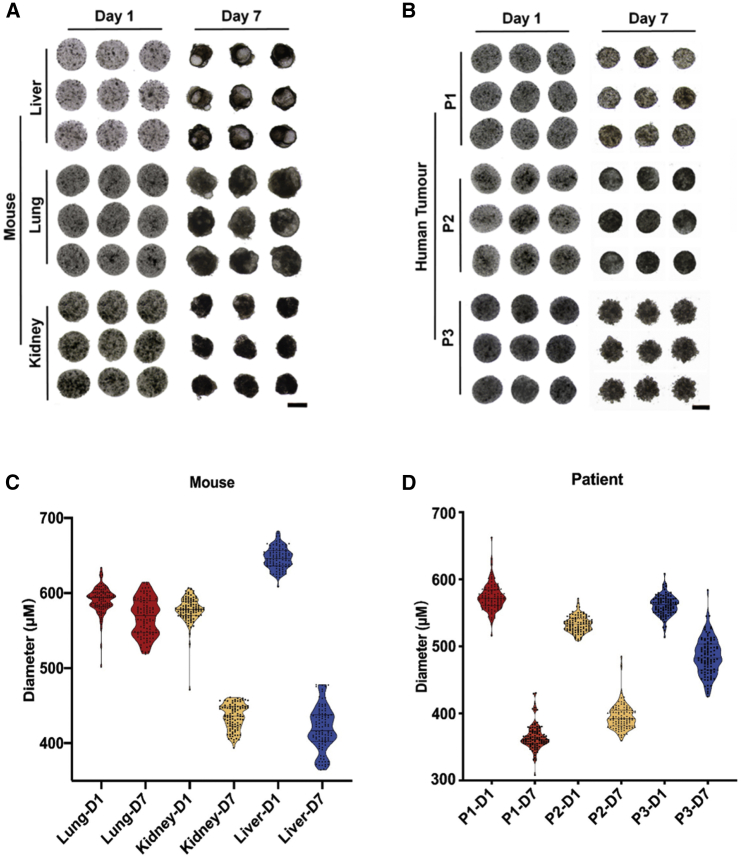
The organoid precursors derived from multiple healthy mouse organs and human tumors were monodisperse in size and shape. After being cultured for 7 days, the precursors developed into organoids possessing the reproducible and demanded constructs as their parental tissues. For example, the mouse lung and liver organoids were grown with semi-transparent buds representing the self-organized epithelium, the typical characteristics of healthy organoids (Figures 2A and 2B; Figure S2). The size counts of the organoid precursors and the developed organoids exhibited narrow distribution, proving the significantly reduced variability of organoids in our platform (Figures 2C and 2D). Notably, the organoid morphology development was heavily correlated with the organs of cell derivation. For example, healthy lung organoids displayed nearly identical sizes as their organoid precursors, but liver and kidney organoids were remarkably shrunk, possibly attributed to the improved cell density after morphology development in these tissues. The liver developed large volumes of epithelium, judging from the appearance of pseudo-transparent tissues that emerged on day 7. The morphological development in tumor organoids was less significant than their healthy counterparts, except for the decrease in sizes. This was in accordance with previous reports.12
Figure 2.
Inter-organoid Homogeneity
(A and B) Bright-field images of identical organoid precursors (day 1) and organoids (day 7) of (A) mouse lung, kidney, liver, and (B) human lung, kidney and stomach tumors from three patients (P1, P2, P3), acquired after they were printed (day 1) and cultured for 7 days (day 7). Scale bar, 400 μm.
(C and D) The violin plots of diameters of the mouse and human tumor organoid precursors (D1) and organoids (D7) at (A) and (B). The initial cell seeding density was 2.0 × 107 cells mL–1 for all panels (n = 100).
See also Figure S2.
Each Matrigel droplet, about 0.08 μL, encapsulated approximately 1,506 ± 13 cells (n = 3), equivalent to a cell density of 2.0 × 107 cells per mL. At this high initial cell density, the organoids developed rapidly from cell-laden Matrigel spheres, recapitulating the organotypic constructs within 5–7 days. Traditionally, it required 4–6 weeks growth time for the organoids to reach large sizes for drug screening (e.g., larger than 400 μm across).4 The rapid organoid development in our system may shorten the duration from patient tumor sampling to accomplishment of drug evaluation and therapy recommendation to 1 week.
Organoid Validation by Histology and Gene Expression
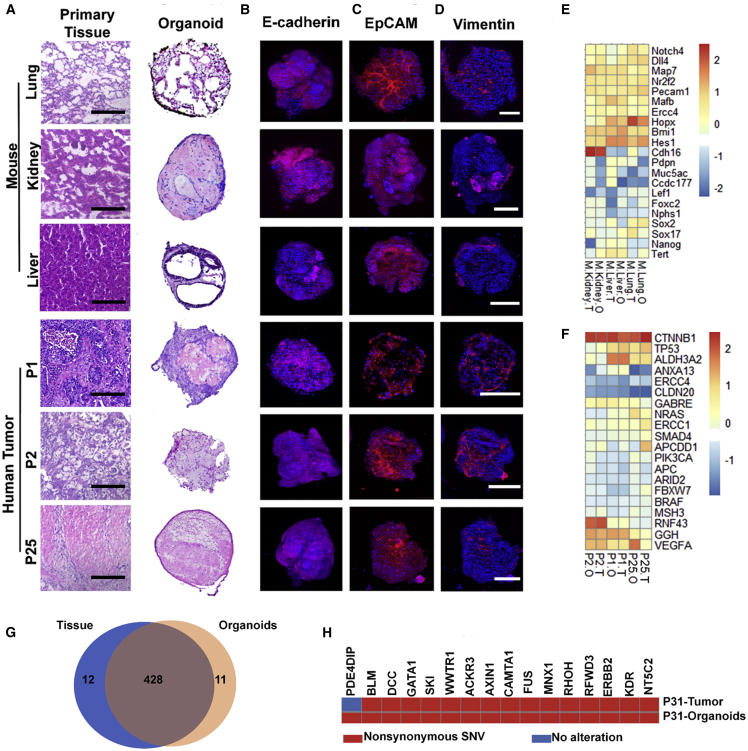
To further assess whether the organoids resembled the characteristics of their parental healthy tissues and tumors, we first performed the histological analysis. The mouse organoids derived from lung, kidney, and liver displayed analogous histology in microscale to the corresponding tissues of cell derivation, with significant recapitulation of epithelial organization. Contrarily, the tumor organoids displayed much reduced epithelium in the volume12 (Figure 3A).
Figure 3.
Histopathological Characterization and Gene-Expression Profiling of Healthy and Tumor Organoids
(A) Histological images of organoids derived from mouse lung, kidney, liver, and human lung (P1), liver (P2), and gastric (P25) tumors and the comparison with their parental tissues/tumors. Scale bar, 200 μm.
(B–D) Immunofluorescence staining for (B) E-cadherin, (C) EpCAM, and (D) vimentin of the organoids in (A). Scale bar, 200 μm.
(E and F) Heatmap of gene expression of the organoids and their parental (E) tissues and (F) tumors by RNA-seq. It profiles (E) the organ-associated genes and chemotherapy-related genes and (F) the cancer genes and chemotherapy-related genes.
(G) Venn diagram showing ∼97% mutational (SNV) overlap between the P31 tumor organoids and the parental tumor tissue.
(H) Overview of the oncogene mutations detected in the P31 tumor organoids and the parental tumor tissue.
See also Figure S3.
The organoids were also investigated by immunofluorescence imaging of their interiors. Figure 3B shows the projects of z stack confocal microscopic immunostaining images for E-cadherin (magenta), the marker for epithelial cadherins, and nuclei (DAPI, blue). The homogeneous and wide distribution of E-cadherin suggested that the organoids had resembled the parental tissue-alike epithelium, and the tumor organoids were dominated with epithelial cells (Figure 3B). The dominant distribution of epithelial cells was further approved by the single confocal image planes displaying wide distribution of the epithelial cell adhesion molecule (EpCAM) (Figure 3C). The organization of epithelial monolayer domains were indicated by the membrane staining in the Figure S3A and the Videos S4 and S5.
It must be noted that the Matrigel droplets encapsulated cell mixtures extracted from the parental tissues, where fibroblasts and epithelial cells were the major components. Fibroblasts grew at higher rates than tumor epithelial cells,12 so we deliberately suppressed the overwhelming growth of fibroblasts by using the human 9 fibroblast growth inhibitor (FibrOut, CHI Scientific). Consequently, the number of fibroblasts, indicated by the distribution of immunostained intermediate filament vimentin (fibroblast marker, red), became less substantial and predominantly located along the organoid peripheries (Figure 3D). Moreover, the immune environment was preliminarily recapitulated in organoids, as indicated by the immunostaining for CD3, the T cell co-receptor (Figure S3B).
The images in Figures 3B–3D and Figure S3 were acquired from non-sectioned organoids, where the light scattering blocked the signal transmission from deep tissues. Penetration of immunostaining reagents were also impeded by the dense matrices. However, these probed virtual planes provided improved acquisition of the regional organo-constructs in intact organoids.
We next assessed whether organoids maintained the gene-expression signatures of their parental healthy tissues and tumors. We performed transcriptomic analysis by bulk RNA sequencing (RNA-seq) across 3 mice and 3 patients (Figures 3E and 3F). The organoids were cultured for 7 days. Heatmap of gene expression compared the organoids (O) and their parental tissues (or tumors, T). The organ-specific marker genes and chemotherapy-related genes composed the library for the mouse organoids, whereas the cancer genes and chemotherapy-related genes composed the comparison set for tumor organoids. In both groups, organoids displayed analogous profiles to their parental healthy tissues or tumors, but the profiles were more distinctive among the organoids derived from different organs and tumors. We also performed the whole exome sequencing. The tumor-derived organoids were found comprising a highly overlapped (over 97%) single nucleotide variants (SNVs) profile with their parental tumor tissue (P31) (Figure 3G). For the oncogene dataset comprising 731 genes, we found 16 genetic mutations in the P31 organoids and 15 genetic mutations in the parental tumor tissue (Figure 3H). It suggests that the organoids recapitulated the genetic information of the parental tumor.
Droplet Circulation Has Minimal Effect on Altering Gene Expression
Matrigel droplets circulated in the long incubation tubing (∼10 m) for around 10 min before reaching the outlet when they gradually phase-transformed into gel spheres. The structure functioned as the templating scaffold for organoid development and contributed to reduce inter-organoid variability. Moving droplets had friction with the tubing wall that induced internal circulation and shear. Our previous studies found that the flow topology in moving microfluidic droplets was dominated by the inner-to-outer viscosity ratio across the droplet interfaces, and the internal circulation and shear were dramatically suppressed by reducing the viscosity of the oil phase.24, 25, 26 The kinematic viscosity of HFE7000 was 0.32 cSt, significantly lower than that of the Matrigel solution. Therefore, the Matrigel droplet volume adopted a uniform flow profile with negligible internal circulation and exerted minimal shear stress on the encapsulated cells throughout the process.
To further verify the elimination of detrimental or misregulation effects from fluid shear on the encapsulated cells, we chose a highly sensitive cell type, human induced pluripotent stem cells (hiPSCs),27 as the cell model to examine the shear effects. The iPSCs were first suspended in the Matrigel solution at 1.0 × 107 cells per mL and formulated into monodisperse cell-laden Matrigel droplets (D = 560 μm). The droplets were circulated and incubated in tubing and cultured for 1 day (S1) and 4 days (S4) after being patterned. The RNA-seq data were compared with the undifferentiated iPSCs (S0) cultured according to the standard culturing protocols (see STAR Methods). The S0 cells were employed as the control to investigate the effects of culture conditions because both the S1 and S4 samples were derived from S0.
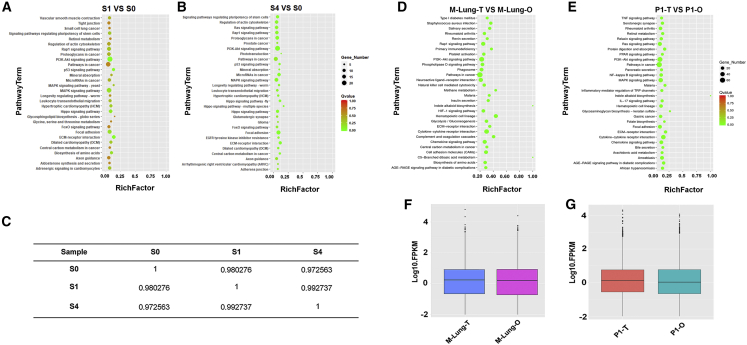
Figures 4A and 4B show the most enriched 30 pathways in S1 and S4, compared with S0, performed by the KEGG pathway enrichment analysis. The alteration was insignificant. The Pearson’s correlation analysis (Figure 4C) provides an overview of all the variation between the three samples (S0, S1, and S4). The Pearson’s correlation coefficient r2 were all over 0.97, indicating that the mechanical interference in microfluidics and droplet templating had negligible mis-regulatory effects on gene expression of encapsulated cells. It paved the way for using synchronized microfluidic droplet printing as a tool to achieve automated organoid production and manipulation.
Figure 4.
Cell-Phenotype Consistency of Organoids and Their Parental Tissues/Tumors Profiled by RNA-Seq
(A and B) Scatterplot of the KEGG pathway enrichment analysis of differentially expressed genes in paired comparisons of (A) S0 versus S1 and (B) S0 versus S4 iPSCs. The Rich factor in the x axis is the ratio of differentially expressed gene numbers annotated in a pathway term to all gene numbers annotated in this pathway term. A greater Rich factor indicates a higher degree of pathway enrichment. The color codes the p values.
(C) Pairwise Spearman’s ρ correlation coefficients between S0, S1, and S4 iPSCs. The iPSCs were loaded in Matrigel at 1.0 × 107 cells mL–1, formulated into droplets, incubated in tubing for 10 min, printed, and cultured in vitro before being sequenced.
(D and E) Scatterplot of the KEGG pathway enrichment analysis of differentially expressed genes in paired comparisons of (D) mouse lung tissue (M-Lung-T) versus mouse lung organoid (M-Lung-O) and (E) patient 1 derived organoid (P1-O) versus patient 1 tumor (P1-T, lung).
(F and G) Boxplot of the log FPKM expression values in (F) M-Lung-T and M-Lung-O, and (G) P1-O and P1-T.
The Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis showed that few of the biological pathways were altered significantly against their parental tissues (Figure 4D) or tumors (Figure 4E). This was further verified by the boxplots of the log FPKM (fragments per kilobase of exon per million fragments mapped) expression values in Figures 4F and 4G. It suggests that the organoids are able to replicate the inter-patient heterogeneity, which determines their feasibility on personalized therapy evaluation.
Tumor Organoids Capture Inter-patient Heterogeneous Responses to Anticancer Drugs
The advent of organoid technology supplements the insufficient coverage of gene-targeted therapies, because most tumor occurrence is associated with hundreds of gene mutations.28 Patient-derived organoids replicate the genomic and transcriptomic information of the parental tumors and capture the patient-specific response to anticancer drugs, including immunotherapy drugs.14,15,29, 30, 31
To assess the potential of our organoids in evaluating heterogeneous drug responses across patients, we conducted the therapeutic profiling on an anticancer drug library composed of 29 chemotherapeutic drugs and 2 targeted drugs, rituximab and cetuximab. The library covered most of the FDA-approved first-line chemodrugs. The tumor organoids were grown from Matrigel spheres embedded with patient tumor cells for 7 days and conditioned with 10 μM single drugs for 2 days. We chose the single concentration approximating the upper level of the typical concentration range for hit discovery, i.e., 1–10 μM,32 as a primary screen33 to find effective drugs for a patient. It could be followed with further rounds of dose-response evaluation of single and combination drugs from a pre-screened library.
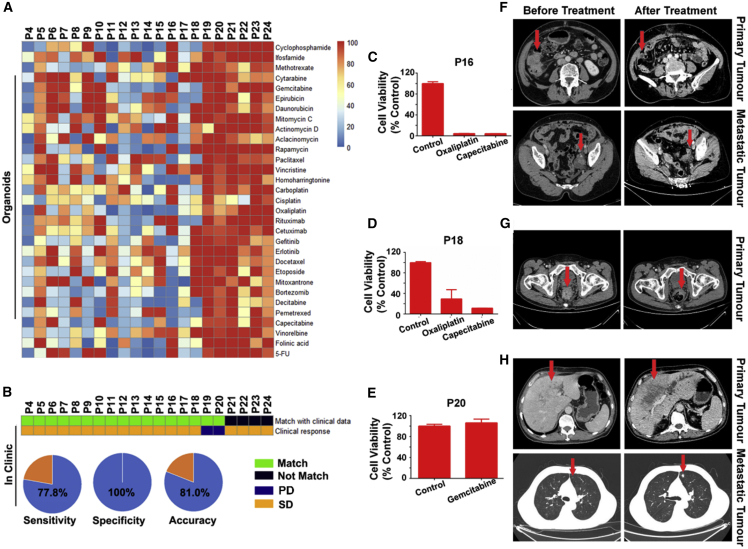
The drug efficacy was evaluated by testing the metabolic levels of at least 5 individual organoids and averaged. The inter-patient heterogeneous sensitivity to certain drugs was reflected in the heatmap of drug screening outcomes and compared with the clinical treatment of 21 patients (Figure 5A; Figure S4). 15 patients responded to a therapy (stable disease [SD]) that included the efficacious drugs as screened by their organoids. 2 patients responded poorly to a therapy not including the screened efficacious drugs. However, 4 patients had a conflict in their clinical outcome with the screening outcome on their organoids, by responding to a drug that was failed to be recommended by the organoid screening. In total, 17 out of 21 patients had matched responses in terms of the organoid screening outcome and the clinical treatment. The screening accuracy rate reached 81.0%, sensitivity 77.8%, and specificity 100% (Figure 5B). A follow-up study on the clinical treatment for prolonged terms shall be conducted to further validate the organoid screening, including evaluation of combination therapies.
Figure 5.
Tumor Organoids Capture Inter-patient Heterogeneous Responses to Anticancer Drugs
(A) Heatmap of cell viability profiled for organoids derived from 21 patients of different tumors conditioned in 31 individual anticancer compounds. The organoids were cultured under the drug-free conditions for 7 days before being dosed with single drugs at 10 μM for 2 days (n = 5).
(B) Heatmap of the organoid response and patients’ clinical outcomes.
(C–H) Drug efficacy profiles (C–E) (n = 5) and computed tomography (CT) (F–H) of patient tumors matching (C, F, and D, G) or mismatching (E, H) the screening outcomes before and after the anticancer treatment for colon (F), rectal (G), and liver (H) tumors.
See also Figures S4–S6 and Tables S1 and S2.
The screening outcome on patient organoids was also checked on three patient tumors examined using computed tomography (CT) before and after 3, 4, and 1 chemotherapy courses, respectively. Two patient tumors exhibited significant volume shrinkage after treatment by the screened highly efficacious drugs (Figures 5C, 5F, 5D, and 5G), whereas another patient failed to achieve tumor shrinkage by using drugs that were screened inefficacious (Figures 5E and 5H).
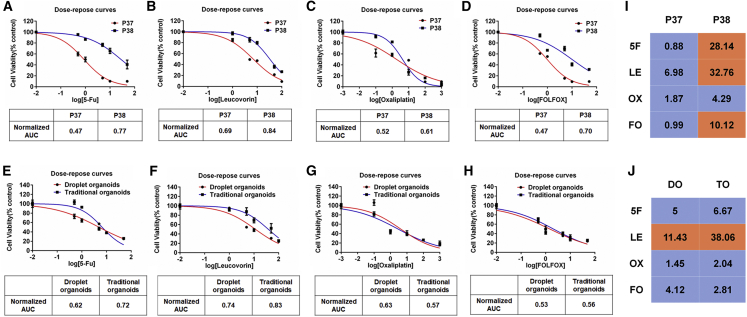
Our droplet-templated organoids were sensitive to drug doses (Figures 6A–6D), including multi-drug combinations (Figures 6E–6H), and displayed inter-patient differences (Figure 6I). We then compared the chemosensitivity evaluation of our droplet-templated organoids (D7) and the organoids (14 days in growth, D14) fabricated using the traditional method34 but derived from the same parental tumor. Both types of organoids produced the same recommendation outcome, though the suggested IC50 values were slightly different (Figure 6J). Single-dose administration at a fixed drug concentration, e.g., 10 μM as performed in Figure 5A and Figures S5 and S6A, reached the same recommendation outcome by choosing from viability rates, though it failed to reflect the whole profile of a drug treatment. The assessment outcome for extending the drug conditions to 6 days was in agreement with that from the 2-day drug condition (Figure S6B). Overall, the droplet organoid technology shortened the incubation period and meanwhile minimize the uses of Matrigel, as only the organoid volume was supplemented with the extracellular scaffold (Table S1).
Figure 6.
Dose Responses to Chemotherapy in Rectal Tumor Organoids
(A–D) Chemosensitivity of P37, P38 organoids to 5-FU (A), leucovorin (B), Oxaliplatin (C), FOLFOX (D) (5-Fu: leucovorin: Oxaliplatin = 25:5:1) in the form of dose-response curves (n = 3). AUC was calculated from the raw dose-response data.
(E–H) Dose responses to chemotherapy in droplet organoids and traditional organoids. Chemosensitivity of P33 organoids to 5-FU (E), leucovorin (F), Oxaliplatin (G), and FOLFOX (H) (5-Fu: Leucovorin: Oxaliplatin = 25:5:1) in the form of dose-response curves (n = 3). AUC was calculated from the raw dose response data.
(I and J) The heatmaps show the values of IC50 of P37 and P38 organoids (I), and responses of P33 droplet organoids (DO) and traditional organoids (TO) to 5-FU (5F), leucovorin (LE), Oxaliplatin (OX), FOLFOX (FO) (J). The colors code sensitivity (blue) and resistance (orange) at an arbitrary cutoff value of 10 μM. See also Table S2.
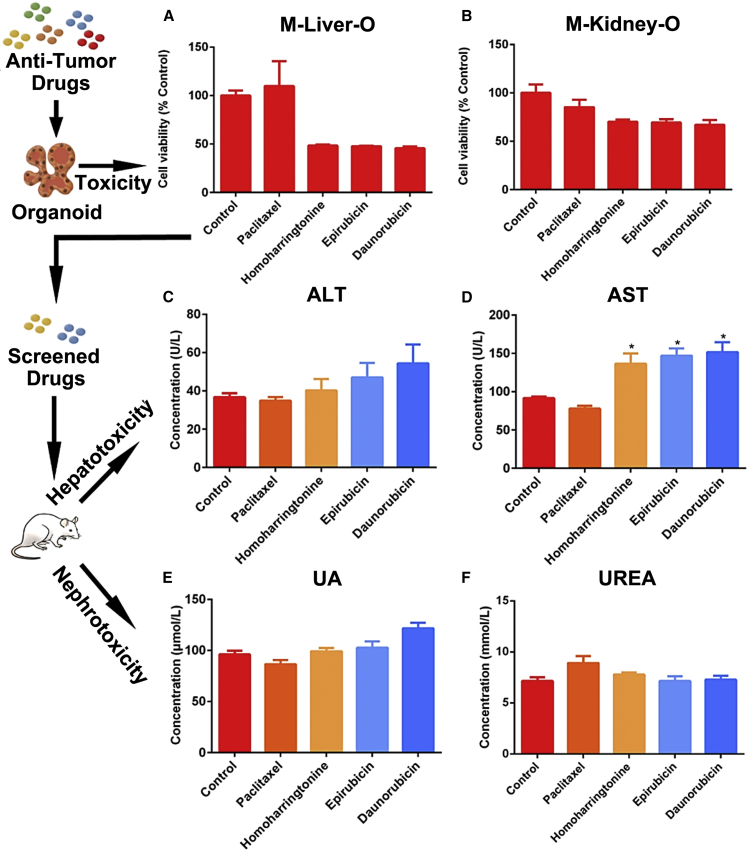
Kidney and Liver Organoid Predict Anticancer Drug Toxicity
Cancer chemotherapy is often associated with drug toxicity to healthy organs, in particular, kidney and liver that function in drug metabolism. Organoids capture the sensitivity of patient tumors to chemodrugs, but their ability to predict the drug toxicity to healthy organs has been less explored.
We examined the toxicity on liver and kidney organoids, by dosing the culturing medium with 4 chemotherapeutic single drugs at 10 μM for 2 days to organoids that had been cultured for 7 days (Figures 7A and 7B; Figure S7). Paclitaxel exhibited non-toxicity to both liver and kidney organoids, whereas Homoharringtonine, Epirubicin, and Daunorubicin exhibited significant liver toxicity. To validate the toxicity screening outcome, we intravenously injected single drugs to BALB/c mice (n = 6 for each drug). To probe the organ-specific function alteration, we performed the alanine aminotransferase (ALT) and aspartate aminotransferase(AST) tests for the liver, and the urinalysis (UA) and urea test for the kidney. It revealed significant increases in the ALT and AST concentrations in mice exposed to Homoharringtonine, Epirubicin, and Daunorubicin for 2 days, compared to the control mice that had no drug exposure (Figures 7C and 7D). No significant increase was observed in the ALT and AST levels in mice exposed to Paclitaxel, or the UA and urea levels in mice exposed to either drug (Figures 7E and 7F).
Figure 7.
Toxicity Evaluation of Anticancer Drugs on Liver and Kidney Organoids
(A and B) The toxicity of 4 anticancer drugs screened on organoids derived from mouse (A) livers and (B) kidneys (n = 4).
(C–F) Validation of the screening outcomes, by testing Paclitaxel, Homoharringtonine, Epirubicin, and Daunorubicin on mice (n = 6).
See also Figure S7.
The matched testing outcome on organoids and mice supported the applications of liver organoids on screening drug toxicity, and the all-in-one screening on drug efficacy, specificity, and toxicity on a particular patient by using co-cultured cancerous and healthy organoids.
Discussion
Unlike deriving organoids by embedding a single or a cluster of cells in Matrigel to grow into tissue structures of variable sizes and cell compositions, in our platform, the organoids are templated from monodisperse Matrigel droplets encapsulated with a certain number of primary tissue cells, e.g., 1,506 ± 13 cells (n = 3). This count of cells is statistically representative of the cell classification in a parental tumor. The derived organoids recapitulate the inter-patient heterogeneity.
It also takes advantage of the droplet-templating that provides uniform and controlled the shaping of ultra-soft scaffolds, such as Matrigel. The microfluidics is re-innovated to be compatible with Matrigel manipulation. The shaped Matrigel provides mechanical support for encapsulated cells and allows them to resemble the primary tissue constructs. The volume of the templating droplet determines the organoid size. The cellular composition remains statistically consistent across individual Matrigel droplets. Consequently, the derived organoids possess the inter-organoid homogeneity.
The unique platform synchronizing Matrigel sphere formation and printing endows uniformity, high-throughputs, and automation in organoid applications. It reduces manual manipulation induced variability, reduces labor costs and organoid application barrier, and, therefore, offers scalability and expansion opportunities for organoid technologies.
Notably, the RNA-seq profiling of organoids and their parental tissues and tumors displays high similarity in the genes relevant to organ functions, cancer, and chemotherapy. We have validated our organoids in the recapitulation of tumor heterogeneity, histology, gene-expression and mutation profiles, and inter-patient heterogeneous responses to anticancer drugs. Minor gene-expression alterations were observed, which might be attributed to the brutal cell extraction from parental tissues by microdissection and enzymatic degradation. Growing organoids from microdissects can avoid the detrimental factors exerted on cells by enzymatic degradation, though maintaining the consistency of cell counts across organoids might be compromised.29
In this study, we employ the serum-conditioned culturing35 to accelerate organoid growth. Together with the Matrigel droplet templating effect, it fulfills the goal of accomplishing drug screening on organoids within 1 week and at a reduced cost. However, the recapitulation of some genetic information might be compromised in the presence of serum. Medium customization by supplementing serum and selective growth factors for specific tumor organoids might be demanded to simultaneously improve the morphogenesis, accelerate organoid growth, and reduce the cost for expansion.
The Matrigel droplet templating strategy also offers a solution toward increasing organoid complexity, e.g., incorporating stromal cells in tumoroids.20,36 It is suggested by our previous unpublished study that, in Matrigel droplets smaller than 1 mm across, the fibroblasts mixed homogeneously with cancer cells quickly migrates to the periphery, to form a stroma “shell” enclosing the “tumor.”
“Organoids on a chip” have been reported, but they failed to replicate the patient tumor heterogeneity and eliminate the organoid-to-organoid variability.5 The marriage between our organoid platform with “organoids on a chip” offers the solution toward high-throughput and low-cost organoid formation, automated organoid culturing, and reduced variability and inconsistent manual inference during repetitive laboratory procedures.4,21 Additionally, parallel testing of drugs on healthy and tumor organoids provides parallel information on both the killing efficiency for cancer cells and toxicity for healthy cells.37
This study has shown the creation of multi-organ, cross-species, healthy, and cancerous organoids in high-throughput, high uniformity, and reduced manual manipulation in our platform. Organoids are also therapeutic reagents for regenerative medicine, as the feasibility of transplanting organoids to stimulate neo-tissue generation and repair damaged organs has also been reported.38 Our high-throughput and automated organoid platform is expected to play increasing roles in personalized cancer medicine, and theranostics innovation for new drug development, and regenerative medicine.
Limitations of Study
The current success rate of printing organoid beads into 96-well plates reached above 95%, i.e., up to 4 missing placements in a 96-well plate. It was mainly attributed to the lack of sufficiently low-temperature control on Matrigel loaded in the infusion syringe, as the cooling setup was a fridge and not customized for this task. For high-throughput production, the M2 module was lack of a robotic arm to transfer and position plates, which had been carried out manually.
The size of human tumor samples is not enough to evaluate the concentration-dependent and/or combination therapies. For example, for concentration-depend drug screening of 31 drugs with 6 concentrations and 6 repeats, it needs 1,116 organoids (or organoid contained wells). In the current study, we were able to produce 300–500 organoids from a single surgery tumor sample. For biopsy samples, the count was even smaller. The cell-extraction techniques from patient samples require improvement.
Further, the current platform achieved 81% accuracy in evaluating personalized chemotherapy, but to be further validated for clinical uses, the evaluation accuracy must to further improve. It might be achieved by optimizing the primary cell-extraction and organoid culturing procedures and enriching the acquired information volumes.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-E-cadherin | Abcam | Cat# ab76055; RRID:AB_1310159 |
| Rabbit monoclonal anti-EpCAM | Abcam | Cat# ab71916; RRID:AB_1603782 |
| Rabbit monoclonal anti-Vimentin | Abcam | Cat# ab92547; RRID:AB_10562134 |
| Rabbit monoclonal anti-CD3 | Abcam | Cat# ab135372; RRID:AB_2884903 |
| Anti-Mouse IgG H&L (Alexa Fluor® 594) | Abcam | Cat# ab150116; RRID:AB_2650601 |
| Anti-Rabbit IgG H&L (Alexa Fluor® 594) | Abcam | Cat# ab150080; RRID:AB_2650602 |
| Biological Samples | ||
| Human tumor tissue samples | This study | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM/F12 | Thermo Fisher Scientific | Cat# 11320033 |
| Fetal bovine serum | invigentech | Cat# A6903FBS |
| GlutaMAX Supplement | Thermo Fisher Scientific | Cat# 35050061 |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 |
| B-27 Supplement | Thermo Fisher Scientific | Cat# 17504044 |
| red blood cell lysis buffer | Roche | Cat# 11814389001 |
| Nicotinamide | Sigma-Aldrich | Cat# N0636 |
| N-Acetylcysteine | Sigma-Aldrich | Cat# A9165 |
| Noggin (Human) | MCE | Cat# HY-P7051A |
| Noggin (Mouse) | MCE | Cat# HY-P7086 |
| R-spondin 1 | MCE | Cat# HY-P7114 |
| SB431542 | MCE | Cat# HY-10431 |
| CHIR99021 | MCE | Cat# HY-10182 |
| FGF4 | MCE | Cat# HY-P7014 |
| FGF-basic(Human) | MCE | Cat# HY-P7004 |
| FGF-basic(Mouse) | MCE | Cat# HY-P7066 |
| FibrOut | Chi scientific | Cat# 4-21547, 4-21565, 4-21502, 4-20520 |
| Y-27632 | Abmole | Cat# M1817 |
| EGF | Peprotech | Cat# AF-100-15 |
| AQIX ® | AQIX LTD | NA |
| cyclophosphamide | MCE | Cat# HY-L025 |
| ifosfamide | MCE | Cat# HY-L025 |
| methotrexate | MCE | Cat# HY-L025 |
| cytarabine | MCE | Cat# HY-L025 |
| gemcitabine | MCE | Cat# HY-L025 |
| epirubicin | MCE | Cat# HY-L025 |
| daunorubicin | MCE | Cat# HY-L025 |
| mitomycin C | MCE | Cat# HY-L025 |
| actinomycin D | MCE | Cat# HY-L025 |
| aclacinomycin | MCE | Cat# HY-L025 |
| rapamycin | MCE | Cat# HY-L025 |
| paclitaxel | MCE | Cat# HY-L025 |
| vincristine | MCE | Cat# HY-L025 |
| homoharringtonine | MCE | Cat# HY-L025 |
| carboplatin | MCE | Cat# HY-L025 |
| cisplatin | MCE | Cat# HY-L025 |
| oxaliplatin | MCE | Cat# HY-L025 |
| rituximab | MCE | Cat# HY-L025 |
| cetuximab | MCE | Cat# HY-L025 |
| erlotinib | MCE | Cat# HY-L025 |
| docetaxel | MCE | Cat# HY-L025 |
| etoposide | MCE | Cat# HY-L025 |
| mitoxantrone | MCE | Cat# HY-L025 |
| bortezomib | MCE | Cat# HY-L025 |
| decitabine | MCE | Cat# HY-L025 |
| vinorelbine | MCE | Cat# HY-L025 |
| folinic acid | MCE | Cat# HY-L025 |
| 5-FU | MCE | Cat# HY-L025 |
| gefitinib | MCE | Cat# HY-L025 |
| pemetrexed | MCE | Cat# HY-L025 |
| capecitabine | MCE | Cat# HY-L025 |
| paraformaldehyde | Beyotime | Cat# P0099 |
| bovine serum Albumin | Sigma-Aldrich | Cat# V900933 |
| sucrose | Sigma-Aldrich | Cat# S7903 |
| urea | Sigma-Aldrich | Cat# U5378 |
| D-sorbitol | Sigma-Aldrich | Cat# S1876 |
| glycerol | Sigma-Aldrich | Cat# G5516 |
| DMSO | Sigma-Aldrich | Cat# D8418 |
| Matrigel | Corning | Cat# 356237 |
| Collagenase | Sigma | Cat# C9407 |
| Critical Commercial Assays | ||
| CellTiter-Glo 3D cell viability assay | Promega | Cat# G9683 |
| Deposited Data | ||
| RNA-seq | This paper | GSE161928 |
| Whole-exome sequencing (WES) | This paper | PRJNA679439 |
| Experimental Models: Organisms/Strains | ||
| Mouse: BALB/c | Guangdong Medical Laboratory Animal Center | NA |
| Software and Algorithms | ||
| Prism Graphpad 6 | GraphPad Software | NA |
| Other | ||
| HFE-7000 | 3M | Cat# 200418AT |
| 1 ml / 10 ml injection syringe | Yuekang | NA |
| SYLGARD 184 Silicone Elastomer Kit | Dow | Cat# 01673921 |
| 24T PTFE tubing, ID = 0.56 mm | Woer | NA |
| Heater | homemade | NA |
| Refrigerator | AUS | Cat# JC-95 |
| 3D printer | Homemade | NA |
| Needle, Blunt end, gauge size 22 | Local supplier | NA |
| Injection Pump | LEAD FLUID | Cat# TYD01 |
| Biopsy punch | Miltex | Cat# 33-31AA |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shaohua Ma (ma.shaohua@sz.tsinghua.edu.cn).
Materials Availability
This study did not generate new unique reagents
Data and Code Availability
The RNA-seq dataset generated during this study is available atGEO: GSE161928. The accession number for the Whole-exome sequencing (WES) data reported in this paper is available at NCBI Trace and Short-Read Archive (SRA): PRJNA679439.
Experimental Model and Subject Details
The collection of patient data and tissue for tumor organoids fabrication has been performed according to the guidelines of The First Affiliated Hospital of Zhengzhou University Ethics Committees following China, following both the national and the local laws. Clinical information is available in Table S2. All animal experiments were approved by the Animal Ethical Committee of Tsinghua University and performed in accordance with the Guidelines for the Care and Use of Laboratory Animals. Adult male BALB/c mice were purchased from Guangdong Medical Laboratory Animal Center and maintained in the specific pathogen-free Tsinghua University Laboratory Animal Center.
Method Details
iPSCs culture
Undifferentiated human iPSCs were maintained in the feeder-free conditions in iPSCeasy human pluripotent stem cells (hESCs / hiPSCs) medium (Cellapy Biological, Beijing, China) on Matrigel & VTN-N coated plates (BD Biosciences). iPSCs were passaged at 60%–80% confluence using 0.5 mM EDTA solution for 5 min and reseeding 1:6 onto pre-coated plates. S0 refers to the undifferentiated iPSC cells cultured according to the standard culturing protocols, i.e., on top of the Matrigel & Vitronectin (VTN-N) coating in a Petri dish. S1 and S4 refer to iPSCs cultured for 1 day (S1) and 4 days (S4)
Human tumor processing
In this study, we employed cancer tissue specimens obtained from patients who had been diagnosed preoperatively as having different cancers and underwent surgical resections, but before the chemotherapy treatment at the First Affiliated Hospital of Zhengzhou University (Henan, China). Details of the patients contributing the tissue specimens are provided in Table S2.
The remnant tissue samples (approximately 10 mm × 10 mm × 5 mm) from surgical resections after clinicopathological diagnosis were utilized for research. The specimens were immersed in an organic preservation solution (AQIX, UK) and transported from the hospital to the laboratory at 4°C. The specimens were used to extract cells for fabricating tumor organoids and extract RNA for transcriptome analysis.
Tumor tissues were washed three times by cold PBS (1 × ) solution with 2.0% penicillin-streptomycin (vol/vol, GIBCO) and then cut into small pieces. The tissues were then digested by 1.0 mg/mL collagenase type I (Sigma-Aldrich) and 2.0% fetal bovine serum (FBS) (vol/vol, Invigentech) on an orbital shaker at 37°C and incubated for 1 - 2 h. After digestion, the tissues were sheared by 10 mL plastic pipettes. The suspension was passed through a 100 μm filter (Falcon), and centrifuged at 1000 rpm for 5 min. The pellet was then resuspended in 5 mL red blood cell lysis buffer (Roche) for 5 min at room temperature. Afterward, the suspension was re-centrifuged at 1000 rpm for 5 min, and the pellet was resuspended in DMEM/F12 medium. The number of extracted cells was dependent on the size of each tumor specimen. For a fingernail-sized tissue, about 4.0 × 106 cells were extracted. Cells were calculated by using a hemocytometer.
Mouse tissue processing
BALB/c mice (male, 6 - 8-weeks old) were euthanized by excessive CO2. Then, the organs (lung, liver, kidney, pancreas, spleen, heart) were isolated and cut into small pieces (1 - 3 mm3 for each piece). After being washed with cold PBS (1 × ) for three times and supplemented with 2.0% penicillin-streptomycin (vol/vol, GIBCO), the tissues were digested by 1.0 mg/mL collagenase type I (Sigma-Aldrich) and 2.0% fetal bovine serum (FBS) (vol/vol, Invigentech) on an orbital shaker at 37°C and incubated for 1 - 2 h. After digestion, the tissues were sheared by 10 mL plastic pipettes. The suspension was passed through a 100 μm filter (Falcon), and centrifuged at 1000 rpm for 5 min. The pellet was resuspended in 5 mL red blood cell lysis buffer (Roche) for 5 min at room temperature. Afterward, the suspension was recentrifuged at 1000 rpm for 5 min, and the pellet was resuspended in DMEM/F12 medium. The cell number was counted for organoid fabrication.
Organoid platform structure
We used SYLGARD 184 Silicone Elastomer Kit (DOW) to fabricate the 3-way polydimethylsiloxane (PDMS) connector. Following the operation manual, we mixed two liquids at the ratio of 10 to 1 in beaker and stirred with a magnetic stirrer for 5min. The mixed liquid was transferred into a 6-cm dish. Afterward, the mixture was degassed under vacuum for 20 min and crosslinked in a drying oven at 65°C for 24 h. Then, the solidified PDMS was cut into small pieces (length: 10 mm, width: 5 mm, height: 10 mm). The T-junction channel was punched by using a 1.0-mm biopsy punch.
Structure of the microfluidics M1 module: a 1 mL plastic injection syringe (Yuekang) was connected to a piece of 24T PTFE tubing (Woer, ID = 0.56 mm), with a blunt end needle (Yuekang) (gauge size 22), and then installed in an infusion syringe pump (LEAD FLUID). Both the Matrigel and oil phases were connected in the same way as described. The other ends of the tubing were connected to the 3-way PDMS connector. The third channel of the connector was connected to a third tubing (∼10 m long), where cell-laden Matrigel droplets were formulated, incubator and transported to the end of this tubing, which was anchored to the 3D droplet printing module, i.e., the M2 module. The pumps and syringes were placed in a refrigerator set at 4°C.
Structure of the M2 module: the tri-axis translational stage was homemade, analogous to a regular 3D printing frame. One end of 10-m 24T PTFE tubing was connected to the 3-way PDMS connector in M1 and the other one was mounted to the printing head in M2. The middle part of 10-m 24T PTFE tubing (approximately 9.5 m long) was warmed by a small volume water bath kettle set at 37°C.
Organoids precursor fabrication
The cells extracted from mouse tissues or human tumors were suspended in growth factor reduced Matrigel (Corning) at the density of 2.0 × 107 cells per mL. Then the Matrigel phase was loaded in a 1 mL syringe and installed in an injection pump. A fluorocarbon oil (HFE-7000, 3M Novec) was loaded in a 10 mL injection syringe and installed in another injection pump. Both pumps were placed in a 4°C refrigerator. The two phases were co-injected through polytetrafluoroethylene (PTFE) tubing (the inner diameter ID = 600 μm, Woer) to a third piece of PTFE tubing via a 3-way hand-made PDMS connector. The Matrigel phase was injected at the flow rate of 20 μL/min and subsequently sheared into monodisperse droplets by the fluorocarbon oil at the flow rate of 30 μL/min. The tubing conducting the Matrigel droplets, about 10 m long, was heated to 37°C by a small water bath kettle. The Matrigel droplets were circulated in the warmed tubing for 10 min before reaching the outlet, which was connected to the droplet printer head. The Matrigel beads, gelled from droplets, were printed into 96-well plate or 10 cm culture dish individually. The printing sequence of the beads matched with their sequence in the tubing. After being printed, the beads were further incubated at 37°C for 20 min to complete solidification. (Figure 1A) 200 μL culturing medium (for human sample: 20% fetal bovine serum, 1% penicillin-streptomycin, 100 ng/mL noggin (MCE), 100 ng/mL R-spondin 1 (MCE), 5 ng/ml EGF (Peprotech), 10 ng/mL FGF-basic (MCE), 1X GlutaMAX Supplement (Thermo Fisher Scientific), 10mM HEPES (Thermo Fisher Scientific), 1X B-27 Supplement (Thermo Fisher Scientific), 5mM Nicotinamide (Sigma-Aldrich), 1.25 mM N-Acetylcysteine (Sigma-Aldrich), 5 μM Y-27632 (Abmole), 1X FibrOut (Chi scientific). For mouse sample: 20% fetal bovine serum, 1% penicillin-streptomycin, 100 ng/mL noggin (MCE), 100 ng/mL R-spondin 1 (MCE), 5 ng/ml EGF (Peprotech), 10 μM SB431542 (MCE), 2 μM CHIR99021 (MCE), 200 ng/mL FGF4 (MCE), 10 ng/mL FGF-basic (MCE), 5 μM Y-27632 (Abmole), 1X FibrOut (Chi scientific)) was added to each well containing one cell-laden Matrigel bead, i.e., organoid precursor, and cultured at 37°C in an incubator, supplied with 5% CO2. Medium was changed every 3 days. Images of organoids were acquired at day 1 and day 7. Afterward, the organoids were harvested for further analysis or conditioned with drugs.
Traditional organoid culture
We followed Nicola Valeri’s protocol (38) to fabricate traditional orgnaoids. After human tumor processing, 30 μl of matrigel containing 5000 cells were seeded in standard 96-well cell culture plates, and solidified in a 37°C and 5% CO2 cell culture incubator for 20 min. Then, 100 μL culturing medium were added into individual culturing wells. Medium was changed for every 3 days. After 2 weeks culture, the orgaoids were used to perform the drug screening assay.
Histology
Tissue and organoids were fixed in 4.0% paraformaldehyde (PFA) (Beyotime) and quickly frozen in liquid nitrogen. Afterward, they were cut into 10 μm slices by a Thermo Scientific NX50 freezing microtome. The sections were performed following the standard HE staining protocol. Images were acquired on a Leica DM1000 inverted microscope.
Immunofluorescence imaging with tissue clearance
After being cultured for 7 days, organoids were fixed with 4.0% PFA and washed for three times using PBS (1 × ). Then, the organoids were permeabilized with 0. 5% Triton X-100 for 5 min at room temperature. After incubation in 5% bovine serum albumin (Sigma-Aldrich) for 1 h, the organoids were incubated with antibodies against E-cadherin, EpCAM or Vimentin (1:200, Abcam) at 4°C for 48 h. After washing with PBS (1 × ), the organoids were further incubated with Alexa Fluor 594 conjugated secondary antibody (1:500, Abcam). Finally, the nuclei were stained with DAPI (1:1000, Beyotime). Then, the organoids were dehydrated in 30% sucrose solution (wt/vol, sigma) at 4°C for 24h, and cleared by using the ultrafast optical clearing regents, composed of 20% urea (wt/vol, sigma), 30% D-sorbitol (wt/vol, sigma), and 5% glycerol (wt/vol, sigma) dissolved in DMSO (sigma), for 5min at room temperature before imaging. The images were captured by a Nikon A1R+ Laser scanning confocal microscopy.
RNA-seq analysis
Standard RNA-seq analyses were performed by Sangon Biotech (Shanghai) Co., Ltd. China (for hiPSC-derived organoids) and GENEWIZ (Suzhou) Co., Ltd. China (for patient tissues and patient-derived organoids). The hiPSCs, tissues and organoids after 7 days in culture were send to GENEWIZ (Suzhou, China) or Sangon Biotech (Shanghai, China) to perform the standard RNA-seq analysis. Total RNA of each sample was extracted using the TRIzol Reagent (Invitrogen) /RNeasy Mini Kit (QIAGEN). 1 μg total RNA with RIN value above 6.5 was used for library preparation. Next generation sequencing library preparations were constructed according to the manufacturer’s protocol. Then libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to manufacturer’s instructions. Sequencing was carried out using a 2 × 150bp paired-end (PE) configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. Differential expression analysis was performed using the DESeq2 Bioconductor package, a model based on the negative binomial distribution. The estimates of dispersion and logarithmic fold changes incorporate data-driven prior distributions. Padj of genes were set < 0.05 to detect differential expressed ones. KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances (https://en.wikipedia.org/wiki/KEGG). We used scripts in house to enrich significant differential expression gene in KEGG pathways.
Whole-exome sequencing (WES) analysis
Whole exome sequencing was performed by Sangon Biotech (Shanghai) Co., Ltd. China (for hiPSC-derived organoids) and GENEWIZ (Suzhou) Co., Ltd. China (for patient tumor tissues and patient-derived organoids). The tumor tissues and the organoids cultured for 7 days were sent to GENEWIZ (Suzhou, China) to perform the whole exome sequencing by Illumina HiSeq. Primary analysis was performed by built-in software, HiSeq Control Software (HCS), RTA 2.3 plus, and demultiplexing was performed by bcl2fastq 2.17. The raw data of exome sequencing were analyzed by bioinformatics analysts of GENEWIZ Inc. The GATK haplotypecaller or samtools was used to call SNV and the variants were annotated by Annovar (Version11Feb2016).
Drug screening
Human tumor organoids or mouse normal tissue organoids were printed on 96-well plates (one organoid per well), followed by exposure to 31 anti-tumor drugs (cyclophosphamide, ifosfamide, methotrexate, cytarabine, gemcitabine, epirubicin, daunorubicin, mitomycin C, actinomycin D, aclacinomycin, rapamycin, paclitaxel, vincristine, homoharringtonine, carboplatin, cisplatin, oxaliplatin, rituximab, cetuximab, gefitinib, erlotinib, docetaxel, etoposide, mitoxantrone, bortezomib, decitabine, pemetrexed, capecitabine, vinorelbine, folinic acid, 5-FU) (MCE) at the fixed concentration of 10 μM, with DMSO exposure as the control, for 48 h or 6 days. For FOLFOX treatments, tumor organoids were cultured with FOLFOX (the molar ratio of the FOLFOX chemotherapy cocktail used was as follows: 5-FU: leucovorin: oxaliplatin = 25: 5: 1) for 48 h. The final 5-FU concentrations were 50, 10, 5, 1, 0.5 and 0 μM. A CellTiter-Glo 3D cell viability assay (Promega)was employed to detect cell viability according to the manufacturer’s instruction. The drugs achieving over 50% cell viability reduction compared with control were regarded as efficacious.
Outcome evaluation for patients
Response Evaluation Criteria in Solid Tumors (RECIST) was the guideline of clinical response to anti-tumors drugs therapy. After Chemotherapy, RECIST was applied by experienced oncologists and pathologists at The First Affiliated Hospital of Zhengzhou University, for evaluating clinical response of patients to anti-tumors drugs, and compared with their conditions before the therapy.
Acute hepatorenal toxicity evaluation in vivo
Adult male BALB/c mice (6 - 8 weeks old) were maintained in the specific pathogen-free Tsinghua University Laboratory Animal Center. The mice (n = 6) were injected with 10 mg/kg paclitaxel, homoharringtonine, epirubicin and daunorubicin for two days. Normal saline (0.9% NaCl) was employed as a control (n = 7). After exposure, all mice were sacrificed through excessive CO2. The livers and kidneys were sectioned fixed at 4.0% PFA. Serum were also collected for hepatic and renal functional analysis. Serum parameters were analyzed with a Mindray BS-240VET system. All animal experiments were approved by the Animal Ethical Committee of Tsinghua University and performed in accordance with the Guidelines for the Care and Use of Laboratory Animals.
Quantification and Statistical Analysis
Unless otherwise indicated, all the data are presented as the mean ± SEM. The statistical comparisons between the different treatments were conducted with two-tailed Student’s t test by Prism Graphpad. The P value less than 0.05 or 0.01 was considered as significant or highly significant difference.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (grant numbers 61971255 and 81872368), Shenzhen Science and Technology Innovation Committee (grant numbers KQJSCX20180327143623167, JCYJ20180508152130899, and GJHZ20180420180754917), Shenzhen Development and Reform Commission Subject Construction Project [2017]1434, and State Key Lab Construction Project (GuoKeFaJi [2018]38).
Author Contributions
S.M. and L.H. conceived this project. S.J., H.Z., and Y.L. performed most of the experiments, including cell extraction, tissue processing, organoid culturing, drug screening, and animal studies. J.W. performed the experiments on iPSCs. W.Z., S.W., Y.Z., and W.W. provided the human samples. W.Z. provided the patient with clinical data and advice. H.Z. processed the images. S.M., L.H., and P.E.L. oversaw the experimental design and progress. S.M., S.J., and H.Z. wrote the manuscript.
Declaration of Interests
S.M., L.H., S.J., and H.Z. are listed as inventors on provisional patent applications based on components of this work.
Published: December 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100161.
Contributor Information
Laiqiang Huang, Email: huanglq@tsinghua.edu.cn.
Shaohua Ma, Email: ma.shaohua@sz.tsinghua.edu.cn.
Supplemental Information
References
- 1.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 2.Keller L., Pantel K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer. 2019;19:553–567. doi: 10.1038/s41568-019-0180-2. [DOI] [PubMed] [Google Scholar]
- 3.Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y., Jin K., Seung L.J., Min S., Suran K., Da-Hee A., Yun-Gon K., Seung-Woo C. Drug Screening: Vascularized Liver Organoids Generated Using Induced Hepatic Tissue and Dynamic Liver-Specific Microenvironment as a Drug Testing Platform. Adv. Funct. Mater. 2018;28:1801954. [Google Scholar]
- 6.Yin S., Xi R., Wu A., Wang S., Xi J.J. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci. Transl. Med. 2020;12:eaaz1723. doi: 10.1126/scitranslmed.aaz1723. [DOI] [PubMed] [Google Scholar]
- 7.Aparicio S., Hidalgo M., Kung A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo M., Amant F., Biankin A.V., Budinská E., Byrne A.T., Caldas C., Clarke R.B., de Jong S., Jonkers J., Mælandsmo G.M. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H., Gehart H., Artegiani B., Löpez-Iglesias C., Dekkers F., Basak O., van Es J., Chuva de Sousa, Lopes S.M., Begthel H., Korving J. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell. 2018;175:1591–1606. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Schutgens F., Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. 2020;15:211–234. doi: 10.1146/annurev-pathmechdis-012419-032611. [DOI] [PubMed] [Google Scholar]
- 11.Schutgens F., Rookmaaker M.B., Margaritis T., Rios A., Ammerlaan C., Jansen J., Gijzen L., Vormann M., Vonk A., Viveen M. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 2019;37:303–313. doi: 10.1038/s41587-019-0048-8. [DOI] [PubMed] [Google Scholar]
- 12.Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 13.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 14.Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.-H., Salahudeen A.A., Smith A.R. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins R.W., Aref A.R., Lizotte P.H., Ivanova E., Stinson S., Zhou C.W., Bowden M., Deng J., Liu H., Miao D. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phipson B., Er P.X., Combes A.N., Forbes T.A., Howden S.E., Zappia L., Yen H.-J., Lawlor K.T., Hale L.J., Sun J. Evaluation of variability in human kidney organoids. Nat. Methods. 2019;16:79–87. doi: 10.1038/s41592-018-0253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbluth J.M., Schackmann R.C.J., Gray G.K., Selfors L.M., Li C.M., Boedicker M., Kuiken H.J., Richardson A., Brock J., Garber J. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 2020;11:1711. doi: 10.1038/s41467-020-15548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H., Chen Y., Shao L., Xie M., Nie J., Qiu J., Zhao P., Ramezani H., Fu J., Ouyang H., He Y. Airflow-Assisted 3D Bioprinting of Human Heterogeneous Microspheroidal Organoids with Microfluidic Nozzle. Small. 2018;14:e1802630. doi: 10.1002/smll.201802630. [DOI] [PubMed] [Google Scholar]
- 20.Takebe T., Wells J.M. Organoids by design. Science. 2019;364:956–959. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S.E., Georgescu A., Huh D. Organoids-on-a-chip. Science. 2019;364:960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 23.Cole R.H., Tang S.Y., Siltanen C.A., Shahi P., Zhang J.Q., Poust S., Gartner Z.J., Abate A.R. Printed droplet microfluidics for on demand dispensing of picoliter droplets and cells. Proc. Natl. Acad. Sci. USA. 2017;114:8728–8733. doi: 10.1073/pnas.1704020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma S., Sherwood J.M., Huck W.T.S., Balabani S. On the flow topology inside droplets moving in rectangular microchannels. Lab Chip. 2014;14:3611–3620. doi: 10.1039/c4lc00671b. [DOI] [PubMed] [Google Scholar]
- 25.Ma S., Sherwood J.M., Huck W.T., Balabani S. The microenvironment of double emulsions in rectangular microchannels. Lab Chip. 2015;15:2327–2334. doi: 10.1039/c5lc00346f. [DOI] [PubMed] [Google Scholar]
- 26.Ma S., Mukherjee N., Mikhailova E., Bayley H. Gel Microrods for 3D Tissue Printing. Adv. Biosyst. 2017;1:e1700075. doi: 10.1002/adbi.201700075. [DOI] [PubMed] [Google Scholar]
- 27.Kanie K., Sakai T., Imai Y., Yoshida K., Sugimoto A., Makino H., Kubo H., Kato R. Effect of mechanical vibration stress in cell culture on human induced pluripotent stem cells. Regen. Ther. 2019;12:27–35. doi: 10.1016/j.reth.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S. A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell. 2020;180:188–204. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopper O., de Witte C.J., Lõhmussaar K., Valle-Inclan J.E., Hami N., Kester L., Balgobind A.V., Korving J., Proost N., Begthel H. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019;25:838–849. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 31.Yao Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torrance C.J., Agrawal V., Vogelstein B., Kinzler K.W. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat. Biotechnol. 2001;19:940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 34.Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernández-Mateos J., Khan K., Lampis A., Eason K., Huntingford I., Burke R. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X., Nadauld L., Ootani A., Corney D.C., Pai R.K., Gevaert O., Cantrell M.A., Rack P.G., Neal J.T., Chan C.W. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014;20:769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shyer A.E., Tallinen T., Nerurkar N.L., Wei Z., Gil E.S., Kaplan D.L., Tabin C.J., Mahadevan L. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., Massa S., Riahi R., Chae S., Hu N. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampaziotis F., Justin A.W., Tysoe O.C., Sawiak S., Godfrey E.M., Upponi S.S., Gieseck R.L., 3rd, de Brito M.C., Berntsen N.L., Gómez-Vázquez M.J. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The play was accelerated by 6 times.
The play was accelerated by 6 times.
Data Availability Statement
The RNA-seq dataset generated during this study is available atGEO: GSE161928. The accession number for the Whole-exome sequencing (WES) data reported in this paper is available at NCBI Trace and Short-Read Archive (SRA): PRJNA679439.