Abstract
Identification of antibody-binding epitopes is crucial to understand immunological mechanisms. It is of particular interest for allergenic proteins with high cross-reactivity as observed in the lipid transfer protein (LTP) syndrome, which is characterized by severe allergic reactions. Art v 3, a pollen LTP from mugwort, is frequently involved in this cross-reactivity, but no antibody-binding epitopes have been determined so far. To reveal human IgE-binding regions of Art v 3, we produced three murine high-affinity mAbs, which showed 70–90% coverage of the allergenic epitopes from mugwort pollen–allergic patients. As reliable methods to determine structural epitopes with tightly interacting intact antibodies under native conditions are lacking, we developed a straightforward NMR approach termed hydrogen/deuterium exchange memory (HDXMEM). It relies on the slow exchange between the invisible antigen-mAb complex and the free 15N-labeled antigen whose 1H-15N correlations are detected. Due to a memory effect, changes of NH protection during antibody binding are measured. Differences in H/D exchange rates and analyses of mAb reactivity to homologous LTPs revealed three structural epitopes: two partially cross-reactive regions around α-helices 2 and 4 as well as a novel Art v 3–specific epitope at the C terminus. Protein variants with exchanged epitope residues confirmed the antibody-binding sites and revealed strongly reduced IgE reactivity. Using the novel HDXMEM for NMR epitope mapping allowed identification of the first structural epitopes of an allergenic pollen LTP. This knowledge enables improved cross-reactivity prediction for patients suffering from LTP allergy and facilitates design of therapeutics.
Keywords: allergen, monoclonal antibody, epitope mapping, NMR spectroscopy, lipid transfer protein, hydrogen/deuterium exchange, Art v 3, mugwort pollen, allergen, epitope mapping, nuclear magnetic resonance (NMR), immunoglobulin E (IgE), immunoglobulin G (IgG)
Interaction of antibodies with their antigen-binding sites (epitopes) is vital to maintain health but may also contribute to immunological diseases. To understand these interactions, determination of binding specificities and precise epitope localization is a crucial but not trivial task (1). A reliable technique to investigate binding of an antigen with its respective antibody is X-ray crystallography. This, however, typically requires production and extensive testing of truncated antibody fragments that need to generate high-quality (co-)crystals (2, 3). More recently, MS-based approaches have used hydrogen/deuterium exchange to reveal binding regions (4, 5). Differences in exchange rates can be measured at the peptide mass level, but resolution is typically poor, because it mostly depends on the accessibility of proteolytic cleavage sites. Another option i.e. screening of immobilized synthetic peptides covering the entire antigen sequence is straightforward but limited to analysis of continuous, linear epitopes (6, 7).
A powerful technique for mapping protein interactions is NMR spectroscopy using chemical shift deviations (8). However, the large molecular size of the mAb-antigen complexes prevents a straightforward application. Analogous to X-ray crystallography, so far Fab fragments of the mAbs obtained by enzymatic cleavage are a typical prerequisite for analyses (9). However, Fab fragments often show low stability, and although the size of the Fab-antigen complex is still large and challenging for NMR measurements, it was successful in several cases. Typically, additional deuteration of the protein and special NMR techniques like TROSY experiments are required (10). Whereas this methodology seems successful for NMR epitope mapping (11–16), it relies on the availability of isolated Fab fragments and works best with interactions under a fast-exchange regime. Whereas weak interactions typically with a fast-exchange regime can be well-analyzed, tight interactions with low koff rates are challenging. Moderately tight interactions involving large proteins can be studied by cross-saturation experiments like chemical exchange saturation transfer or dark-state exchange saturation transfer as long as there is sufficient exchange during the NMR experiment, ideally with exchange time scales between 10 ms and 1 s (17, 18). However, these methods rely on exchange between the free and the bound form, and they fail if the koff rates are much slower than a scan of the NMR experiment. For interactions with antibodies, deuteration of the 15N-labeled binding partner largely improves the performance (16). Another strategy for tight interactions involves weakening of affinity e.g. by the use of detergents (15). However, such additions might influence the results, as the fold and stability of the antibody and antigen can be affected. NMR-detected H/D exchange has also been used for interaction mapping with antibodies, either based on directly observable fragments (19, 20) or on a tedious protocol involving quenching the H/D exchange, separating the antigen from the mAb and detecting the amount of NH in spectra of the separated antigen for each time point (20, 21). Considering all mentioned applications, we currently lack a straightforward, high-resolution method for determination of structural epitopes of antigens interacting very tightly with intact antibodies under native conditions.
Identification of antibody-binding epitopes is required to efficiently study IgE cross-reactivity, design novel vaccines, and monitor allergen immunotherapies (22). At present, more than 100 tertiary structures of allergens have been solved, but information on their antibody-binding epitopes is very limited (23). This also applies to nonspecific lipid transfer proteins (LTPs) that were identified as allergens in food, pollen, and latex (6, 24). LTPs are small, nonglycosylated proteins with a typical α-helical fold stabilized by disulfide bonds (25). Due to their high thermal and proteolytic stability, they are able to trigger severe allergic reactions. Clinical manifestations ranging from mild oral symptoms to anaphylaxis are described as LTP syndrome (26–29). The complex clinical picture is based on IgE cross-reactivity, which can cause allergic reactions to multiple LTP-containing sources because of high structural similarities of the underlying allergens (30–34). However, besides cross-reactive LTP epitopes, additional source-specific IgE epitopes do exist (35, 36), which contributes to the complexity of this syndrome. The only pollen LTP with IgE cross-reactivity relevant to this syndrome is Art v 3 from mugwort pollen. Our recent study revealed that patients' IgE and murine IgG recognize exclusively conformational antibody-binding epitopes (25), suggesting a folded protein during allergic sensitization (37, 38). So far, no information on the localization of antibody-binding sites accounting for the IgE (cross)-reactive epitopes of Art v 3 is available.
Thus, Art v 3 serves as an excellent representative to study structural epitopes using high-affinity antibodies. For this purpose, we developed a novel NMR-based approach termed hydrogen/deuterium-exchange memory (HDXMEM). This method utilizes the equilibrium of free and mAb-bound protein, which results in reduced H/D exchange rates of antibody-bound regions due to a memory effect. To reveal antigen-antibody interaction at the residue level, murine monoclonal antibodies that covered relevant human IgE-binding epitopes of Art v 3 were used for mapping. Together with reduced antibody reactivity to Art v 3 epitope variants, distinct residues relevant for IgE binding and LTP cross-reactivity were determined.
Results
Art v 3–specific murine mAbs recognize structural epitopes
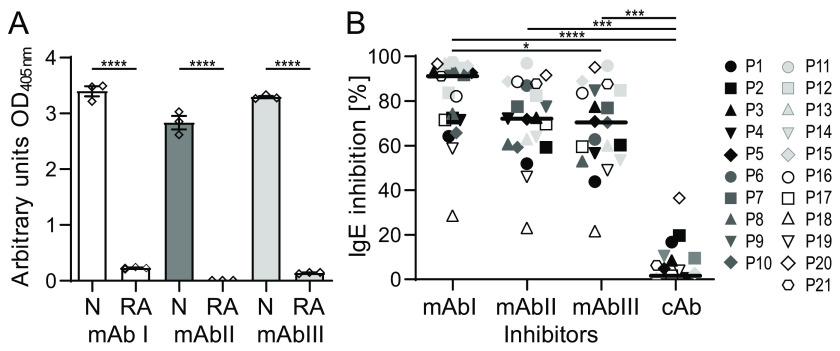
To generate mAbs, mice were immunized with recombinant Art v 3.0201 (for simplicity termed Art v 3). We produced the nontagged allergen using Escherichia coli Rosetta-gami cells enabling disulfide bond formation relevant for the LTP fold (25). The protein showed the correct identity and high purity (>98%) as verified by gel electrophoresis and MS (Fig. S1, A and B). Recombinant Art v 3 was subcutaneously administered six times, and murine spleens were harvested after 56 days (Fig. S1C). After generating hybridoma cells, single B cells were obtained by limiting dilution. Three clones with high affinity to Art v 3 were selected, and mAbs were purified using protein G columns (Fig. S1D). We finally obtained 10–12 mg of each Art v 3–specific IgG1 antibody (i.e. mAb I, mAb II, and mAb III). All mAbs efficiently bound the native protein in ELISA, whereas no reactivity was observed when Art v 3 was reduced and alkylated (Fig. 1A). This suggests that the obtained mAbs recognize structural epitopes on the disulfide bond–stabilized structure.
Figure 1.
Characterization and IgE inhibition capacities of Art v 3–specific mAbs. A, binding of mAbs I, II, and III to native Art v 3 (N) or reduced and alkylated Art v 3 (RA) was determined by ELISA. Measurements were performed in triplicates; means and S.E. (error bars) are given. B, IgE binding of allergic patients (P1–P21) to immobilized Art v 3 inhibited by the respective monoclonal antibodies was determined by ELISA. An unrelated anti-Amb a 1 antibody was used as control Ab (cAb). Black lines, medians. ****, p ≤ 0.0001; ***, p ≤ 0.001; *, p ≤ 0.05.
Affinity and driving forces of allergen recognition by individual mAbs
To characterize the binding affinity between Art v 3 and the mAbs, we used isothermal titration calorimetry (ITC) and surface acoustic wave (SAW) measurements. Highly comparable binding affinities ranging from 5.5 to 75.6 nm were observed with both techniques (Fig. 2). Recognition by mAb I generated a large enthalpy gain, but the binding had to overcome an entropic barrier (positive −TΔS). Interaction with mAb II again generated a smaller enthalpy gain than entropy gain, resulting in a very similar ΔG and thus KD compared with mAb I. Interaction with mAb III showed a slightly lower affinity and was mainly entropy-driven. These results suggest that despite similar affinities, the antibody-binding mechanisms on the molecular level must be quite different. Interestingly, the SAW data revealed extremely slow binding kinetics (Fig. S2A). Dissociation rate constants (koff) extracted from fitting the decay phases were in the range of 10−5 to 10−4 s−1 for the three mAbs, corresponding to half-lives on the order of several hours (Fig. S2B).
Figure 2.
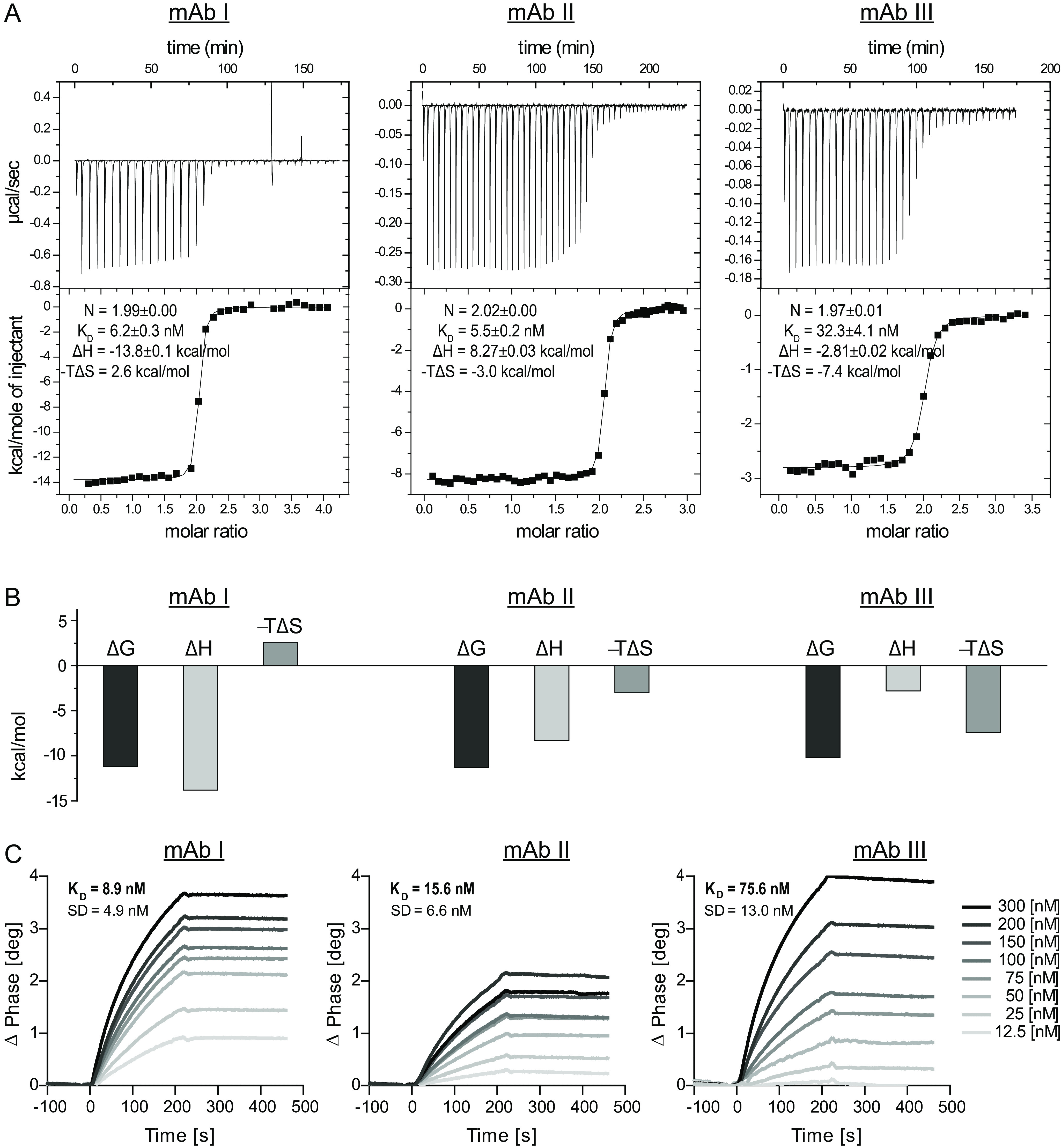
Isothermal titration calorimetry and surface acoustic wave data of the interaction between Art v 3 and mAb I, mAb II and mAb III. A, raw data and integrated heats as a function of the molar ratio with the best fit. B, illustration of the thermodynamic components ΔG, ΔH, and −TΔS for each of the interactions. C, representative binding curves between the respective mAb and Art v 3 monitored by surface acoustic wave technology. Average KD values and S.D. of 12 channels are given.
Binding of mAb overlaps with human IgE epitopes from mugwort pollen–allergic patients
To determine whether purified murine mAbs cover relevant human IgE-binding epitopes, we performed ELISAs using sera of 21 mugwort pollen–allergic patients. All sera were positive to Art v 3, whereas IgE binding to homologous LTPs revealed diverse and patient-specific sensitization patterns (Table S1). In ELISA, preincubation with mAb I reduced on average 90% (range 29–98%) of IgE binding. Both, mAb II and mAb III led to a decrease in IgE reactivity of around 70%. The specificity of the assay was verified using an unrelated mAb as control (Fig. 1B). Based on these data, our mAbs qualify as surrogate antibodies for mapping structural IgE-binding regions of Art v 3.
Confirming the structural integrity of Art v 3 in solution by sequence-specific NMR assignment
To verify whether Art v 3 adopts the typical LTP fold in solution and to subsequently map epitopes on the amino acid level, we used NMR spectroscopy to sequence-specifically assign the backbone chemical shifts. This was achieved using recombinant 13C15N-labeled Art v 3 (Fig. S1, A and B) and standard triple-resonance experiments HNCA, HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO (39) at 278 and 298 K. An extensive buffer exchange to phosphate buffer, pH 6.0, was required to obtain a homogeneous protein preparation, as judged from the 15N HSQC spectrum (Fig. S3) providing a protein fingerprint, in which every amino acid is represented by one signal. The chemical shift assignment determined at 298 K was used to predict the secondary structure of Art v 3 in solution (Fig. S4). Localization of α-helices is in agreement with regions determined in the crystal structure (PDB entry 6FRR), thus confirming the correct folding of this Art v 3 preparation in solution.
Evaluating traditional NMR approaches for mapping the binding sites
To evaluate whether traditional NMR titration studies are feasible to map binding epitopes, as suggested by Razzera et al. (40), a series of 15N HSQC spectra was used to monitor the binding of the individual mAbs to the 15N-labeled allergen. This resulted in a gradual disappearance of Art v 3 signals, indicative of a slow exchange regime and a formed complex that was beyond the size limit of NMR spectroscopy, resulting in the disappearance of all NMR signals. Neither changes in peak positions nor changes in line widths were observed (Fig. S5), which could have been used for epitope mapping. Instead, the signal intensity of all NH signals decreased uniformly for all residues with increasing mAb concentration. In summary, interactions of Art v 3 with mAbs could not be mapped by traditional two-dimensional NMR titrations because no chemical shift perturbations or line broadening effects were observed.
HDXMEM identified three structural epitopes on distinct surface regions of Art v 3
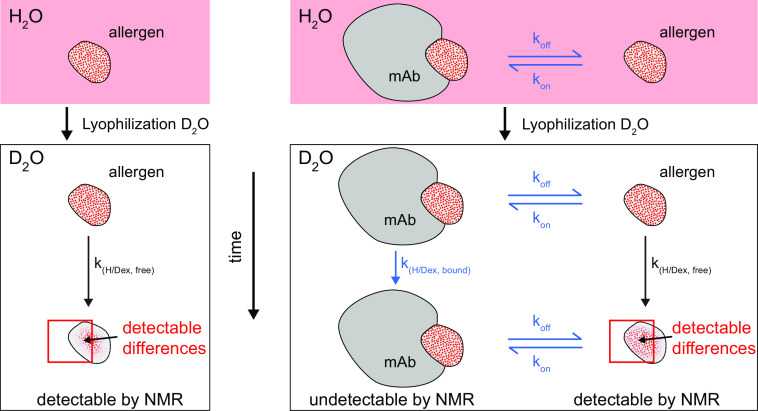
To map the functional epitopes of Art v 3 with the three intact mAbs in situ despite the tight binding and extremely slow koff rates, we developed an innovative approach based on H/D exchange. This overcomes the limitations of conventional H/D exchange measurements because such tight interactions between antigens and intact mAbs that are not directly detectable by NMR spectroscopy due to their large size, or rely on a tedious protocol of quenching the exchange and separating the antigen from the mAb for each time point (21). Our novel approach, the HDXMEM, uses the memory of the H/D exchange in the bound, invisible form and the exchange between free and bound allergen. We therefore measured H/D exchange times of the free allergen in the absence and presence of 0.25 eq of mAb in which ∼50% of Art v 3 was free and ∼50% was bound to mAb (Fig. 3). This method exploits the exchange of the small protein binding partner between the free and bound state, so that some information of the bound state can be picked up by the signals of the free allergen. A comparison between measurements with and without mAb enables determination of differences originating from altered H/D exchange rates in the bound form. The H/D exchange can be measured in situ over a time range of minutes up to months, which makes it likely that a memory of the bound state can be detected.
Figure 3.
Schematic presentation of the HDXMEM NMR methodology. The 15N-labeled allergen is dissolved in H2O and contains nearly 100% amide protons schematically indicated by red dots. After lyophilization and dissolving in D2O, the amide protons start slowly exchanging with deuterium (white dots) and become invisible in the 1H-detected NMR spectra. For each observable proton, the exchange rates are measured. A similar experiment is done in the presence of a mAb at a ratio at which the allergen is present as a mixture of free and bound states (1:1). Only the amide signals of the free allergen are detectable, but because there is an equilibrium between free and bound allergen, a mixture of H/D exchange rates is observed containing contributions of the free and bound state. Regions that are bound to the mAb are expected to be protected from H/D exchange and therefore show a lower exchange rate compared with the free allergen.
In our experimental setup, H/D exchange of unbound 15N-labeled Art v 3 was monitored at 278 K using 1H-15N HSQC spectra over a period of 5–9 days (Fig. 4). Most exchange curves could be fit well (Fig. S6), and precise exchange rates/times could be extracted, ranging from 1 × 104 to >2 × 106 s (Fig. S7), corresponding to 16 min to 12 weeks. However, for the very slowly exchanging amides, whose intensity did not decrease significantly, curve fitting delivered only inaccurate exchange times. For these signals, H/D exchange was additionally measured at 298 K, speeding up exchange times to 4 days maximum, resulting in exchange curves that could be fit well (Fig. S8). Analog measurements in the presence of the three different mAbs, using samples containing ∼50% free and 50% bound Art v 3 revealed corresponding exchange times (Fig. S9–S14). H/D exchange times could be extracted for 36% of all amino acids at 278 K for the free protein and for 33–36% for the samples containing mAbs.
Figure 4.
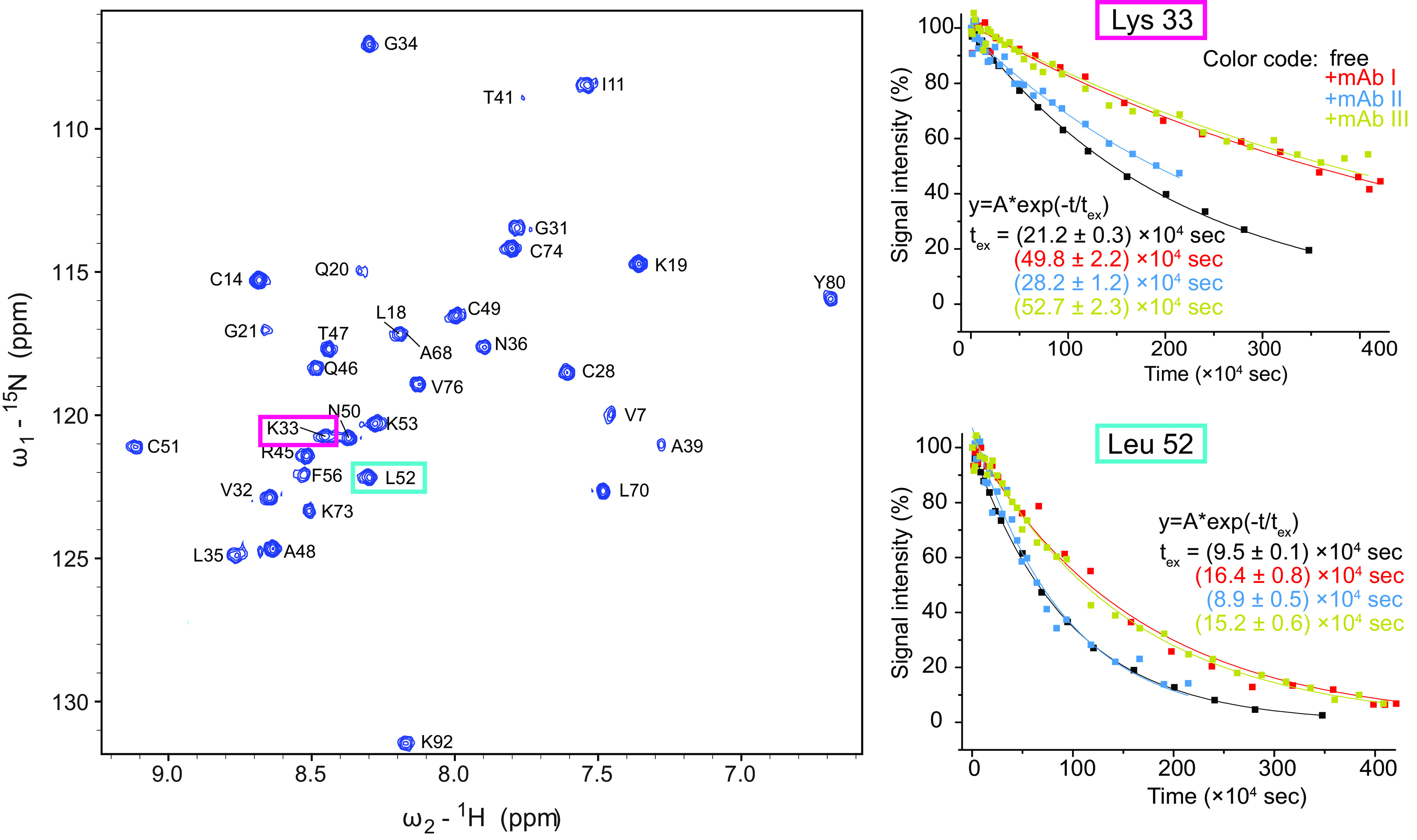
HDXMEM data of Art v 3 in the presence and absence of mAbs. Left, 15N-1H HSQC of Art v 3 in the absence of a mAb 10 min after dissolving the sample in D2O. The signals of the two amino acids Lys-33 and Leu-52 are highlighted. Right, the intensities of the two example residues (Lys-33 and Leu-52) are plotted over time in the absence and presence of mAb I, II, and III measured at 298 K with an exponential fit and the extracted exchange times as fitting parameter.
To map the binding epitope, the H/D exchange times of the allergen were compared between the measurements with and without mAb (Figs. 2 and 3). An increase in H/D exchange times indicates a local protection by the mAb. Indeed, several residues displayed (up to 4-fold) increased H/D exchange times (Fig. 5). Each mAb showed a distinct profile in exchange time changes and affected different residues. In the case of mAb I, changes clustered on a surface-exposed region in the vicinity of Gly-34 and Ala-39. For mAb II, the largest changes were detected for residues Arg-45, Gln-46, Tyr-80, and Lys-92. Regarding mAb III, a cluster of the largest changes was observed around Cys-28 (not surface-exposed), Gly-31, Lys-73, Cys-74, and Val-76 (Fig. 4). The differences in localization of antibody-binding residues are better comprehensible by mapping them on a surface presentation of the Art v 3 structure and taking the surface accessibility of each residue into account (Fig. 4). In summary, each mAb recognized a distinct surface epitope demonstrating the power of the presented HDXMEM approach.
Figure 5.
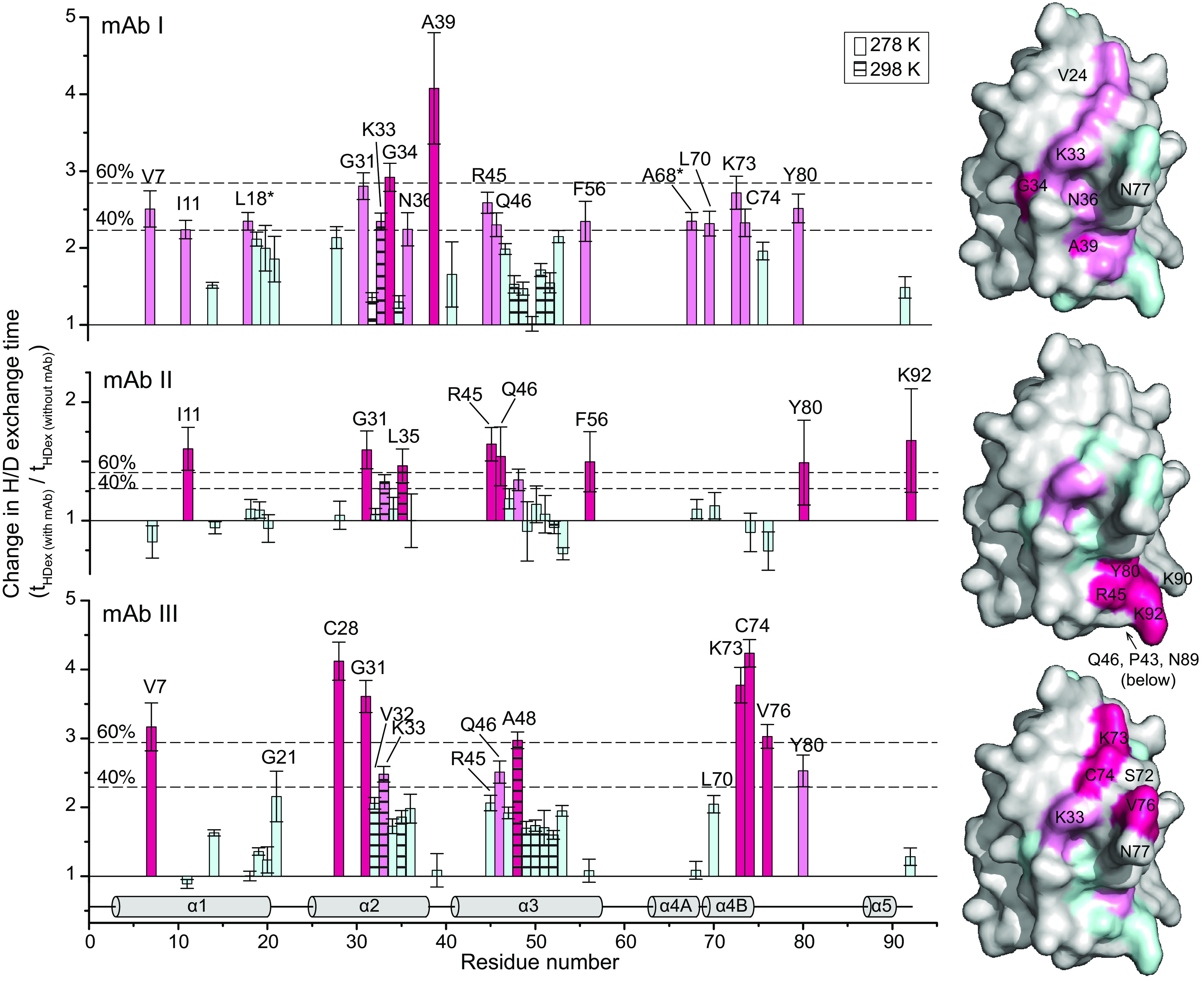
Effects of mAb binding on the H/D exchange times with residue resolution. Shown are the factors at which the H/D exchange times for each Art v 3 residue changed in the presence of 0.25 equivalents mAb compared with free Art v 3. Hatched bars, data recorded at 298 K; all other bars, data obtained at 278 K. Thresholds at 40 and 60% of the largest change are indicated as dotted lines. The secondary structure is shown at the bottom, and the surface presentations on the right are based on the crystal structure (PDB entry 6FRR, chain A). Changes in the H/D exchange rates are color-coded in hot pink (>60%), moderate changes in pink (>40%), and changes below the lowest threshold in pale cyan. Residues without a detectable NH group are depicted in gray. Error bars, S.D.
Antibody binding revealed a distinct recognition pattern toward homologous LTPs
To reveal reactivity profiles, antibody-binding titers of the three mAbs to another Art v 3 isoform and six other LTPs were determined. Homologous LTPs represent relevant allergens from pollen and food with sequence identities to Art v 3 ranging from 36 to 54% (Fig. S15). Among the mAbs, the broadest reactivity, recognizing six of eight proteins, was observed for mAb I. In contrast, mAb II and mAb III were more specific for Art v 3, with additional reactivity solely for Fra e 3 and Mal d 3/Api g 2, respectively (Fig. 6A). Amb a 6 was not detected by any of the mAbs due to a very low sequence identity of 36%. The individual reactivity pattern toward the panel of LTPs corroborates the fact that all mAbs recognize distinct surface-located epitopes.
Figure 6.
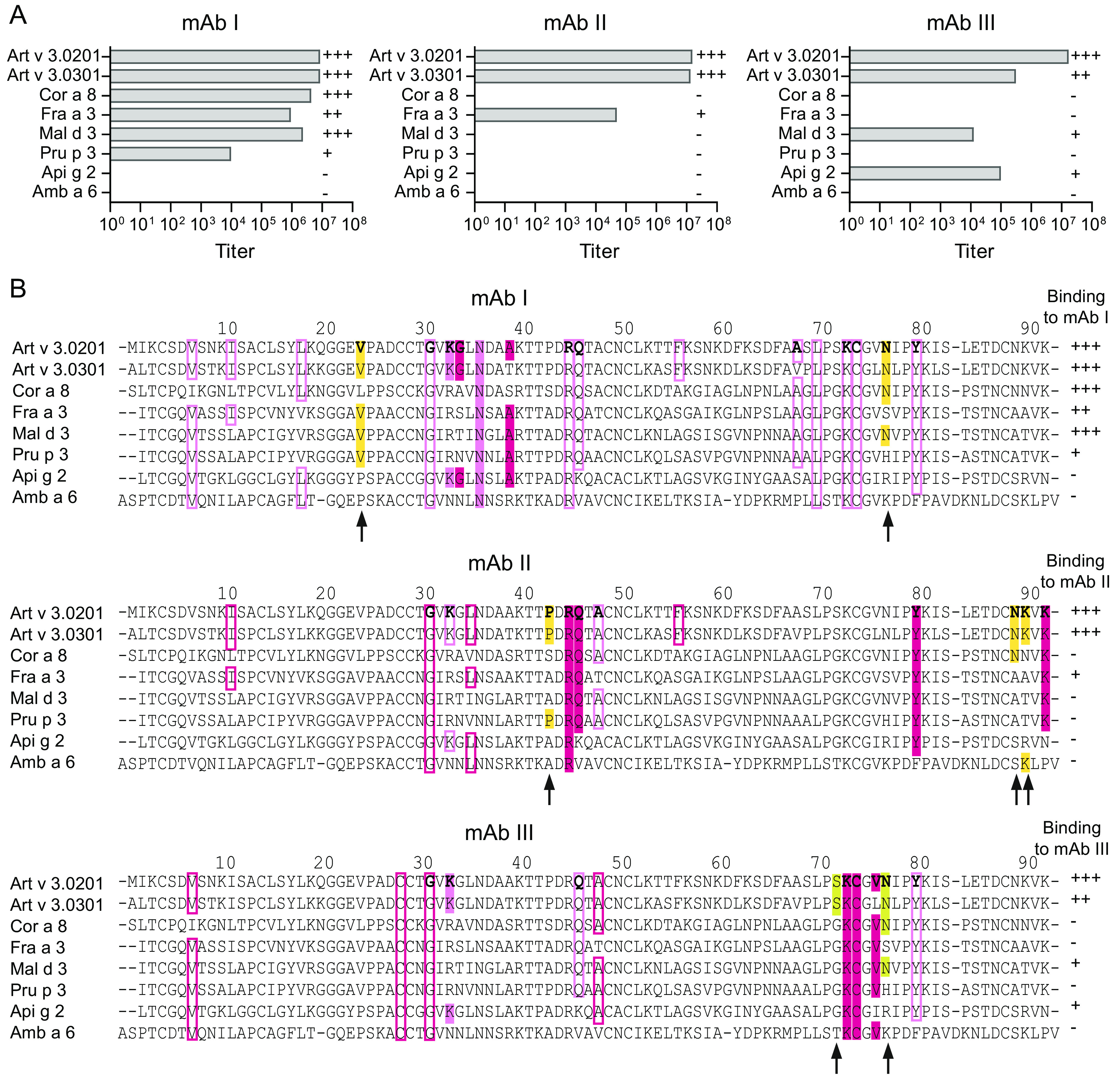
Recognition pattern of mAbs to Art v 3 and homologous LTPs. A, titers of mAb I, mAb II, and mAb III for Art v 3 and six other LTPs were measured by ELISA. Binding strengths of mAbs are depicted as follows: titers >106 (+++), titers between 105 and 106 (++), titers between 104 and 105 (+), and titers <104 (−). B, sequence alignment of Art v 3 and homologs to refine binding patterns of individual mAbs. Color-coded hotspots observed during the interaction with mAbs are used in accordance with Fig. 4 (filled boxes, amino acids involved in the epitope; rimmed boxes, amino acids that have been identified by HDXMEM NMR and are thus influenced by binding but not part of the epitope). Yellow boxes, amino acids invisible by HDXMEM NMR that potentially contribute to the epitope. Surface-exposed residues involved in mAb binding of Art v 3.0201 are shown in boldface type. Binding strengths of different homologs to the mAbs are indicated on the right. Residues that correlate with the mAb-binding pattern are highlighted with an arrow.
For epitope refinement, we additionally evaluated the antibody recognition pattern together with the individual amino acid sequences of homologous LTPs. This considers potential residues involved in epitope formation that were not detectable in NMR due to very fast H/D exchange rates. For mAb I, the main interaction as determined by NMR is observed in the central part of Art v 3 (as displayed in Fig. 5), in which Lys-33 and Asn-36 are flanked by strong perturbations of Gly-34 and Ala-39. Considering the antibody recognition pattern from negative (−) to strongly positive (+++), residues, Val-24 and Asn-77 are mostly conserved in sequences with positive reactivity and might thus contribute to epitope formation (Fig. 6B). The surface region for the interaction with mAb II clustered around the C terminus, including Arg-45, Gln-46, Tyr-80, and Lys-92. Residues in the vicinity, which were not detectable by NMR but show coherence in strongly recognizing only the two Art v 3 isoallergens, are Pro-43, Asn-89, and Lys-90. The epitope recognized by mAb III localizes to another part of the molecule around residues Lys-33, Lys-73, Cys-74, and Val-76. In addition, Ser-72 and Asn-77 could be relevant to establish the antibody-binding site due to structural proximity and the antibody-binding pattern of the LTP homologs. The individual mAb-binding profiles and amino acid sequences of all investigated LTPs enabled us to propose additional residues involved in the binding epitope.
Art v 3 epitope variants present considerable differences in antibody-binding reactivity
To further analyze the identified epitopes, we generated four Art v 3 variants targeting epitopes of mAb I (V1), mAb II (V2), and mAb III (V3A and V3B) using a computational approach to maintain protein stability (Fig. 7A and Fig. S16A). As V3A shared two exchanged residues with V1, another variant termed V3B lacking Lys-33 and Asn-77 was generated. Recombinant Art v 3 variants were obtained from E. coli expression. Protein purity, identity, and formation of disulfide bonds were confirmed by gel electrophoresis and MS, respectively (Fig. S16, B and C). In ELISA experiments, recognition of V1 was selectively disrupted when using mAb I (Fig. 7B). Unexpectedly, V2 maintained its antibody reactivity with mAb II (Fig. 7C). Recognition by mAb III was completely abrogated for V3A and partially for V3B (Fig. 7D). Interestingly, amino acid exchanges targeting the mAb III epitope also affected antibody binding by the other antibodies.
Figure 7.
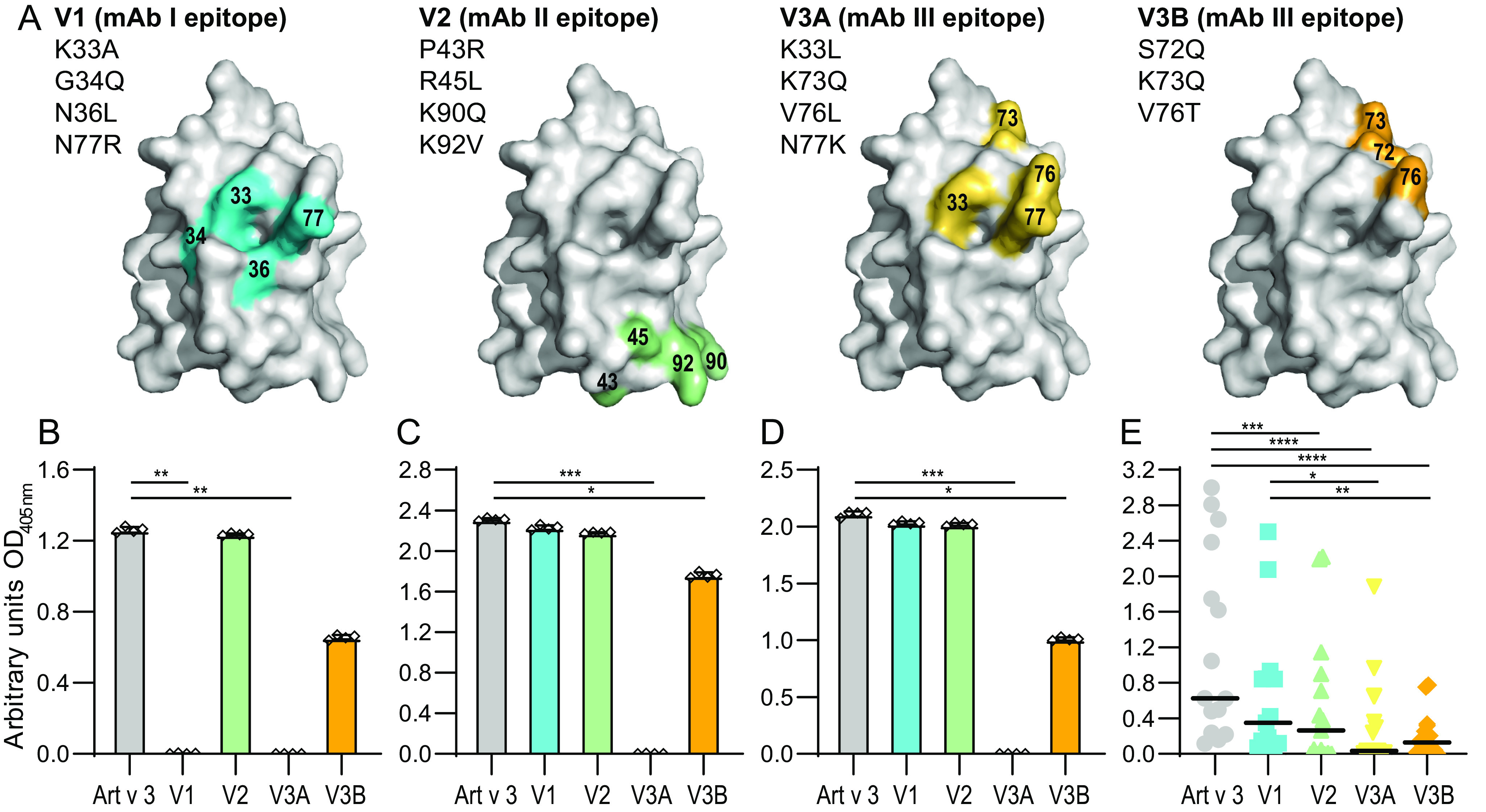
Art v 3 epitope variants and recognition by mAbs and patients' serum IgE. A, surface representation of residues exchanged for epitope variants V1, V2, V3A, and V3B based on the crystal structure (PDB entry 6FRR, chain A). Shown is ELISA antibody reactivity to Art v 3 and epitope variants using mAb I (B), mAb II (C), and mAb III (D). Bars, mean of four technical replicates; whiskers, S.D. E, IgE reactivity of allergic patients' sera (n = 15) to Art v 3 and epitope variants. Black lines, medians. ****, p ≤ 0.0001; ***, p ≤ 0.001; **, p ≤ 0.01; *, p ≤ 0.05.
Testing all epitope variants with serum from allergic patients, IgE reactivity to V2, V3A, and V3B was significantly reduced compared with Art v 3 (Fig. 7E), with median declines ranging from 88.2 to 83.4% when calculated for each patient individually. In addition, V1 also showed changes in IgE reactivity (i.e. 47.7% lower reactivity), which was, however, not statistically significant. It is noteworthy that patients presented highly individual IgE reactivity profiles toward the epitope variants (Fig. S17). In summary, all variants showed reduced IgE-binding capacities, and thus relevant IgE-binding epitopes were successfully targeted by the residue exchanges.
Discussion
Identification of antibody-binding epitopes and determination of IgE cross-reactivity is crucial to better understand the mechanisms of allergic responses. However, so far, we lack straightforward methods for determination of structural epitopes using intact antibodies. To overcome this limitation, we developed HDXMEM, a novel NMR method, and elucidated three clinically relevant IgE-binding epitopes of the mugwort pollen allergen Art v 3.
Recombinant Art v 3 was used to immunize mice for subsequent generation of mAbs. Using this approach, we obtained large amounts of three high-affinity IgG1 antibodies with different binding mechanisms at the molecular level. Reflecting the fact that human IgE antibodies from allergic patients solely recognize structural epitopes of Art v 3 (25), we selected mAbs with strong binding to the native protein but lacking reactivity to the reduced and alkylated protein. Such in vitro–generated mAbs have several advantages, as they are highly specific for a single epitope (with affinities in the nanomolar range) and can be produced in large quantities as needed for interaction studies. In addition, they can be used to overcome the low bioavailability of human antibodies, especially IgE (15, 40–42). A key feature of our surrogate mAbs for mapping clinically relevant allergenic epitopes is their overlap with human IgE-binding regions from allergic patients. The majority of investigated patients showed a high overlap with mean inhibitions ranging from 70 to 90%. Such a high overlap is remarkable, as human IgE is polyclonal, and patient-tailored reactivity profiles are frequently observed within the LTP syndrome (31). This supports the assumption that antibodies recognize similar regions on allergens irrespective of antibody subtypes or even organisms and emphasizes the use of surrogate antibodies for subsequent analyses (22).
Concerning epitope mapping of allergen-antibody interactions, only some allergen-antibody co-crystals have been elucidated so far, whereas the majority of binding regions were indirectly determined by in vitro mutagenesis (22, 43, 44). Besides, H/D exchange monitored by MS can be explored for determination of interaction sites at the peptide level. However, analysis requires sufficient accessible proteolytic protein cleavage sites to narrow down resolution and ensure suitable display of discontinuous epitopes on the surface (45, 46). With regard to epitope mapping of Art v 3, neither co-crystallization experiments with Fab fragments nor H/D exchange coupled with MS were successful in our previously conducted studies.
We therefore exploited NMR spectroscopy for epitope mapping studies. First, we confirmed the typical α-helical LTP fold as previously revealed by X-ray crystallography (PDB entry 6FRR) (25) using secondary structure chemical shift analysis of Art v 3. So far, NMR spectroscopy has been used for mapping of structural epitopes of two house dust mite allergens using either Fab fragments (14) or intact antibodies in combination with a detergent to weaken the interactions (15). However, when studying tight interactions between allergens and an intact mAb, traditional chemical shift deviation approaches typically fail. Moreover, cross-saturation transfer experiments are usually unsuccessful due to slow koff values. As such, the traditional NMR approaches were not applicable for analyzing the high-affinity binding to Art v 3.
Therefore, we developed a combination of H/D exchange and the indirect observation of the invisible bound form for epitope mapping. In contrast to previous H/D exchange studies, which relied on a very demanding protocol involving quenching the H/D exchange, separating the antigen from the mAb and detecting the amount of NH in spectra of the separated antigen for each time point (21), our approach requires only one sample per mAb in addition to the measurement of the free antigen. The novel HDXMEM approach exploits the exchange between free and bound antigen in samples in which 50% of free and 50% of bound antigen is present. Although only the free form is detected, the signals show a memory effect reflecting to a certain degree the H/D exchange in the otherwise invisible bound form. Even though we measure a mixture of the H/D exchange in the bound form and the H/D exchange in the free form, this is sufficient to detect changes in the effective exchange rates when compared with antigen in the absence of mAb. Because HDXMEM critically depends on the observation of the amide protons and thus exchange times longer than the dead time of the experiment, the H/D exchange was slowed down by measuring at low temperature. At 278 K, the lowest limit for measurements in D2O, we could measure H/D exchange rates for 33–36% of the residues, representing an equivalent or enhanced coverage compared with other H/D exchange–based epitope mapping studies measured with the traditional tedious approach (20, 21, 41). Although the coverage is incomplete and sometimes an increase in protection occurs in regions remote from the interaction site (20, 47), the binding region can typically be still localized. We thus conclude that HDXMEM is a suitable method for epitope mapping of antigens interacting tightly with intact antibodies.
Using HDXMEM and mAb-binding pattern, three IgE epitopes of Art v 3 involving Val-24, Lys-33, Gly-34, Asn-36, Ala-39, and Asn-77 (mAb I); Pro-43, Arg-45, Gln-46, Tyr-80, Asn-89, Lys-90, and Lys-92 (mAb II); and Lys-33, Ser-72, Lys-73, Cys-74, Val-76, and Asn-77 (mAb III) were identified. These IgE-binding regions represent the first structural epitopes of a pollen LTP with relevance in the LTP syndrome. Analogous to other structural epitopes, residues involved in binding are not continuous in the sequence but only assemble in the three-dimensional structure (37). Previous studies revealed charged residues, in particular lysines, are crucial for binding (3, 14, 48, 49). In accordance with these findings, we determined Lys-33, Arg-45, Lys-73, Lys-90, and Lys-92 to be involved in epitope formation of Art v 3. As indicated by different binding mechanisms and patterns, the investigated mAbs recognize three distinct regions on the protein surface. The epitope of mAb I is central (according to the illustration in Fig. 5) and mostly involves residues of the α2-helix. The other epitopes localize to the outer edges thereof, with the mAb II comprising of residues in α3 as well as the C terminus and the mAb III epitope predominately localizing at the end of α4.
To corroborate the identified epitopes, we tested antibody binding to four Art v 3 epitope variants, each presenting 3 or 4 amino acid exchanges. Variants were computationally designed to change surface properties of each mAb epitope while maintaining the protein stability. Using mAb I and mAb III, the expected reduction in antibody binding toward the respective variants was observed. Interestingly, V3A did not interact with any of the mAbs, suggesting that residue exchanges also influenced other binding regions due to epitope proximity and/or local structural changes. Although V2 did not lead to changes in mAb recognition, the majority of patients showed a strongly reduced or completely lacking IgE reactivity to this variant, suggesting the exchange of residues highly relevant for human IgE antibody binding. Despite the fact that all variants showed a reduced IgE-binding capacity, the reactivity profile was highly distinct for each patient. Similarly, the binding pattern of mAbs toward the investigated LTP homologs ranging from broad cross-reactivity (mAb I) to higher specificity (mAb II and mAb III) was noted. This observation illustrates the complex and patient-specific IgE (cross-)reactivity as observed in the LTP syndrome (30–34, 50).
So far, several linear and some conformational IgE-binding epitopes of peach and wheat LTP had been identified (6, 7, 35). Those conformational epitopes were, however, determined by mimotope mapping followed by three-dimensional modeling, which is not based on direct antibody-allergen binding experiments as in our setup. Residues within 31–43 and 71–80 were suggested as relevant epitopes for peach and wheat LTP (6, 35), which are in accordance with our mAb I and mAb III epitopes. The epitope recognized by mAb I seems to be widely conserved in other allergenic LTPs. We therefore propose this region to be part of an IgE cross-reactive epitope involved in the LTP syndrome.
Even though the mAb III epitope co-localizes with a previously determined IgE-binding region of LTPs (6, 35), our antibody seems to have a high specificity that prevented cross-reactivity with peach LTP in this case. On the other hand, residue exchanges of V3A led to a dramatic decrease in antibody recognition by all mAbs as well as IgE of most patients. We therefore confirm that this region also constitutes an important antibody-binding epitope of Art v 3. Considering all data, a certain overlap between the mAb I and mAb III epitope seems plausible. This is also noticeable in the individual patients' IgE reactivity profiles, where mostly V1, V3A, and V3B follow a congruency in antibody recognition, but there were also several exceptions to this pattern. Notably, the epitope revealed by mAb II involving the C terminus of the protein is a novel epitope within the LTP family. IgE reactivity to V2 was highly reduced or completely abrogated in the majority of patients' sera tested. Both the mAb-binding pattern and the testing of variants suggest that this epitope is highly distinct in localization.
So far, the generation of low IgE-binding LTP variants for allergen immunotherapy proved to be challenging, as molecules either lost their α-helical structure and thus stability or showed only moderate reduction in IgE reactivity (51, 52). Based on the herein identified antibody binding residues, design of a therapeutic variant lacking all relevant IgE-binding epitopes while maintaining immunogenicity seems reasonable (53).
In conclusion, we herein describe HDXMEM as an innovative approach to map clinically relevant epitopes on Art v 3 using intact antibodies. This method is broadly applicable to all sorts of tight interactions, in particular with large binding partners like mAbs, and could be widely used for biopharmaceutical characterization and epitope mapping. HDXMEM for the first time enables determination of conformational IgE-binding regions on a pollen LTP. The obtained results have a direct implication on the clinical interpretation of IgE cross-reactivity in the LTP syndrome and development of novel therapeutics.
Experimental procedures
Details about the materials and methods are provided in the supporting information.
Recombinant production of allergens
Recombinant Art v 3.0201, was produced as described previously (25). For protein isotope labeling, bacteria were grown in M9 minimal medium with 15N or 15N and 13C as sole nitrogen and carbon source (54). Proteins were purified as described previously and analyzed by reducing gel electrophoresis (25). Another isoform (i.e. Art v 3.0301) as well as Api g 2, Cor a 8, Pru p 3, Fra a 3, Mal d 3, and Amb a 6 were obtained as recombinant proteins as published and described in the supporting information (33, 55–60). Art v 3 epitope variants V1, V2, V3A, and V3B (Fig. 7 and Fig. S16) were obtained using the stability predictor MAESTRO (61) to the crystal structure of Art v 3 (PDB entry 6FRR). Recombinant production and purification were performed analogous to Art v 3.
MS
MS analyses of Art v 3, isotopically labeled proteins, and epitope variants were conducted on the intact protein level using the Thermo ScientificTM Q ExactiveTM and Thermo ScientificTM Q ExactiveTM Plus Hybrid Quadrupole-OrbitrapTM mass spectrometers (Thermo Fisher Scientific). The extent of isotope labeling was calculated manually based on the shift of isotope distribution of the most intense charge state.
Generation and purification of monoclonal anti-Art v 3 antibodies
For immunization, three female BALB/c mice (Charles River Laboratories) were immunized subcutaneously with 10 µg of recombinant Art v 3 adsorbed to Alu-gel S in sterile PBS. After six immunizations, B cells from the spleens were collected and used for hybridoma production (62). Secreted antibodies in the supernatant were tested for IgG (IgG1, IgG2a, IgG2b, and IgG3) and IgE reactivity in an ELISA. For purification, secreted antibodies were obtained from B-cell hybridoma cultures and purified using protein G affinity chromatography. Animal experiments were approved by the Austrian Ministry of Science (permission no. GZ 66.012/0047-II/3b/2017).
Affinity determination
ITC was performed on a VP-ITC microcalorimeter (MicroCal), loading the sample cell with respective mAbs and the microsyringe with Art v 3. Raw data were analyzed using the MicroCal ITC module of Origin 7.0, applying a 1:2 binding model. For SAW measurements, Art v 3 was immobilized on the surface of a SAW CM-Dextran three-dimensional sensor chip and incubated with serial dilutions of mAbs. SAW phase changes were recorded, and Trace Drawer 1.7 software was used to calculate the affinities.
mAb ELISA
mAb reactivities to native and reduced/alkylated Art v 3 as well as antibody titers to Art v 3, homologous LTPs, and epitope variants were determined by ELISA. Briefly, 200 ng of protein/well was coated overnight to ELISA plates. Plates were incubated with anti-Art v 3–specific monoclonal antibodies (in serial dilution for titer calculation), and an alkaline phosphatase–labeled rabbit anti-mouse IgG/IgM antibody was used for subsequent colorimetric detection.
Patients' IgE ELISA
Twenty-one patients with allergic symptoms to mugwort pollen and positive in vitro reactivity to Art v 3 were included (Table S1). The study was approved by the Institutional Review Board (approval no. 106-CE-2005). Sera of those patients were used to determine IgE reactivity to Art v 3, homologous LTPs, and epitope variants. Briefly, 200 ng of protein/well was coated overnight to ELISA plates. Plates were incubated with individual patients' sera, and an alkaline phosphatase–labeled mouse anti-human IgE antibody was used for subsequent colorimetric detection. For cross-inhibition studies, coated proteins were first incubated with respective mAbs. After washing, individual patients' sera were applied and detected as described above. Decline of IgE reactivity compared with measurements without mAbs was calculated as percentage of inhibition.
NMR spectroscopy measurements
All spectra were recorded on a 600-MHz Bruker Avance III HD spectrometer equipped with a QXI quadruple-resonance probe (1H/13C/15N/13C) at 278 or 298 K. Standard three-dimensional triple-resonance experiment spectra were recorded for sequential assignment.
Data availability
Chemical shift assignments were deposited in the BioMagRes data bank under accession number 28092. All remaining data are contained within the article and the supporting information.
Supplementary Material
Acknowledgments
We thank Reinhard Schwarz, Gabriele Schmutzler, and Frauke Schocker for providing materials and protocols. We appreciate beneficial discussions and valuable comments on the manuscript from Fred Damberger, Martin Tollinger, Therese Wohlschlager, Peter Schmieder, Lawrence McIntosh, and Arthur Hinterholzer.
This article contains supporting information.
Author contributions—M. D. M., S. W., S. H., E. V., J. L., M. S., and G. G. formal analysis; M. D. M., S. W., E. V., M. S., and G. G. validation; M. D. M., S. W., S. H., M. H., E. V., W. A., C. R., J. L., D. Z., P. L., and M. S. investigation; M. D. M., S. W., M. S., and G. G. visualization; M. D. M., S. W., S. H., M. H., E. V., W. A., C. R., D. Z., N. W., and M. S. methodology; S. W., M. S., and G. G. writing-original draft; M. H., N. W., C. G. H., R. v. R., A. M., P. L., F. F., M. S., and G. G. supervision; J. L. and P. L. software; C. G. H., F. F., M. S., and G. G. funding acquisition; R. v. R., A. M., and F. F. resources; M. S. and G. G. conceptualization; M. S. and G. G. project administration; M. S. and G. G. writing-review and editing.
Funding and additional information—This work was supported by the Austrian Federal Ministry for Digital and Economic Affairs; the National Foundation for Research, Technology, and Development; a Start-up Grant of the Province of Salzburg; and Austrian Science Fund (FWF) Grant P26125.
Conflict of interest—R. v. R. reports consultancies from HAL Allergy BV, Citeq BV, and Amgany Inc. and speaker's fees from HAL Allergy BV, and Thermo Fisher Scientific. F. F. is a member of the Scientific Advisory Boards of HAL Allergy BV, AllergenOnline, and SIAF. G. G. is member of the Scientific Advisory Board of Bencard.
- LTP
- lipid transfer protein
- HDXMEM
- H/D exchange memory
- ITC
- isothermal titration calorimetry
- SAW
- surface acoustic wave
- HSQC
- heteronuclear single quantum coherence
- PDB
- Protein Data Bank.
References
- 1. Abbott W. M., Damschroder M. M., and Lowe D. C. (2014) Current approaches to fine mapping of antigen-antibody interactions. Immunology 142, 526–535 10.1111/imm.12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mirza O., Henriksen A., Ipsen H., Larsen J. N., Wissenbach M., Spangfort M. D., and Gajhede M. (2000) Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1. J. Immunol. 165, 331–338 10.4049/jimmunol.165.1.331 [DOI] [PubMed] [Google Scholar]
- 3. Padavattan S., Flicker S., Schirmer T., Madritsch C., Randow S., Reese G., Vieths S., Lupinek C., Ebner C., Valenta R., and Markovic-Housley Z. (2009) High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J. Immunol. 182, 2141–2151 10.4049/jimmunol.0803018 [DOI] [PubMed] [Google Scholar]
- 4. Willison L. N., Zhang Q., Su M., Teuber S. S., Sathe S. K., and Roux K. H. (2013) Conformational epitope mapping of Pru du 6, a major allergen from almond nut. Mol. Immunol. 55, 253–263 10.1016/j.molimm.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 5. Zhang Q., Yang J., Bautista J., Badithe A., Olson W., and Liu Y. (2018) Epitope mapping by HDX-MS elucidates the surface coverage of antigens associated with high blocking efficiency of antibodies to birch pollen allergen. Anal. Chem. 90, 11315–11323 10.1021/acs.analchem.8b01864 [DOI] [PubMed] [Google Scholar]
- 6. Garcia-Casado G., Pacios L. F., Diaz-Perales A., Sanchez-Monge R., Lombardero M., Garcia-Selles F. J., Polo F., Barber D., and Salcedo G. (2003) Identification of IgE-binding epitopes of the major peach allergen Pru p 3. J. Allergy Clin. Immunol. 112, 599–605 10.1016/S0091-6749(03)01605-1 [DOI] [PubMed] [Google Scholar]
- 7. Pacios L. F., Tordesillas L., Cuesta-Herranz J., Compes E., Sánchez-Monge R., Palacín A., Salcedo G., and Díaz-Perales A. (2008) Mimotope mapping as a complementary strategy to define allergen IgE-epitopes: peach Pru p 3 allergen as a model. Mol. Immunol. 45, 2269–2276 10.1016/j.molimm.2007.11.022 [DOI] [PubMed] [Google Scholar]
- 8. Williamson M. P. (2013) Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 10.1016/j.pnmrs.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 9. Simonelli L., Pedotti M., Bardelli M., Jurt S., Zerbe O., and Varani L. (2018) Mapping antibody epitopes by solution NMR spectroscopy: practical considerations. Methods Mol. Biol. 1785, 29–51 10.1007/978-1-4939-7841-0_3 [DOI] [PubMed] [Google Scholar]
- 10. Pervushin K., Riek R., Wider G., and Wüthrich K. (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U. S. A. 94, 12366–12371 10.1073/pnas.94.23.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Veverka V., Henry A. J., Slocombe P. M., Ventom A., Mulloy B., Muskett F. W., Muzylak M., Greenslade K., Moore A., Zhang L., Gong J., Qian X., Paszty C., Taylor R. J., Robinson M. K., et al. (2009) Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J. Biol. Chem. 284, 10890–10900 10.1074/jbc.M807994200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boschert V., Frisch C., Back J. W., van Pee K., Weidauer S. E., Muth E. M., Schmieder P., Beerbaum M., Knappik A., Timmerman P., and Mueller T. D. (2016) The sclerostin-neutralizing antibody AbD09097 recognizes an epitope adjacent to sclerostin's binding site for the Wnt co-receptor LRP6. Open Biol. 6, 160120 10.1098/rsob.160120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scarselli M., Cantini F., Santini L., Veggi D., Dragonetti S., Donati C., Savino S., Giuliani M. M., Comanducci M., Di Marcello F., Romagnoli G., Pizza M., Banci L., and Rappuoli R. (2009) Epitope mapping of a bactericidal monoclonal antibody against the factor H binding protein of Neisseria meningitidis. J. Mol. Biol. 386, 97–108 10.1016/j.jmb.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 14. Naik M. T., Chang C. F., Kuo I. C., Kung C. C., Yi F. C., Chua K. Y., and Huang T. H. (2008) Roles of structure and structural dynamics in the antibody recognition of the allergen proteins: an NMR study on Blomia tropicalis major allergen. Structure 16, 125–136 10.1016/j.str.2007.10.022 [DOI] [PubMed] [Google Scholar]
- 15. Ichikawa S., Takai T., Inoue T., Yuuki T., Okumura Y., Ogura K., Inagaki F., and Hatanaka H. (2005) NMR study on the major mite allergen Der f 2: its refined tertiary structure, epitopes for monoclonal antibodies and characteristics shared by ML protein group members. J. Biochem. 137, 255–263 10.1093/jb/mvi039 [DOI] [PubMed] [Google Scholar]
- 16. Takahashi H., Nakanishi T., Kami K., Arata Y., and Shimada I. (2000) A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 7, 220–223 10.1038/73331 [DOI] [PubMed] [Google Scholar]
- 17. Fawzi N. L., Ying J., Ghirlando R., Torchia D. A., and Clore G. M. (2011) Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature 480, 268–272 10.1038/nature10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vallurupalli P., Bouvignies G., and Kay L. E. (2012) Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 10.1021/ja3001419 [DOI] [PubMed] [Google Scholar]
- 19. Gouda H., Shiraishi M., Takahashi H., Kato K., Torigoe H., Arata Y., and Shimada I. (1998) NMR study of the interaction between the B domain of staphylococcal protein A and the Fc portion of immunoglobulin G. Biochemistry 37, 129–136 10.1021/bi970923f [DOI] [PubMed] [Google Scholar]
- 20. Williams D. C. Jr., Benjamin D. C., Poljak R. J., and Rule G. S. (1996) Global changes in amide hydrogen exchange rates for a protein antigen in complex with three different antibodies. J. Mol. Biol. 257, 866–876 10.1006/jmbi.1996.0207 [DOI] [PubMed] [Google Scholar]
- 21. Paterson Y., Englander S. W., and Roder H. (1990) An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science 249, 755–759 10.1126/science.1697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pomes A. (2010) Relevant B cell epitopes in allergic disease. Int. Arch. Allergy Immunol. 152, 1–11 10.1159/000260078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pomes A., Chruszcz M., Gustchina A., Minor W., Mueller G. A., Pedersen L. C., Wlodawer A., and Chapman M. D. (2015) 100 years later: celebrating the contributions of X-ray crystallography to allergy and clinical immunology. J. Allergy Clin. Immunol. 136, 29–37.e10 10.1016/j.jaci.2015.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asturias J. A., Gómez-Bayón N., Eseverri J. L., and Martínez A. (2003) Par j 1 and Par j 2, the major allergens from Parietaria judaica pollen, have similar immunoglobulin E epitopes. Clin. Exp. Allergy 33, 518–524 10.1046/j.1365-2222.2003.01631.x [DOI] [PubMed] [Google Scholar]
- 25. Wildner S., Griessner I., Stemeseder T., Regl C., Soh W. T., Stock L. G., Völker T., Alessandri C., Mari A., Huber C. G., Stutz H., Brandstetter H., and Gadermaier G. (2019) Boiling down the cysteine-stabilized LTP fold—loss of structural and immunological integrity of allergenic Art v 3 and Pru p 3 as a consequence of irreversible lanthionine formation. Mol. Immunol. 116, 140–150 10.1016/j.molimm.2019.10.012 [DOI] [PubMed] [Google Scholar]
- 26. Scala E., Till S. J., Asero R., Abeni D., Guerra E. C., Pirrotta L., Paganelli R., Pomponi D., Giani M., De Pità O., and Cecchi L. (2015) Lipid transfer protein sensitization: reactivity profiles and clinical risk assessment in an Italian cohort. Allergy 70, 933–943 10.1111/all.12635 [DOI] [PubMed] [Google Scholar]
- 27. Azofra J., Berroa F., Gastaminza G., Saiz N., Gamboa P. M., Vela C., García B. E., Lizarza S., Echenagusia M. A., Joral A., Aranzabal M. A., Quiñones M. D., Jauregui I., Madera J. F., Navarro J. A., et al. (2016) Lipid transfer protein syndrome in a non-Mediterranean area. Int. Arch. Allergy Immunol. 169, 181–188 10.1159/000445893 [DOI] [PubMed] [Google Scholar]
- 28. Asero R., Piantanida M., and Pravettoni V. (2018) Allergy to LTP: to eat or not to eat sensitizing foods? A follow-up study. Eur. Ann. Allergy Clin. Immunol. 50, 156–162 10.23822/EurAnnACI.1764-1489.57 [DOI] [PubMed] [Google Scholar]
- 29. Asero R., Piantanida M., Pinter E., and Pravettoni V. (2018) The clinical relevance of lipid transfer protein. Clin. Exp. Allergy 48, 6–12 10.1111/cea.13053 [DOI] [PubMed] [Google Scholar]
- 30. Gao Z. S., Yang Z. W., Wu S. D., Wang H. Y., Liu M. L., Mao W. L., Wang J., Gadermaier G., Ferreira F., Zheng M., and van Ree R. (2013) Peach allergy in China: a dominant role for mugwort pollen lipid transfer protein as a primary sensitizer. J. Allergy Clin. Immunol. 131, 224–226.e3 10.1016/j.jaci.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 31. Egger M., Hauser M., Mari A., Ferreira F., and Gadermaier G. (2010) The role of lipid transfer proteins in allergic diseases. Curr. Allergy Asthma Rep. 10, 326–335 10.1007/s11882-010-0128-9 [DOI] [PubMed] [Google Scholar]
- 32. Salcedo G., Sanchez-Monge R., Diaz-Perales A., Garcia-Casado G., and Barber D. (2004) Plant non-specific lipid transfer proteins as food and pollen allergens. Clin. Exp. Allergy 34, 1336–1341 10.1111/j.1365-2222.2004.02018.x [DOI] [PubMed] [Google Scholar]
- 33. Gadermaier G., Egger M., Girbl T., Erler A., Harrer A., Vejvar E., Liso M., Richter K., Zuidmeer L., Mari A., and Ferreira F. (2011) Molecular characterization of Api g 2, a novel allergenic member of the lipid-transfer protein 1 family from celery stalks. Mol. Nutr. Food Res. 55, 568–577 10.1002/mnfr.201000443 [DOI] [PubMed] [Google Scholar]
- 34. Palacín A., Gómez-Casado C., Rivas L. A., Aguirre J., Tordesillas L., Bartra J., Blanco C., Carrillo T., Cuesta-Herranz J., de Frutos C., Alvarez-Eire G. G., Fernández F. J., Gamboa P., Muñoz R., Sánchez-Monge R., et al. (2012) Graph based study of allergen cross-reactivity of plant lipid transfer proteins (LTPs) using microarray in a multicenter study. PLoS ONE 7, e50799 10.1371/journal.pone.0050799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tordesillas L., Pacios L. F., Palacin A., Quirce S., Armentia A., Barber D., Salcedo G., and Diaz-Perales A. (2009) Molecular basis of allergen cross-reactivity: non-specific lipid transfer proteins from wheat flour and peach fruit as models. Mol. Immunol. 47, 534–540 10.1016/j.molimm.2009.07.028 [DOI] [PubMed] [Google Scholar]
- 36. Gadermaier G., Hauser M., Egger M., Ferrara R., Briza P., Santos K. S., Zennaro D., Girbl T., Zuidmeer-Jongejan L., Mari A., and Ferreira F. (2011) Sensitization prevalence, antibody cross-reactivity and immunogenic peptide profile of Api g 2, the non-specific lipid transfer protein 1 of celery. PLoS ONE 6, e24150 10.1371/journal.pone.0024150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pomés A., Chruszcz M., Gustchina A., and Wlodawer A. (2015) Interfaces between allergen structure and diagnosis: know your epitopes. Curr. Allergy Asthma Rep. 15, 506 10.1007/s11882-014-0506-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanchez-Trincado J. L., Gomez-Perosanz M., and Reche P. A. (2017) Fundamentals and methods for T- and B-cell epitope prediction. J. Immunol. Res. 2017, 2680160 10.1155/2017/2680160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sattler M., Schleucher J., and Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 10.1016/S0079-6565(98)00025-9 [DOI] [Google Scholar]
- 40. Razzera G., Gadermaier G., de Paula V., Almeida M. S., Egger M., Jahn-Schmid B., Almeida F. C., Ferreira F., and Valente A. P. (2010) Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure 18, 1011–1021 10.1016/j.str.2010.05.012 [DOI] [PubMed] [Google Scholar]
- 41. Mueller G. A., Smith A. M., Chapman M. D., Rule G. S., and Benjamin D. C. (2001) Hydrogen exchange nuclear magnetic resonance spectroscopy mapping of antibody epitopes on the house dust mite allergen Der p 2. J. Biol. Chem. 276, 9359–9365 10.1074/jbc.M010812200 [DOI] [PubMed] [Google Scholar]
- 42. Asam C., Batista A. L., Moraes A. H., de Paula V. S., Almeida F. C., Aglas L., Kitzmüller C., Bohle B., Ebner C., Ferreira F., Wallner M., and Valente A. P. (2014) Bet v 1–a Trojan horse for small ligands boosting allergic sensitization? Clin. Exp. Allergy 44, 1083–1093 10.1111/cea.12361 [DOI] [PubMed] [Google Scholar]
- 43. Mueller G. A., Min J., Foo A. C. Y., Pomés A., and Pedersen L. C. (2019) Structural analysis of recent allergen-antibody complexes and future directions. Curr. Allergy Asthma Rep. 19, 17 10.1007/s11882-019-0848-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu C., and Sathe S. K. (2018) Food allergen epitope mapping. J. Agric. Food Chem. 66, 7238–7248 10.1021/acs.jafc.8b01967 [DOI] [PubMed] [Google Scholar]
- 45. Gessner C., Steinchen W., Bédard S., Skinner J. J., Woods V. L., Walsh T. J., Bange G., and Pantazatos D. P. (2017) Computational method allowing hydrogen-deuterium exchange mass spectrometry at single amide resolution. Sci. Rep. 7, 3789 10.1038/s41598-017-03922-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Percy A. J., Rey M., Burns K. M., and Schriemer D. C. (2012) Probing protein interactions with hydrogen/deuterium exchange and mass spectrometry—a review. Anal. Chim. Acta 721, 7–21 10.1016/j.aca.2012.01.037 [DOI] [PubMed] [Google Scholar]
- 47. Benjamin D. C., Williams D. C. Jr., Smith-Gill S. J., and Rule G. S. (1992) Long-range changes in a protein antigen due to antigen-antibody interaction. Biochemistry 31, 9539–9545 10.1021/bi00155a005 [DOI] [PubMed] [Google Scholar]
- 48. Oezguen N., Zhou B., Negi S. S., Ivanciuc O., Schein C. H., Labesse G., and Braun W. (2008) Comprehensive 3D-modeling of allergenic proteins and amino acid composition of potential conformational IgE epitopes. Mol. Immunol. 45, 3740–3747 10.1016/j.molimm.2008.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kringelum J. V., Nielsen M., Padkjær S. B., and Lund O. (2013) Structural analysis of B-cell epitopes in antibody:protein complexes. Mol. Immunol. 53, 24–34 10.1016/j.molimm.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tordesillas L., Sirvent S., Díaz-Perales A., Villalba M., Cuesta-Herranz J., Rodríguez R., and Salcedo G. (2011) Plant lipid transfer protein allergens: no cross-reactivity between those from foods and olive and Parietaria pollen. Int. Arch. Allergy Immunol. 156, 291–296 10.1159/000323503 [DOI] [PubMed] [Google Scholar]
- 51. Gómez-Casado C., Garrido-Arandia M., Gamboa P., Blanca-López N., Canto G., Varela J., Cuesta-Herranz J., Pacios L. F., Díaz-Perales A., and Tordesillas L. (2013) Allergenic characterization of new mutant forms of Pru p 3 as new immunotherapy vaccines. Clin. Dev. Immunol. 2013, 385615 10.1155/2013/385615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eichhorn S., Hörschlager A., Steiner M., Laimer J., Jensen B. M., Versteeg S. A., Pablos I., Briza P., Jongejan L., Rigby N., Asturias J. A., Portolés A., Fernandez-Rivas M., Papadopoulos N. G., Mari A., et al. (2019) Rational design, structure-activity relationship, and immunogenicity of hypoallergenic Pru p 3 variants. Mol. Nutr. Food Res. 63, e1900336 10.1002/mnfr.201900336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tscheppe A., and Breiteneder H. (2017) Recombinant allergens in structural biology, diagnosis, and immunotherapy. Int. Arch. Allergy Immunol. 172, 187–202 10.1159/000464104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sambrook J., and Russell D. W. (eds) (2001) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 55. Zuidmeer L., van Leeuwen W. A., Budde I. K., Cornelissen J., Bulder I., Rafalska I., Besolí N. T., Akkerdaas J. H., Asero R., Fernandez Rivas M., Rivas M. F., Gonzalez Mancebo E., Mancebo E. G., and van Ree R. (2005) Lipid transfer proteins from fruit: cloning, expression and quantification. Int. Arch. Allergy Immunol. 137, 273–281 10.1159/000086420 [DOI] [PubMed] [Google Scholar]
- 56. Schocker F., Lüttkopf D., Scheurer S., Petersen A., Cisteró-Bahima A., Enrique E., San Miguel-Moncín M., Akkerdaas J., van Ree R., Vieths S., and Becker W. M. (2004) Recombinant lipid transfer protein Cor a 8 from hazelnut: a new tool for in vitro diagnosis of potentially severe hazelnut allergy. J. Allergy Clin. Immunol. 113, 141–147 10.1016/j.jaci.2003.09.013 [DOI] [PubMed] [Google Scholar]
- 57. Schmutzler G. (2005) Recombinant Production of Allergens of the nsLTP Family in E. coli. Master's thesis, University of Salzburg [Google Scholar]
- 58. Schwarz R. (2004) Celery-mugwort Spice Syndrome: A Possible Involvement of Non-specific Lipid Transfer Proteins. Master's thesis, University of Salzburg [Google Scholar]
- 59. Gadermaier G., Harrer A., Girbl T., Palazzo P., Himly M., Vogel L., Briza P., Mari A., and Ferreira F. (2009) Isoform identification and characterization of Art v 3, the lipid-transfer protein of mugwort pollen. Mol. Immunol. 46, 1919–1924 10.1016/j.molimm.2009.03.021 [DOI] [PubMed] [Google Scholar]
- 60. Gadermaier G., Wopfner N., Wallner M., Egger M., Didierlaurent A., Regl G., Aberger F., Lang R., Ferreira F., and Hawranek T. (2008) Array-based profiling of ragweed and mugwort pollen allergens. Allergy 63, 1543–1549 10.1111/j.1398-9995.2008.01780.x [DOI] [PubMed] [Google Scholar]
- 61. Laimer J., Hofer H., Fritz M., Wegenkittl S., and Lackner P. (2015) MAESTRO–multi agent stability prediction upon point mutations. BMC Bioinformatics 16, 116 10.1186/s12859-015-0548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Köhler G., and Milstein C. (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256, 495–497 10.1038/256495a0 [DOI] [PubMed] [Google Scholar]
- 63. Peri S., Steen H., and Pandey A. (2001) GPMAW—a software tool for analyzing proteins and peptides. Trends Biochem. Sci. 26, 687–689 10.1016/s0968-0004(01)01954-5 [DOI] [PubMed] [Google Scholar]
- 64. Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R. F., Sykes B. D., and Wishart D. S. (2003) VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31, 3316–3319 10.1093/nar/gkg565 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Chemical shift assignments were deposited in the BioMagRes data bank under accession number 28092. All remaining data are contained within the article and the supporting information.