Abstract
Pseudomonas aeruginosa biofilm infections in the cystic fibrosis (CF) lung are highly resistant to current antimicrobial treatments and are associated with increased mortality rates. The existing models for such infections are not able to reliably mimic the clinical biofilms observed. We aimed to further optimise an ex vivo pig lung (EVPL) model for P. aeruginosa CF lung infection that can be used to increase understanding of chronic CF biofilm infection. The EVPL model will facilitate discovery of novel infection prevention methods and treatments, and enhanced exploration of biofilm architecture. We investigated purine metabolism and biofilm formation in the model using transposon insertion mutants in P. aeruginosa PA14 for key genes: purD, gacA and pelA. Our results demonstrate that EVPL recapitulates a key aspect of in vivo P. aeruginosa infection metabolism, and that the pathogen forms a biofilm with a clinically realistic structure not seen in other in vitro studies. Two pathways known to be required for in vivo biofilm infection - the Gac regulatory pathway and production of the Pel exopolysaccharide - are essential to the formation of this mature, structured biofilm on EVPL tissue. We propose the high-throughput EVPL model as a validated biofilm platform to bridge the gap between in vitro work and CF lung infection.
Keywords: 3R’s, Biofilm, Chronic infection, Cystic fibrosis, ex vivo model, Pseudomonas aeruginosa
Introduction
Biofilms are microbial aggregates that consist of bacterial cells and a self-produced matrix primarily composed of proteins, lipids, polysaccharides and nucleic acids [1]. The biofilm matrix provides the microbial population with increased antibiotic tolerance and protection against the host immune response [2]. The persistent, biofilm-associated, lung infections that affect people with the genetic condition Cystic Fibrosis (CF) are highly resistant to antibiotic treatment and, despite being well studied, continue to have a large impact on health and life expectancy. More than 70,000 people worldwide are believed to have CF, though it is thought to be widely underdiagnosed across Asia as epidemiological reports are largely derived from data collected in western countries [[3], [4], [5]]. Despite the rising average life expectancy of people with CF [6], it still has one of the highest mortality rates among human genetic conditions [5].
CF lung infections are known to be polymicrobial [7]. However, Pseudomonas aeruginosa is the most abundant CF pathogen in the UK. It is estimated that 75% of adults with CF are either chronically or transiently infected with P. aeruginosa during their lifetime [8,9]. By 5 years old approximately 53% of children with CF are infected with P. aeruginosa [10,11]. Detection of P. aeruginosa infection is associated with worsened disease outcomes including rapid lung function decline and higher mortality rates [[12], [13], [14]].
P. aeruginosa is able to switch from the initial acute stage of infection, where virulence is high, infection is transient and biofilms are not formed [15], to chronic biofilm infection. This chronic infection is characterised by embedding of P. aeruginosa biofilm aggregates in the mucus plugs typically found in the airways of people with CF [16]. Chronic infection is traditionally difficult to replicate in vitro and so the ability to study P. aeruginosa virulence, persistence and treatment response at this stage of infection is limited. Once established, chronic P. aeruginosa infection is almost impossible to eradicate, and there is a poor correlation between antibiotic susceptibility testing and the clinical outcomes of antibiotic treatment [17,18]. This is largely a result of altered resistance phenotypes in in vivo biofilms compared with in vitro [19]. Without the development of novel models to study clinically realistic, CF-associated chronic biofilm infections in the laboratory, development of more effective treatments to prevent or treat these infections will continue to be hindered.
There are a number of models currently used to study P. aeruginosa in mono- and polymicrobial CF biofilm infections. These include, but are not limited to, artificial sputum media (ASM) [[20], [21], [22], [23]], in vitro surface-attached biofilm models [24] and murine models with cystic fibrosis transmembrane conductance regulator mutations that mimic the mutations causative of CF in humans [25]. Limitations with such model systems have been identified. There is a distinct difference in the P. aeruginosa transcriptomic profiles between isolates grown in ASM and direct transcriptomic analysis of CF sputum samples. It has been shown that P. aeruginosa grown in laboratory media, including ASM, demonstrates upregulation of key genes involved in amino acid and carbohydrate biosynthesis, the TCA cycle and quorum sensing regulation, in comparison to human infection [26]. These differences indicate that although advancements have been made for laboratory media that more closely reflects the CF lung environment, it is still missing key aspects of chronic infection such as metabolism.
It is also important to consider the structure of P. aeruginosa biofilms. While this is hard to quantify, there are some distinct qualitative differences in the structure of biofilms grown in different in vitro platforms. For example the dense, homogenous mass of cells and matrix seen in microtitre plates versus the “mushroom” structures and open channels observed in flow cells [27,28]. Comparison of these systems with in vivo biofilms highlights further differences in structure. In particular, lung biopsies of people with CF reveals biofilms suspended in bronchial mucus with a ‘sponge’-like appearance. This mass of bacterial cells is punctuated by gaps which may be filled with mucus, alginate or lung fluid [16,29,30]. Hence, it is likely in vitro biofilms do not accurately mimic CF biofilms or the aspects of infection that are structure dependent, such as nutrient gradients in the biofilm matrix and resultant pockets of dormant cells, which affect quorum sensing [2]. In addition, mouse models are also unable to reliably mimic the P. aeruginosa infections that occur in people with CF [31]. The airway secretions from CF mice have been shown to be very different from human airway secretions [32], which could explain the significant differences in P. aeruginosa transcriptome that have found between mouse and human CF infection [26].
We previously published brief details of a novel ex vivo pig lung (EVPL) model for P. aeruginosa biofilm infection of CF bronchioles. The model combines one formulation of ASM with sections of porcine bronchiole [33]. The EVPL bronchiolar tissue demonstrated potential in recapitulating clinically relevant chronic infection [33,34]. In comparison to more commonly used model organisms, including rats and mice, pig lungs demonstrate closer similarity to human lungs. The two are similar in their metabolic composition and overall physiology, anatomy and immunology [32,35]. The lungs used for the model are obtained from pigs slaughtered for meat, thus the EVPL model offers a replacement for live animal (rodent) infection models, which is clinically realistic, economical and ethical.
Here, we provide detailed validation of EVPL as a model for chronic P. aeruginosa biofilm infection in CF. We have built on our previous work to demonstrate that the model is able to successfully mimic clinically relevant aspects of in vivo CF P. aeruginosa metabolism and biofilm formation. We show key pathways required for CF P. aeruginosa biofilm formation and maturation in vivo are similarly essential in EVPL. Our findings highlight the importance of visualising biofilms in order to understand chronic infection in a way that in vitro models of bacterial growth cannot achieve. We have also described the antibiotic tolerance of P. aeruginosa CF isolates grown as biofilms in this model [36]. EVPL has the potential to provide a platform to aid in the development of novel drugs and treatment regimens, which can be used to target the growing incidence of highly antibiotic-tolerant biofilms in the unique environment of CF lungs.
Materials and methods
Bacterial strains
Two isolates of P. aeruginosa (SED42, SED43), from a chronically-colonised person with CF, were used as example clinical isolates. These strains have previously been phenotypically characterised [37]. P. aeruginosa PA14 transposon insertion mutant strains (Table 1) and the isogenic wild type (WT) used in this work were obtained from the confirmed PA14 Non-Redundant Transposon Insertion Mutant Set [38,39]. The transposon’s presence in the correct locus of the putative PA14 mutants for leuB, purD, gacA and pelA was confirmed by arbitrary PCR and Sanger sequencing [38,39]. Auxotrophy of the leuB and
Table 1.
Description of the Pseudomonas aeruginosa PA14 transposon insertion mutants used for our work [38,39]. Information for each gene is provided and the associated CF infection phenotype following loss of gene function [23,40,41].
| Mutant ID | Active Gene ID | Active Gene Name | Active Gene Description | Predicted Infection Phenotype in EVPL |
|---|---|---|---|---|
| 31800 | GID1896 | leuB | 3-isopropylmalate dehydrogenase | No effect |
| 23405 | GID1574 | metX | Homoserine O-acetyltransferase |
No effect |
| 29794 | GID1301 | purD | Phosphoribosylamine--glycine ligase | No effect |
| 54630 | GID3719 | gacA | Response regulator GacA | Biofilm maturation defects |
| 26187 | GID86 | pelA | Conserved hypothetical protein | Unable to form biofilm |
purD mutants was confirmed by absence of growth in Agrobacterium (AB) medium + citrate and AB + glucose minimal media (data not shown). No sequence was obtained from three repeated PCRs for the metX mutant hence locus-specific primers for metX were designed (5′-ATCGGGTCCTCACACGCTG-3′ and 5′-TCCGACCCTGAGTTCCTCG-3′). PCR and gel electrophoresis confirmed amplification of a fragment in the putative metX mutant of ~1.5 kb. This is consistent with the presence of the transposon (994 bp); the expected fragment in the absence of a transposon is ~500 bp.
Artificial sputum media
The first artificial sputum medium (ASM) version, ASM1, was prepared following a published recipe, also referred to as SCFM [22]. The second ASM version: ASM2, is a slightly modified version of ASM1 and was similarly prepared following a published method, also referred to as SCFM2 [23]. For instances where glucose was removed from these recipes, it was substituted with an equivalent volume of deionised water.
In vitro P. aeruginosa growth and biofilm formation in ASM
Colonies of PA14, SED42 and SED43 were taken from Luria-Bertani (LB) agar plates (Melford) and grown overnight at 37 °C. Colonies were used to inoculate replicate 100 μl aliquots of ASM1 or ASM2, ±3 μM glucose, in a 96-well microplate with a peg lid (Innovotech), n = 4 per growth condition. Cultures were incubated at 37 °C for 24 h. Growth of planktonic fractions of the populations was assessed by briefly shaking the well plate then reading absorbance at 600 nm using a Tecan Spark 10M plate reader. Biofilm formation on pegs was quantified using a crystal violet assay [42]. Briefly, pegs were rinsed with 150 μl phosphate-buffered saline (PBS) in a fresh 96-well plate to remove loosely-adhering cells. The peg lid was then transferred to a further 96-well plate with 150 μl 0.1% (w/v) crystal violet (Vickers Laboratories) per well. It was incubated at room temperature for 15 min and the pegs rinsed twice in PBS as above, before drying in a laminar flow hood for 30 min. The pegs were transferred to another 96-well plate containing 150 μl 95% (v/v) ethanol per well to solubilise the crystal violet. Absorbance of the solubilised crystal violet was read at 590 nm in a Tecan Spark 10M. The relative allocation of bacterial cells to the biofilm per well was calculated by dividing A590nm of crystal violet by A600nm of corresponding planktonic subpopulation.
In vitro growth of P. aeruginosa metabolic mutants
Cultures were grown for 6 h in LB at 37 °C. Bacterial cells were washed and starved in PBS for 2 h, then resuspended in ASM1 or ASM2. One set of ASM cultures were left as per the original method, and the other supplemented with either 20 μg ml−1 leucine, 20 μg ml−1 methionine or 30 μg ml−1 each of guanine, adenine, xanthine and hypoxanthine as relevant for each mutant. Purines had to be dissolved in NaOH, so the comparative original ASM was supplemented with an equivalent amount of NaOH. Subsequently, 150 μl of each resuspended culture was transferred to a 96-well plate and grown for a further 24 h at 37 °C in a Tecan Spark 10M. Every 20 min, the plate was shaken for 5 s and the absorbance read at 600 nm.
EVPL dissection and infection
EVPL was prepared as previously described [33]. Briefly, porcine lungs were obtained from a local butcher (Quigley and Sons, Cubbington) and dissected on the day of delivery under sterile conditions. The pleura of the ventral surface was heat sterilised using a hot pallet knife. A sterile razor blade was then used to make an incision in the lung, exposing the bronchiole. A section of the bronchiole was extracted and the exterior alveolar tissue removed using dissection scissors. Bronchiolar sections were washed once in a 1:1 mix of Dulbecco’s modified Eagle medium (DMEM) and RPMI 1640 supplemented with 50 μg ml−1 ampicillin (Sigma-Aldrich) then cut into ~5 mm wide longitudinal strips. The bronchiolar strips were placed in a second 1:1 DMEM, RPMI 1640 + 50 μg ml−1 ampicillin wash and cut into squares (~5 × 5 mm). The tissue squares were washed for a third time in 1:1 DMEM, RPMI 1640 + 50 μg ml−1 ampicillin. Bronchiolar pieces were then further washed in ASM, UV sterilised for 5 min and transferred to individual wells of a 24-well plate containing 400 μl ASM (+20 μg ml−1 ampicillin) solidified with 0.8% (w/v) agarose per well.
A sterile 29G hypodermic needle (Becton Dickinson Medical) was touched to the surface of a P. aeruginosa colony grown on LB agar overnight at 37 °C and used to pierce an individual piece of bronchiolar tissue. Uninfected control tissue sections were mock inoculated with a fresh, sterile needle. Following infection of all tissue pieces, 500 μl of ASM +20 μg ml−1 ampicillin was added to each well. Tissue was incubated at 37 °C with a Breathe-Easier® membrane (Diversified Biotech) for the desired length of time.
EVPL biofilm recovery and assessment of bacterial load
Bronchiolar tissue pieces were removed from the 24-well plate following incubation, and each briefly washed in 500 μl PBS in a fresh 24-well plate. Tissue pieces were then transferred into sterile homogenisation tubes (Fisherbrand) containing eighteen 2.38 mm metal beads (Fisherbrand) and 1 ml PBS. Tissue was bead beaten in a FastPrep-24 5G (MP Biomedicals) for 40 s at 4 m s−1 to recover the bacteria and virulence factors from the tissue-associated biofilm. To determine the bacterial load, the homogenate was serially diluted in PBS and plated on LB agar. Plates were incubated overnight at 37 °C and colony counts used to calculate colony forming units (CFU) per tissue piece.
Determination of P. aeruginosa virulence factor production in EVPL biofilms
To quantify virulence factors produced by the P. aeruginosa biofilm population, the homogenate was diluted 1:4 in PBS to obtain a sufficient volume for further experiments. The diluted homogenate was passed through a 0.2 μm filter to remove bacterial cells and tissue debris.
To assay the amount of the siderophores pyoverdine and pyochelin present in homogenates, 100 μl aliquots of diluted sterile homogenate were transferred to a black 96-well plate. Fluorescence was measured with excitation/emission of 400±20/460±20 nm for pyoverdine [43] and 360±35/460±20 nm for pyochelin [44] in a Tecan Spark 10M.
To measure total protease, 100 μl of diluted sterile homogenate was mixed with 5 mg azocasein (Sigma-Aldrich) dissolved in 900 μl 100 mM Tris-HCl + 1 mM CaCl2. The reaction mixture was incubated with 170 rpm shaking for 2 h at 37 °C, then stopped by the addition of 100 μl 120 mM EDTA. The mixture was centrifuged at 13,000 rpm for 1 min and the absorbance of the supernatant read at 400 nm.
The amount of two quorum sensing (QS) signals in the diluted sterile homogenate was assayed using Escherichia coli biosensors [45]. Expression of a luxCDABE reporter in response to either 3-oxo-C12-HSL or C4-HSL was measured as relative light units (RLU) for E. coli strains carrying the reporter plasmids pSB1075 or pSB406 respectively.
Extracellular DNA detection in EVPL biofilms
Following 2 d and 7 d post-infection (PI), homogenate from EVPL cultures of PA14 and SED43 and uninfected control tissue were centrifuged at 13,000 rpm for 1 min (n = 3). The pellet was run through a Promega Wizard® Genomic DNA extraction kit. Recovered DNA was eluted in 50 μl sterile water and 10 μl of the eluate was loaded onto a 0.7% (w/v) agarose + SYBR™ Safe stain (Invitrogen) gel. Following electrophoresis, the resulting gel was visualised under blue light.
Crystal violet assay on EVPL biofilms
Infected and uninfected control bronchiolar tissue pieces were placed in a 24-well plate with 500 μl PBS per well. The plate was briefly shaken to remove any loosely-adhering P. aeruginosa cells from the tissue. Tissue pieces were then moved to another 24-well plate with 500 μl 0.1% (v/v) crystal violet per well, and incubated at room temperature for 15 min with 170 rpm shaking. Tissue sections were transferred into a fresh 24-well plate and the crystal violet treatment and incubation was repeated. The tissue was then transferred to a further 24-well plate and left to dry in a laminar flow hood for 30 min. The tissue and biofilm bound crystal violet was solubilised in 500 μl 95% (v/v) ethanol and incubated at room temperature for 15 min with 170 rpm shaking. The tissue was removed from the wells and absorbance read at 590 nm in a Tecan Spark 10M.
Congo red binding assay on EVPL biofilms
A Congo red binding assay to measure exopolysaccharide Pel production was tested and adapted [46] for use with the EVPL model. Infected EVPL bronchiolar tissue pieces were placed in individual 2 ml Eppendorf tubes containing 1 ml of 2 mg ml−1 Congo red (Vickers Laboratories Ltd., UK) and uninfected tissue pieces used as negative assay controls. Tubes were incubated for 90 min at 37 °C with 170 rpm shaking. Following incubation, the samples were centrifuged at 13,000 rpm for 5 min and the supernatant transferred to a transparent 96-well plate. Each supernatant was diluted 1 in 10 in PBS to a final volume of 200 μl to be within the limit of detection. The absorbance was read at 490 nm in a Tecan Spark 10M. The bound Congo red absorbance was determined by subtracting the A490nm of the infected tissue sample from the mean average uninfected tissue A490nm.
Hematoxylin & eosin staining
P. aeruginosa infected EVPL tissue pieces and uninfected control tissue were fixed in 10% (v/v) neutral buffered formalin (VWR Chemicals). The tissue samples were sent to the University of Manchester’s Histology Core Facility for paraffin wax embedding, sectioning and mounting. Mounted tissue sections were de-paraffinized in xylene for 20 min. To re-hydrate the tissue, slides were transferred to 95% (v/v) ethanol followed by 70% (v/v) ethanol. Any residual ethanol was removed by washing slides in distilled water. Samples were stained in Mayer’s hemalum solution (Merck Millipore) then washed in running ‘tap water’ (1 L tap water with 2 tsp sodium carbonate). Samples were counterstained in eosin Y solution (Merck Millipore) for then dehydrated by dipping the slides in 95% (v/v) ethanol. Samples were transferred to fresh 95% (v/v) ethanol then placed in 100% (v/v) ethanol. The samples were then placed in xylene. The samples were mounted using DPX mounting fluid and images taken using a Zeiss Axio Scope. A1 light microscope with the Zeiss Axiocam Erc 5s and Zeiss Zen 2.3 pro software.
Gram staining
P. aeruginosa infected EVPL tissue pieces, plus uninfected controls, were fixed and paraffin-embedded as described above for H & E staining. The tissue sections were de-paraffinized in xylene. Samples were re-hydrated in 100% (v/v) ethanol for then moved to fresh 100% (v/v) ethanol. Slides were placed in 95% (v/v) isopropanol then 70% (v/v) isopropanol. Residual alcohol was removed by washing slides in distilled water. Crystal violet (Pro-Lab Diagnostics) was applied to the samples and rinsed with water, then iodine (Pro-Lab Diagnostics) was applied. The iodine was then rinsed from the samples using tap water and acetone added for decolourisation. Tap water was used to rinse off the acetone and 1% (v/v) neutral red (Pro-Lab Diagnostics) applied to counterstain. The slides were again rinsed with tap water and blotted dry with filter paper. The samples were dehydrated in 100% (v/v) isopropanol then placed in two fresh changes of 100% (v/v) xylene. Samples were mounted using DPX mounting fluid and images taken using a Zeiss Axio Scope. A1 light microscope with the Zeiss Axiocam Erc 5s and Zeiss Zen 2.3 pro software.
Alcian blue staining
EVPL tissue pieces infected with P. aeruginosa, and uninfected tissue negative controls, were fixed, paraffin-embedded, de-paraffinized and re-hydrated as described for H & E staining. Slides were stained in 1% Alcian blue solution (pH 2.5) (Sigma-Aldrich) then rinsed using distilled water. The samples were counterstained in nuclear fast red solution (Alfa Aesar) and rinsed briefly in distilled water. Tissue sections were dehydrated in increasing ethanol concentrations and finally placed in xylene, as performed for H & E staining. Samples were mounted with DPX mounting fluid and images were taken of stained tissue sections using a Zeiss Axio Scope. A1 light microscope with the Zeiss Axiocam Erc 5s and Zeiss Zen 2.3 pro software.
Statistical analysis
Data were analysed using either R v3.5.1 or RStudio (v1.2.1335 on Mac OS Mojave v10.14.6) [47], with P < 0.05 considered significant. The car package [48] was used to conduct ANOVAs with type II SS when missing values caused non-orthogonality (i.e. analyses of C4-HSL and total protease). The multcomp package [49] was used for post-hoc comparisons. The FactoMineR package [50] was used for principal components analysis. All raw data and R code used for analyses, including data transformations used to meet model assumptions, are provided in the Data Supplement.
Results
Simple and complex formulations of artificial CF sputum lead to comparable P. aeruginosa growth and virulence in EVPL
Various formulations of artificial CF sputum media have been published [[20], [21], [22], [23]]. Here we focus on two formulations, which we term ASM1 and ASM2. ASM1 is the original formula we chose for use with the EVPL model [33,34,51,52] and ASM2 is an updated version that has since been published by the same laboratory [23]. ASM1 contains concentrations of free amino acids, cations, anions and lactate that are representative of the average concentrations found in a selection of sputum samples from people with CF. ASM1 was initially chosen for use with EVPL as it was shown to cue comparable carbon-usage pathways and expression of quorum sensing signals by P. aeruginosa PA14 to growth in medium made from lyophilised patient sputum [22]. ASM2 adds bovine maxillary mucin, membrane lipid 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), N-acetyl glucosamine (NAG) and free DNA. These additions were made to better represent the presence of host mucins, bacterial membrane and wall components, and the extracellular DNA (eDNA) found in the biofilm plugs formed in the bronchioles of people with CF. ASM2 has been shown to result in a comparable essential genome for P. aeruginosa PA14 to lyophilised patient sputum media in vitro [23]. We aimed to determine whether the EVPL model required the more complex ASM2, or whether EVPL + ASM1 would provide a sufficient platform for laboratory growth of P. aeruginosa with more clinically realistic characteristics.
In our previous work with EVPL, we removed glucose from the original ASM1 formulation as it appeared to enhance the growth of endogenous bacteria from the porcine lungs [34]. We have tested the effects of glucose removal from both versions of ASM on P. aeruginosa growth and biofilm formation in vitro (Fig. S1). Our results show small and inconsistent effects of ASM version and glucose removal on PA14 and two clinical CF isolates. The limited effect of glucose removal is consistent with the expectation that P. aeruginosa growth in the CF lung environment mostly depends on using amino acids and short-chain fatty acids as carbon sources [22,23,53,54]. Hence ASM without glucose is an appropriate approach for use with EVPL, so glucose was omitted from ASM for all further work.
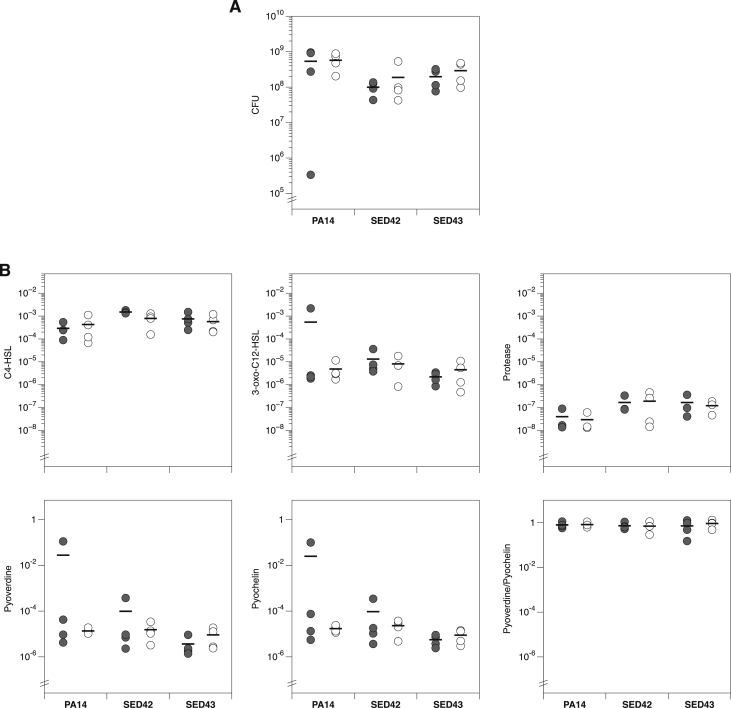
Our previous work also showed typical morphology of P. aeruginosa growing as extensive biofilms on EVPL + ASM1 4 d post-infection (PI) [33]. We have now further investigated the effect of ASM version on growth and virulence in EVPL. P. aeruginosa PA14 and clinical isolates SED42 and SED43 were used to infect EVPL with either ASM1 or ASM2, and biofilm was recovered 2 d PI. The biofilm load did not differ between EVPL + ASM1 and EVPL + ASM2 for PA14 or the exemplar clinical isolates (Fig. 1A). To confirm that ASM version did not impact on other aspects of P. aeruginosa biology, we measured the production of a selection of exoproducts associated with virulence: the quorum sensing signals C4-HSL and 3-oxo-C12 HSL, total protease and the siderophores pyoverdine and pyochelin (Fig. 1B). ANOVAs confirmed that there was not a significant effect of ASM version on any of these variables, as either a main effect or interaction with strain (Table 2). Principal component analysis did not reveal any clear clustering of phenotypes by ASM version (Fig. S2). We also found that EVPL + ASM1 was able to maintain extended P. aeruginosa growth over 14 d (Fig. S3), with detectable amounts of eDNA present in the biofilms formed 7 d PI (Fig. S4). It is not clear whether this results from bacterial lysis or host cell damage, but as the purD- mutant grows well in the presence of lung tissue it is likely that a proportion of this eDNA is tissue-derived. These results indicate the addition of EVPL removes the requirement of further additions made to ASM1 in the form of ASM2, and enhances the suitability of the simpler artificial media to mimic CF infection.
Fig. 1.
(A) Growth of the laboratory isolate Pseudomonas aeruginosa PA14 and two exemplar P. aeruginosa Cystic Fibrosis (CF) isolates: SED42 and SED43, on four pieces of ex vivo pig lung (EVPL) bronchiole plus either artificial sputum media 1 (ASM1) (closed circles) or ASM2 (open circles). The colony forming units (CFU) were retrieved from biofilm after 2 d growth. Bars denote means per ASM version. Strains varied in their biofilm load (ANOVA: F1,18 = 3.64, p = 0.047), but it was not affected by ASM version (main effect F1,18 = 0.84, p = 0.37; interaction with strain F2,18 = 0.01, p = 0.99). Results were unaffected if the outlier in PA14 + ASM1 was excluded (p-values of 0.002, 0.70 and 0.64, respectively). (B) Production of selected virulence factors by P. aeruginosa isolates in EVPL + ASM1 (closed circles) or EVPL + ASM2 (open circles). Bars denote means per ASM version. Units are as follows: for C4-HSL and 3-oxo-C12-HSL, Relative Light Units (RLU) of Escherichia coli biosensor per CFU of P. aeruginosa; for total protease, A400 following azocasein assay per CFU of P. aeruginosa; for pyoverdine and pyochelin, fluorescence per CFU of P. aeruginosa individually and as a ratio.
Table 2.
Results of ANOVAs for the data shown in Fig. 1B. None of the analyses show a significant effect of ASM version, either as a main effect or as an interaction with strain. Raw data and R code are provided in the Data Supplement.
| Strain |
ASM |
Strain∗ASM |
||||
|---|---|---|---|---|---|---|
| F2,18 | p | F1,18 | p | F2,18 | p | |
| C4-HSL | 6.00 | 0.011∗ | 2.27 | 0.150 | 1.86 | 0.185 |
| 3-oxo-C12-HSL | 1.41 | 0.342 | 0.49 | 0.493 | 0.35 | 0.709 |
| Total protease | 2.07 | 0.155 | 0.37 | 0.553 | 0.95 | 0.406 |
| Pyoverdine (PVD) | 2.07 | 0.155 | 0.37 | 0.553 | 0.95 | 0.406 |
| Pyochelin (PCH) | 2.14 | 0.147 | 0.65 | 0.430 | 0.89 | 0.429 |
| PVD/PCH | 0.25 | 0.781 | 0.26 | 0.619 | 0.28 | 0.758 |
EVPL removes the essentiality of purine biosynthesis for P. aeruginosa growth in both simple and complex formulations of ASM
To further confirm the optimal ASM version for use in all subsequent work, and the effect on P. aeruginosa of adding EVPL tissue to ASM, we compared one key aspect of primary metabolism in EVPL + ASM1 and EVPL + ASM2. Purine biosynthesis was selected as it was previously found to be essential in ASM2 in vitro but not in sputum taken from people with CF [23]. This lack of consistency with true infection highlights a specific area where the composition of ASM2 appears sub-optimal as a model for CF.
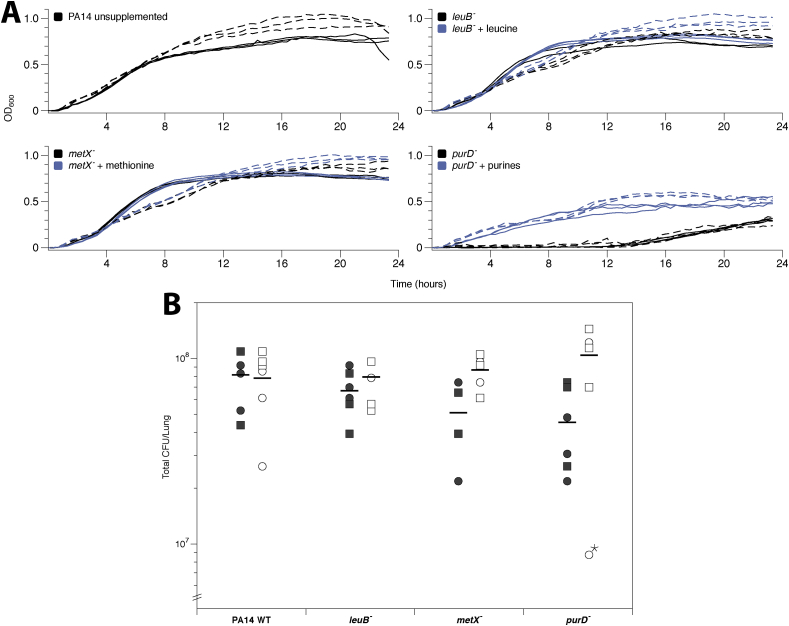
To confirm this previous finding, we performed in vitro growth curves in ASM1 and ASM2 of the laboratory strain wild type (WT) P. aeruginosa PA14 and a PA14 transposon insertion mutant for the purine auxotroph gene purD. We also measured growth of two isogenic mutants auxotrophic for amino acids abundant in ASM: leuB and metX, as positive controls. We found that in both versions of ASM purine supplementation is required for purD mutant growth in vitro (Fig. 2A).
Fig. 2.
(A) Growth of Pseudomonas aeruginosa PA14 and transposon insertion metabolic mutants in vitro in artificial sputum media 1 (ASM1) (solid) and ASM2 (dashed). Black lines are ASM alone, blue lines show ASM cultures supplemented with 20 μg ml−1 leucine, 20 μg ml−1 methionine or 30 μg ml−1 each of purines (guanine, adenine, xanthine and hypoxanthine) as appropriate. All treatments were replicated in triplicate and lines show individual replica cultures. (B) Growth of P. aeruginosa PA14 and transposon insertion metabolic mutants on three pieces of ex vivo pig lung (EVPL) bronchiole from each of two independent lungs (circles vs. squares) plus ASM1 (closed symbols) or ASM2 (open symbols). The graph shows colony-forming units (CFU) retrieved from individual EVPL biofilms after 48 h growth. The bars denote means per ASM version. An ANOVA was run to test for effects of lung identity, genotype, version of ASM used and the interaction between genotype and version of ASM used, on biofilm load. One clear outlier (purD-, ASM2, denoted with a star) was identified and excluded from analysis of this data. There was no main effect of lung identity or genotype on biofilm load (ANOVA: F3,38 = 0.56, p = 0.448 and F1,38 = 0.46, p = 0.712). Biofilm load was affected by the version of ASM used (main effect F1,38 = 12.1, p = 0.001; interaction with genotype F3,38 = 3.50, p = 0.024). A Tukey test showed statistical interaction was driven by the purD mutant (p < 0.01). Planned contrasts by conducting t-tests where t = (mutant CFU – WT CFU in the same medium)/SEdiff, where SEdiff is the standard error of the difference, calculated using the residual mean square from the ANOVA, showed no significant difference in CFU between individual mutants and WT in the same version of ASM (2-tailed p ≥ 0.126). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To identify whether the presence of EVPL could compensate for this shortcoming we infected two independent lungs with the four P. aeruginosa PA14 strains used in vitro (WT; purD, leuB and metX mutants). Bacterial growth was recovered at 2 d PI to determine CFU per lung (Fig. 2B). This was repeated using either ASM1 or ASM2 with the EVPL. Fig. 2B highlights the consistency in CFU per lung of the WT between EVPL + ASM1 and EVPL + ASM2. Statistical analysis found no significant difference in CFU between any individual mutant and the WT in the same version of ASM (Fig. 2B). Crucially, the purD mutant demonstrated no reduction in bacterial load compared with the WT in the corresponding ASM version. It can be concluded that addition of EVPL removes the essentiality of purine biosynthesis in ASM1, making it consistent with clinical infection. It is also worth noting that the slight growth defect of the WT observed in ASM1 in vitro compared to ASM2 (Fig. 2A) is not observable when EVPL is added (Fig. 2B).
In conclusion, there does not appear to be any significant differences in P. aeruginosa growth, virulence or purine metabolism in EVPL + ASM1 and EVPL + ASM2. We therefore used EVPL + ASM1 to explore the architecture of P. aeruginosa biofilms formed.
P. aeruginosa biofilm formation on EVPL with ASM recapitulates the morphology of in vivo CF biofilms, and is dependent on the Gac regulatory system and Pel polysaccharide
Biofilm formation is arguably the defining phenotype of chronic P. aeruginosa lung infection in individuals with CF. Following identification of optimal EVPL model conditions, we explored the structure and mechanisms of biofilm formation by P. aeruginosa PA14 on EVPL + ASM1. Initially, we confirmed that we were observing PA14 growth in the model and not simply maintaining inoculation CFU by measuring growth across 7 days in two independent lungs, and in ASM1 in vitro. Similar growth curves were obtained in lungs and ASM1 (Fig. S5). We then compared the biofilm formed by WT PA14 with those formed by transposon insertion mutants for the genes gacA and pelA. Both gacA and pelA are important in biofilm infection by P. aeruginosa PA14; either by direct regulation (gacA) or production of a structural matrix polysaccharide (pelA). GacA is a response regulator from the global activator of antibiotic and cyanide synthesis (Gac) two-component regulatory system. The Gac system is well-studied and known to be largely responsible for the switch from acute to chronic infection; in particular it regulates biofilm maturation [55,56]. PelA is a periplasmic protein required for biosynthesis of the biofilm matrix exopolysaccharide Pel. It is one of three exopolysaccharides associated with P. aeruginosa biofilm formation: Pel, Psl and alginate. In PA14, alginate is non-essential for biofilm formation and PA14 is Psl deficient. Hence, biofilm formation in this strain requires the Pel exopolysaccharide. Pel-deficient mutants have been shown to arrest in an early stage of biofilm development, forming thin, flat monolayers [41,57].
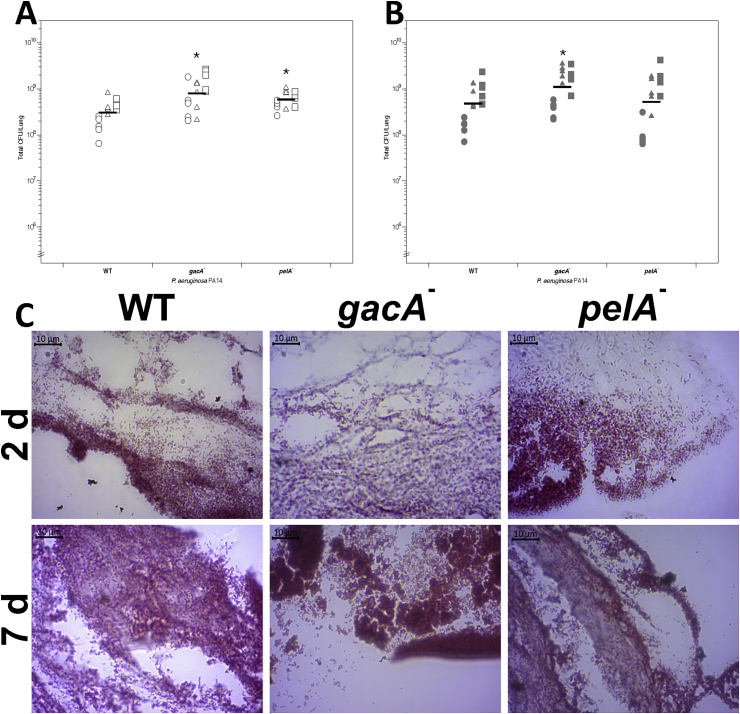
To investigate biofilm formation by P. aeruginosa PA14 WT and the gacA and pelA mutants, strains were inoculated individually into replica lung pieces from three independent lungs. Uninfected tissue was used as the negative control. We recovered the biofilm at 2 d and 7 d PI to determine CFU per EVPL section across an extended time period (Fig. 3A). ANOVAs identified a significant difference in CFU per EVPL tissue section between both the gacA and pelA mutants and the WT at 2 d PI (Fig. 3A). However, there was only a significant difference in CFU per EVPL section between the gacA mutant and WT 7 d PI (Fig. 3A). The interaction term between replicate lungs and infection strain was found to be significant, reflecting the natural variation between lungs rather than true differences in WT and mutant growth. This is not dissimilar to the patient-to-patient variation expected clinically.
Fig. 3.
(A, B) Colony forming units (CFU) per ex vivo pig lung (EVPL) bronchiolar tissue section infected with Pseudomonas aeruginosa PA14 wild type (WT) and transposon mutants at (A) 2 d and (B) 7 d post-infection (PI). Five replicate pieces of tissue were infected per strain from three independent lungs. Bars denote mean for each genotype across all three lungs, and asterisks denote a significant difference from the WT under that condition (post-hoc Dunnett’s test after ANOVA). ANOVA found a significant difference between strains, lung and their interaction at 2 d PI (strain F4,60 = 10.27, P < 0.01; lung F2,60 = 26.05, P < 0.01; interaction F8,60 = 1.99, P = 0.06). Post-hoc analysis showed that there was significantly higher CFU recovered of the gacA mutant compared to the WT (P < 0.01), and higher CFU of pelA mutant than the WT (P = 0.01). At 7 d PI a significant difference in bacterial recovery was found between strains and between lungs, as well as a significant interaction between strain and lung (ANOVA: strain F4,60 = 5.40, P < 0.01; lung F2,60 = 78.98, P < 0.01; interaction F8,60 = 2.01, P = 0.06). There was no longer a significant difference in CFU per lung between the WT and pelA mutant (P = 0.69). A significantly greater CFU per lung was recovered from the gacA mutant than the WT (P = 0.047) 7 d PI. (C) Micrographs of Gram-stained PA14 WT and selected transposon mutants in EVPL + ASM1 at 2 d and 7 d PI. All images are x100 magnification.
Gram staining confirmed that the uninfected tissue was clear of bacterial growth at 2 d and 7 d PI (data not shown). Aggregates of Gram-negative rods were visible on the surface of EVPL tissue infected with each strain at both time points (Fig. 3B). The Gram stain images also demonstrate the increase in density of bacterial cells from 2 d PI to 7 d PI for both mutants and the WT.
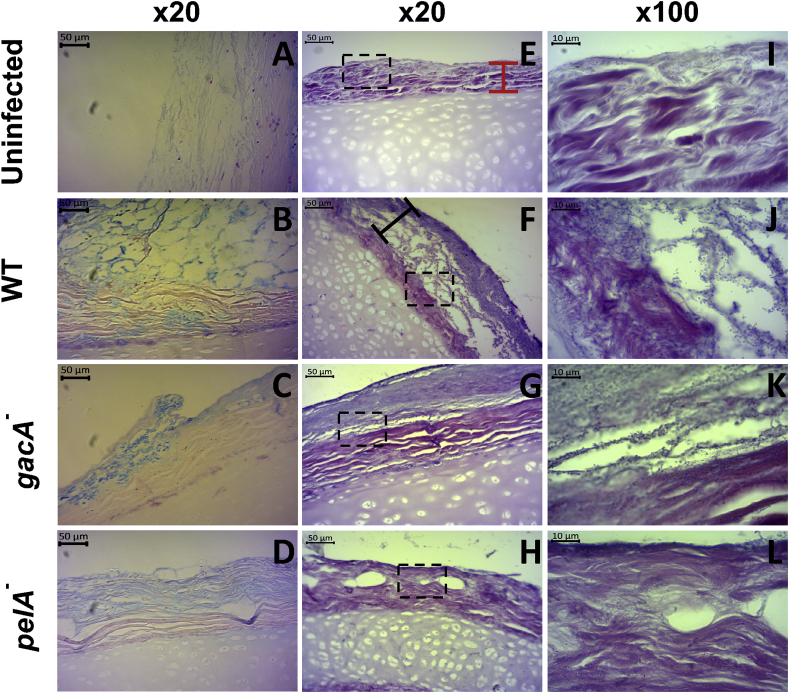
H & E staining is a histological technique traditionally used to visualise mammalian tissue architecture. However there are a number of studies that have adopted this technique to detect biofilms associated with host tissue for use as a clinical diagnostic tool [[58], [59], [60], [61]]. Although it is a non-specific stain that cannot distinguish between different bacterial species, when performed alongside Gram staining (Fig. 3C), it is a quick, cost-effective approach to detect bacteria, their biofilms and the structure of associated mammalian tissue. It has been reported as a reliable method for biofilm staining by cross-referencing H & E stained tissue sections with sections imaged using fluorescence in situ hybridisation (FISH) and confocal laser scanning microscopy [58,60]. We found that the standard crystal violet biofilm assay is not suitable for use on ex vivo tissue samples. As shown in Fig. S6A, the lung tissue preferentially binds crystal violet obscuring any signal from dye bound to the biofilm matrix. A similar issue was found with the Congo red binding assay for Pel production (Fig. S6B). Hence, H & E staining was used to visualise the P. aeruginosa biofilms on EVPL tissue as it stains both the bacteria and extracellular polymeric substances of the biofilm, as well as the porcine tissue. Alcian blue staining was also performed as a standard technique for identifying the biofilm matrix [16,62,63]. Replica sets of infected lung pieces from two independent lungs were fixed at 2 d PI and 7 d PI and sections stained with either Gram strain to identify bacteria (Fig. 3C), Alcian blue to stain the biofilm polysaccharide matrix (Fig. 4A–D), or H & E to visualise the total biofilm mass and tissue architecture (Fig. 4E-L).
Fig. 4.
Sections of ex vivo pig lung (EVPL) bronchiolar tissue with artificial sputum media 1 (ASM1) infected with Pseudomonas aeruginosa at 7 d post infection (PI). EVPL was infected with Pseudomonas aeruginosa PA14 wild type (WT) and selected PA14 transposon mutants, with uninfected tissue as a negative control. (A–D) Alcian blue stained sections counterstained with nuclear fast red. The biofilm matrix is stained blue and the EVPL tissue and cartilage stained red/pink. (E–L) Hematoxylin and eosin (H & E) stained sections. The cartilage and tissue surface (red bar) stain pink and the bacterial biofilm (black bar) purple, including the bacterial cells and biofilm matrix. A detailed diagram of the biofilm structure as shown by H & E staining is provided in the supplementary information. The x100 magnification images are taken from the same section as shown in x20 (denoted by the dashed boxes). The P. aeruginosa rods are visible, which make up the structures evident in the lower magnification images. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
H & E images for each strain at 7 d PI are shown in Fig. 4: the basophilic bacteria and biofilm matrix are stained purple by the hematoxylin, and the connective tissue of the bronchiole is stained pink by the eosin (replicate images from a second lung are provided in Fig. S7). Whilst Gram stain images identified Gram-negative rods present 2 d and 7 d PI (Fig. 3C), H & E staining gave a clear image of all aspects of the EVPL biofilm model: the tissue, bacteria and biofilm matrix. A diagram explaining each aspect of the tissue and the biofilm evident in the H & E images is shown in Fig. S8. This was further complemented with Alcian blue staining (Fig. 4) showing the biofilm matrix associated with the tissue.
There were clear qualitative differences in the biofilm observed between the mutant strains and WT 7 d PI using Alcian blue (Fig. 4A–D) and H & E staining (Fig. 4E-L). As shown in Fig. 4F,J the WT formed a thick structure on the surface of the tissue, comprising of layers of bacteria and matrix punctuated with spaces, comparable to CF biofilms observed in vivo using H & E or Alcian blue staining (see Figure 4 in Ref. [16] and Figure 8 in Ref. [29]). Alcian blue staining confirmed the spaces are truly open with a comparable structure visible in the blue stained WT biofilm (Fig. 4B), indicating there is no matrix polysaccharide occupying the empty space visualised with H & E staining. This structure was not present in the biofilm formed by the gacA mutant: the purple stained region on the surface of the EVPL tissue demonstrates the homogenous, dense biofilm matrix formed by the gacA mutant (Fig. 4G,K). It was supported by dense Alcian blue staining of the biofilm matrix (Fig. 4C) compared to the WT (Fig. 4B). Images of the pelA mutant biofilm are comparable to the uninfected tissue: we identified only a thin layer of purple stain at 7 d PI (Fig. 4H,L). Although the pelA mutant was able to grow on EVPL tissue (Fig. 3), it was unable to form a biofilm. There was evidence of aspects of a biofilm matrix produced by the pelA mutant on the Alcian blue images (Fig. 4D). This is consistent with work on PA14 in in vitro biofilm systems, in which pelA mutants can attach to a surface to form a thin monolayer, but do not develop further into biofilms reminiscent of the WT [41]. Biofilm depth measurements taken from the images are provided in Fig. S10, supporting visual findings. At 2 d PI, these differences although visible were not as clearly defined with Alcian blue or H & E staining (Fig. S9).
The difference between WT growth and mutants associated with pathways known to be essential for biofilm matrix production (Pel polysaccharide) and biofilm maturation (GacAS regulatory system) confirmed that the structures formed by P. aeruginosa PA14 on the surface of EVPL + ASM1 are biofilm. Comparison with published images from CF lung biopsies show that the biofilm structure of PA14 WT on EVPL is in vivo-like.
Discussion
CF can dramatically impact an individual’s quality of life and has a high mortality rate. Alongside this, the condition also has a major impact on healthcare systems worldwide. Throughout their lifetime people with CF continually require treatment for lung infections. In the US, the estimated median annual costs for treatment of pulmonary exacerbations, a period of acute-like infection that is highly virulent, ranges from $9,456 (USD) to $48,263 (USD) per person [64]. There is a limited choice of effective antibiotics to treat CF lung infections currently available to clinicians, and the antibiotic tolerance observed in vivo means that standard antibiotic susceptibility testing does not accurately predict whether drugs will actually work [17,18]. As individuals with CF are living longer, there is a growing medical and economic need for novel and improved treatments for lung infections that are more effective and less rigorous. The rising incidence of antimicrobial resistance (AMR) is only adding to this pressure. Prescribing cannot be improved, nor can a true insight into the impact of potential novel treatment agents (e.g. anti-virulence drugs) be achieved, until there is a better understanding of the fundamental microbiology underpinning these chronic biofilm infections. We aimed to develop a clinically-relevant ex vivo model for the predominant cause of chronic CF lung infections: P. aeruginosa. We have demonstrated that addition of EVPL tissue to a simple formulation of ASM results in a clinically realistic P. aeruginosa biofilm, overcoming a number of obstacles associated with current laboratory models of CF infection.
We initially aimed to define the optimal P. aeruginosa culture conditions for the EVPL model to ensure we are closely modelling clinical CF lung infections. Investigation of P. aeruginosa growth, virulence and purine biosynthesis showed that addition of the porcine bronchiolar tissue overcomes issues associated with in vitro growth in the laboratory media ASM1 and ASM2.
ASM1 is simpler and cheaper to make than ASM2, thus our conclusion that ASM1 is sufficiently realistic when used with EVPL makes the system readily accessible and tractable for use in research and potentially diagnostic laboratories. It is plausible that ASM1 is sufficient with the addition of EVPL for two primary reasons. Firstly, mucin is unlikely to be an important physiological cue in vivo and in EVPL as it is a poorly used carbon source for P. aeruginosa [54,65]. In vitro it is likely that mucin acts as a structural cue for P. aeruginosa aggregation and the switch to biofilm lifestyle, however we observed biofilm formation on EVPL in the absence of exogenous mucin. Additionally, the prolonged growth periods and high biofilm densities possible with EVPL mean it is likely that membrane lipids, NAG and eDNA will be released from cells within the tissue and/or maturing biofilm. Our findings suggest that EVPL tissue with ASM1 provides key compounds that are lacking from the media alone, creating a closer match with authentic sputum from people with CF.
Visualisation of the biofilm is key to comparing laboratory models with in vivo biofilms. We used H & E staining as this stains both the EVPL tissue and the bacterial cells [58,60], which is not possible with other more bacteria specific microscopy techniques. This allowed us to detect the formation of a biofilm and its location on the tissue, as well as any potential tissue damage. This approach is relatively simple to perform and cost-effective [58,59], thus beneficial to address our aim of detecting the biofilm visualising its structure using approaches that will make the model tractable for diagnostics where time and cost savings are necessary. We also performed Alcian blue staining to visualise the biofilm matrix [16,63] to confirm the H & E images, which supported our conclusions.
However, unlike techniques such as FISH, H & E staining does not provide taxonomic bacterial identification [58]. It is suitable for work focusing on mono-species biofilm, in which we know the species we have introduced. More specific approaches must be used in work focused on polymicrobial biofilms. Our confirmatory work using H & E and P. aeruginosa paves the way for the use of EVPL to conduct more complex microscopic analysis of single and mixed-species lung biofilms using FISH or scanning electron microscopy to gain further insight into infection.
We found that the laboratory P. aeruginosa strain PA14 forms a biofilm on the surface of EVPL tissue, with a lace-like structure of bacterial aggregates surrounding air or fluid spaces. On a structural level it is comparable to P. aeruginosa biofilms that have been observed in explanted lung tissue from people with CF (see Figure 8 in Ref. [29], Figure 4 in Ref. [30] and Figure 4 in Ref. [16]). The EVPL biofilm appears more structurally heterogeneous than has been previously observed in vitro [28], and distinct from the “mushroom” type typically seen in flow cell biofilms (see Figure 8 in Ref. [2]). Our findings support the conclusion that EVPL mimics a clinically realistic P. aeruginosa biofilm infection. The porcine tissue provides the structure and physiological cues that facilitate the formation of biofilms representative of CF infection.
To explore the mechanisms responsible for biofilm formation in the EVPL model, we also imaged the biofilm formed by two PA14 transposon mutants: pelA and gacA. We identified clear differences in growth in EVPL. The pelA gene encodes a deacetylase and hydrolase enzyme involved in the synthesis of the biofilm exopolysaccharide pellicle: Pel [57]. Previous studies have shown that the Pel polysaccharide is necessary for cell-cell interactions and development of P. aeruginosa PA14 biofilms [41]. This is consistent with our images showing that the pelA mutant was unable to form a biofilm on the surface of the EVPL bronchiolar tissue. Interestingly, whilst the pelA mutant formed visually less biofilm than the WT, similar CFU were recovered from tissues infected by both genotypes. This may suggest that a large portion of cells visible in WT biofilms are dead or viable but not culturable. Alcian blue stained the bacterial biofilm on the surface of the tissue infected with the pelA mutant 7 d PI. As the mutant is Pel-deficient and PA14 is Psl-deficient, this is likely to be staining of lipopolysaccharide and/or alginate. The presence of alginate could explain the mutant’s ability to attach to the tissue surface despite being unable to form a biofilm.
The gacA mutant however was found to form a biofilm, but it lacked the structure that was observed for the WT by H & E staining. This was similarly observed with Alcian blue staining, and the biofilm matrix components appeared to be densely packed in comparison to the WT. GacA has been shown to be essential for P. aeruginosa PA14 biofilm maturation, and lack of gacA function results in a reduction in biofilm formation capacity and structure, and reduced antibiotic resistance [55]. The development of the WT biofilm architecture is dependent on the GacAS system so may be considered a “mature” state. Our observations thus confirm the importance of established biofilm regulatory pathways in the formation of the structured communities observed on EVPL tissue. This supports the conclusion that P. aeruginosa is forming a mature biofilm in the EVPL model.
Further analysis of EVPL infection at a genotypic level will be required to fully conclude which aspects of chronic CF infection dynamics the model is able to recapitulate. However, we present EVPL + ASM1 as a cheap, tractable and high-throughput platform for growing P. aeruginosa biofilms. The EVPL infections we observed exhibit some of the key morphological and metabolic characteristics observed for this pathogen in CF. Our results clearly demonstrate the importance of visualising biofilms grown in laboratory models. When we compared the CFU per lung of WT, pelA and gacA mutants, there were no clear differences in bacterial recovery – but there were obvious differences in the structure of the biofilms they formed. Visualisation of biofilms formed on EVPL tissue are a valuable addition to future work, with the aim to understand infection dynamics, antibiotic tolerance, polymicrobial interactions and the ability of novel drugs to penetrate and destroy these biofilms.
Conclusions
We have shown, in extension to our previous work [30,31], that the EVPL model is able to successfully replicate a phenotypically realistic P. aeruginosa CF lung biofilm. Our work can now be expanded to further understanding of other key aspects of infection, as well as a basis for development of ex vivo models for other biofilm infection contexts. The model provides a high-throughput approach to bridge the gap between in vitro models and true human infection.
CRediT authorship contribution statement
Niamh E. Harrington: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Visualization. Esther Sweeney: Methodology, Investigation, Writing - review & editing. Freya Harrison: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
We thank all at Steve Quigley & Sons butchers for donating pig lungs; Leo Eberl, Steve Higgins, Sophie Darch, Steve Diggle and Paul Williams for bacterial strains; and John Moat and Ian Hands-Portman for help setting up imaging. The authors would also like to acknowledge the help of the Media Preparation Facility in the School of Life Sciences at the University of Warwick, with special thanks to Cerith Harries and Caroline Stewart. The Histology Facility equipment used in this study was purchased with grants from The University of Manchester Strategic Fund. Special thanks to Peter Walker and Grace Bako for their help with the histology. Finally, we thank Ákos Kovacs and two anonymous reviewers for comments on the manuscript. This work was supported by an MRC New Investigator Research Grant [grant number MR/R001898/1] awarded to FH and by a PhD studentship from the BBSRC Midlands Integrative Biosciences Training Partnership (MIBTP) awarded to NEH.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2020.100024.
Contributor Information
Niamh E. Harrington, Email: N.Harrington@warwick.ac.uk.
Esther Sweeney, Email: E.Sweeney@warwick.ac.uk.
Freya Harrison, Email: F.Harrison@warwick.ac.uk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Flemming H.C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 2.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heltshe S.L., Cogen J., Ramos K.J., Goss C.H. Cystic fibrosis: the dawn of a New therapeutic era. Am J Respir Crit Care Med. 2017;195:979–984. doi: 10.1164/rccm.201606-1250PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aali M., Caldwell A., House K., Zhou J., Chappe V., Lehmann C. Iron chelation as novel treatment for lung inflammation in cystic fibrosis. Med Hypotheses. 2017;104:86–88. doi: 10.1016/J.MEHY.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Taylor-Robinson D., Archangelidi O., Carr S.B., Cosgriff R., Gunn E., Keogh R.H. Data resource profile: the UK cystic fibrosis registry. Int J Epidemiol. 2018;47:9–10e. doi: 10.1093/ije/dyx196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magalhães A.P., Lopes S.P., Pereira M.O. Insights into cystic fibrosis polymicrobial consortia: the role of species interactions in biofilm development, phenotype, and response to in-use antibiotics. Front Microbiol. 2017;7 doi: 10.3389/fmicb.2016.02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surette M.G. The cystic fibrosis lung microbiome. Ann. Am. Thorac. Soc. 2014;11:S61–S65. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 9.Sanders D.B., Fink A.K. Background and epidemiology. Pediatr Clin. 2016;63:567–584. doi: 10.1016/j.pcl.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd T.J., Ramsay K.A., Vidmar S., Carlin J.B., Bell S.C., Wainwright C.E. Pseudomonas aeruginosa genotypes acquired by children with cystic fibrosis by age 5-years. J Cyst Fibros. 2015;14:361–369. doi: 10.1016/j.jcf.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Smith W.D., Bardin E., Cameron L., Edmondson C.L., Farrant K.V., Martin I. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnx121. [DOI] [PubMed] [Google Scholar]
- 12.Nixon G.M., Armstrong D.S., Carzino R., Carlin J.B., Olinsky A., Robertson C.F. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J Pediatr. 2001;138:699–704. doi: 10.1067/MPD.2001.112897. [DOI] [PubMed] [Google Scholar]
- 13.Konstan M.W., Morgan W.J., Butler S.M., Pasta D.J., Craib M.L., Silva S.J. Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J Pediatr. 2007;151:134–139.e1. doi: 10.1016/J.JPEDS.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 14.Lund-Palau H., Turnbull A.R., Bush A., Bardin E., Cameron L., Soren O. Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expet Rev Respir Med. 2016;10:685–697. doi: 10.1080/17476348.2016.1177460. [DOI] [PubMed] [Google Scholar]
- 15.Broder U.N., Jaeger T., Jenal U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2017;2:16184. doi: 10.1038/nmicrobiol.2016.184. [DOI] [PubMed] [Google Scholar]
- 16.Bjarnsholt T., Østrup Jensen P., Fiandaca M.J., Pedersen J., Rønne Hansen C., Bøgelund Andersen C. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 17.Smith A.L., Fiel S.B., Mayer-Hamblett N., Ramsey B., Burns J.L. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration. Chest. 2003;123:1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 18.Hurley M.N., Ariff A.H.A., Bertenshaw C., Bhatt J., Smyth A.R. Results of antibiotic susceptibility testing do not influence clinical outcome in children with cystic fibrosis. J Cyst Fibros. 2012;11:288–292. doi: 10.1016/j.jcf.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müsken M., Klimmek K., Sauer-Heilborn A., Donnert M., Sedlacek L., Suerbaum S. Towards individualized diagnostics of biofilm-associated infections: a case study. npj Biofilms Microbiomes. 2017;3:22. doi: 10.1038/s41522-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sriramulu D.D., Lünsdorf H., Lam J.S., Römling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- 21.Fung C., Naughton S., Turnbull L., Tingpej P., Rose B., Arthur J. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol. 2010;59:1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 22.Palmer K.L., Aye L.M., Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189 doi: 10.1128/JB.01138-07. 8079–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner K.H., Wessel A.K., Palmer G.C., Murray J.L., Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternberg C., Bjarnsholt T., Shirtliff M. Methods for dynamic investigations of surface-attached in vitro bacterial and fungal biofilms. Methods Mol Biol. 2014;1147:3–22. doi: 10.1007/978-1-4939-0467-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson D.J., Rolfe M. Mouse models of cystic fibrosis. Trends Genet. 2001;17:S29–S37. doi: 10.1016/S0168-9525(01)02452-0. [DOI] [PubMed] [Google Scholar]
- 26.Cornforth D.M., Dees J.L., Ibberson C.B., Huse H.K., Mathiesen I.H., Kirketerp-Møller K. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E5125–E5134. doi: 10.1073/pnas.1717525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts A.E.L., Kragh K.N., Bjarnsholt T., Diggle S.P. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Kragh K.N., Alhede M., Kvich L., Bjarnsholt T. Into the well—a close look at the complex structures of a microtiter biofilm and the crystal violet assay. Biofilms. 2019;1 doi: 10.1016/j.bioflm.2019.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson A.G., Ehre C., Button B., Abdullah L.H., Cai L.-H., Leigh M.W. Cystic fibrosis airway secretions exhibit mucin hyperconcentration and increased osmotic pressure. J Clin Invest. 2014;124:3047–3060. doi: 10.1172/JCI73469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baltimore R.S., Christie C.D.C., Walker Smith G.J. Immunohistopathologic localization of Pseudomonas aeruginosa in lungs from patients with cystic fibrosis. Implications for the pathogenesis of progressive lung deterioration. Am Rev Respir Dis. 1989;140:1650–1661. doi: 10.1164/ajrccm/140.6.1650. [DOI] [PubMed] [Google Scholar]
- 31.Bayes H.K., Ritchie N., Irvine S., Evans T.J. A murine model of early Pseudomonas aeruginosa lung disease with transition to chronic infection. Sci Rep. 2016;6:35838. doi: 10.1038/srep35838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benahmed M.A., Elbayed K., Daubeuf F., Santelmo N., Frossard N., Namer I.J. NMR HRMAS spectroscopy of lung biopsy samples: comparison study between human, pig, rat, and mouse metabolomics. Magn Reson Med. 2014;71:35–43. doi: 10.1002/mrm.24658. [DOI] [PubMed] [Google Scholar]
- 33.Harrison F., Diggle S.P. An ex vivo lung model to study bronchioles infected with Pseudomonas aeruginosa biofilms. Microbiology. 2016;162:1755–1760. doi: 10.1099/mic.0.000352. [DOI] [PubMed] [Google Scholar]
- 34.Harrison F., Muruli A., Higgins S., Diggle S.P. Development of an ex vivo porcine lung model for studying growth, virulence, and signaling of Pseudomonas aeruginosa. Infect Immun. 2014;82:3312–3323. doi: 10.1128/IAI.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meurens F., Summerfield A., Nauwynck H., Saif L., Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/J.TIM.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan M.M., Harrington N.E., Sweeney E., Harrison F. Predicting antibiotic-associated virulence of Pseudomonas aeruginosa using an ex-vivo lung biofilm model. BioRxiv. 2020 doi: 10.1101/2020.02.24.963173. 2020.02.24.963173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darch S.E., McNally A., Harrison F., Corander J., Barr H.L., Paszkiewicz K. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep. 2015;5:7649. doi: 10.1038/srep07649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci Unit States Am. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G. PA14 transposon insertion mutant library. 2006. http://Pa14.Mgh.Harvard.Edu/Cgi-Bin/Pa14/Home.Cgi [DOI] [PMC free article] [PubMed]
- 40.Parkins M.D., Ceri H., Storey D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 41.Colvin K.M., Gordon V.D., Murakami K., Borlee B.R., Wozniak D.J., Wong G.C.L. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moskowitz S.M., Foster J.M., Emerson J., Burns J.L. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 2004;42:1915–1922. doi: 10.1128/JCM.42.5.1915-1922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiricny N., Diggle S.P., West S.A., Evans B.A., Ballantyne G., Ross-Gillespie A. Fitness correlates with the extent of cheating in a bacterium. J Evol Biol. 2010;23:738–747. doi: 10.1111/j.1420-9101.2010.01939.x. [DOI] [PubMed] [Google Scholar]
- 44.Dumas Z., Ross-Gillespie A., Kümmerli R. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. R. Soc. B Biol. Sci. 2013;280 doi: 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winson M.K., Swift S., Fish L., Throup J.P., Jørgensen F., Chhabra S.R. Construction and analysis of luxCDABE -based plasmid sensors for investigating N -acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 46.Ghafoor A., Hay I.D., Rehm B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Core Team . R Found. Stat. Comput.; 2018. R: a language and environment for statistical computing.https://www.r-project.org/ [Google Scholar]
- 48.Fox J., Weisberg S. Second. Sage; Thousand Oaks CA: 2011. An R companion to applied regression.http://socserv.socsci.mcmaster.ca/jfox/Books/Companion [Google Scholar]
- 49.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 50.Lê S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. J Stat Software. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 51.Davies S., Fearn S., Allsopp L., Harrison F., Ware E., Diggle S. Visualizing antimicrobials in bacterial biofilms: three-dimensional biochemical imaging using TOF-SIMS. mSphere. 2017;2:e00211–e00217. doi: 10.1128/mSphere.00211-17. https://core.ac.uk/download/pdf/84340786.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison F., McNally A., da Silva A.C., Heeb S., Diggle S.P. Optimised chronic infection models demonstrate that siderophore ‘cheating’ in Pseudomonas aeruginosa is context specific. ISME J. 2017;11:2492–2509. doi: 10.1038/ismej.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmer K.L., Mashburn L.M., Singh P.K., Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flynn J.M., Niccum D., Dunitz J.M., Hunter R.C. Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkins M.D., Ceri H., Storey D.G. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 56.Jimenez P.N., Koch G., Thompson J.A., Xavier K.B., Cool R.H., Quax W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marmont L.S., Whitfield G.B., Rich J.D., Yip P., Giesbrecht L.B., Stremick C.A. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biol Chem. 2017;292:19411–19422. doi: 10.1074/jbc.M117.812842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hochstim C.J., Choi J.Y., Lowe D., Masood R., Rice D.H. Biofilm detection with hematoxylin-eosin staining. Arch Otolaryngol - Head Neck Surg. 2010;136:453–456. doi: 10.1001/archoto.2010.62. [DOI] [PubMed] [Google Scholar]
- 59.Tóth L., Csomor P., Sziklai I., Karosi T. Biofilm detection in chronic rhinosinusitis by combined application of hematoxylin-eosin and gram staining. Eur Arch Oto-Rhino-Laryngol. 2011;268:1455–1462. doi: 10.1007/s00405-011-1623-x. [DOI] [PubMed] [Google Scholar]
- 60.Hong S.D., Dhong H.-J., Chung S.-K., Kim H.Y., Park J., Ha S.Y. Hematoxylin and eosin staining for detecting biofilms: practical and cost-effective methods for predicting worse outcomes after endoscopic sinus surgery. Clin. Exp. Otorhinolaryngol. 2014;7:193–197. doi: 10.3342/ceo.2014.7.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Becerra S.C., Roy D.C., Sanchez C.J., Christy R.J., Burmeister D.M. An optimized staining technique for the detection of Gram positive and Gram negative bacteria within tissue. BMC Res Notes. 2016;9:216. doi: 10.1186/s13104-016-1902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hengzhuang W., Song Z., Ciofu O., Onsøyen E., Rye P.D., Høiby N. OligoG CF-5/20 disruption of mucoid Pseudomonas aeruginosa biofilm in a murine lung infection model. Antimicrob Agents Chemother. 2016;60:2620–2626. doi: 10.1128/AAC.01721-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Høiby N., Bjarnsholt T., Moser C., Jensen P.Ø., Kolpen M., Qvist T. Diagnosis of biofilm infections in cystic fibrosis patients. APMIS. 2017;125:339–343. doi: 10.1111/APM.12689. [DOI] [PubMed] [Google Scholar]
- 64.Rubin J.L., Thayer S., Watkins A., Wagener J.S., Hodgkins P.S., Schechter M.S. Frequency and costs of pulmonary exacerbations in patients with cystic fibrosis in the United States. Curr Med Res Opin. 2017;33:667–674. doi: 10.1080/03007995.2016.1277196. [DOI] [PubMed] [Google Scholar]
- 65.Flynn J.M., Phan C., Hunter R.C. Genome-wide survey of Pseudomonas aeruginosa PA14 reveals a role for the glyoxylate pathway and extracellular proteases in the utilization of mucin. Infect Immun. 2017;85 doi: 10.1128/IAI.00182-17. e00182-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.