Abstract
Recent CYP2D6 phenotype standardization efforts by CYP2D6 activity score (AS) are based on limited pharmacokinetic (PK) and pharmacodynamic (PD) data. Using data from two independent clinical trials of metoprolol, we compared metoprolol PK and PD across CYP2D6 AS with the goal of determining whether the PK and PD data support the new phenotype classification. S‐metoprolol apparent oral clearance (CLo), adjusted for clinical factors, was correlated with CYP2D6 AS (P < 0.001). The natural log of CLo was lower with an AS of 1 (7.6 ± 0.4 mL/minute) vs. 2–2.25 (8.3 ± 0.6 mL/minute; P = 0.012), similar between an AS of 1 and 1.25–1.5 (7.8 ± 0.5 mL/minute; P = 0.702), and lower with an AS of 1.25–1.5 vs. 2–2.25 (P = 0.03). There was also a greater reduction in heart rate with metoprolol among study participants with AS of 1 (−10.8 ± 5.5) vs. 2–2.25 (−7.1 ± 5.6; P < 0.001) and no significant difference between those with an AS of 1 and 1.25–1.5 (−9.2 ± 4.7; P = 0.095). These data highlight linear trends among CYP2D6 AS and metoprolol PK and PD, but inconsistencies with the phenotypes assigned by AS based on the current standards. Overall, this case study with metoprolol suggests that utilizing CYP2D6 AS, instead of collapsing AS into phenotype categories, may be the most precise approach for utilizing CYP2D6 pharmacogenomics in clinical practice.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
CYP2D6 genotype‐derived activity score (AS) is used to assign CYP2D6 phenotype. However, recent approaches to standardize CYP2D6 genotype‐to‐phenotype translation are based on limited pharmacokinetic (PK) or pharmacodynamic (PD) data.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study compared metoprolol apparent oral clearance and change in heart rate across CYP2D6 genotype‐derived AS and evaluated the performance of an AS‐exclusive method compared with consensus CYP2D6 genotype‐to‐phenotype translation.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study characterized PK and PD variability for metoprolol across the AS continuum. Metoprolol PK and PD differed between AS of 1 and 2–2.25, supporting initiatives to separate these scores into different phenotypes. However, metoprolol PK also differed between AS of 1.25–1.5 and 2, suggesting they should not share phenotype assignment.
HOW MIGHT THIS CHANGE DRUG DISCOVERY, DEVELOPMENT, AND/OR THERAPEUTICS ?
These data overall suggest an AS‐exclusive approach for CYP2D6 genotype translation may provide better granularity for clinical implementation of CYP2D6 pharmacogenomics.
Cytochrome P450 2D6 (CYP2D6) plays an important role in biotransformation of ~ 25% of drugs, including metoprolol. 1 CYP2D6 is highly polymorphic, with > 140 alleles described (https://www.pharmvar.org/gene/CYP2D6). The highly polymorphic properties of CYP2D6 contribute to wide interpatient variation in CYP2D6 catabolic activity and drug response.
The CYP2D6 Activity Score (AS), determined based on genotype and drug interactions (i.e., use of CYP2D6 inhibitors), is used to assign CYP2D6 phenotype as referenced in Clinical Pharmacogenetic Implementation Consortium (CPIC) guidelines. 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 The CYP2D6 AS system assigns each gene allele an activity value, and the sum of the individual allele values forms the AS. 3 Then, if applicable, the AS is multiplied by a factor of 0.5 or 0 to account for phenoconversion with use of a moderate or strong CYP2D6 inhibitor, respectively. 2 , 3 The CYP2D6 AS system is used clinically to predict CYP2D6 metabolizer phenotype. 2 , 5 , 6 , 7 , 8 , 9 For example, an individual with an AS of 0 is classified as a poor metabolizer (PM) with no expected enzyme activity, whereas an individual with CYP2D6 copy number variation leading to an AS > 2.25 would be assigned the ultra‐rapid metabolizer (UM) phenotype. As clinical pharmacogenetic test results become more readily available, appropriate assignment of phenotype based on the CYP2D6 AS is critical to optimally predict drug response and tailor therapy accordingly.
Based on pharmacokinetic (PK) data specific for sparteine, debrisoquine, and dextromethorphan, a CYP2D6 genotype‐derived AS of 1 to 2, reflecting the equivalent of 1 to 2 fully functional CYP2D6 alleles, was originally considered by CPIC as the normal metabolizer (NM) phenotype. This contrasted with phenotype assignment by the Dutch Pharmacogenetics Working Group (DPWG), which classified an AS of 1 as the intermediate metabolizer (IM) phenotype. Inconsistencies in CYP2D6 phenotype classification between CPIC and DPWG created confusion in the pharmacogenetic community and manifested clinically as patients with the same CYP2D6 genotype receiving different genotype‐guided pharmacotherapy recommendations depending on whether the CPIC or DPWG guideline was followed. 10 , 11
Recently, CPIC and DPWG collaboratively engaged a panel of CYP2D6 experts who were administered a series of surveys using a modified Delphi approach in an effort to reach consensus on CYP2D6 phenotype assignment. This resulted in revision of the CYP2D6 genotype to phenotype translation so that an AS of 0.25 to < 1.25 denotes the IM phenotype, and an AS of 1.25–2.25 denotes the NM phenotype, which was recommended for universal adoption for both research and clinical practice. 12 However, these consensus revisions were based on expert opinion, 12 not on PK and pharmacodynamic (PD) data by CYP2D6 AS. Thus, whether the new genotype to phenotype translation more accurately predicts the PK and PD of CYP2D6 substrates compared with the historical nomenclature remains unclear.
Metoprolol, a beta‐blocker commonly prescribed for treating hypertension, heart failure, myocardial infarction, and other cardiovascular diseases, serves as a model CYP2D6 substrate by which to compare PK by CYP2D6 AS because 70–80% of metoprolol metabolism is mediated via CYP2D6. 13 Several studies have shown that metoprolol PK is influenced by CYP2D6 genotype‐inferred metabolizer phenotype, consistently demonstrating a gene‐dose effect between CYP2D6 phenotype and metoprolol disposition. 13 , 14 , 15 , 16 These studies specifically show that metoprolol oral clearance increases as the number of partially or fully functional alleles increases. Compared with the CYP2D6 NM phenotype, this translates to increased metoprolol oral clearance in UMs and decreased clearance in PMs and IMs. However, none of these studies examined metoprolol PK or PD by CYP2D6 AS, and phenotype translations for AS of 1 were variable. Variability in metoprolol plasma concentrations by CYP2D6 phenotype translates to PD differences in heart rate (HR) response, with greater HR reductions observed in PMs and IMs compared with NMs; however, no differences in blood pressure (BP) response or risk for adverse effects with metoprolol were shown. 14 , 16 , 17 , 18 , 19 , 20 , 21
The objective of this study was to evaluate the PK and PD of metoprolol, as a case drug, across CYP2D6 genotype‐derived activity scores with the goal of determining whether the PK and PD data support the new phenotype classification. We were especially interested in whether metoprolol PK and PD differed across AS of 1 to 2 given the recent reclassification of an AS of 1 from NM to an IM. Additionally, we explored whether consideration of the rs5758850 single nucleotide polymorphism (SNP), located in the CYP2D6 enhancer region and shown to increase gene expression, improved the association between AS and metoprolol PK and PD parameters. 22 , 23 , 24
METHODS
Study cohorts
The data for analyses were generated from two clinical trials. The PK cohort was from an open‐label study that compared the PK and cardiovascular effects of the brand metoprolol succinate product and two generic formulations in adults with hypertension (clinicaltrials.gov identifier: NCT02417246). Trial exclusion criteria are available as Supplementary Information . The trial population was intentionally enriched for CYP2D6 PMs and IMs. As part of the trial, all participants underwent a 24‐hour PK study after at least 5 days of treatment with the brand name formulation. On the day of PK study, 10‐mL blood samples were collected in heparinized tubes immediately before and 0.5, 1, 2, 3, 4, 6, 8, 12, 16, 20, and 24 hours after the metoprolol dose. Data from 37 hypertensive study participants who were treated with the reference formulation of metoprolol succinate at doses of 50 mg/day (n = 30), 100 mg/day (n = 5), or 150 mg/day (n = 2) were included in the current PK analysis.
The PD cohort came from the Pharmacogenomic Evaluation of Antihypertensive Responses 2 (PEAR‐2) trial (clinicaltrials.gov identifier: NCT01203852), which has been previously described. 16 Briefly, PEAR‐2 was a prospective, multicenter, clinical trial conducted to assess the genomic variability attributed to BP and adverse metabolic effects following sequential monotherapy treatment with metoprolol tartrate and chlorthalidone. 16 Participants were started on metoprolol tartrate at an initial dose of 50 mg twice daily, with up‐titration after 2 weeks to 100 mg twice daily unless BP was < 120/70 mmHg or HR < 55 beats/minute. For the current analysis, change in HR (beats/minute (bpm)), as assessed after 2 weeks of metoprolol tartrate 50 mg twice daily, was selected as the PD end point because of its previously observed association with CYP2D6 genotype‐derived phenotype. 16
The PK study protocol was approved by the University of Florida institutional review board. The PEAR‐2 study protocol was approved by the institutional review board of each participating location (University of Florida, Gainesville, FL; Emory University, Atlanta, GA; and the Mayo Clinic, Rochester, MN). Patients in both studies provided voluntary, written informed consent.
CYP2D6 genotyping and AS assignment
In both the PK and PD studies, genomic DNA from study participants was isolated from lymphocytes using the FlexiGene DNA kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Genotyping for CYP2D6*2, *3, *4, *5, *6, *10, *17, *29, *40, and *41 variant alleles was performed by polymerase chain reaction and Pyrosequencing or Sanger sequencing. If no sequence variation was detected, then the allele was, by default, assigned as *1. The genotyping for rs5758550 (CYP2D6 enhancer SNP) was done by Pyrosequencing and TaqMan genotyping methods. The CYP2D6 copy number variations were estimated by the TaqMan Copy Number Assay (Life Technologies, Foster City, CA) and quantification‐based pyrosequencing genotyping method. 25
The AS method was used to assign CYP2D6 metabolizer phenotype from genotype data and use of CYP2D6 inhibitors. 2 , 3 , 26 Without considering the enhancer SNP, a value of 1 was assigned for each normal function allele (i.e., *1 and *2), 0.5 for each decreased function allele (i.e., *17, *29, and *41), 0.25 for each *10 allele, and 0 for each no function allele (i.e., *3, *4, *5, *6, and *40). 2 , 3 , 10 To test the associations between CYP2D6 AS and metoprolol PK and PD parameters after including the enhancer SNP (rs5758550), CYP2D6 alleles containing rs16947 T plus rs5758550 A were considered to have an activity value of 0.5, whereas alleles containing rs16947 T plus rs5758550 G were considered to have an activity value of 1. 22 , 23 , 24 Phenotypes were assigned based on the sum of allele values (i.e., AS) and were concordant with CPIC/DPWG recommendations: 0, PM; 0.25–1, IM; 1.25–2.25, NM; and > 2.25, UM. 12
Metoprolol analytical assay
A detailed explanation is available as Supplementary Information .
Data analyses
Data are presented as mean ± SD, median (interquartile range) for continuous data, or count (%) for categorical data. S‐metoprolol apparent oral clearance (CLo (mL/minute)) was calculated for each CYP2D6 AS group by dividing metoprolol dose (mg) by S‐metoprolol area under the curve (AUC0–24; min*mg/mL). S‐metoprolol CLo data were not normally distributed, and the variable was natural log transformed (ln(CLo) (mL/minute)) prior to analysis. A multiple linear regression analysis with contrast and estimate procedures for multiple comparisons was performed to compare ln(CLo) by CYP2D6 AS after controlling for the clinical factors: age, race, sex, and body mass index (BMI). Two multiple regression models were constructed; one model contained the conventional CYP2D6 AS without considering rs5758550, whereas the other included an AS that considered the functional effects of the enhancer SNP. Both multiple regressions were performed using CYP2D6 AS, age, race, sex, and BMI as explanatory variables, and ln(CLo) as the outcome. A sensitivity power analysis was conducted for the PK study. With a sample size of 37 and an alpha of 0.05, this study had 80% power to detect an effect size of 1.37 between activity scores of 1 vs. 2–2.25 and an effect size of 1.63 between activity scores of 1 vs. 1.25–1.5. 27
For the PD end point, the analysis of variance test was used to compare mean change in HR from baseline to 2‐weeks after metoprolol tartrate 50 mg twice daily was started between CYP2D6 AS groups. Given the sample size and an alpha of 0.05, the PD study had 80% power to detect an effect size of 0.46 and 0.57 between AS of 1 vs. 2–2.25 and AS of 1.25–1.5, respectively. 27 Similar to the PK analysis, to control for the effects of baseline characteristics (age, race, sex, and BMI) on metoprolol response, multiple regression was performed to evaluate the differences in HR changes across CYP2D6 AS groups. This method was used for both the conventional and enhancer SNP‐derived CYP2D6 AS.
Changes in CYP2D6 AS after considering the enhancer SNP were further compared between participants of African (hereafter referred to as black) and European (hereafter referred to as white) ancestry because of differing linkage disequilibrium between the *2 allele and enhancer SNP by race, with the continuity adjusted χ2 test or Fisher’s exact test as appropriate. To assess model fit for PK and PD outcomes, Akaike information criterion (AIC) 28 and adjusted R 2 values were generated from multiple linear regression models of CYP2D6 phenotype, as assigned by CPIC/DPWG consensus recommendations, and CYP2D6 AS on a continuous scale after adjusting for clinical factors (age, race, sex, and BMI). P values < 0.05 were considered statistically significant. The data analysis for this paper was generated using SAS software (SAS Institute, Cary, NC).
RESULTS
Study participant characteristics and CYP2D6 activity score
Clinical characteristics and AS for study participants included in the PK and PD analyses are summarized in Table 1 . Blacks comprised 32% of the PK and 37% of the PD cohorts; most of the remaining participants were white. Clinical characteristics were similar across CYP2D6 AS in both studies (data not shown). CYP2D6 and rs5758550 allele frequencies are shown in Supplementary Table S1 . One participant in the PK study with the CYP2D6 *1/*1 diplotype was taking bupropion, a strong CYP2D6 inhibitor that phenoconverts the CYP2D6 AS to 0. 29
Table 1.
Characteristics of study participants from two clinical trials
| Pharmacokinetic (n = 37) | Pharmacodynamic (n = 227) | |
|---|---|---|
| Age, years | 53.3 ± 11.7 | 50.4 ± 9.4 |
| Males | 19 (51.4) | 104 (45.8) |
| Race | ||
| Black/African American | 12 (32.4) | 84 (37.0) |
| Asian | 1 (2.7) | 1 (0.4) |
| White/European American | 24 (64.9) | 139 (61.2) |
| Other | 0 | 3 (1.3) |
| BMI, kg/m2 | 30.4 ± 4.7 | 30.6 ± 5.3 |
| CYP2D6 ASa | ||
| 0 | 4 (10.8) | 12 (5.3) |
| 0.25–0.75 | 3 (8.1) | 17 (7.5) |
| 1 | 7 (18.9) | 69 (30.4) |
| 1.25–1.5 | 7 (18.9) | 38 (16.7) |
| 2–2.25 | 14 (37.8) | 82 (36.1) |
| > 2.25 | 2 (5.4) | 9 (4.0) |
| CYP2D6 AS + rs5758550b | ||
| 0 | 4 (10.8) | 12 (5.3) |
| 0.25–0.75 | 5 (13.5) | 25 (11.0) |
| 1 | 8 (21.6) | 66 (29.1) |
| 1.25–1.5 | 7 (18.9) | 43 (18.9) |
| 2–2.25 | 12 (32.4) | 68 (30.0) |
| > 2.25 | 1 (2.7) | 8 (3.5) |
| Unknown | 0 | 5 (2.2) |
Data are presented as mean ± SD or n (%).
AS, activity score; BMI, body mass index.
AS derived from CYP2D6 genotype and phenoconversion.
AS derived from CYP2D6 genotype, phenoconversion, and rs5758550.
Metoprolol pharmacokinetics
S‐metoprolol ln(CLo), adjusted for clinical covariates, increased with increasing CYP2D6 AS (Figure 1 ; adjusted P < 0.001). The ln(CLo) was higher among those with an AS of 2–2.25 (8.3 ± 0.6 mL/minute) vs. an AS of 1 (7.6 ± 0.4 mL/minute; P = 0.012) or 1.25–1.5 (7.8 ± 0.5 mL/minute; P = 0.030). The ln(CLo) was significantly higher with an AS of 1 vs. an AS of 0 (6.6 ± 0.6 mL/minute; P = 0.010) and numerically higher with an AS of 0.25–0.75 vs. an AS of 0 (7.3 ± 0.2 vs. 6.6 ± 0.6 mL/minute), although small sample sizes made this comparison underpowered to show a statistical difference. The ln(CLo) was similar between AS of 1.25–1.5 (7.8 ± 0.5) and 1 (7.6 ± 0.4), and between AS of 1 (7.6 ± 0.4) and 0.25–0.75 (7.3 ± 0.2). The adjusted R 2 value for ln(CLo) in the CYP2D6 phenotype model was 48.1% and the AIC was −36.1. When evaluating CYP2D6 AS as a continuous variable vs. grouping scores into discrete phenotype categories, the AS remained associated with ln(CLo) (adjusted P < 0.001), and model performance improved, with an increase in the adjusted R 2 value and decrease in the AIC (Table 2 ).
Figure 1.
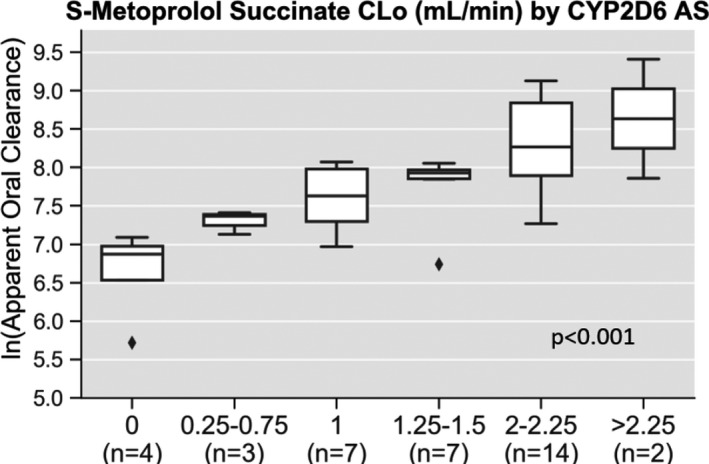
S‐metoprolol succinate apparent oral clearance by CYP2D6 activity score. Pharmacokinetic data are shown for the natural log of S‐metoprolol apparent oral clearance (ln(CLo)) by CYP2D6 AS. After adjusting for other clinical predictors (sex, race, age, and body mass index) CYP2D6 AS was significantly associated with S‐metoprolol ln(CLo) (adjusted P < 0.001).
Table 2.
CYP2D6 activity score model performance across pharmacokinetic and pharmacodynamic parameters
| Pharmacokinetic study outcome: S‐metoprolol ln(CLo) | Pharmacodynamic study outcome: change in heart rate | |||||||
|---|---|---|---|---|---|---|---|---|
| Adjusted R 2 | AIC | Adjusted R 2 | AIC | |||||
| CYP2D6 Phenotype | 48.1% | ‐36.1 | 16.1% | 770.81 | ||||
|
Black 9.4% |
White 55.2% |
Black −8.11 |
White −22.66 |
Black 19.1% |
White 15.9% |
Black 297.07 |
White 464.27 |
|
| Continuous AS | 53.8% | −41.83 | 18.1% | 763.33 | ||||
|
Black 29.8% |
White 59.3% |
Black −11.17 |
White −26.27 |
Black 26.4% |
White 14.8% |
Black 287.35 |
White 464.18 |
|
| CYP2D6 Phenotype + rs5758550 | 51.9% | −38.88 | 14.0% | 759.62 | ||||
|
Black 26.5% |
White 57.1% |
Black −10.63 |
White −23.67 |
Black 16.5% |
White 15.0% |
Black 296.95 |
White 451.98 |
|
| Continuous AS + rs5758550 | 63.0% | −47.49 | 16.5% | 751.23 | ||||
|
Black 43.5% |
White 71.0% |
Black −13.78 |
White −31.84 |
Black 23.0% |
White 14.7% |
Black 288.37 |
White 450.5 |
|
AIC, Akaike information criterion; AS, CYP2D6 activity score; ln(CLo), natural log of apparent oral clearance.
CYP2D6 phenotype definitions: 0, poor metabolizer; 0.25–1, intermediate metabolizer; 1.25–2.25, normal metabolizer; and > 2.25, ultra‐rapid metabolizer. All models were adjusted for participant: age, race, sex, and body mass index.
After considering the CYP2D6 enhancer SNP (rs5758550), 8 of the 37 study participants (22%), including 4 of 12 (33%) blacks and 4 of 24 (17%) whites, had a change in their AS. This led to phenotype reclassification in three (8%) participants; two changed from the NM to IM phenotype, and one shifted from the UM to a NM‐UM ranged phenotype (Table 3 ). CYP2D6 AS remained associated with ln(CLo) in the regression model, including the enhancer SNP AS (Figure 2 ; adjusted P < 0.001), with a higher adjusted R 2 value (51.9%) and lower AIC (−38.88) compared with the conventional CYP2D6 phenotype model. The greatest model fit was observed when the AS was evaluated as a continuous variable in the enhancer SNP model (adjusted P < 0.001; Table 2 ).
Table 3.
Diplotypes with CYP2D6 activity score change after including enhancer SNP
| Diplotype | Race | Conventional ASa | rs5758550: Genotype at Enhancer SNP locus | AS with Enhancerb | Phenotype Reclassification |
|---|---|---|---|---|---|
| Pharmacokinetic study (n = 8) | |||||
| *1/*2 | Black | 2 | T/T | 1.5 | No |
| *2/*17 | Black | 1.5 | C/T | 1 | No |
| *2/*2 | Black | 2 | T/T | 1 | No |
| *2/*2 | Black | 2 | C/T | 1.5 | No |
| *2/*2x2 | White | 3 | C/T | 2–2.5 | Yes |
| *2/*4 | White | 1 | T/T | 0.5 | Yes |
| 2*/*4x2 | White | 1 | T/T | 0.5 | Yes |
| *2/*41 | White | 1.5 | T/T | 1 | No |
| Pharmacodynamic study (n = 24) | |||||
| *1/*2 (n = 3) | Black | 2 | T/T | 1.5 | No |
| *1/*2 (n = 4) | White | 2 | T/T | 1.5 | No |
| *2/*10 | Black | 1.25 | T/T | 0.75 | Yes |
| *2/*17 (n = 2) | Black | 1.5 | C/T | 1 | No |
| *2/*2 (n = 3) | Black | 2 | T/T | 1 | No |
| *2/*2 | Black | 2 | C/T | 1.5 | No |
| *2/*2 | White | 2 | C/T | 1.5 | No |
| *2/*4 (n = 3) | Black | 1 | T/T | 0.5 | Yes |
| *2/*41 | White | 1.5 | T/T | 1 | No |
| *2/*5 (n = 3) | Black | 1 | T/T | 0.5 | Yes |
| *2/*5 | White | 1 | T/T | 0.5 | Yes |
| *2x2/*10 | Black | 2.25 | T/T | 1.25 | No |
AS, activity score; SNP, single nucleotide polymorphism.
Conventional CYP2D6 AS does not consider effects of the enhancer SNP (rs5758550).
CYP2D6 AS after considering functional effects of enhancer SNP (rs5758550).
Figure 2.
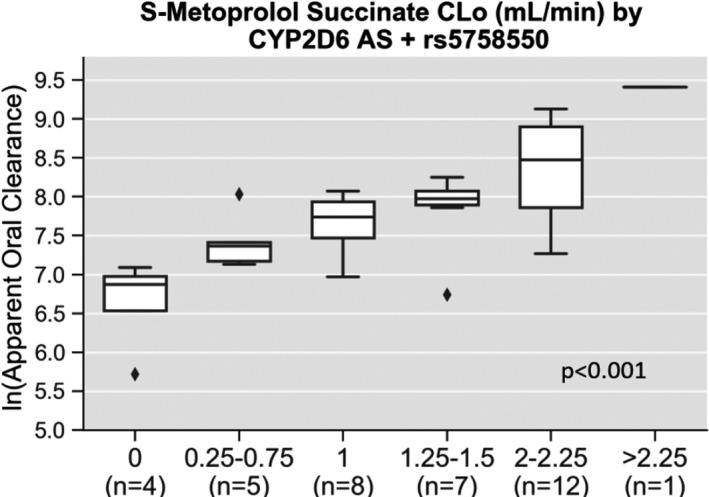
S‐metoprolol succinate apparent oral clearance by CYP2D6 activity score after considering the functional effects of rs5758550. Pharmacokinetic data are shown for the natural log of S‐metoprolol apparent oral clearance (ln(CLo)) by CYP2D6 AS after accounting for rs5758550, a CYP2D6 enhancer single nucleotide polymorphism. After adjusting for other clinical predictors (sex, race, age, and body mass index) CYP2D6 AS was significantly associated with metoprolol ln(CLo) (adjusted P < 0.001).
Metoprolol pharmacodynamics
Change in HR with metoprolol tartrate 50 mg twice daily was associated with CYP2D6 AS across the AS continuum (Figure 3 ; adjusted P < 0.001). There was a greater reduction in HR among those with AS of 1 (−10.8 ± 5.5 bpm) compared to an AS of 2–2.25 (−7.1 ± 5.6 bpm; P < 0.001). Compared with AS of 1, there was a greater reduction in HR with an AS of 0.25–0.75 (−15.9 ± 4.5 bpm; P < 0.001) and tended to be a greater reduction with an AS of 0 (−13.7 ± 4.7 bpm; P = 0.063). There were no significant differences in HR changes with metoprolol between those with an AS of 1.25–1.5 (−9.2 ± 4.7 bpm) and 2–2.25 (P = 0.102), between those with an AS of 1.25–1.5 and 1 (P = 0.095) or between those with an AS of 0 and 0.25–0.75 (P = 0.363). The adjusted R 2 value for change in HR with the CYP2D6 phenotype multiple regression model was 16.1%, and the AIC was 770.81. When evaluating AS as a continuous variable vs. grouping into phenotype categories, it remained associated with change in HR (adjusted P < 0.001), and the model improved as indicated by the increased adjusted R 2 and decreased AIC (Table 2 ).
Figure 3.
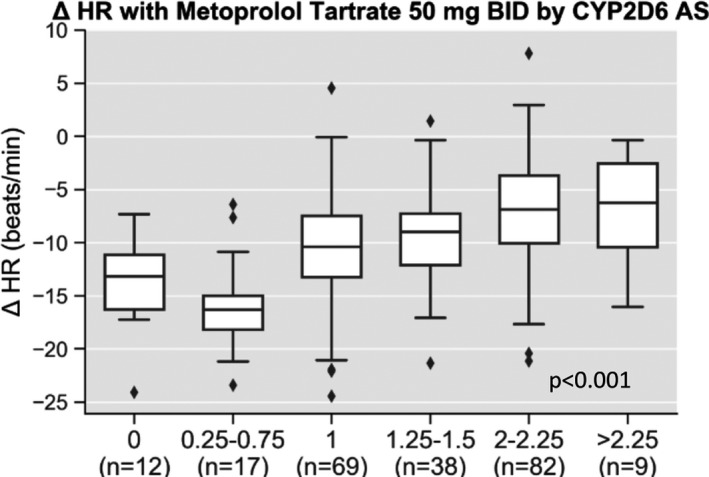
Change in heart rate with metoprolol tartrate by CYP2D6 activity score. Pharmacodynamic data are shown for change in heart rate (HR) after 2 weeks of metoprolol tartrate by CYP2D6 AS. After adjusting for other clinical predictors (sex, race, age, and body mass index) CYP2D6 AS was significantly associated with change in HR (adjusted P < 0.001).
After considering the CYP2D6 enhancer SNP, 24 of the 227 study participants (10.6%) had a change in their AS (Table 3 ). Changes occurred more often in blacks (20.5%) compared with whites (5.4%, P = 0.002); 17 of the 24 participants (70.8%) with a change in AS were black. In eight (4%) cases, consideration of the enhancer SNP led to CYP2D6 phenotype reclassification. CYP2D6 AS remained associated with change in HR in the enhancer SNP AS models whether considered in phenotype categories (Figure 4 ; adjusted P < 0.001) or as a continuous variable (adjusted P < 0.001). The adjusted R 2 and AIC indicated better model fit for the continuous AS variable, regardless of whether the enhancer SNP was considered (Table 2 ).
Figure 4.
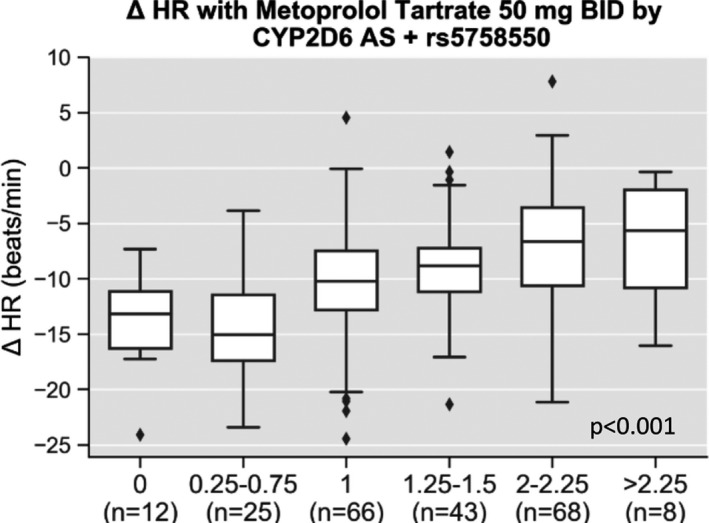
Change in heart rate with metoprolol tartrate by CYP2D6 activity score (AS) after considering the functional effects of rs5758550. Pharmacodynamic data are shown for change in heart rate (HR) after 2 weeks of metoprolol tartrate by CYP2D6 AS after accounting for rs5758550, a CYP2D6 enhancer single nucleotide polymorphism. After adjusting for other clinical predictors (sex, race, age, and body mass index) CYP2D6 AS was significantly associated with change in HR (adjusted P < 0.001).
DISCUSSION
This study demonstrated that CYP2D6 AS is significantly associated with S‐metoprolol succinate oral clearance and change in HR after initiating metoprolol tartrate. There were apparent differences in metoprolol PK and PD parameters in those with an AS of 1 vs. an AS of 2–2.25. However, metoprolol PK and PD did not significantly differ across the CYP2D6 AS range of 1 to 1.25–1.5. Expected differences in PK and PD were observed between an AS of 0 and 1. Although S‐metoprolol ln(CLo) was similar between AS of 0.25–0.75 and 1, likely secondary to the small size of the PK cohort, there were greater reductions in HR with an AS of 0.25–0.75 compared with 1. PK, but not PD, was different between AS of 1.25–1.5 compared with AS of 2–2.25, which might be explained by the sigmoidal concentration‐response relationship with beta‐blockers. Overall, these data suggest a CYP2D6 paradigm shift from considering metabolizer status nomenclature (e.g., PM, IM, NM, and UM) to using an AS‐based system may improve CYP2D6‐guided pharmacotherapy.
The new CPIC/DPWG consensus CYP2D6 genotype to phenotype translation method expands the definition of IM to include those with an AS of 1; this is in contrast to the prior nomenclature where NMs encompassed the AS range of 1–2. 12 Although some aspects of our data are consistent with the standardized CYP2D6 genotype to phenotype translation, there are other results that are not. Our data showing apparent differences in metoprolol PK and PD parameters in those with an AS of 1 vs. an AS of 2–2.25 support different phenotype categories for CYP2D6 AS of 1 and 2, consistent with the new CYP2D6 phenotype classification system. 12 However, S‐metoprolol ln(CLo) and HR changes were similar between AS of 1 and 1.25–1.5, suggesting these CYP2D6 AS should share the same CYP2D6 phenotype designation, which is not the case with the current classification system. The current data highlight the complexities of clinical CYP2D6 phenotype translation based on CYP2D6 AS and provide insight on the PK and PD differences between AS of 1 and 2.
When CYP2D6 AS was considered for metoprolol oral clearance and change in HR, the AS‐exclusive model outperformed CYP2D6 genotype to phenotype translation, and a linear relationship between metoprolol clearance and AS was observed. An AS‐exclusive method may be preferred to collapsing AS into predicted phenotypes, particularly because for some drugs with CPIC guidance, an AS of 1 may translate to an IM, whereas for others it is closer to an NM. 2 , 7 , 11 An AS‐exclusive paradigm dissolves the debate regarding phenotype status for an AS of 1 and permits substrate‐specific pharmacotherapy recommendations across the AS continuum. Potential benefits of removing the translation of AS into metabolizer phenotype for CYP2D6 include increased flexibility of CPIC prescribing recommendations for CYP2D6 substrates, improved precision for associations between PK/PD, and improved ability to characterize differences in drug response between the AS range from 0.25 to 2.25. From a pragmatic standpoint, an AS‐exclusive approach could occupy a discrete field within the electronic health record and support the current landscape of pharmacogenomic implementation that utilizes clinical decision support to facilitate prescribing decisions.
We focused on metoprolol as the case CYP2D6 substrate for which to examine PK and PD by genotype‐derived activity score based on the significant role of CYP2D6 in metoprolol metabolism and evidence from multiple studies that its disposition is altered across the CYP2D6 phenotype spectrum; as the number of active alleles increases, so does metoprolol metabolism. 14 , 15 , 21 , 30 , 31 , 32 , 33 Although, up to sixfold higher apparent oral clearance of metoprolol has been observed in PMs vs. NMs, PD responses are less predictable given the drug’s wide therapeutic index and sigmoidal response curve. This translates to PD consequences (e.g., HR response) of variable metoprolol plasma concentrations being less evident at higher doses. 33 CYP2D6 has not been associated with BP lowering or adverse effects with metoprolol, 14 , 16 , 21 and as such, CYP2D6 is not recommended to guide metoprolol use in hypertension management. 16 , 33 However, multiple studies have demonstrated variable HR response to metoprolol therapy across CYP2D6 phenotypes, with greater HR reductions in PM/IMs compared with NMs. 16 , 17 , 18 , 19 , 20 , 33 Thus, notwithstanding the limitations of clinical pharmacogenetic implementation, metoprolol is an ideal substrate for examining PK effects and their potential translation to PD end points (e.g., HR response). Given CYP2D6 is subject to substrate‐dependent effects, 34 whether the findings herein are generalizable to other substrates remains to be determined.
The uncertainties surrounding CYP2D6 AS and phenotype assignment may be partially explained by additional CYP2D6 variants that influence CYP2D6 activity but are not included on most genotyping platforms. The CYP2D6*2 haplotype is generally considered to display similar activity to the CYP2D6*1 allele. However, Wang et al. reported rs16947, the CYP2D6*2 allele defining SNP, alters exon splicing and is associated with reduced expression and CYP2D6 activity. 23 , 24 The rs5758550 SNP is a CYP2D6 regulatory SNP located in a distant downstream enhancer region (> 100 kb) and interacts with the CYP2D6 promoter to increase CYP2D6 mRNA expression. 24 Increased gene expression conferred by the rs5758850 SNP overcomes the reduced expression of rs16947 resulting in overall “normal” CYP2D6 mRNA expression. 22 , 23 , 24 The rs16947 and rs5758850 SNPs are in high linkage disequilibrium (D′ = 0.9507) in whites. 35 However, there is lower linkage disequilibrium between the two SNPs in blacks (D′ = 0.4645). 35 This presents a potential obstacle for interpreting several rs16947 containing alleles because characterization of functional status in the absence of interrogating the enhancer SNP may lead to overestimation of CYP2D6 activity, particularly in blacks who are less likely to have the enhancer SNP in conjunction with the rs16947 SNP.
As reported herein, consideration of the CYP2D6 enhancer SNP decreases allele AS relative to conventional AS assignment that does not take the enhancer into account, especially in blacks. Current proposals to incorporate rs5758550 into CYP2D6 genotyping panels are limited by the absence of PK and PD data with CYP2D6 substrates. 22 , 23 , 36 In the current study, taking the CYP2D6 enhancer SNP into account when assigning AS with the *2 allele led to more variability being explained in metoprolol CLo, but not PD, across AS compared with conventional CYP2D6 AS assignment methods. Regarding metoprolol PK, superior model fit was observed when CYP2D6 AS, either collapsed into phenotype categories or used as a continuous variable, was considered in the context of the enhancer SNP. Further assessment of the enhancer SNP effects may be warranted, particularly in those of African ancestry, given the potential for overestimating substrate clearance in the absence of its consideration.
The current investigation utilized metoprolol PK and PD data from two prospective clinical trials with similar enrollment criteria among participants with essential hypertension to better elucidate phenotype classifications across the CYP2D6 AS continuum. Notably, a limitation of the study is that PK and PD data were not available within the same study population, and thus, we cannot definitively conclude that the association between reduced CYP2D6 AS and greater HR response to metoprolol was secondary to higher metoprolol concentrations. Another limitation is that while there were no significant differences in ln(CLo) and HR changes between AS of 1 and 1.25–1.5 and no difference in HR between an AS of 1.25–1.5 and 2–2.25, we recognize that these observations may be subject to type 2 error, which cannot be ruled out given the small sample sizes in the individual AS groups. Additionally, the metoprolol total daily dose was inconsistent between studies. Most participants in the PK study received 50 mg total daily dose of metoprolol compared with all PEAR‐2 participants receiving 100 mg total daily dose. To account for this difference, S‐metoprolol apparent oral clearance was selected as the primary PK outcome given its determination is a function of metoprolol dose and AUC.
In conclusion, this study highlights differences in metoprolol PK and PD between an AS of 1 and 2, thereby supporting standardization efforts to dissolve the NM phenotype consisting of AS in the 1–2 range. However, PK and PD parameters for metoprolol were similar between an AS of 1 and 1.25–1.5 and differed between an AS of 1.25–1.5 and 2, suggesting that the AS of 1.25–1.5 results in CYP2D6 metabolic activity toward metoprolol that is more similar to AS of 1 than an AS of 2. Treating CYP2D6 AS as a continuous variable, as opposed to collapsing into metabolizer phenotypes, resulted in a better model fit and explained more interpatient variability in metoprolol oral clearance and change in HR. Additionally, including the enhancer SNP as part of AS determination explained greater variability in metoprolol ln(CLo) compared with conventional‐based AS methods. The data from these two prospective clinical trials reiterates the complex nature of inferring clinical CYP2D6 phenotype from genotype and supports initiatives to implement a CYP2D6 AS‐exclusive method of phenotype determination.
Funding
Funding for this research was made possible, in part, by the Food and Drug Administration through grant 1U01 FD005235. Views expressed do not necessarily reflect the official policies of the Department of Health and Human Services; nor does any mention of drug names or organization imply endorsement by the United States Government. Additional funding support was provided by NIH U01 GM074492, NIH NHGRI 1T32HG008958‐01A1, and NIH NCATS UL1TR001427.
Conflict of Interest
All authors declared no competing interests for this work.
Author Contributions
C.D.T., S.A.M., N.E., T.L., Y.G., D.W., S.S., R.F.F., J.A.J., and L.H.C. wrote the manuscript. T.L., S.O.S., P.B., D.S.E., K.F., H.K., M.K., Z.L., L.F., A.C., R.M.C.D., J.G., L.Z., S.S., R.F.F., J.A.J., and L.H.C. designed the research. C.D.T., S.A.M., N.E., and I.S.H. performed the research. C.D.T., S.A.M., S.K., K.L., N.E., T.L., Y.G., D.W., S.O.S., I.S.H., S.S., R.F.F., J.A.J., and L.H.C. analyzed the data.
Supporting information
Supplementary Material
Table S1
Supplementary Material
Acknowledgments
The authors greatly appreciate Evan Waters and Diane Biernacki for their efforts in coordinating study activities and recruiting study participants. Dr. Thomas is supported by T32 HG008958.
References
- 1. Eichelbaum, M. & Gross, A.S. The genetic polymorphism of debrisoquine/sparteine metabolism–clinical aspects. Pharmacol. Ther. 46, 377–394 (1990). [DOI] [PubMed] [Google Scholar]
- 2. Crews, K.R. et al Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther. 95, 376–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaedigk, A. , Simon, S.D. , Pearce, R.E. , Bradford, L.D. , Kennedy, M.J. & Leeder, J.S. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 83, 234–242 (2008). [DOI] [PubMed] [Google Scholar]
- 4. Swen, J.J. et al Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 5. Bell, G.C. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 102, 213–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown, J.T. et al Clinical Pharmacogenetics Implementation Consortium Guideline for cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin. Pharmacol. Ther. 106, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goetz, M.P. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 103, 770–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hicks, J.K. et al Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hicks, J.K. , Swen, J.J. & Gaedigk, A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metab. 15, 218–232 (2014). [DOI] [PubMed] [Google Scholar]
- 11. Bank, P.C.D. et al Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 103, 599–618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caudle, K.E. et al Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 13, 116–124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson, J.A. & Burlew, B.S. Metoprolol metabolism via cytochrome P4502D6 in ethnic populations. Drug Metab. Dispos. 24, 350–355 (1996). [PubMed] [Google Scholar]
- 14. Blake, C.M. , Kharasch, E.D. , Schwab, M. & Nagele, P. A meta-analysis of CYP2D6 metabolizer phenotype and metoprolol pharmacokinetics. Clin. Pharmacol. Ther. 94, 394–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nozawa, T. et al Influence of CYP2D6 genotype on metoprolol plasma concentration and beta-adrenergic inhibition during long-term treatment: a comparison with bisoprolol. J. Cardiovasc. Pharmacol. 46, 713–720 (2005). [DOI] [PubMed] [Google Scholar]
- 16. Hamadeh, I.S. et al Impact of CYP2D6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin. Pharmacol. Ther. 96, 175–181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Batty, J.A. et al An investigation of CYP2D6 genotype and response to metoprolol CR/XL during dose titration in patients with heart failure: a MERIT-HF substudy. Clin. Pharmacol. Ther. 95, 321–330 (2014). [DOI] [PubMed] [Google Scholar]
- 18. Goryachkina, K. , Burbello, A. , Boldueva, S. , Babak, S. , Bergman, U. & Bertilsson, L. CYP2D6 is a major determinant of metoprolol disposition and effects in hospitalized Russian patients treated for acute myocardial infarction. Eur. J. Clin. Pharmacol. 64, 1163–1173 (2008). [DOI] [PubMed] [Google Scholar]
- 19. Kirchheiner, J. et al Impact of the ultrarapid metabolizer genotype of cytochrome P450 2D6 on metoprolol pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 76, 302–312 (2004). [DOI] [PubMed] [Google Scholar]
- 20. Koytchev, R. et al Influence of the cytochrome P4502D6*4 allele on the pharmacokinetics of controlled-release metoprolol. Eur. J. Clin. Pharmacol. 54, 469–474 (1998). [DOI] [PubMed] [Google Scholar]
- 21. Zineh, I. et al Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin. Pharmacol. Ther. 76, 536–544 (2004). [DOI] [PubMed] [Google Scholar]
- 22. Ray, B. , Ozcagli, E. , Sadee, W. & Wang, D. CYP2D6 haplotypes with enhancer single-nucleotide polymorphism rs5758550 and rs16947 (*2 allele): implications for CYP2D6 genotyping panels. Pharmacogenet. Genomics 29, 39–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang, D. , Papp, A.C. & Sun, X. Functional characterization of CYP2D6 enhancer polymorphisms. Hum. Mol. Genet. 24, 1556–1562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang, D. , Poi, M.J. , Sun, X. , Gaedigk, A. , Leeder, J.S. & Sadee, W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 23, 268–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langaee, T. , Hamadeh, I. , Chapman, A.B. , Gums, J.G. & Johnson, J.A. A novel simple method for determining CYP2D6 gene copy number and identifying allele (s) with duplication/multiplication. PLoS One 10, e0113808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borges, S. et al Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J. Clin. Pharmacol. 50, 450–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Faul, F. , Erdfelder, E. , Lang, A.G. & Buchner, A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007). [DOI] [PubMed] [Google Scholar]
- 28. Yamaoka, K. , Nakagawa, T. & Uno, T. Application of Akaike's information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J. Pharmacokinet. Biopharm. 6, 165–175 (1978). [DOI] [PubMed] [Google Scholar]
- 29. Kim, K.M. et al Pharmacogenetics and healthcare outcomes in patients with chronic heart failure. Eur. J. Clin. Pharmacol. 68, 1483–1491 (2012). [DOI] [PubMed] [Google Scholar]
- 30. Fux, R. et al Impact of CYP2D6 genotype on adverse effects during treatment with metoprolol: a prospective clinical study. Clin. Pharmacol. Ther. 78, 378–387 (2005). [DOI] [PubMed] [Google Scholar]
- 31. Lefebvre, J. , Poirier, L. , Poirier, P. , Turgeon, J. & Lacourciere, Y. The influence of CYP2D6 phenotype on the clinical response of nebivolol in patients with essential hypertension. Br. J. Clin. Pharmacol. 63, 575–582 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rau, T. et al Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics 12, 465–472 (2002). [DOI] [PubMed] [Google Scholar]
- 33. Thomas, C.D. & Johnson, J.A. Pharmacogenetic factors affecting β-blocker metabolism and response. Expert. Opin. Drug Metab. Toxicol.https://doi.org/10.1080/17425255.2020.1803279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou, S.F. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin. Pharmacokinet. 48, 689–723 (2009). [DOI] [PubMed] [Google Scholar]
- 35. Machiela, M.J. & Chanock, S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hahn, J.-Y. et al Effect of P2Y12 inhibitor monotherapy vs dual antiplatelet therapy on cardiovascular events in patients undergoing percutaneous coronary intervention: the SMART-CHOICE randomized clinical TrialP2Y12 monotherapy vs DAPT and cardiovascular events in patients undergoing PCIP2Y12 monotherapy vs DAPT and cardiovascular events in patients undergoing PCI. JAMA 321, 2428–2437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1
Supplementary Material


