Abstract
RecQ family helicases are highly conserved from bacteria to humans and have essential roles in maintaining genome stability. Mutations in three human RecQ helicases cause severe diseases with the main features of premature aging and cancer predisposition. Most RecQ helicases shared a conserved domain arrangement which comprises a helicase core, an RecQ C-terminal domain, and an auxiliary element helicase and RNaseD C-terminal (HRDC) domain, the functions of which are poorly understood. In this study, we systematically characterized the roles of the HRDC domain in E. coli RecQ in various DNA transactions by single-molecule FRET. We found that RecQ repetitively unwinds the 3′-partial duplex and fork DNA with a moderate processivity and periodically patrols on the ssDNA in the 5′-partial duplex by translocation. The HRDC domain significantly suppresses RecQ activities in the above transactions. In sharp contrast, the HRDC domain is essential for the deep and long-time unfolding of the G4 DNA structure by RecQ. Based on the observations that the HRDC domain dynamically switches between RecA core- and ssDNA-binding modes after RecQ association with DNA, we proposed a model to explain the modulation mechanism of the HRDC domain. Our findings not only provide new insights into the activities of RecQ on different substrates but also highlight the novel functions of the HRDC domain in DNA metabolisms.
Keywords: DNA repair, E. coli, RecQ, G-quadruplex, helicase, unwinding, single-molecule
RecQ family helicases play essential roles in genome integrity maintenance by processing a wide variety of DNA structures generated during DNA replication, repair, and recombination (1–3). These proteins are conserved in both prokaryotes and eukaryotes (4). In humans, there are five RecQ helicases: RecQ1, BLM, WRN, RecQ4, and RecQ5. Importantly, mutations in BLM, WRN, and RecQ4 genes cause Bloom, Werner, and Rothmund-Thompson syndromes, which are linked to profound developmental abnormalities and increased cancer risk (5). Meanwhile, the latter two syndromes are also characterized by premature aging. In Escherichia coli, RecQ functions in the RecF recombination pathway to repair the ssDNA gaps and dsDNA breaks (6). E. coli RecQ is also involved in suppressing illegitimate recombination (7), repairing stalled replication forks, and promoting the induction of SOS response (8).
The ability of RecQ family helicases to resolve complex DNA structures is associated with their architecture consisting of evolutionarily conserved domains (9). First, a conserved helicase core is formed by two RecA domains that harbor the ATPase cleft and drive the 3′-5′ directed translocation on ssDNA. Second, the helicase core is followed by a RecQ C-terminal (RQC) domain, which contains a Zn2+ binding domain and a β-hairpin winged-helix domain. RQC is primarily responsible for substrate recognition and DNA unwinding. In addition, an auxiliary element, the helicase, and the RNaseD C-terminal (HRDC) domain, which is connected to RQC by a flexible linker, exist in some RecQ helicases, including E. coli RecQ, human BLM, and WRN. The primary sequences and surface properties of the HRDC domain vary remarkably among different species, and HRDC is even absent in two human RecQ helicases (10–15). These evidences then raise important questions about the functions of HRDC in the various DNA transactions of RecQ family helicases, such as translocating on ssDNA, unwinding dsDNA, and resolving G-quadruplex (G4).
In recent years, the duplex DNA unwinding activities of RecQ family helicases have been widely investigated. Single-molecule studies showed that human BLM unwinds duplex DNA in a highly repetitive fashion by switching between unwinding and rewinding modes (16, 17). Similar phenomena have also been reported in other RecQ helicases, such as human or chicken WRN (18, 19), Arabidopsis thaliana RecQ2 (20), and Caenorhabditis elegans HIM-6 (21), implying that these helicases may use a complex mechanism to unwind duplex DNA rather than the simple unidirectional strand separation. Recently, a very complex dsDNA unwinding behavior with frequent pause, shuttling (22), and co-existence of two unwinding modes (23) has been reported for E. coli RecQ using magnetic tweezers. In addition, HRDC was shown to suppress the rate of DNA-activated ATPase activity in E. coli RecQ (24). However, how E. coli RecQ unwinds duplex DNA when there is no external force, and particularly the modulation mechanism of HRDC, needs further investigation.
In addition to processing duplex DNA, bubble DNA, displacement loops (D-loops), and Holliday junctions, helicases in the RecQ family, such as E. coli RecQ, Cronobacter sakazakii RecQ, yeast sgs1, human BLM, and WRN, also participate in resolving G4 DNA structure (25). As noncanonical nucleic acid structures, G4s can be formed in guanine-rich DNA regions and are implicated in several critical cellular processes, including genomic DNA recombination, replication, and telomere maintenance (26). Both BLM and WRN helicases operate on a wide range of G4 structures with repetitive cycles of unfolding and refolding (18, 27–29). Recently, the widespread existence and potential regulatory roles of G4s have been reported in the E. coli genome (30). However, the detailed mechanism of how E. coli RecQ acts on G4 structures, and particularly how HRDC affects the G4 unwinding activity of RecQ, remains to be determined.
In this study, we characterized the activity of E. coli RecQ on different DNA substrates, including 3′-, 5′-partial duplexes, fork DNA, and G4 DNA, and the roles of HRDC by single-molecule FRET (fluorescence resonance efficiency transfer). Our results indicate that RecQ repetitively unwinds the 3′-partial duplex and fork DNA with moderate processivity. RecQ is also able to periodically patrol on the ssDNA in 5′-partial duplex, extruding DNA loops. In addition, RecQ unfolds G4 structure in a stepwise and repetitive fashion and maintains G4 DNA in an unfolded state for a long time. More importantly, we discovered that HRDC oppositely modulates the unwinding or patrolling activity of RecQ on duplex DNA (suppressing) and G4 structure (enhancing). Based on these results, we proposed models to explain the different modulation mechanism of HRDC in different DNA transactions.
Results
The atomic structures of the E. coli RecQ catalytic core (31), C. sakazakii RecQ catalytic core bound to DNA (32), and human BLM bound to DNA with HRDC (12, 33) were previously resolved (Fig. 1A). The HRDC domain is missing in both RecQ structures; however, in BLM, it folds back onto the core and interacts with both RecA domains (12, 33), suggesting that E. coli and C. sakazakii RecQ may undergo similar interactions. To comprehensively address the role of HRDC in the enzymatic activity of RecQ, in this study, we examined and compared the activities of WT E. coli RecQ (referred to as RecQ herein) and RecQ523 which lacks the HRDC domain on a series of DNA substrates including 3′-, 5′-partial duplexes, fork DNA, and G4 DNA. RecQY555A, which abolished the ssDNA-binding ability of HRDC with a Y555A mutation, was also used to further dissect the effect of the interaction between HRDC and ssDNA (Fig. 1B) (10).
Figure 1.
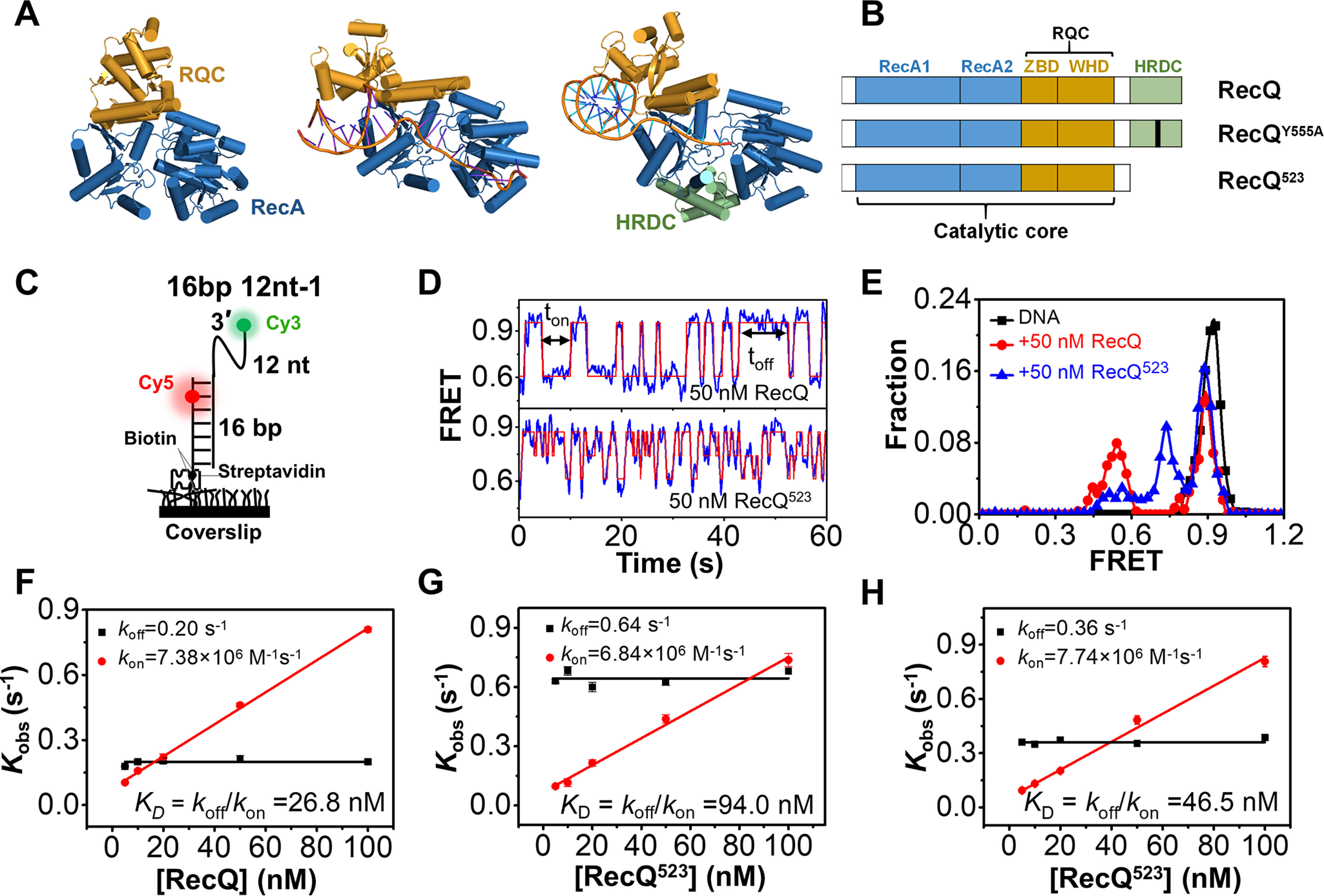
RecQ helicase repetitively associates with and dissociates from the 3′-partial duplex DNA. A, crystal structures of an HRDC-deleted E. coli RecQ (PDB: 1OYW), HRDC-deleted C. sakazakii RecQ in complex with DNA (PDB: 4TMU), and HRDC-containing human BLM in complex with DNA (4O3M). B, domain map of E. coli RecQ constructs used in this study. C, schematic set-up of the smFRET experiment. D, the typical smFRET traces of 16 bp 12 nt-1 in 50 nm RecQ and RecQ523. An automated step-finding algorithm was used to identify the different FRET states (red line). E, FRET histograms of 16 bp 12 nt-1 in 50 nm RecQ or RecQ523 based on the fitted FRET traces. In all the following figures, the FRET histograms were collected from more than 200 traces. F–H, the dissociation rate (koff =1/ton′, red) and the binding rate (kon=1/toff′, black) as a function of protein concentrations. As expected for a binary reaction, the dissociation rate is independent of protein concentration whereas the binding rate has a linear dependence on it. The dissociation constant is thus determined as KD = koff/kon. Error bars denote the standard deviations.
HRDC dynamically interacts with the RecA core and ssDNA overhangs and significantly reinforces RecQ binding on DNA
Before delving into the specific unwinding mechanism of RecQ, we first investigated its DNA-binding activity and the influences of HRDC at the single-molecule level. The substrate (referred to as 16 bp 12 nt-1) contained a 12-nt ssDNA at the 3′ end of a 16-bp duplex and was anchored onto coverslip by the biotin-streptavidin link (Fig. 1C). Cy3 and Cy5 were labeled at the end of the 3′-ssDNA and the fourth nucleotide inside the duplex. 16 bp 12 nt-1 displayed stable FRET efficiency at ∼0.92 (Fig. S1A and Fig. 1E). Upon the addition of RecQ, the FRET trace oscillated frequently between ∼E0.9 and ∼E0.55 in abrupt steps (Fig. S1B; Fig. 1D, upper panel; and Fig. 1E), reflecting the repetitive association and dissociation of RecQ.
HRDC is connected to the remaining portion of RecQ through a long and flexible loop, raising the possibility of dynamic interactions with the RecA core and/or the ssDNA regions of the DNA substrate. In our previous report, we directly labeled the HRDC domain in RecQ by a Cy5 (referred to as Cy5-RecQ) and examined its interaction with different DNA substrates by smFRET (34). We found that the Cy5-labeled HRDC domain can directly interact with the helicase core (Fig. S2, A and B), the 5′-overhang in fork DNA (Fig. S2C), and the free 3′-ssDNA beyond the helicase core (Fig. S2D) (34).
We next addressed the influence of HRDC on the DNA-binding affinity of RecQ. The equilibrium DNA binding assay shown in Fig. S3 indicates that, although HRDC itself can negligibly associate with the partial duplex, the deletion of HRDC significantly attenuated in the binding affinity of RecQ, with the KD value increasing from 16.1 ± 0.4 nm to 91.6 ± 5.8 nm. RecQY555A had an intermediate KD value of 31.3 ± 1.3 nm. Then, we used smFRET to further dissect the differences. Fig. 1D shows that the FRET trace of 16 bp 12 nt-1 in RecQ523oscillated more frequently than that in RecQ. In addition, the RecQ523 association mainly led to an intermediate at E0.74, which may reflect an unstable binding state on DNA (Fig. 1E). We also compared the dwell time ton when helicase remains bound to DNA and the time interval toff between two successive binding events of RecQ, RecQ523, and the mixture of RecQ523 and free HRDC. Both ton and toff followed the single-exponential decay with the average time ton′ and toff′ (Fig. S1, C–E). Although toff′ is almost the same among these three proteins, ton′ of RecQ is much longer, suggesting that RecQ binds to the DNA substrate more tightly than RecQ523. The equilibrium dissociation constant KD can also be obtained based on kon (1/toff′) and koff (1/ton′) (35). Indeed, the WT RecQ had a much lower KD than RecQ523 (Fig. 1, F–H), consistent with the results from fluorescence polarization measurement shown in Fig. S3.
Taken together, the above findings suggest that RecQ523 associates with DNA in an unstable manner and therefore dissociates from the DNA more easily. HRDC can significantly reinforce RecQ binding on DNA by interacting with RecA core and ssDNA overhangs. Indeed, adding excess free HRDC to RecQ523 restored the binding activity to some extent (Fig. S1E, Fig. S3A, and Fig. 1H), confirming the positive effect of the interaction between HRDC and the helicase core in DNA binding.
RecQ repetitively unwinds the 3′-partial duplex and fork DNA with moderate processivity
After examining the DNA-binding activity, we characterized the duplex DNA unwinding mechanism of RecQ and the influences of HRDC. Initially, 16 bp 12 nt-1 was used (Fig. 2A, left panel), and upon addition of 5 nm RecQ and 20 μm ATP, two different types of traces were observed. First, the FRET value dropped from ∼E0.9 to ∼E0.5, representing the binding of RecQ to DNA; after an ∼2.5-s dwell time, the signals of Cy3 and Cy5 disappear almost simultaneously, reflecting the one-step separation of the 16-bp duplex (Fig. 2A and Fig. S4A). Second, the FRET level oscillates until the signals of Cy3 and Cy5 disappear, reflecting complete duplex unwinding (Fig. 2B). The initiation time between RecQ binding and duplex unwinding is referred to as t1 and has an average value of 2.4 s in 20 μm ATP (Fig. 2C). The existence of t1 suggests that, instead of separating the duplex immediately upon arriving at the junction, RecQ may require several seconds to switch into the active unwinding state.
Figure 2.
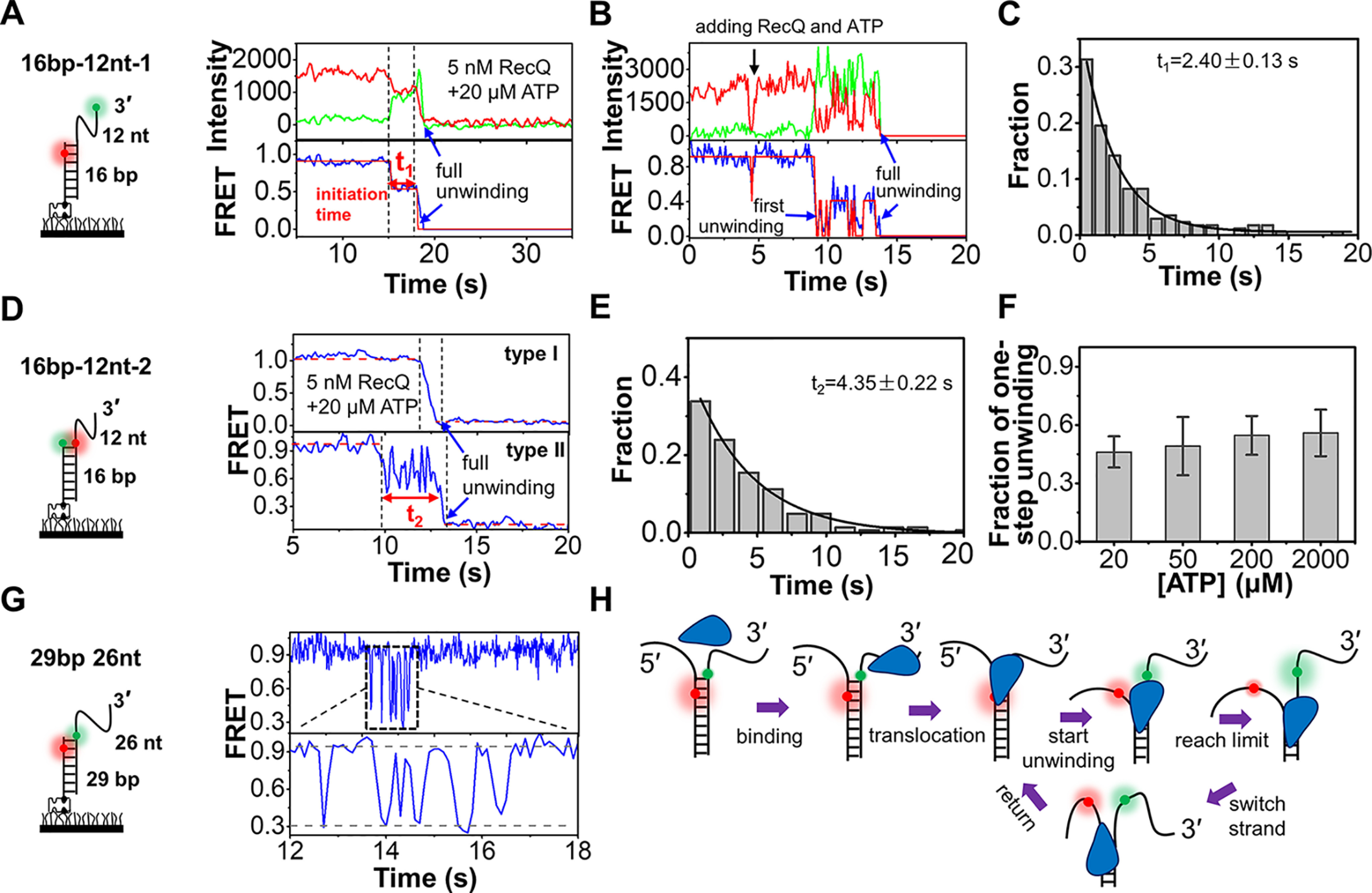
Unwinding of duplex DNA by RecQ. A and B, the typical fluorescence and FRET traces of 16 bp 12 nt-1 in 5 nm RecQ and 20 µm ATP. More reaction traces were exhibited in Fig. S4A. t1 represented the initiation time before duplex unwinding. C, distribution of t1 obtained from single-exponential fitting. In all the following figures, the time histograms were collected from more than 200 traces. D, two typical FRET traces of 16 bp 12 nt-2. Type I represents one-step unwinding, and type II represents repetitive unwinding with total unwinding time t2. E, distribution of t2 obtained from single-exponential fitting. F, the fractions of type I increase with the increase in ATP concentrations. G, repetitive unwinding can be observed in the substrate 29 bp 26 nt. H, a proposed model for RecQ-catalyzed duplex unwinding.
To observe the separation of two DNA strands in the duplex more directly, another substrate 16 bp 12 nt-2 was designed, in which both the donor and acceptor were labeled near the ss/dsDNA junction (Fig. 2D, left panel). Fig. S4B confirmed that the fluorophore on the translocation strand has little effect on RecQ. Upon addition of 5 nm RecQ and 20 μm ATP, both one-step unwinding (type I) and repetitive unwinding (type II) were observed (Fig. 2D), consistent with the phenomena associated with 16 bp 12 nt-1. We used t2 to represent the time taken in the repetitive unwinding. With increases in ATP concentration, the fractions of one-step unwinding were slightly increased (Fig. 2F). The co-existence of the two types may be attributed to the duplex length of 16 bp 12 nt-1 and 16 bp 12 nt-2 being close to the unwinding limit of RecQ. Therefore, in some cases, RecQ can unwind the duplex in one-step abruptly, whereas in other cases, RecQ may reach the limit and reverse the direction. To verify our speculation, another 3′-partial duplex with a 29-bp stem was designed (Fig. 2G). In 5 nm RecQ and 20 μm ATP, continuous FRET fluctuations were observed in most cases, reflecting the repetitive unwinding by RecQ. With the increases in ATP concentration (Fig. S4C), the unwinding fractions were significantly increased. Nevertheless, most traces still displayed the repetitive unwinding by RecQ before the complete unwinding (Fig. S4D), reflecting the moderate processivity of RecQ even at 2 mm ATP. In addition, we discovered that fork DNA was also repetitively unwound by RecQ (Fig. S5A). The processivity was over 20 bp in most cases, and RecQ may only occasionally arrive at the position beyond 28 bp (Fig. S5, B–E).
Fig. 2H demonstrates the proposed mechanism of RecQ-catalyzed duplex unwinding. First, RecQ associates with the 3′-partial duplex or fork DNA at the 3′-ssDNA. Driven by ATP, RecQ translocates to the ss/dsDNA junction. Then, after a short initiation time, RecQ starts to unwind the duplex at a rapid speed. Once RecQ reaches the limit, it may loosen the tracking strand and switch to the 5′-ssDNA (23), translocate or slide back with the reannealing of the two strands, and then repetitively unwind the duplex.
HRDC domain suppresses the duplex DNA unwinding activity of RecQ
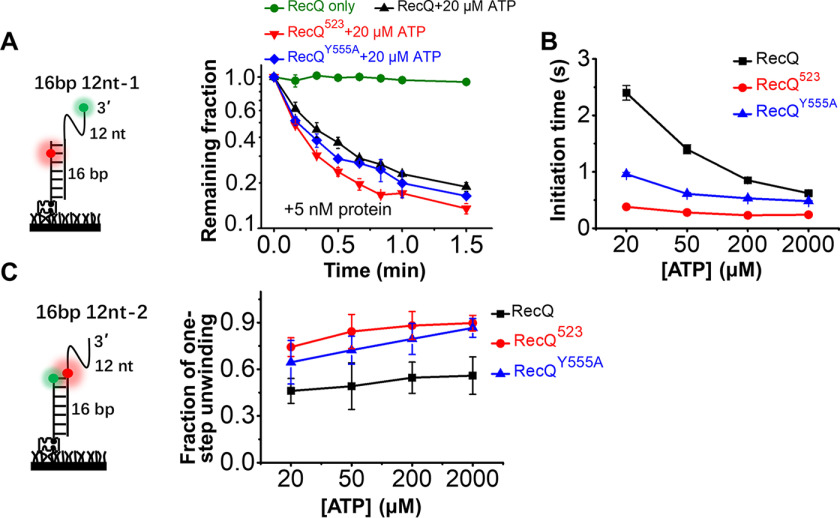
To address the influence of HRDC on the duplex unwinding activity of RecQ, we directly measured the unwinding fractions of 16 bp 12 nt-1 and 16 bp 12 nt-2 by counting the number of Cy5 spots over time, as previously described (36). The remaining fractions with time in Fig. 3A and Fig. S6A both reflect that RecQ523 displays a higher efficiency than RecQ in unwinding the 16-bp duplex.
Figure 3.
HRDC suppresses the duplex unwinding activity of RecQ. A, fractions of remaining 16 bp 12 nt-1 molecules on coverslip versus time after addition of 5 nm RecQ and 20 µm ATP. Lines are the simple connections of the individual data points by Origin 8.0. B, the initiation time of 16 bp 12 nt-1 in 5 nm RecQ, RecQ523, and RecQY555A in different concentrations of ATP. C, the fractions of 16 bp 12 nt-2 traces showing one-step unwinding in 5 nm RecQ, RecQ523, and RecQY555A in different concentrations of ATP.
We next analyzed the FRET traces of 16 bp 12 nt-1 and 16 bp 12 nt-2 in the three types of RecQ. Under our experimental conditions, HRDC has a negative effect on RecQ unwinding initiation (Fig. 3B). For instance, the initiation time at 20 μm ATP is much longer for RecQ than for RecQ523 (2.40 ± 0.13 s versus 0.39 ± 0.05 s; Fig. 2C and Fig. S6B). In addition, the fractions of one-step unwinding in 16 bp 12 nt-2 by RecQ523 are much higher than by RecQ (Fig. 3C), i.e. the number of traces showing repetitive unwinding was greatly reduced without HRDC. The duration of repetitive unwinding in 16 bp 12 nt-2 by RecQ523 was also significantly reduced compared with that by RecQ (1.13 s ± 0.03 versus 4.35 ± 0.22 s, Fig. S6C and Fig. 2E).
Taken together, the above evidence indicates that the HRDC domain suppresses RecQ unwinding activity on duplex DNA mainly by increasing the unwinding initiation time and promoting repetitive unwinding. Importantly, RecQY555A, which abolishes the ssDNA-binding ability of HRDC, displays an activity level between that of the WT RecQ and RecQ523 (Fig. 3). Therefore, we speculate that the interactions of HRDC with the helicase core and with the ssDNA overhang both contribute to the weakened unwinding activity of RecQ by suppressing the ATP hydrolysis rate (24) and promoting RecQ switching to the displaced strand (as indicated by a comparison of the fractions from the one-step unwinding by RecQ and RecQY555A).
RecQ periodically patrols on the 5′-ssDNA overhang in 5′-partial duplex
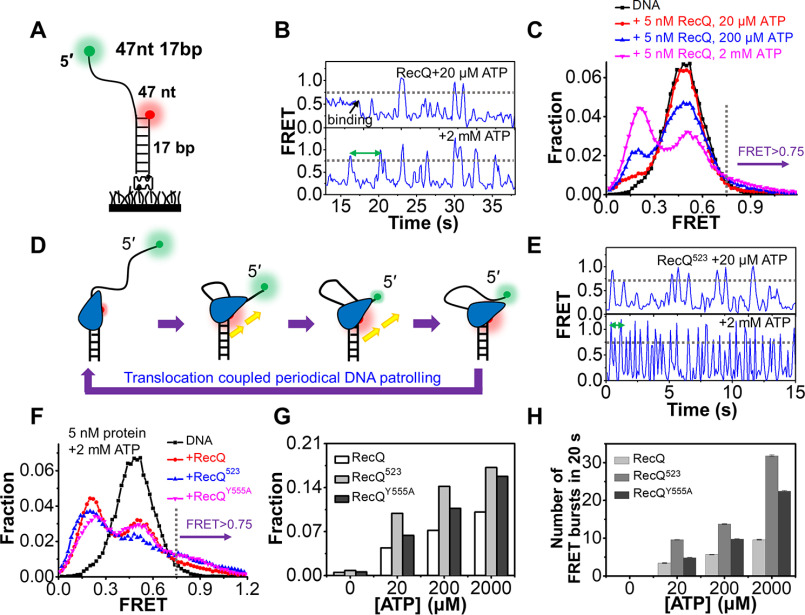
After systematically characterizing the unwinding of 3′-partial duplex and fork DNA by RecQ, we further examined the activity of RecQ on the 5′-partial duplex with the 47 nt 17 bp substrate (Fig. 4A), in which Cy3 and Cy5 were labeled at the end of the 5′-overhang and the junction. Our previous studies have detected that the RecQ family helicases BLM (37) and WRN (38) both possess the activity of anchoring at the 3′ ss/dsDNA junction and reeling in ssDNA by its 3′-5′ translocation activity with the 47 nt 17 bp substrate. DNA alone showed a stable EFRET at ∼0.5 (Fig. 4B, upper panel, and Fig. 4C). Upon addition of 5 nm RecQ and 20 μm ATP, the FRET value first dropped to ∼0.2, reflecting the stretching of the ssDNA by RecQ, and then it occasionally increased to the values greater than 0.75, reflecting the looping of the 5′-ssDNA. At 2 mm ATP, the FRET bursts occurred more frequently (Fig. 4B, lower panel). Fig. S7 further suggests that the repetitive FRET fluctuations should be caused by the same helicase because excess protein was removed in the chamber. The FRET distributions of 47 nt 17 bp in 5 nm RecQ and 20 μm-2 mm ATP are shown in Fig. 4C. Compared with DNA substrate alone, a new population at E0.21 emerged, consistent with the FRET decrease caused by RecQ binding in Fig. 4B. There were also additional populations at higher FRET values, which were likely caused by the transient looping of the ssDNA.
Figure 4.
RecQ periodically patrols on the 5′-ssDNA. A, the 5′-partial duplex 47 nt 17 bp contains a 17 bp duplex and a 47 nt 5′-ssDNA. B, the typical FRET traces of 47 nt 17 bp in 5 nm RecQ and indicated ATP concentrations. C, FRET distributions of 47 nt 17 bp only and in 5 nm RecQ and different concentrations of ATP. D, a proposed action of RecQ on the 5′-partial duplex. E, the typical FRET traces of 47 nt 17 bp in 5 nm RecQ523 and indicated ATP concentrations. F, FRET distributions of 47 nt 17 bp in 5 nm RecQ, RecQ523, and RecQY555A with 2 mm ATP. G, the fractions of DNA at looping states (FRET > 0.75) in different reaction conditions. H, the number of FRET bursts within the 20-s time window in 5 nm RecQ, RecQ523, and RecQY555A with indicated ATP concentration.
Based on the above observations, we hypothesized that RecQ may anchor at the ss/dsDNA junction while translocating on the 5′-overhang in the 3′-5′ direction, thereby extruding an ssDNA loop. Upon arrival at the end of the 5′-ssDNA, RecQ may release the strand and restart a new cycle of translocation (Fig. 4D). In addition, there is a 3–5-s interval between each FRET burst at 2 mm ATP (indicated by the green arrow in Fig. 4B), suggesting that, after releasing ssDNA, RecQ requires a short time to restart a new cycle of ssDNA scanning.
HRDC domain significantly suppresses the periodical patrolling of RecQ on 5′-partial duplex
Next, we examined the influence of HRDC on the periodical patrolling activity of RecQ on the 5′-partial duplex. In 5 nm RecQ523 and 20 μm or 2 mm ATP, the FRET traces show similar bursts as that induced by RecQ, though with a much higher frequency (Fig. 4E). The FRET values may increase to different levels possibly because of the release of ssDNA by RecQ before it reaches the 5′-end; however, most of the bursts were greater than E0.75 (Fig. 4, B and E). Therefore, to better quantify the fractions of DNA in the looping state as shown in Fig. 4F, an artificial threshold was set at E0.75, above which loops were presumed to be extruded. Fig. 4G shows that with increases in ATP concentration, the fractions of DNA with EFRET values above 0.75 increased significantly. More importantly, they were highest in RecQ523 and lowest in RecQ at each ATP concentration (Fig. 4G), reflecting the much stronger periodical patrolling activity of RecQ523. The number of FRET bursts in the 20-s time window was also quantified (Fig. 4H). The frequency of looping events increased significantly with the increases in ATP concentration. Moreover, the frequencies were much higher in RecQ523 than in RecQ, highlighting the extraordinary patrolling activity of RecQ523.
By comparing the FRET traces in Fig. 4, B and E, we noticed that the time intervals between individual FRET bursts were much lower in RecQ523 than in RecQ at both low and high ATP concentrations. This evidence indicates that HRDC significantly prolongs the time for RecQ to restart the next cycle of translocation, consistent with the negative effect of HRDC on the initiation of duplex DNA unwinding, as shown in Fig. 3B. Because both the initiation time in duplex unwinding and time interval in periodical patrolling depend on ATP concentration, the existence and duration of these times may be related to ATP hydrolysis, because HRDC suppresses ATP hydrolysis by interacting with the RecA core (24), leading to a longer unwinding initiation and patrolling restart time. RecQY555A displays a medium level of activity between RecQ and RecQ523 (Fig. 4, F–H), suggesting that the interaction of HRDC with the 5′-ssDNA also contributes to the decrease in the periodical patrolling activity of RecQ. It is possible that HRDC dynamically binds onto the 5′-ssDNA ahead of RecQ, inhibiting RecQ translocation on the strand to some extent.
RecQ unfolds G4 structure in a stepwise and repetitive fashion and maintains G4 DNA in an unfolded state for a long time
Because the E. coli genome indeed includes considerable amounts of G4s that may play important roles in critical cellular processes (30, 39), here we further investigated RecQ-catalyzed G4 unfolding with or without the HRDC domain. Unexpectedly, RecQ displayed a very poor affinity toward the G4 structure with the KD value of 658.2 ± 79 nm (Fig. S8A). Fig. S8A further shows that a 3′-ssDNA overhang is indispensable for RecQ to efficiently associate with the G4 DNA. Therefore, we speculated that RecA1 and RecA2 domains in RecQ may first bind to the 3′-ssDNA region (∼10 nt), anchoring the helicase onto the substrate, and then RQC can interact with the G4 structure, consistent with the structure of C. sakazakii RecQ in complex with G4 DNA (25).
Next, we carried out smFRET unwinding assay with the substrate 29 bp-G4 12 nt, in which the G4 motif was linked with a 29-bp duplex at its 5′ end and a 12-nt ssDNA at its 3′ end (Fig. 5A) as previously reported (29). Cy3 was labeled at the nucleotide between the G4 motif and 3′ tail, and Cy5 was labeled 6 bp inside the duplex. The fluorophores were so spaced that the FRET signal can sensitively report the conformational change of G4 (29). In 100 mm KCl, the FRET value of 29 bp-G4 12 nt remained at a stable level at ∼0.9 (Fig. S8B), reflecting the well-folding of the G4 structure. Therefore, this buffer condition was used for further RecQ-catalyzed unfolding experiments.
Figure 5.
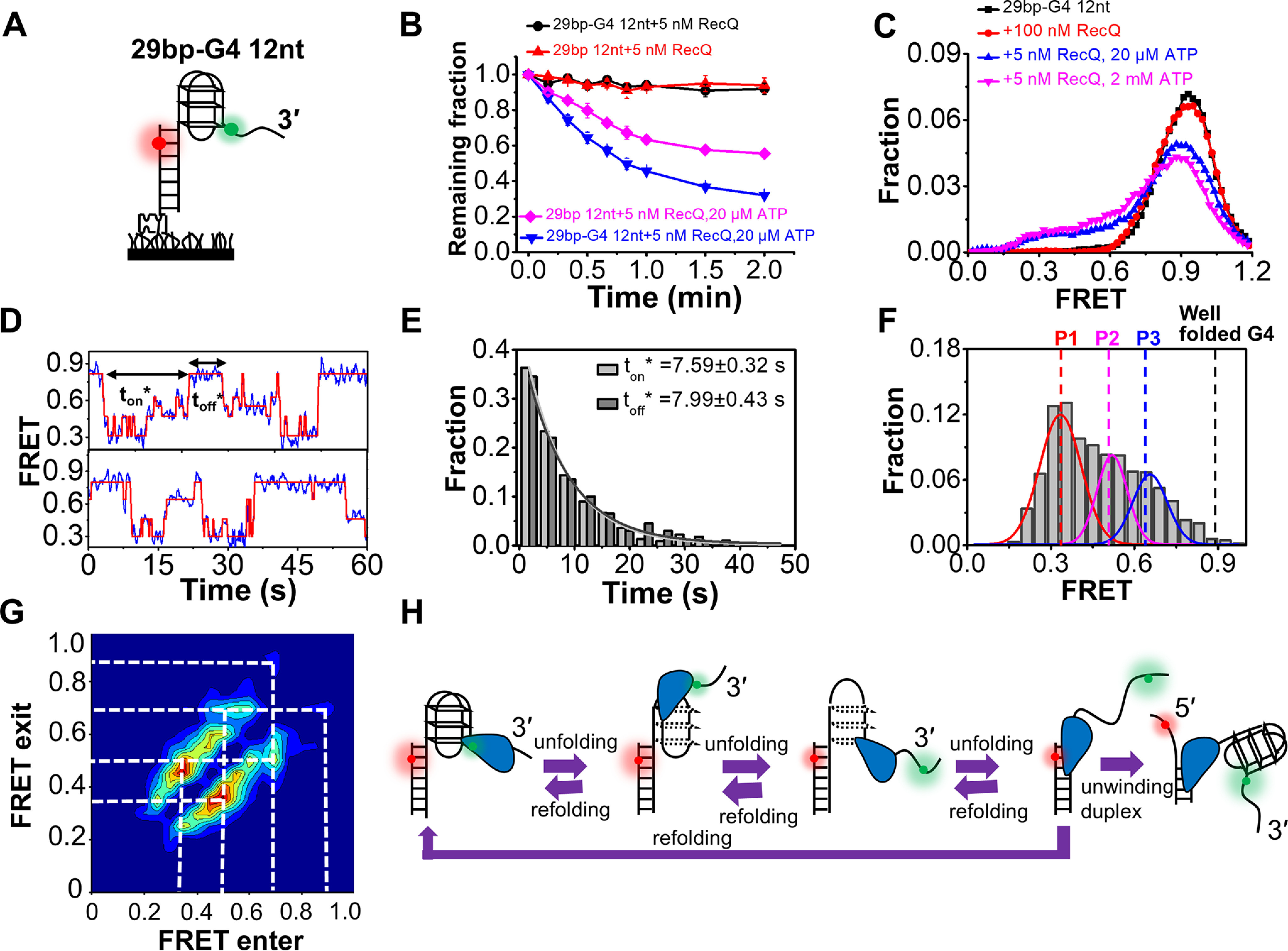
RecQ can unfold the G4 DNA structure. A, the G4-containing DNA substrate 29 bp-G4 12 nt. B, fractions of remaining DNA molecules on coverslip versus time after the addition of RecQ and ATP. C, FRET distributions of G4 substrate in different concentrations of RecQ and ATP. D, in 5 nm RecQ and 20 µm ATP, the FRET values of 29 bp-G4 12 nt fluctuate between different levels. The automated step-finding algorithm was used to identify the individual steps (red line) during G4 unfolding and refolding. The time for the continuous FRET oscillations was defined as ton*, and the time interval between two lasting oscillations was defined as toff*. E, distributions of ton* and toff*. F, distributions of the FRET oscillation regions of 29 bp-G4 12 nt in 5 nm RecQ and 20 µm ATP. G, transition density plot for oscillation regions of 29 bp-G4 12 nt from ∼200 FRET oscillation regions. H, schematic diagram of our model to explain how RecQ unfolds the G4 structure repetitively.
The fractions of remaining DNA molecules versus the time after the addition of 5 nm RecQ and different concentrations of ATP were determined. Fig. 5B indicates that RecQ should be able to unfold the G4 structure in the presence of ATP; otherwise, the downstream duplex cannot be unwound (Fig. 5B). Then, the FRET traces of 29 bp-G4 12 nt after the addition of different concentrations of RecQ and ATP were recorded and analyzed (Fig. 5C). In the absence of ATP, no change was detected in the FRET distributions even at 100 nm RecQ. However, at 5 nm RecQ and 20 μm ATP, the population at ∼E0.9 decreased significantly, accompanied by an increase in low-range FRET populations, reflecting the disruption of the G4 structure. Both pieces of evidence indicate that RecQ-catalyzed G4 unfolding was ATP-dependent. Fig. 5D shows that the FRET traces in 5 nm RecQ and 20 μm ATP switch among at least four states, suggesting the disruption and dynamic conversion of G4 structure between different folding states (the duration time was defined as ton*). Then, the FRET value returns to the original level, likely because of the dissociation of RecQ. After a short interval, toff*, another cycle of similar FRET fluctuation begins. The distributions of both ton* and toff* follow the single-exponential decay with an average time of ∼8 s (Fig. 5E). To further understand the different unfolding states, we selected the fluctuation regions within ton* from ∼200 traces and plotted the FRET histograms (Fig. 5F). Three peaks can be discriminated: the leftmost peak at E0.33 could be the ssDNA (29), whereas the other two peaks, at E0.52 and E0.65, may represent the proposed G-hairpin and G-triplex structures, respectively (29, 40–42). The transition density plot in Fig. 5G further indicates that the transitions between the above states are reversible; i.e. the completely or partially unfolded G4 can refold back to a more complete state whereas RecQ remains associated with the G4 motif. Fig. S9 further confirms the G4 unfolding activity of RecQ with a substrate in which the G4 motif was at the 5′ end of the partial duplex.
A reasonable interpretation of the observed FRET oscillation is presented in Fig. 5H. RecQ unfolds the G4 structure into ssDNA in a stepwise manner with at least two intermediate states, G-triplex and G-hairpin. It is worth noting that this is a simplified model, highlighting that there are multiple states in the dynamic interaction between RecQ and G4; however, the specific structures of those states still need further ascertainment by other methods. In addition, our results further show that RecQ can maintain the G4 structures in unfolded states for a relatively long time (∼8 s at 20 μm ATP), and it may be alike to that observed from FANCJ helicase, which can recognize G-quadruplexes and mediate their longstanding stepwise unfolding in repeating cycles (43). However, according to previous reports, the G4 structure was transiently unfolded by Pif1 (∼1 s) (40) and DHX36, BLM, WRN (∼2 s) (27, 44) during the quick and frequent switching between well-folded and unfolded states.
The HRDC domain is essential for the complete and long-time unfolding of the G4 DNA structure by RecQ
We next examined the influence of HRDC on the G4-unfolding activity of RecQ. First, the fractions of 29 bp-G4 12 nt on coverslip versus the time after the addition of 5 nm helicase and 2 mm ATP were determined (Fig. 6A). Although RecQ displayed the lowest duplex unwinding activity among the three proteins (Fig. 3A), the unwinding of 29 bp-G4 12 nt by RecQ was more efficient than by RecQ523, suggesting that HRDC plays a positive role in G4 unfolding.
Figure 6.
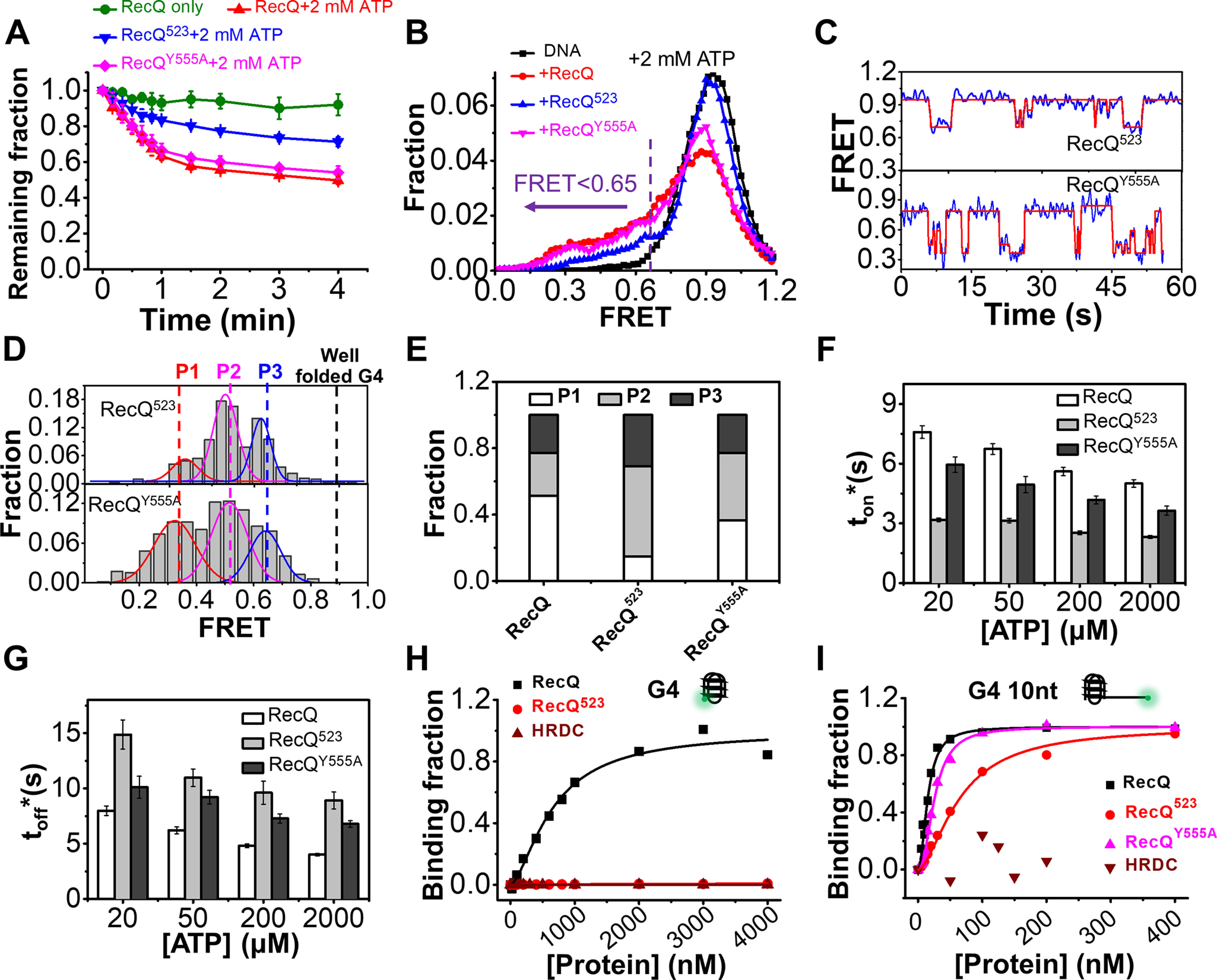
The HRDC domain of RecQ is necessary for the efficient unfolding of the G4 structure. A, fractions of remaining 29 bp-G4 12 nt molecules on coverslip versus time after the addition of 5 nm protein and 2 mm ATP. B, FRET distributions of 29 bp-G4 12 nt in 2 mm ATP and different RecQ constructs. According to the FRET peaks in Fig. 5F, a criterion at E0.65 was set artificially, below which the G4 structure was recognized as being disrupted. C, in 5 nm RecQ523 or RecQY555A and 20 µm ATP, the FRET values of 29 bp-G4 12 nt fluctuate between different levels. D, distributions of the FRET oscillation regions of the G4 substrate from ∼100 traces. E, the fractions of P1, P2, and P3 in the FRET histograms of G4 substrate in different types of RecQ. F and G, histograms of ton* and toff* in RecQ, RecQ523, and RecQY555A. H and I, RecQ constructs binding to G4 (H) or G4 10 nt (I) measured by equilibrium DNA binding assay. The dissociation constant (KD) of RecQ bound to G4 is 658.2 ± 79 nm; KD of RecQ523 and HRDC bound to G4 were both not available. KD of RecQ, RecQ523, and RecQY555A bound to G4 10 nt were 15.1 ± 1.1 nm, 63.7 ± 4.1 nm, and 25.6 ± 1 nm, respectively; KD of HRDC bound to G4 was not available.
Afterward, FRET traces of 29 bp-G4 12 nt after the addition of 5 nm RecQ and 2 mm ATP were analyzed. In all three types of RecQs, the FRET distributions of 29 bp-G4 12 nt showed shifts to the lower band (Fig. 6B), indicating the unfolding of G4. To quantify the differences between them, an artificial threshold at E0.65 corresponding to the G-triplex state in Fig. 5F was set, below which G4 structure is considered as partially or completely unfolded. Fig. S8C shows that the unfolding fractions by RecQ are much higher than by RecQ523, highlighting the importance of HRDC in G4 unfolding. The FRET traces in 5 nm RecQ523 and 20 μm ATP were consistent with that in RecQ, with the FRET value switching between different states (Fig. 6C). Then, we selected the regions showing oscillations and constructed FRET histograms (Fig. 6D). The P1 state corresponding to ssDNA is the lowest in RecQ523 (Fig. 6E, 14% versus 52% in RecQ). Instead, in RecQ523, most of the molecules are at P2 (55%) and P3 (41%) state, which may correspond to G-hairpin and G-triplex; i.e. RecQ523 most likely disrupts G4 into partially unfolded states and can barely disrupt G4 completely.
We also compared the unwinding time ton* and the time interval toff* of FRET traces in the three types of RecQs (Fig. 6, F and G). Under the same experimental conditions, ton* in RecQ was at least 2-fold longer than that in RecQ523, indicating that RecQ523 was more prone to dissociate from the G4 substrate after partially disrupting G4. On the other hand, toff* in RecQ was shorter than that in RecQ523; i.e. RecQ523 takes a longer time to reassociate with the G4 substrate and restart the unfolding.
The differences between RecQ and RecQ523 reflect that the HRDC domain substantially promotes G4 unfolding by increasing the duration time of each unfolding event in parallel with increasing the degree of G4 disruption. Unexpectedly, the G4 unfolding activity of RecQY555A is very similar to that of the WT RecQ (Fig. 6), implying that the interaction between HRDC and ssDNA may have very little impact. To further dissect whether HRDC directly interacts with the G4 structure and whether RecQ523 binds G4 differently than RecQ, we measured the binding affinity between those proteins and the G4 structure. Fig. 6, H and I indicates that, although HRDC itself cannot interact with the G4 structure, both RecQ and RecQY555A bind to the G4 substrate with 3′-ssDNA much stronger than RecQ523. Therefore, the reinforcement of the association of RecQ on the G4 substrate may be mainly caused by the interaction between HRDC and the helicase core.
Discussion
Proposed roles of the HRDC domain in regulating the helicase activity of E. coli RecQ on different nucleic acid substrates
The functions of HRDC in duplex DNA unwinding have been studied previously. Harami et al. (24) reported that HRDC in E. coli RecQ suppresses the rate of DNA-activated ATPase activity in parallel with those of ssDNA translocation and dsDNA unwinding. Using magnetic tweezers, the same group then discovered that HRDC mediates pausing and shuttling during hairpin DNA unwinding (22). Later, Bagchi et al. (23) reported that RecQ unwinds hairpin DNA using a fast mode of continuous unwinding and a slow mode of persistent random walking, and the deletion of HRDC diminished the slow mode. In our current smFRET study without external forces, RecQ mainly displays repetitive unwinding/rewinding activity on 3′-tailed duplex and fork DNA; therefore, we focused on the influence of HRDC on the repetitive unwinding behavior of RecQ. More importantly, we also carried out an in-depth analysis of the effect of HRDC on the repetitive ssDNA patrolling and G4 unfolding activities of RecQ.
We suggest that HRDC suppresses the duplex DNA unwinding activity of RecQ with the proposed role as shown in Fig. 7A. First, HRDC increases the initiation time of RecQ for duplex unwinding (Fig. 3B). The increase in initiation time with the existence of HRDC is possibly because HRDC dynamically interacts with the RecA core, thereby inhibiting ATP hydrolysis (24). Because the HRDC binding site is near the ATP binding cleft of RecA domains, it is also possible that ATP binding or ADP and/or Pi release is inhibited (24). Second, more frequent repetitive unwinding was observed with RecQ whereas more unidirectional unwinding was observed with RecQ523 or RecQY555A (Fig. 3C). This is attributed to HRDC's dynamic interaction with the 5′-ssDNA, which promotes the strand-switching activity of RecQ.
Figure 7.
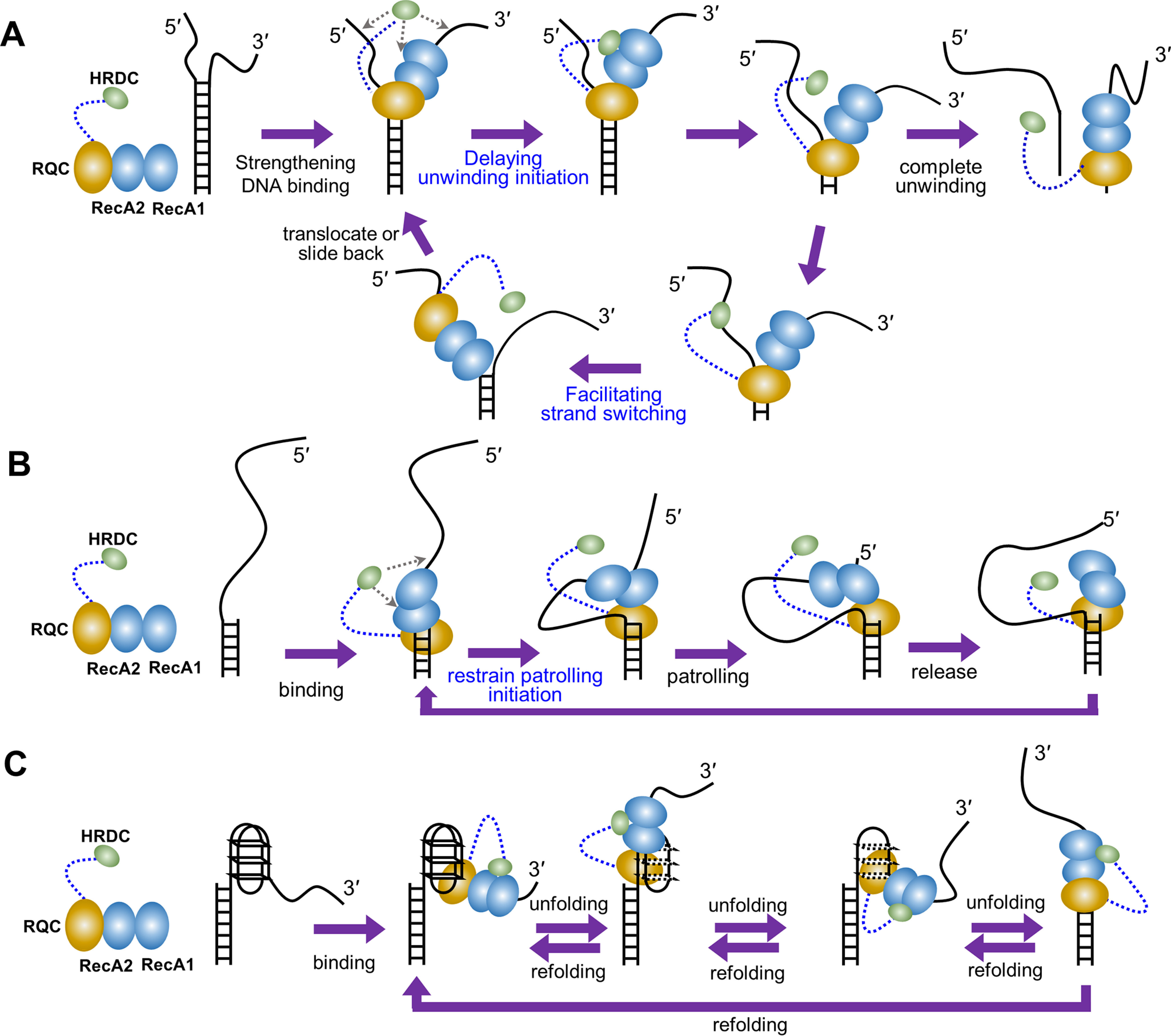
The proposed modulation mechanism of the HRDC domain. A, in duplex DNA unwinding, HRDC delays the unwinding initiation of RecQ possibly by inhibiting ATP hydrolysis, ATP binding, and ADP and/or Pi release due to the interaction with the RecA core. Moreover, HRDC promotes the strand-switch of RecQ by dynamically binding to the 5′-ssDNA, thereby inhibiting the unidirectional unwinding. B, in the periodically patrolling process, HRDC restrains the patrolling initiation of RecQ possibly by inhibiting ATP hydrolysis, ATP binding, and ADP and/or Pi release due to the interaction with the RecA core. C, in G4 unfolding, HRDC reinforces the association of RecQ on the DNA by interacting with the RecA core, leading to the complete and long-time G4 unfolding.
HRDC also significantly suppresses the repetitive ssDNA patrolling activity of RecQ on a 5′-tailed duplex. The major difference between RecQ and RecQ523 is in the patrolling frequency and burst width (Fig. 4), which are related to ATP concentration, suggesting that HRDC may slow down the switch of RecQ into an active translocation state and the translocation rate by inhibiting ATP binding, ATP hydrolysis, or ADP and/or Pi release (24), as mentioned above. It is also possible that the dynamic binding of HRDC to the ssDNA ahead of RecQ inhibits its translocation initiation, thus partially contributing to the reduced patrolling frequency (Fig. 7B).
In sharp contrast with the inhibiting effect of HRDC on duplex unwinding and repetitive ssDNA patrolling, our results demonstrate that HRDC is essential for the complete and long-time unfolding of G-quadruplex DNA by RecQ. Considering that the G4 unfolding activity of RecQY555A is very similar to that of RecQ but more efficient than RecQ523 (Fig. 6), the positive effect of HRDC during G4 unfolding should be mainly caused by the interaction of HRDC with the RecA core. Therefore, we speculate that HRDC is crucial for RecQ to proceed with G4 unfolding by reinforcing the association of RecQ with the DNA substrate through interacting with the RecA core, thus ensuring the complete unfolding of the G4 structure (Fig. 7C).
It is worth noting that the HRDC domain in Neisseria gonorrhoeae RecQ has been previously reported to improve G4 unfolding (45); however, this RecQ helicase has three tandem HRDCs, leading to very complex and different functions compared with other RecQ helicases. Indeed, both N. gonorrhoeae RecQ and its version with two HRDCs deleted bind to G4 relatively well with KD = 55.1 and 86.5 nm (45), respectively; however, E. coli RecQ binds to G4 very poorly with KD = 658.2 nm (RecQ523 can barely bind to G4), and a 3′-ssDNA is required for the RecA domains to bind first, anchoring the helicase onto the substrate. Moreover, N. gonorrhoeae RecQ takes ∼211 s to unfold 50% of the G4 structures, reflecting a very poor G4 unfolding activity (45). Altogether, there are essential differences between these two helicases, and we think that E. coli RecQ is an ideal helicase to address the in-depth functional mechanism of HRDC because most RecQ family members only have one HRDC.
HRDC may play a role in mediating the cooperative binding of RecQ onto DNA substrates
Different RecQ family members may exhibit different oligomeric states in solution. As for E. coli RecQ, our previous studies (46, 47) found that it is monomeric in solution up to a concentration of 20 μm; this property is not affected by the presence of ATP. Although RecQ unwinds DNA as a monomer, our further study (48) and Kowalczykowski group's research (22) both found that multiple E. coli RecQ monomers can cooperate to unwind long DNA substrates, dependent on the protein concentration. In this study, we mainly used 5 nm protein concentration to detect the binding initiation, dsDNA unwinding, 5′-ssDNA translocation, and G4 unwinding process; therefore, we treated RecQ as a monomer, similarly to the previous single-molecule studies (22, 23).
We have noticed that the HRDC domain not only significantly enhances the binding affinity between RecQ and DNA substrate by interacting with the RecA core and ssDNA overhangs but also increases the cooperativity between different RecQ molecules in DNA binding. As shown in Fig. S3, the Hill coefficient is the lowest for RecQ523 among the three helicases (RecQ, RecQ523, and RecQY555A). Meanwhile, adding free HRDC to RecQ523 has little effect on the Hill coefficient (Fig. S3). Therefore, we speculate that, because of the long flexible linker, HRDC might also be able to dynamically interact with the RecA core of another RecQ nearby, leading to the relatively high cooperativity of RecQ molecules in DNA binding. It is also possible that multiple RecQ monomers may unwind the duplex DNA and G4 DNA with the cooperative translocation at high protein concentration.
Potential biological significances of HRDC in modulating RecQ activities in DNA repair
Our results indicate that HRDC suppresses the dsDNA unwinding activity of RecQ although it reinforces RecQ binding to DNA. In addition, HRDC significantly promotes the repetitive unwinding of the duplex by RecQ with a moderate processivity. Because E. coli RecQ is a central DNA recombination and repair helicase, the above observations suggest that HRDC may play a positive role in improving the precision and efficiency when RecQ removes the short duplex invasions in D-loops formed in the illegitimate homologous recombination (7). What's more, because HRDC can significantly strengthen RecQ binding to the G4 substrate and ensure complete and long-time unfolding of G4 structure, we speculate that HRDC may be crucial for some key biological processes mediated by RecQ such as repairing stalled replication forks induced by G4 (30, 39). In brief, our study reveals that the auxiliary structural component HRDC differentially modulates the activities of E. coli RecQ in processing different DNA structures by dynamically interacting with the RecA core and ssDNA and provides new insights into the functioning of RecQ during DNA replication, recombination, and repair.
Experimental procedures
Buffers
The RecQ reaction buffer contained 50 mm KCl, 2 mm MgCl2, 1 mm DTT, and 0.1 mg/ml BSA in 20 mm Tris-HCl, pH 7.5, unless otherwise specified. For single-molecule measurements, 0.8% D-glucose, 1 mg/ml glucose oxidase (266,600 units/g, Sigma-Aldrich), 0.4 mg/ml catalase (2,000-5,000 units/mg, Sigma-Aldrich), and 1 mm Trolox (Sigma-Aldrich) were added to the reaction buffer.
DNA constructs
All oligonucleotides required to generate DNA substrates were purchased from Sangon Biotech (Shanghai, China). The sequences and labeling positions of all the oligonucleotides are listed in Table S1. For DNA constructs used in single-molecule measurements, DNA was annealed with a 1:3 mixture of stem and ssDNA or G4 strands by incubating the mixture at 95 °C for 5 min, then slowly cooling down to room temperature in about 7 h. The strand without biotin was used in excess to reduce the possibility of having nonannealed strands anchored to the coverslip surface. The concentration of the stem strand was 2.5 μm, and all annealing was carried out in the annealing buffer containing 100 mm KCl and 20 mm Tris-HCl, pH 7.5. The partial duplex DNA used in the equilibrium DNA binding assay was annealed with a 1:1 mixture of the two strands.
Protein expression, purification, and labeling
The expression and purification of E. coli RecQ, RecQ523, and RecQY555A were carried out as described previously (49). For simplicity, we refer to E. coli RecQ as RecQ herein. In brief, each RecQ construct was cloned into the pET15b-SUMO vector and expressed in BL21 (DE3) induced by 0.3 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 16 h, respectively. Then, the recombinant protein was severally purified by Ni affinity chromatography; after being digested by SUMO protease at 4 °C overnight, each RecQ construct was purified again by Ni affinity chromatography to remove His6 and SUMO tag. The protein purity was more than 95% determined by SDS-PAGE as described previously (49). The protein concentration was more than 5 mg/ml determined by UV280 using Thermo Fisher Scientific Nanodrop 2000c. When measuring the protein concentration, the molar extinction coefficients of RecQ, RecQY555A, RecQ516, and HRDC were 48,820 m−1cm−1, 47,330 m−1cm−1, 44,560 m−1cm−1, and 2,560 m−1cm−1, respectively.
Because RecQ contains 11 cysteine residues, it is difficult to specifically label the HRDC domain with a single fluorophore. To avoid nonspecific labeling, we depended on the flexible linker (∼22 aa) between the RQC and HRDC domains to establish a scheme based on sortase A ligation (50), as in our previous report (34). In brief, recombinant protein NH2-GGG-HRDCE610C labeled with a single Cy5 fluorophore (Lumiprobe Corporation, Hunt Valley, MD, USA) and RecQ1–516-LPETG were separately prepared and then ligated by sortase A in ligation buffer (50 mm Tris-HCl, pH 7.0, 150 mm NaCl, and 20 mm CaCl2) at 34 °C for 1 h. The final ligated protein was purified and stored at −80 °C.
Single-molecule fluorescence data acquisition
The smFRET assay was performed as described previously (51). 5 nm protein concentration was mainly used in our experiment unless otherwise specified. Imaging was initiated before RecQ and ATP were flowed into the chamber. We used an exposure time of 100 ms for all measurements at a constant temperature of 22 °C. To determine the fractions of unwound DNA with time, a series of 1-s movies were recorded at different times, and the Cy5 spots were counted to represent the number of remaining DNA molecules.
FRET data analysis
The FRET efficiency was calculated using E = IA/(ID+IA), where ID and IA represent the intensities of the donor and acceptor, respectively. Basic data analysis including transition density plot was carried out by scripts written in MATLAB. All data fitting was conducted with Origin 8.0. An automated step-finding method (from http://bio.physics.illinois.edu/HaMMy.asp) was used to characterize RecQ's association with and dissociation from partial duplex DNAs and the stepwise patterns observed in G4 unfolding. The histograms of FRET efficiency and dwell time from more than 300 molecules were fitted with multi-peak Gaussian distribution and single-exponential decay, respectively.
Equilibrium DNA binding assay
Binding of RecQ to DNA was analyzed by fluorescence polarization assay using Infinite F200 PRO (Tecan, Männedorf, Switzerland) at a constant temperature of 25 °C (52). DNA labeled with FAM was used in this study (Table S1). Varying amounts of protein were added to a 150-μl aliquot of binding buffer containing 5 nm DNA. Each sample was allowed to equilibrate for 5 min, and the fluorescence polarization value was then measured. The binding curve was fitted by the Hill equation: y = [RecQ]n/(KDn + [RecQ]n), where y is the binding fraction, n is the Hill coefficient, and KD is the apparent dissociation constant.
Data availability
All data are contained within this article and in the supporting information.
Supplementary Material
Acknowledgments
We thank Dr. Wen-Qiang Wu at Henan University for insightful discussions.
This article contains supporting information.
Funding and additional information—This work was supported by the National Natural Science Foundation of China Grants 32071225, 31870788, and 32071291, the Research Startup Funding of the Affiliated Hospital of Southwest Medical University Grant 18102, Scientific Research Funding of Luzhou-Southwest Medical University Grants 2019LZXNYDJ06 and 2018-ZRZD-003, and the Chinese Universities Scientific Fund Grant Z109021718. The research was conducted within the context of the International Associated Laboratory “Helicase-mediated G-quadruplex DNA unwinding and Genome Stability.”
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- RQC
- RecQ C-terminal
- HRDC
- helicase and RNaseD C-terminal
- smFRET
- single-molecule fluorescence resonance energy transfer
- G4
- G-quadruplex.
References
- 1. Brosh R. M. (2013) DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer 13, 542–558 10.1038/nrc3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vindigni A., Marino F., and Gileadi O. (2010) Probing the structural basis of RecQ helicase function. Biophys. Chem. 149, 67–77 10.1016/j.bpc.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 3. Croteau D. L., Popuri V., Opresko P. L., and Bohr V. A. (2014) Human RecQ helicases in DNA repair, recombination, and replication. Ann. Rev. Biochem. 83 83, 519–552 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Larsen N. B., and Hickson I. D. (2013) RecQ helicases: Conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 973, 161–184 10.1007/978-1-4614-5037-5_8 [DOI] [PubMed] [Google Scholar]
- 5. Mohaghegh P., Karow J. K., Brosh R. M., Bohr V. A., and Hickson I. D. (2001) The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849 10.1093/nar/29.13.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Handa N., Morimatsu K., Lovett S. T., and Kowalczykowski S. C. (2009) Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 23, 1234–1245 10.1101/gad.1780709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanada K., Ukita T., Kohno Y., Saito K., Kato J., and Ikeda H. (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94, 3860–3865 10.1073/pnas.94.8.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hishida T., Han Y. W., Shibata T., Kubota Y., Ishino Y., Iwasaki H., and Shinagawa H. (2004) Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 18, 1886–1897 10.1101/gad.1223804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vindigni A., and Hickson I. D. (2009) RecQ helicases: multiple structures for multiple functions? HFSP J. 3, 153–164 10.2976/1.3079540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernstein D. A., and Keck J. L. (2005) Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13, 1173–1182 10.1016/j.str.2005.04.018 [DOI] [PubMed] [Google Scholar]
- 11. Kim Y. M., and Choi B. S. (2010) Structure and function of the regulatory HRDC domain from human Bloom syndrome protein. Nucleic Acids Res. 38, 7764–7777 10.1093/nar/gkq586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Newman J. A., Savitsky P., Allerston C. K., Bizard A. H., Özer O., Sarlós K., Liu Y., Pardon E., Steyaert J., Hickson I. D., and Gileadi O. (2015) Crystal structure of the Bloom's syndrome helicase indicates a role for the HRDC domain in conformational changes. Nucleic Acids. 43, 5221–5235 10.1093/nar/gkv373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato A., Mishima M., Nagai A., Kim S. Y., Ito Y., Hakoshima T., Jee J. G., and Kitano K. (2010) Solution structure of the HRDC domain of human Bloom syndrome protein BLM. J. Biochem. 148, 517–525 10.1093/jb/mvq097 [DOI] [PubMed] [Google Scholar]
- 14. Kitano K., Yoshihara N., and Hakoshima T. (2007) Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J. Biol. Chem. 282, 2717–2728 10.1074/jbc.M610142200 [DOI] [PubMed] [Google Scholar]
- 15. Liu Z., Macias M. J., Bottomley M. J., Stier G., Linge J. P., Nilges M., Bork P., and Sattler M. (1999) The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure 7, 1557–1566 10.1016/S0969-2126(00)88346-X [DOI] [PubMed] [Google Scholar]
- 16. Yodh J. G., Stevens B. C., Kanagaraj R., Janscak P., and Ha T. (2009) BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. Embo J. 28, 405–416 10.1038/emboj.2008.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S., Qin W., Li J. H., Lu Y., Lu K. Y., Nong D. G., Dou S. X., Xu C. H., Xi X. G., and Li M. (2015) Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase. Nucleic Acids Res. 43, 3736–3746 10.1093/nar/gkv209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu W. Q., Hou X. M., Zhang B., Fossé P., René B., Mauffret O., Li M., Dou S. X., and Xi X. G. (2017) Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 7, 43954 10.1038/srep43954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee M., Shin S., Uhm H., Hong H., Kirk J., Hyun K., Kulikowicz T., Kim J., Ahn B., Bohr V. A., and Hohng S. (2018) Multiple RPAs make WRN syndrome protein a superhelicase. Nucleic Acids Res. 46, 4689–4698 10.1093/nar/gky272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klaue D., Kobbe D., Kemmerich F., Kozikowska A., Puchta H., and Seidel R. (2013) Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat. Commun. 4, 2024 10.1038/ncomms3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi S., Lee S. W., Kim H., and Ahn B. (2019) Molecular characteristics of reiterative DNA unwinding by the Caenorhabditis elegans RecQ helicase. Nucleic Acids Res. 47, 9708–9720 10.1093/nar/gkz708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harami G. M., Seol Y., In J., Ferencziová V., Martina M., Gyimesi M., Sarlós K., Kovács Z. J., Nagy N. T., Sun Y., Vellai T., Neuman K. C., and Kovács M. (2017) Shuttling along DNA and directed processing of D-loops by RecQ helicase support quality control of homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 114, E466–E475 10.1073/pnas.1615439114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bagchi D., Manosas M., Zhang W., Manthei K. A., Hodeib S., Ducos B., Keck J. L., and Croquette V. (2018) Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB. Nucleic Acids Res. 46, 8500–8515 10.1093/nar/gky647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harami G. M., Nagy N. T., Martina M., Neuman K. C., and Kovács M. (2015) The HRDC domain of E-coli RecQ helicase controls single-stranded DNA translocation and double-stranded DNA unwinding rates without affecting mechanoenzymatic coupling. Sci. Rep. 5, 11091 10.1038/srep11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voter A. F., Qiu Y., Tippana R., Myong S., and Keck J. L. (2018) A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat. Commun. 9, 4201 10.1038/s41467-018-06751-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bochman M. L., Paeschke K., and Zakian V. A. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tippana R., Hwang H., Opresko P. L., Bohr V. A., and Myong S. (2016) Single-molecule imaging reveals a common mechanism shared by G-quadruplex-resolving helicases. Proc. Natl. Acad. Sci. U. S. A. 113, 8448–8453 10.1073/pnas.1603724113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chatterjee S., Zagelbaum J., Savitsky P., Sturzenegger A., Huttner D., Janscak P., Hickson I. D., Gileadi O., and Rothenberg E. (2014) Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 5, 5556 10.1038/ncomms6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu W. Q., Hou X. M., Li M., Dou S. X., and Xi X. G. (2015) BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 43, 4614–4626 10.1093/nar/gkv361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaplan O. I., Berber B., Hekim N., and Doluca O. (2016) G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-hairpin-duplex switch. Nucleic Acids Res. 44, 9083–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernstein D. A., Zittel M. C., and Keck J. L. (2003) High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 22, 4910–4921 10.1093/emboj/cdg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manthei K. A., Hill M. C., Burke J. E., Butcher S. E., and Keck J. L. (2015) Structural mechanisms of DNA binding and unwinding in bacterial RecQ helicases. Proc. Natl. Acad. Sci. U. S. A. 112, 4292–4297 10.1073/pnas.1416746112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swan M. K., Legris V., Tanner A., Reaper P. M., Vial S., Bordas R., Pollard J. R., Charlton P. A., Golec J. M., and Bertrand J. A. (2014) Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA. Acta Crystallogr. D. Biol. Crystallogr. 70, 1465–1475 10.1107/S139900471400501X [DOI] [PubMed] [Google Scholar]
- 34. Teng F.-Y., Jiang Z.-Z., Huang L.-Y., Guo M., Chen F., Hou X.-M., Xi X.-G., and Xu Y. (2020) A toolbox for site-specific labeling of RecQ helicase with a single fluorophore used in the single-molecule assay. Front. Mol. Biosci. 7, 586450 10.3389/fmolb.2020.586450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Markiewicz R. P., Vrtis K. B., Rueda D., and Romano L. J. (2012) Single-molecule microscopy reveals new insights into nucleotide selection by DNA polymerase I. Nucleic Acids Res. 40, 7975–7984 10.1093/nar/gks523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang B., Wu W. Q., Liu N. N., Duan X. L., Li M., Dou S. X., Hou X. M., and Xi X. G. (2016) G-quadruplex and G-rich sequence stimulate Pif1p-catalyzed downstream duplex DNA unwinding through reducing waiting time at ss/dsDNA junction. Nucleic Acids Res. 44, 8385–8394 10.1093/nar/gkw669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu W. Q., Hou X. M., Li M., Dou S. X., and Xi X. G. (2015) BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 43, 4614–4626 10.1093/nar/gkv361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu W. Q., Hou X. M., Zhang B., Fossé P., René B., Mauffret O., Li M., Dou S. X., and Xi X. G. (2017) Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci. Rep. 7, 43954 10.1038/srep43954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teng F. Y., Hou X. M., Fan S. H., Rety S., Dou S. X., and Xi X. G. (2017) Escherichia coli DNA polymerase I can disrupt G-quadruplex structures during DNA replication. FEBS J. 284, 4051–4065 10.1111/febs.14290 [DOI] [PubMed] [Google Scholar]
- 40. Hou X. M., Wu W. Q., Duan X. L., Liu N. N., Li H. H., Fu J., Dou S. X., Li M., and Xi X. G. (2015) Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 466, 189–199 10.1042/BJ20140997 [DOI] [PubMed] [Google Scholar]
- 41. Zhou R., Zhang J., Bochman M. L., Zakian V. A., and Ha T. (2014) Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 3, e02190 10.7554/eLife.02190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hou X. M., Fu Y. B., Wu W. Q., Wang L., Teng F. Y., Xie P., Wang P. Y., and Xi X. G. (2017) Involvement of G-triplex and G-hairpin in the multi-pathway folding of human telomeric G-quadruplex. Nucleic Acids Res. 45, 11401–11412 10.1093/nar/gkx766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu C. G., and Spies M. (2016) G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 44, 8742–8753 10.1093/nar/gkw574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen M. C., Tippana R., Demeshkina N. A., Murat P., Balasubramanian S., Myong S., and Ferré-D'Amaré A. R. (2018) Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469 10.1038/s41586-018-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cahoon L. A., Manthei K. A., Rotman E., Keck J. L., and Seifert H. S. (2013) Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J. Bacteriol. 195, 2255–2261 10.1128/JB.02217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu H. Q., Deprez E., Zhang A. H., Tauc P., Ladjimi M. M., Brochon J. C., Auclair C., and Xi X. G. (2003) The Escherichia coli RecQ helicase functions as a monomer. J. Biol. Chem. 278, 34925–34933 10.1074/jbc.M303581200 [DOI] [PubMed] [Google Scholar]
- 47. Zhang X. D., Dou S. X., Xie P., Hu J. S., Wang P. Y., and Xi X. G. (2006) Escherichia coli RecQ is a rapid, efficient, and monomeric helicase. J. Biol. Chem. 281, 12655–12663 10.1074/jbc.M513089200 [DOI] [PubMed] [Google Scholar]
- 48. Li N., Henry E., Guiot E., Rigolet P., Brochon J. C., Xi X. G., and Deprez E. (2010) Multiple Escherichia coli RecQ helicase monomers cooperate to unwind long DNA substrates: a fluorescence cross-correlation spectroscopy study. J. Biol. Chem. 285, 6922–6936 10.1074/jbc.M109.069286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kocsis Z. S., Sarlós K., Harami G. M., Martina M., and Kovács M. (2014) A nucleotide-dependent and HRDC domain-dependent structural transition in DNA-bound RecQ helicase. J. Biol. Chem. 289, 5938–5949 10.1074/jbc.M113.530741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mao H., Hart S. A., Schink A., and Pollok B. A. (2004) Sortase-mediated protein ligation: a new method for protein engineering. J. Am. Chem. Soc. 126, 2670–2671 10.1021/ja039915e [DOI] [PubMed] [Google Scholar]
- 51. Wu W. Q., Zhu X., and Song C. P. (2019) Single-molecule technique: a revolutionary approach to exploring fundamental questions in plant science. New Phytol. 223, 508–510 10.1111/nph.15928 [DOI] [PubMed] [Google Scholar]
- 52. Dou S. X., Wang P. Y., Xu H. Q., and Xi X. G. (2004) The DNA binding properties of the Escherichia coli RecQ helicase. J. Biol. Chem. 279, 6354–6363 10.1074/jbc.M311272200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article and in the supporting information.