Abstract
Faithful replication of the mitochondrial genome is carried out by a set of key nuclear-encoded proteins. DNA polymerase γ is a core component of the mtDNA replisome and the only replicative DNA polymerase localized to mitochondria. The asynchronous mechanism of mtDNA replication predicts that the replication machinery encounters dsDNA and unique physical barriers such as structured genes, G-quadruplexes, and other obstacles. In vitro experiments here provide evidence that the polymerase γ heterotrimer is well-adapted to efficiently synthesize DNA, despite the presence of many naturally occurring roadblocks. However, we identified a specific G-quadruplex–forming sequence at the heavy-strand promoter (HSP1) that has the potential to cause significant stalling of mtDNA replication. Furthermore, this structured region of DNA corresponds to the break site for a large (3,895 bp) deletion observed in mitochondrial disease patients. The presence of this deletion in humans correlates with UV exposure, and we have found that efficiency of polymerase γ DNA synthesis is reduced after this quadruplex is exposed to UV in vitro.
Keywords: DNA structure, DNA polymerase γ, G-quadruplex, mtDNA deletion, DNA replication, heavy-strand promoter, mitochondrial DNA (mtDNA), mitochondrial DNA damage
Mitochondria are membrane-enclosed organelles that are present in every multicellular eukaryote. Within the mitochondrial membrane are the protein complexes of the electron transport chain, where oxidative phosphorylation leads to over 90% of the cellular ATP production. Eukaryotic cells can contain up to thousands of mitochondrial organelles, and each mitochondrion is home to multiple copies of the mitochondrial genome (mtDNA) found in the matrix of the mitochondrial network (1). The mtDNA encodes 13 polypeptides that make up subunits of the electron transport chain, as well as 22 tRNA genes and two ribosomal genes required for the translation of these 13 polypeptides (2). In humans, these mitochondrial genomes are copied and maintained by the minimal replisome components DNA polymerase γ (Pol γ), Twinkle helicase, and the mitochondrial ssDNA binding protein (mtSSB) (3). Polymerase γ is a heterotrimeric complex composed of a large catalytic subunit, p140, and a homodimer of the accessory subunit, p55 (4, 5). Whereas the p140 catalytic subunit contains the DNA polymerase and 3′–5′ exonucleolytic proofreading functions, the accessory subunit p55 functions to tighten DNA binding and increase processivity during DNA synthesis (6–8).
Replication of the mtDNA is thought to occur via an asynchronous strand displacement model (9, 10). In this model, replication of the genome begins at the heavy chain origin (OriH), in the noncoding region (NCR). Replication proceeds in one direction, as the light (L) strand templates the synthesis of nascent heavy (H) strand. After replication proceeds about two-thirds of the way around the genome, the L-strand origin of replication (OriL) becomes exposed and nascent L-strand replication begins. A unique feature of the strand displacement model is the significant presence of H-strand ssDNA. Transiently exposed ssDNA H-strand potentially increases the likelihood of DNA to adopt secondary structures. The binding of mtSSB proteins may protect and prevent formation of some secondary structures; however, our previous study showed that mtSSB binding to DNA is not cooperative (11), leaving the ssDNA free to fold. Regions of mtDNA that may have a propensity to form secondary structures or roadblocks to DNA replication include the 22 tRNA genes, a stem-loop structure that forms at OriL, and more generally, guanine-rich stretches of the H-strand.
Replication roadblocks present opportunities for the mtDNA replisome to stall or decouple from the helicase, which are believed to be major factors in forming mtDNA deletions. We and others have previously shown that environmental and chemical factors can cause Pol γ to stall (12–16). Furthermore, in patients with mitochondrial disease, multiple deletion events are believed to initiate from stalling of Pol γ as a result of substitutions that render the enzyme partially inactive (17–19).
One of the natural roadblocks that has gained much attention is G-quadruplex (G4) structures (20, 21). The mitochondrial helicase Twinkle has been shown to have significantly reduced efficiency when unwinding some of these structures (21). Although it has been demonstrated that DNA Pol γ displays a significant decrease in efficiency synthesizing past damaged and adducted DNA (13, 14), the effect of structured ssDNA on Pol γ DNA synthesis is largely unknown.
To understand how these natural barriers or roadblocks specifically affect the mitochondrial DNA polymerase, we constructed several artificial DNA substrates to emulate those found in the mtDNA and assessed the ability of human DNA Pol γ to extend primers through natural barriers in the template strand. To our surprise, Pol γ proved capable of replicating through the majority of obstacles tested here, including structured tRNA genes, stem-loops, and G-quadruplexes. Notably, a single, specific G-quadruplex at HSP1 caused significant blockage to primer extension catalyzed by Pol γ. Characterization of Pol γ activity on this substrate, G4_5, allowed us to identify a correlation between this G-quadruplex and a common mtDNA deletion observed in patient samples.
Results
DNA synthesis by Pol γ through stem-loop structures
During mtDNA replication, progressive displacement of the H-strand leads to a significant accumulation of ssDNA. As synthesis of the nascent H-strand passes the OriL, the displaced parental H-strand folds into a stem-loop structure at OriL. This structure functions to recruit the mitochondrial RNA polymerase, which primes synthesis of the nascent L-strand (22). As synthesis of the nascent L-strand nears completion, Pol γ reencounters the OriL stem-loop and must faithfully copy through this structure to complete synthesis. To understand the implications of this stem-loop structure on DNA synthesis by Pol γ, we created an artificial OriL template. We then assessed the activity of the isolated catalytic subunit (p140 alone) and the heterotrimeric form (p140 and the dimeric p55 accessory subunit) of Pol γ on this substrate in primer extension reactions in vitro (Fig. 1). Polymerase activity and the ability to synthesize DNA through the stem-loop structure were measured at both “low” (100 mm) and “high” (200 mm) sodium chloride concentrations to match the activity optima of both enzyme forms (6). The products of primer extension reactions were resolved by urea-PAGE, and DNA synthesis both up to and past the structured region was determined by quantifying DNA substrates, reaction intermediates, and products by fluorescence image scanning.
Figure 1.
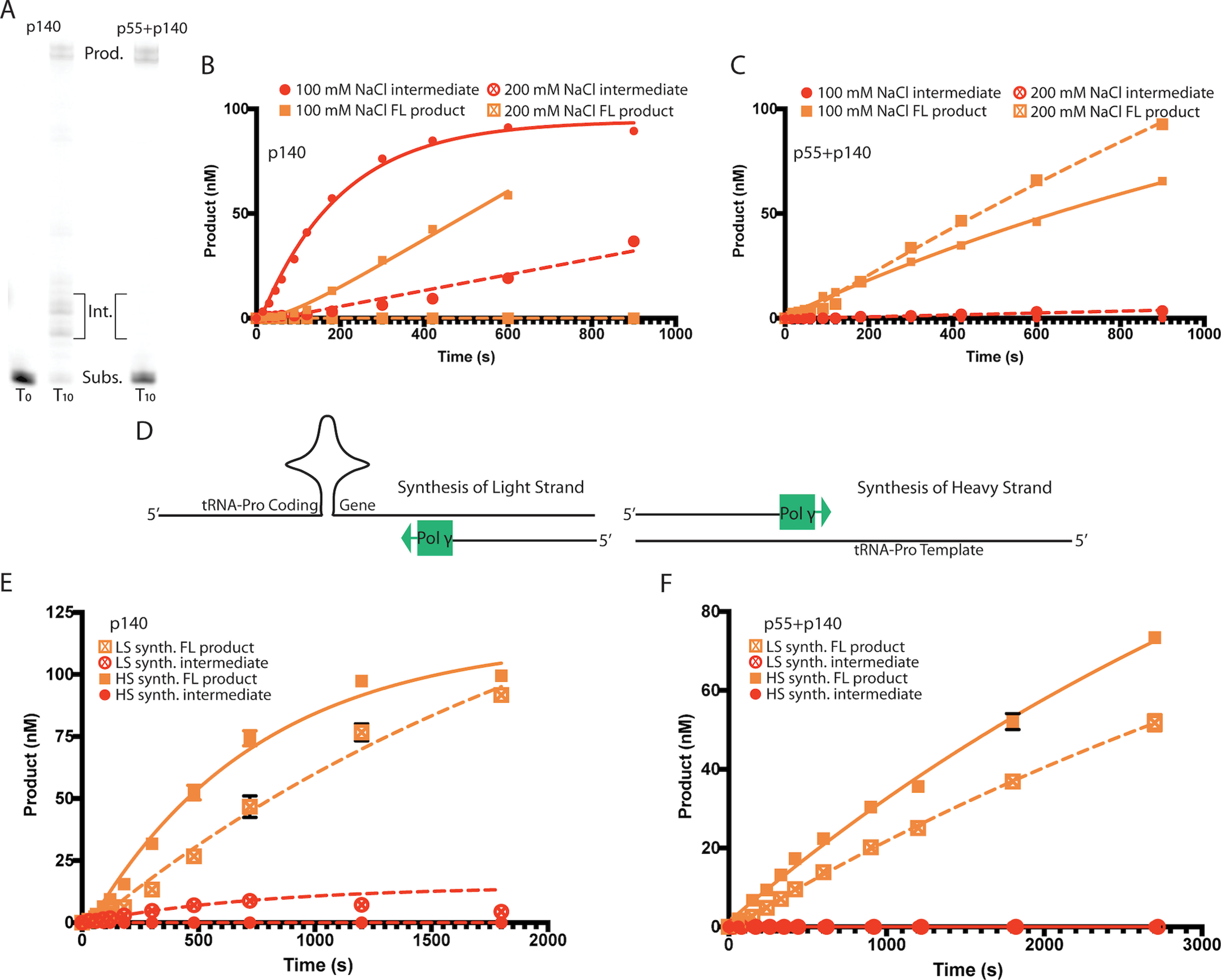
Pol γ primer extension with structured oligo templates. A, illustrative denaturing PAGE gel slices, at time = 0 or 10 min, showing Pol γ catalytic subunit pausing to form intermediate (left) or Pol γ heterotrimer catalyzing only full-length primer extension (right) during primer extension across the OriL template oligo. Full-time course gel is shown in Fig. S1. B, primer extension time course for Pol γ catalytic subunit alone. Substrate template strand is an OriL mimic capable of adopting a stem-loop fold. Formation of reaction intermediates (red) and full-length product (orange) are shown. Reactions were carried out in 100 mm NaCl (closed symbols) and 200 mm NaCl (open symbols). In low salt, p140 (30 nm) catalyzed primer extension to form intermediate species at an initial rate of 0.51 nm/s and full-length product at 0.12 nm/s. At high salt, p140 catalyzed extension to intermediate-length products at 0.04 nm/s. C, primer extension time courses as described in (B) for the Pol γ heterotrimer with an OriL substrate template. The Pol γ complex catalyzed primer extension to produce full-length products in low and high salt at 0.09 nm/s and 0.12 nm/s, respectively. D, schematic representation of tRNA-coding oligo substrates, with Pol γ shown as a green box. The mtDNA gene encoding tRNA-Pro is templated on the light strand and coded by the heavy strand. E and F, reaction progress curves for Pol γ-catalyzed primer extension using either the catalytic subunit alone (E) or the heterotrimer (F) and the tRNA-Pro oligos. Synthesis of the mtDNA light strand is denoted by open symbols, whereas heavy-strand synthesis is denoted by closed symbols. Red circles represent formation of intermediate species and orange squares represent formation of full-length product. When synthesizing L-strand (E), p140 alone (5 nm) catalyzed primer extension to form intermediate species at a rate of 0.02 nm/s and full-length product at 0.08 nm/s. Alternatively, during H-strand synthesis, p140 alone catalyzed primer extension to full-length product at a rate of 0.17 nm/s. The Pol γ complex (F) extended primers synthesizing both L-strand and H-strand to full-length products at 0.02 nm/s and 0.04 nm/s, respectively. Error bars, when larger than data symbols, represent standard error of two replicates.
The catalytic subunit p140 was able to extend the DNA primer at both high and low salt concentrations (Fig. S1, lanes 1–23 and Fig. 1, A–C). Consistent with previous results, primer extension by p140 occurred at a higher rate in the low salt conditions (Fig. 1B). The initial rate of product formation was 0.10 nm/s at low salt and more than an order of magnitude slower at high salt. Interestingly, this rate for forming full-length products was considerably lower than the initial rate for p140 to form intermediate-length products at low salt (0.51 nm/s). Visual inspection of the bands in the urea-PAGE gels indicates accumulation of reaction intermediates 4–6 nucleotides (nt) longer than the original primer, suggesting that DNA synthesis proceeds immediately up to the beginning of the stem-loop structure prior to the enzyme stalling. In contrast, the Pol γ complex (p55+p140) catalyzes DNA synthesis across the stem-loop template to form full-length products under both low and high salt conditions (Fig. S1, lanes 24–46 and Fig. 1C). Thus, addition of the p55 accessory subunit fully alleviates Pol γ stalling on the stem-loop substrate in vitro.
tDNA (structured gene encoding tRNA-Pro)
The secondary structure that forms at OriL during mtDNA replication is well-established (23, 24). However, mtDNA also encodes 22 tRNA genes, and the tDNA sequences for some of these tRNAs may also adopt secondary structures when rendered transiently single-stranded during mtDNA replication. To test the possibility that Pol γ may stall during replication of such tDNA sites, we designed a substrate containing the full tDNA-proline sequence. This tDNA encoding tRNA proline (tRNA-Pro) is the first gene encountered by Pol γ during synthesis of the nascent H-strand in the strand displacement model of mtDNA replication (10, 25). tRNA-Pro is templated on the mtDNA L-strand and coded on the H-strand (Fig. 1D). Therefore, we hypothesized that any replication stalling would exhibit a strand bias, wherein synthesis of the L-strand, but not of the H-strand, may be affected by the tRNA-Pro gene. Indeed, reactions utilizing only p140 exhibited brief stalling when extending the L-strand at tDNA-Pro as judged by the accumulation of replication intermediates (Fig. 1E). Initial rates for formation of the intermediate-length product were 4-fold lower than the rate of generating the full-length product (Table 1). Synthesis of full-length tDNA-Pro H-strand was twice as efficient as L-strand synthesis, and p140 exhibited initial rates for full-length product formation of 0.17 nm/s and 0.08 nm/s, respectively (Table 1). This increased apparent efficiency is likely due to the absence of detectable stalling by p140 when replicating the H-strand sequence (Fig. 1E).
Table 1.
Pol γ synthesis across tRNA-Pro gene
Initial rates of intermediate and product formation catalyzed by Pol γ. The substrate column represents the template strand of DNA.
| Protein(s) | Substrate | Vo,int (nm/s) | Vo,prod (nm/s) |
|---|---|---|---|
| p140 | tRNA-Pro coding | 0.02 | 0.08 |
| p140 | tRNA-Pro template | <0.01 | 0.17 |
| p55+p140 | tRNA-Pro coding | <0.01 | 0.02 |
| p55+p140 | tRNA-Pro template | <0.01 | 0.04 |
Similar to the results with the OriL-based substrate, the addition of accessory subunit p55 ablates the accumulation of any intermediate-length species during either H-strand or L-strand synthesis on the tDNA-based substrate (Fig. 1F). Synthesis of the L-strand catalyzed by p55+p140 occurs at a slower initial rate than synthesis of the H-strand (Table 1). These data are consistent with Pol γ–catalyzed primer extension slowing during DNA synthesis across potentially structured DNA templates but also suggest that Pol γ is able to competently synthesize DNA across tRNA-coding genes. Along with the results using the OriL substrate, these data continue a pattern of replication challenges affecting L-strand synthesis by p140 that are relieved by addition of the accessory subunit p55.
Displacement (encountering 5′ DNA or RNA)
Whereas mtDNA is densely packed with genes, ∼1 kb between tRNA-Phe and tRNA-Pro is notably barren. This noncoding span of the mtDNA (the NCR) is considered a “control region.” Synthesis of the mtDNA H-strand at OriH is initiated by an RNA priming event in the NCR (9). After replicating the entire H-strand of the circular mtDNA genome, Pol γ will encounter either the original RNA primer or the 5′-end of the nascent H-strand DNA in the mtDNA control region. Accordingly, we designed a hairpin-containing oligonucleotide substrate to test the response of Pol γ upon encountering downstream RNA or DNA 5′-ends. This substrate consists of a primer-template DNA hairpin, a 5-nt gap, and downstream RNA and DNA oligos annealed to the hairpin (Fig. 2A). The three oligonucleotide components were labeled with distinct fluorophores to facilitate concurrent monitoring of each by urea-PAGE and fluorescence imaging.
Figure 2.
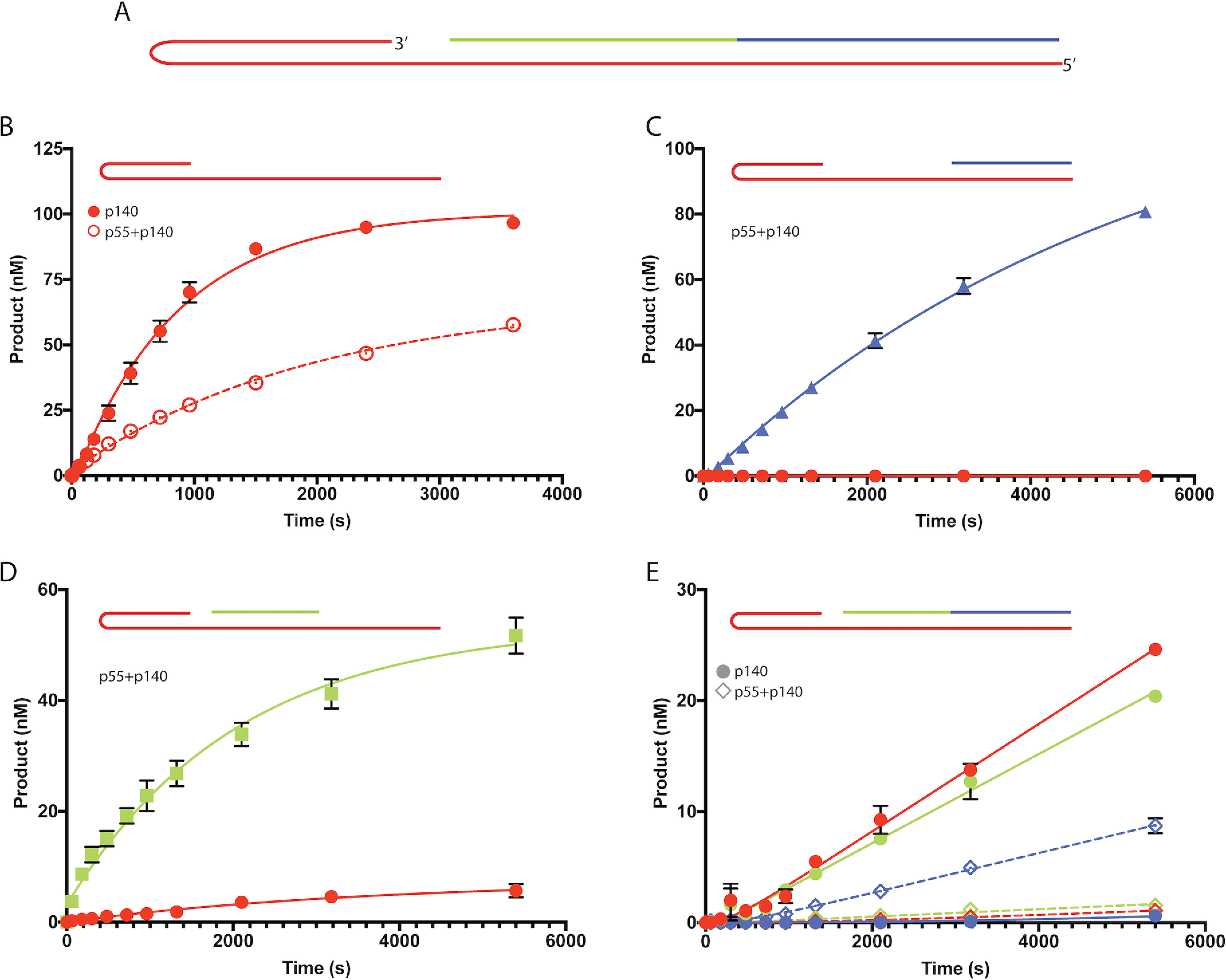
Displacement of downstream oligonucleotides. A, hairpin substrate used to measure the strand displacement activity of Pol γ. The red oligo is a Cy5-labeled 100-mer hairpin and provides both the template and the 3′-OH site for Pol γ extension. The green strand is a 25-mer RNA with an internal Cy3 label. The blue strand, which anneals immediately adjacent to the RNA, is a 28-mer DNA with an internal FAM label. The three fluorophores excite at unique wavelengths, allowing for visualization of each oligo component after PAGE. B, reaction progress curves showing activity of Pol γ catalytic subunit alone (closed circles) and heterotrimer (open circles) on the hairpin substrate lacking downstream oligonucleotides. Pol γ catalyzes primer extension to full-length product. C and D, reaction progress curves for Pol γ heterotrimer with the hairpin substrate and either the downstream 28-mer DNA (C) or 25-mer RNA (D). Displacement of the downstream oligo is represented by the matching colored symbols (displaced DNA is blue and displaced RNA is green. Red symbols represent products from gap-filling that stalled at the downstream oligo). E, reaction progress curves for Pol γ activity on the hairpin substrate with both downstream oligos annealed. Closed circles represent activity by the Pol γ catalytic subunit alone and open diamonds represent the heterotrimer. Red symbols correspond to intermediate products formed through gap-filling activity, green symbols correspond to higher molecular weight intermediate products formed through displacement of the RNA strand, and blue symbols correspond to full-length products formed through displacement of both RNA and DNA strands. In all graphs, error bars, when larger than data symbols, represent standard error of up to six replicates in (B), triplicates in (C) and (D), and duplicates in (E).
In control reactions using only the hairpin-containing oligo, both p140 and p55+p140 catalyzed primer extension to the full length of the hairpin template (Fig. 2B). In the absence of any downstream nucleic acid (either RNA or DNA), p140 synthesized full-length product with a rate constant for total product oligo (kcat = 0.024 s−1) about 4× higher than that of the Pol γ heterotrimer (kcat = 0.006 s−1).
To test the ability of Pol γ to perform strand displacement DNA synthesis, the 28-mer DNA oligo was annealed at the downstream end of the hairpin template, leaving a 30-nt gap between the 3′-OH primer and the 5′-end of the downstream barrier strand. Pol γ heterotrimer (p55+p140) catalyzed extension of the primer across the full length of the DNA template, effectively displacing the downstream DNA strand (Fig. 2C). No detectable intermediate-length species were formed, suggesting that pausing of the polymerase did not occur to an appreciable level. However, the kinetic parameters show that displacement of the downstream DNA comes with an apparent 2–3-fold decrease in kcat (Table 2). Using this same methodology, the 25-mer RNA was annealed to the hairpin substrate to leave a 5-nt gap between the primer 3′-OH and the RNA 5′-end. Consistent with the result for the downstream 28-mer DNA, the enzyme displaced the RNA and catalyzed synthesis of full-length product (Fig. 2D) with the same 2–3-fold decrease in kcat relative to that for the hairpin substrate lacking downstream barriers (Table 2). Interestingly, this substrate also presents a second 3′-OH from which Pol γ could extend: the 3′-end of the 25-mer RNA strand. Consistent with previous studies, Pol γ extends the RNA primer at a reduced efficiency compared with extension of a DNA primer (Table 2) (26, 27).
Table 2.
Displacement of downstream oligos
Rate constants for oligo displacement using a hairpin oligo primer and template. Rate constants are per molecule of product formed. Standard errors are indicated and calculated based on 2–6 reaction replicates as noted in Fig. 2, B–E.
| Protein(s) | Substrate present |
kcat × 10−4 (s−1) |
||||
|---|---|---|---|---|---|---|
| Hairpin | 25-mer RNA | 28-mer DNA | Gap-filling | Displacement of 25-mer RNA | Full-length product | |
| p140 | + | − | − | n/a* | n/a* | 240 ± 14 |
| p55+p140 | + | − | − | n/a* | n/a* | 60 ± 2 |
| p55+p140 | + | +‡ | − | <0.1† | n/a* | 25 ± 2 |
| p55+p140 | + | − | + | <0.1† | n/a* | 24 ± 1 |
| p140 | + | + | + | 9.6 ± 0.4 | 8 ± 0.2 | <0.1† |
| p55+p140 | + | + | + | 0.6 ± 0.1 | 0.7 ± 0.1 | 3.6 ± 0.1 |
*Data points are not applicable for this enzyme/substrate combination.
†Activity was below measurable threshold.
‡kcat for extension from this exposed ribonucleotide primer 3′-OH is 0.5 × 10−4 s−1.
Finally, both the 28-mer DNA and 25-mer RNA oligos were annealed with the hairpin substrate concurrently (Fig. 2E). The p140 catalytic subunit alone was able to catalyze synthesis across the 5-nt gap (kcat = 9.6 × 10−4 s−1) and displace the RNA to form a higher molecular weight intermediate (kcat = 8.0 × 10−4 s−1). However, p140 alone was unable to displace the 28-mer DNA downstream from the RNA oligo, so no full-length product was formed (Fig. 2E and Table 2). In contrast, the Pol γ heterotrimer (p55+p140) catalyzed displacement of both the RNA and DNA to generate full-length product. Although the full-length product was the dominant species observed, the heterotrimer also generated small amounts of reaction intermediates that corresponded to filling the 5-nt gap and displacing the RNA strand. Although displacement of either the downstream RNA or DNA oligonucleotide resulted in a 2- to 3-fold decrease in kcat (see above), the presence of both downstream oligos decreased enzyme turnover 17-fold (from 60 × 10−4 s−1 to 3.6 × 10−4 s−1) relative to the hairpin lacking all barriers (Table 2).
G-quadruplexes
DNA synthesis by the nuclear replicative polymerases α, δ, and ε, and the repair polymerase κ, stalls when the enzymes encounter G4 structures in the template DNA strand (28–30). Because the mtDNA H-strand is enriched for guanine relative to the L-strand and has a naturally higher propensity to form G4 structures (31), we hypothesized that G4 structures in mtDNA would be a barrier to primer extension by Pol γ. To gain a preliminary understanding of the behavior of Pol γ upon encountering a G-quadruplex during DNA synthesis, we designed a pair of oligonucleotide substrates modeled after a canonical G4 motif found by analyzing human nuclear DNA (32, 33). The positive control substrate (G4_Cntrl) follows a GxLaGxLbGxLcGx pattern in which guanine runs (Gx runs of 3–5 nt) are separated by loops (La–c loops <7 nt.) in the template DNA strand. The corresponding negative control substrate (G4_Neg) substitutes nonguanine residues within the Gx runs on the template strand (Table S1).
In primer extension reactions utilizing the G4_Neg substrate, Pol γ was able to synthesize full-length products in the presence or absence of the accessory subunit (Fig. 3, A–C). In contrast, when utilizing the G4_Cntrl substrate, both forms of Pol γ rapidly synthesized just 6 nt before pausing at the start of the G4 structure (Fig. 3, D–F). The intermediate species reach a concentration greater than the enzyme, consistent with the enzyme dissociating from the oligo substrates after stalling at the G4 structure. However, the quadruplex is not a complete block to DNA synthesis, and Pol γ was able to efficiently synthesize full-length product following the lag period spent creating the intermediate and pausing or dissociating. The transient accumulation of intermediate species is more pronounced with p140 alone (Fig. 3, E and F), suggesting that the p55 accessory subunit facilitates synthesis through the G4 structure on the template strand. Furthermore, because intermediate accumulates to concentrations greater than enzyme before declining, these data suggest that Pol γ reassociates with aborted substrates to convert intermediate to product.
Figure 3.

Primer extension by Pol γ pauses at G4 structures. A, time course of primer extension by Pol γ heterotrimer on the G4_Neg substrate was assessed by denaturing PAGE. B and C, activity of the Pol γ catalytic subunit alone and the Pol γ heterotrimer, respectively, on the G4_Neg oligo substrate. Pol γ catalyzes primer extension to form only full-length product (orange squares). No intermediate species are observed (red circles). D, time course of primer extension by Pol γ heterotrimer with the G4_Cntrl substrate was assessed by denaturing PAGE. Both intermediate-length and full-length products are observed. E and F, activity of the Pol γ catalytic subunit alone and the Pol γ heterotrimer, respectively, on the G4_Cntrl oligo substrate. Pol γ catalyzes primer extension to form both full-length product (orange squares) and intermediate species (red circles). Total primer extension of both intermediate- and full-length products is shown (green diamonds).
With these data in hand, we sought to examine the effects of G-quadruplexes on replication of mtDNA sequences. The web tools G4-Hunter (34) and QGRS (35) were used to map potential G4 structures in the human mitochondrial genome (NC_012920). Over 90 putative quadruplex-forming sequences were found, with a majority identified by both predictive algorithms (File SA). From these predictions, a small G4 oligo library was assembled. A series of substrates was designed to share a common primer and complementary strand. The complementary strand was extended with variable sequences that mimic native G4 sites in mtDNA to act as ssDNA templates. In total, eight G4 sequences were chosen (Table S1) because of their strong predictive scores for structure formation, or, in the case of G4_3, because of its unusual location in the mtDNA light strand (Fig. 4).
Figure 4.
G4 substrate library. MtDNA was screened for potential G-quadruplex structures with the G4 Hunter and QGRS mapping algorithms. Eight specific G-quadruplex–forming sequences were selected as substrates to assess primer extension activity of Pol γ in vitro. A, mapped location of the eight mtDNA sequences chosen as quadruplex formations. B, schematic of the library design for G4 substrates. The substrates share a common primer/template combination. The primer (red) has a 5′ Cy5 label. The conserved template (black) is extended by a 9-nt spacer (orange), separate mtDNA sequences containing putative G-quadruplexes (green), and a 6-nt flanking tail (orange).
The secondary structure of each G4 oligonucleotide was analyzed by CD spectroscopy. Changes in the CD spectra of G-quadruplexes provide information on specific topology (parallel, antiparallel, or mixed). For example, parallel G-quadruplexes tend to have CD maxima near 265 nm and minima near 240 nm, whereas antiparallel G4s have CD maxima near 295 nm and minima near 265 nm. G4 structures with mixed topology often show CD maxima at both 265 nm and 295 nm and a minima near 240 nm. Conversely, unstructured ssDNA typically shows local maxima and minima at 280 nm and 250 nm, respectively. All eight selected G4 sequences demonstrate formation of secondary structure, and each oligonucleotide substrate shows a CD spectrum consistent with either parallel or mixed topologies (Fig. S2). No antiparallel G4s were observed. These eight substrates (G4_1–G4_8) provide a biologically relevant sampling of G4-forming mtDNA from varying regions of the genome (Fig. 4A).
As before, the efficiency of Pol γ to utilize each G4 mtDNA substrate (Fig. 4B) was assessed in primer extension reactions. The initial rates for generation of intermediate-length (Vo,I) and full-length (Vo,P) products were compared to quantify the degree of blockage posed by each of the various G4 substrates. The activity of the Pol γ catalytic subunit alone varied between the substrates (Table 3). One of the substrates, G4_3, caused little or no blockage to synthesis of full-length product (Vo,I/Vo,p < 0.1), whereas four substrates, G4_2, G4_4, G4_6, and G4_8, caused “moderate” blockage (defined here by 0.1 ≤ Vo,I/Vo,p ≤ 1.0), and three substrates, G4_1, G4_5 and G4_7, led to a “severe” blockage of primer extension (Vo,I/Vo,p > 1.0). The Pol γ heterotrimer efficiently utilized most of these substrates. Five substrates posed little or no blockage, two substrates posed only moderate blockage, and only one substrate (G4_5) presented a severe blockage to primer extension (Table 3). Because K+ can have a greater stabilizing effect on G-quadruplexes than Na+ (36), we also measured primer extension in the presence of 100 mm KCl on three substrates that exhibited low (G4_8), moderate (G4_1), and strong (G4_5) blockage in standard reactions with NaCl. Although potassium ions appear to provide moderate stabilization, the primer extension patterns and substrate ranking for the Pol γ heterotrimer remain consistent for these quadruplex structures (Table 3, see footnote).
Table 3.
G-quadruplex substrate library
Summary of eight G4-forming substrates used for in vitro activity assays with Pol γ. The Vo column is the ratio of initial rates of formation for intermediate-length and full-length products. Values less than 1 favor formation of full-length product, whereas values greater than 1 favor formation of intermediate-length products. Nucleotide sequences of these substrates are listed in Table S1.
| Substrate | Heavy or light strand | nt position | p140 | p55/p140 |
|---|---|---|---|---|
| G4_1‡ | HS | 3595–3556 | 12.3 | 0.6 |
| G4_2 | HS | 9268–9234 | 0.1 | 0† |
| G4_3 | LS | 16058–16027 | 0† | 0† |
| G4_4 | HS | 2085–2048 | 0.1 | 0† |
| G4_5‡ | HS | 581–525 | 71.1 | 28.2 |
| G4_6 | HS | 10220–10169 | 0.6 | 0† |
| G4_7 | HS | 13067–13017 | 2.8 | 1 |
| G4_8‡ | HS | 8290–8243 | 0.6 | 0† |
†Value zero denotes no detectable intermediate formation.
‡In reactions substituting 100 mm KCl for NaCl, ratios of initial rates with the p55/p140 heterotrimeric complex were 2.0 for G4_1, 63.7 for G4_5, and 0 for G4_8 (intermediate products were undetectable).
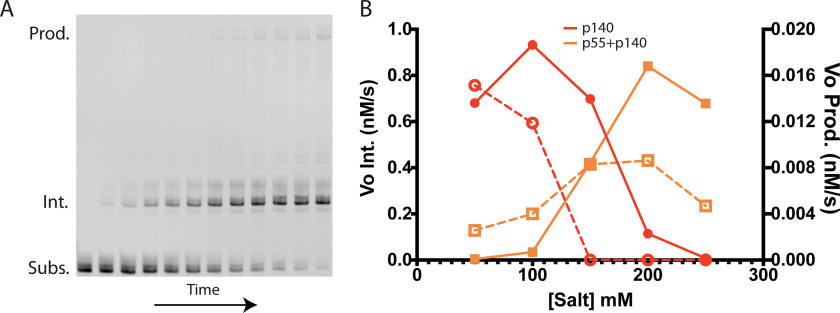
The G4_5 quadruplex substrate was the only structure that blocked primer extension for both the isolated catalytic subunit and the Pol γ complex. This unexpected result prompted us to examine utilization of the G4_5 quadruplex substrate more closely under reaction conditions known to minimize enzyme disassociation from DNA and to favor processive DNA synthesis. For example, monovalent salt can modulate the primer extension activity of Pol γ in vitro (37, 38). Also, the presence of the dimeric p55 accessory subunit raises the DNA binding affinity of the Pol γ complex, which allows the complex both to remain bound to DNA at higher salt concentrations and to synthesize significantly longer DNA products prior to disassociation from the DNA primer template (6–8). Of particular interest is whether or not monovalent salt ions differentially affect stalling and dissociation of Pol γ at the G4_5 structure. Accordingly, we measured the DNA polymerase activity of the isolated catalytic subunit and the heterotrimeric Pol γ complex in primer extension reactions with salt concentrations ranging from 50 to 250 mm NaCl (Fig. 5A). The catalytic subunit displayed maximum activity at 100 mm NaCl. Higher salt concentrations reduced activity, and initial reaction rates dropped more than 10-fold at 250 mm NaCl (Fig. 5B). In contrast, the activity of the heterotrimeric Pol γ complex is positively associated with salt concentrations increasing from 50 to 200 mm NaCl, and activity begins to decline with >200 mm NaCl. Although these activity profiles are not unusual, both forms of the enzyme clearly generated intermediate-length products far more rapidly than full-length products at all salt concentrations. The inability to relieve this potent block to DNA synthesis by optimizing the concentration of monovalent salt suggests that the G4_5 quadruplex structure promotes both stalling and disassociation of Pol γ.
Figure 5.
Pol γ catalytic activity with G4_5 substrate is salt-dependent. A, sample PAGE gel depicting a Pol γ time course from left to right. The Pol γ heterotrimer was used for this time course, with salt at 200 mm. Substrate is primarily extended to the intermediate species, though it occasionally reaches full-length product. B, initial rates for formation of the intermediate (Vo Int., closed symbols) and full-length product (Vo Prod., open symbols) by either the isolated catalytic subunit (red circles) or the heterotrimeric Pol γ complex (orange squares) are plotted.
Correlation between the G4_5 structure and a UV-induced deletion breakpoint
The variable region of the G4_5 quadruplex substrate matches H-strand nucleotides 581–525 of the human mtDNA sequence (Table 3 and Fig. 4A). This region also contains the HSP1 sequence (initiating at nt 561) that is essential for transcription of the two rRNA sequences in vivo (39, 40). Previously, Moraes et al. (41, 42) described a replication-competent human mtDNA bearing a 3,895-bp deletion (mtDNA: 547:4443, MITOMAP ID:RRID:SCR_002996) in two patients with progressive external ophthalmoplegia. The deletion was flanked by 13-bp direct repeat sequences and resulted in loss of both rRNA sequences and the HSP1 at the first deletion breakpoint (Fig. 6A). Interestingly, we show here that the G4_5 quadruplex sequence also coincides with first 13-bp direct repeat associated with this large mtDNA deletion (Fig. 6B). The conspicuous overlap of these features with the mtDNA breakpoint suggests that they may be contributing factors to generation of this deletion. Subsequently, the frequency of occurrence of the 3,895-bp deletion was also found to show a significant correlation with increasing UV exposure in samples collected from human skin (43). Because UV light has been shown to crosslink human telomeric G-quadruplexes by formation of cyclobutane pyrimidine dimers between thymine bases on adjacent loops (44–46), we hypothesized that UV crosslinking of the G4_5 oligonucleotide substrate stabilizes this quadruplex structure and enhances stalling of DNA synthesis by Pol γ in vitro.
Figure 6.
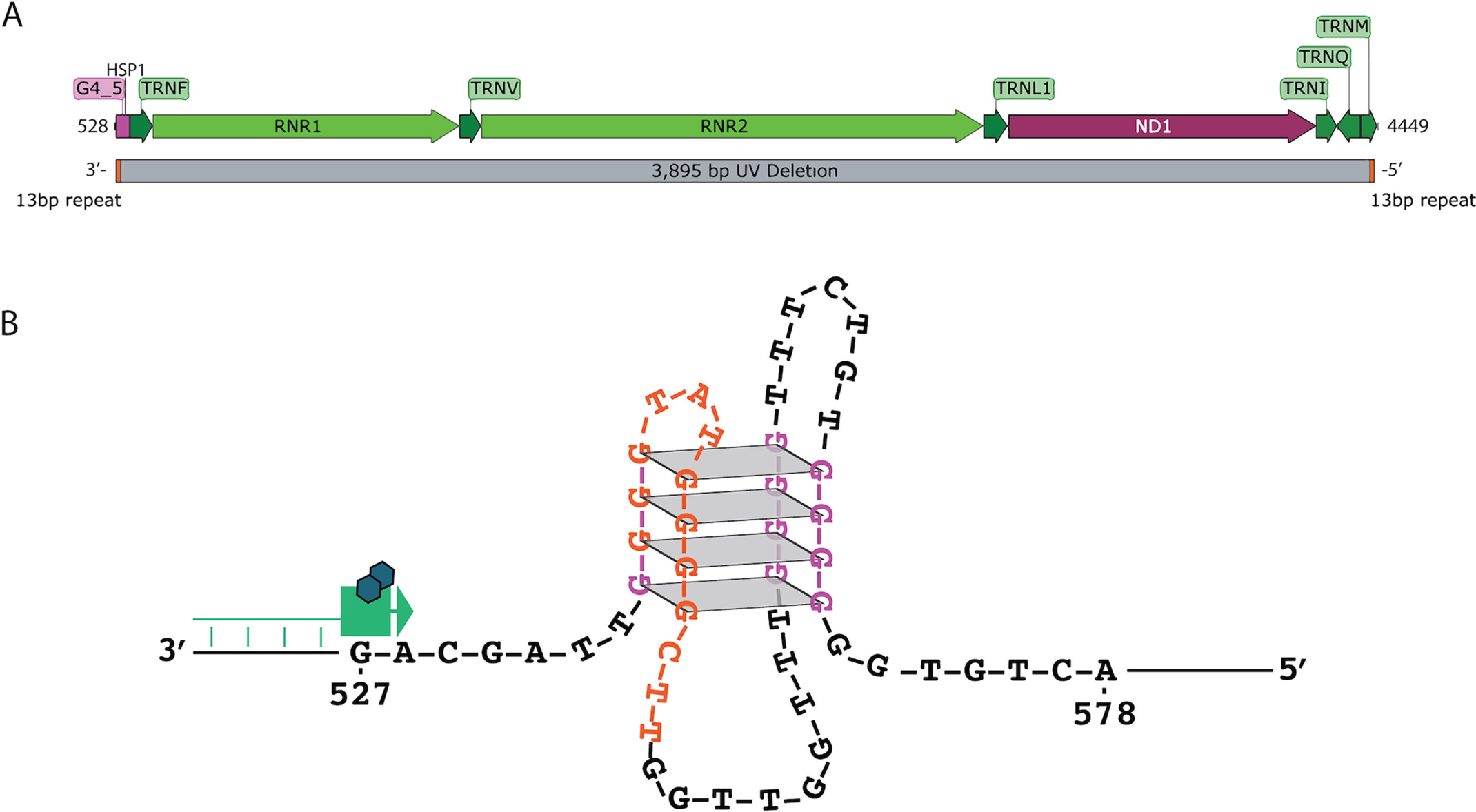
Schematic of G4_5 positioned at UV breakpoint. A, portion of mtDNA that has been reported to be deleted in UV-exposed human skin samples. Mapped along the deletion are the genes lost. The deletion is flanked by 13-bp repeats (orange). The quadruplex formed by G4_5 is shown in purple. B, predicted mixed topology structure of G4_5 based on CD spectra. The 13-bp repeat, shown in orange, overlaps with the first and second stretch of tetrad guanines. In our in vitro experiments, DNA synthesis by Pol γ (green) pauses at the G-quadruplex structure. HSP1 corresponds to nucleotides 561–570, where position 561 is the 5′-end of the transcript.
To approach this idea, we exposed samples of the tRNA-Pro and G4_5 quadruplex substrates to varying doses of 254-nm UV radiation. We first exposed the tRNA-Pro substrate to either 10,000 J/m2 or 30,000 J/m2 of UV irradiation to control for UV dosage. The ability of the Pol γ complex to utilize the treated substrates was tested in primer extension reactions in vitro, and reaction products were resolved by urea-PAGE and quantified as before. Replication through UV crosslinks was blocked in a dose-dependent manner (Fig. S3), as previously shown by Kasiviswanathan et al. (13). On both UV-treated and untreated G4_5 quadruplex substrates, the Pol γ complex could extend primers to generate three distinct length classes of reaction intermediates (intermediates A, B, and C in Fig. 7A) and full-length product. The overall efficiency of primer utilization, judged as the sum of all intermediate- and full-length product species, was equivalent for both UV-treated and untreated G4_5 substrates (Fig. 7B). However, the accumulation of certain intermediate-length species changed with irradiation of the G4_5 template (Fig. 7C). For example, initial rates for forming intermediate A (circles) were similar for both treated and untreated template strands, but this shortest reaction intermediate was slowly consumed at longer incubation times only when the template was unirradiated. Conversely, intermediate B (squares) formed slightly more quickly with the unirradiated template, suggesting that intermediate B may be formed by extension of intermediate A on this substrate. The most significant difference in formation of intermediate species occurs with intermediate C, the longest class of reaction intermediates. Whereas accumulation of intermediate C was barely detectable with the untreated substrate, irradiation of the G4_5 template led to a significant increase in the accumulation of intermediate C (diamonds). Taken together, these data suggest that the decreased formation of full-length primer extension product in reactions utilizing the UV irradiated G4_5 template is attributable to the greater accumulation and stability of intermediate species.
Figure 7.
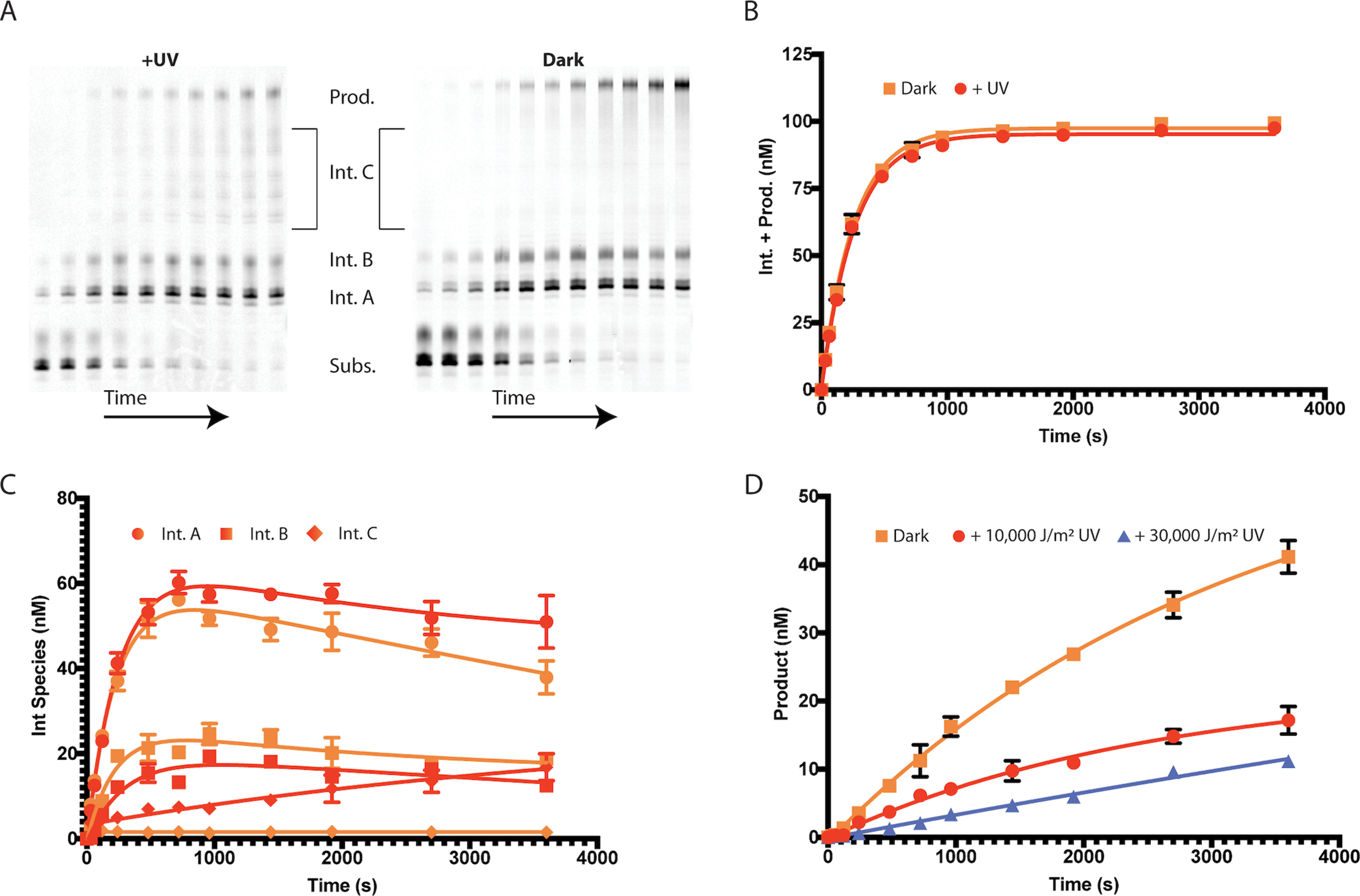
UV irradiation of G4_5 exacerbates stalling by Pol γ. The G4_5 quadruplex substrate was exposed to UV radiation, as described under “Experimental procedures.” A, the activity of Pol γ complex (p140+p55) on the G4_5 quadruplex substrates that had been either exposed to 10,000 J/m2 (+UV) or shielded from (Dark) 254 nm UV light was assessed in primer extension reactions. Products generated as a function of time were analyzed by denaturing PAGE. The positions of substrate, full-length product, and three main regions of intermediate-length products (intermediates A, B, and C) are labeled. B, the sum of all intermediate- and full-length product species generated at the indicated reaction times from +UV (red) and dark (orange) G4_5 quadruplex substrates was quantified. C, intermediate species formed by Pol γ on G4_5 quadruplex substrates that were exposed to (red) or shielded from (orange) UV were quantified for intermediates A (circles), B (squares), and C (diamonds) for the indicated reaction times. D, full-length product species formed by Pol γ on G4_5 quadruplex substrates that were shielded (orange squares) or exposed to either 10,000 J/m2 (red circles) or 30,000 J/m2 (blue triangles) of UV irradiation were quantified for the indicated reaction times. Error bars, when larger than data symbols, represent standard error of two replicates.
Next, we examined whether increased irradiation of the template strand would further decrease the rate for Pol γ to generate full-length primer extension products. Pol γ efficiently synthesized full-length product on untreated G4_5 substrates (kcat = 19.1 × 10−4 s−1). When the G4_5 substrates had been treated with 10,000 J/m2 UV, the reaction rate dropped 2-fold (kcat = 8.9 × 10−4 s−1) (Fig. 7D). We also examined reactions with the G4_5 substrate irradiated with 30,000 J/m2 UV, and the reaction rate decreased a further 2.5-fold (kcat = 3.5 × 10−4 s−1). Collectively, the decrease in efficiency of creating full-length products was roughly proportional to the UV dose utilized to pretreat the G4_5 quadruplex substrate. Accordingly, we infer that UV radiation may be stabilizing the G4_5 quadruplex structure, thereby enhancing the barrier to primer extension by Pol γ.
Discussion
Because of the asynchronous method of DNA replication in mitochondria, the mtDNA replication machinery is likely to encounter several potential roadblocks. Here, we examined the ability of the mtDNA polymerase γ to replicate through several of these obstacles by relaxing secondary structure in the templating ssDNA. Efficiency of DNA replication past these physical barriers was assessed via primer extension assays in vitro using both the Pol γ heterotrimer and the catalytic subunit alone.
Previous work by our laboratory demonstrated that DNA synthesis by Pol γ is severely impaired by large DNA adducts and UV-induced cyclobutane dimers of thymine (13, 14). Specifically, the bulky exocyclic adduct γ-hydroxy-1,N2-propano-2′-deoxyguanosine, formed through the reaction of acrolein with DNA, was found to cause a nearly complete block to incorporation of incoming dCTP. Likewise, DNA templates containing thymine dimers were replicated with an 800-fold decrease in efficiency compared with nondamaged DNA. In the current study, we sought to understand how Pol γ synthesizes DNA on templates containing naturally occurring barriers to replication.
Stem-loops and structured genes
During mtDNA replication, the light-chain origin of replication, OriL, is exposed as ssDNA. This region has been shown to adopt a stem-loop structure required for recruitment of the mitochondrial RNA polymerase (24, 47). When performing synthesis alone, the catalytic subunit p140 paused during replication across the stem-loop structure in the template strand. Interestingly, the addition of the accessory subunit p55 to the reaction led to a decrease in stalling because the heterotrimeric Pol γ favored formation of full-length product. Similar behavior was observed for Pol γ on DNA templates encoding the tRNA-Pro gene, and this tRNA-encoding substrate provided additional, strand-specific information. Both the catalytic subunit and the heterotrimeric form of Pol γ exhibited higher DNA synthesis rates with the noncoding L-strand as the DNA template compared with the rates with the complementary coding H-strand as template. These data are consistent with the interpretation that the Pol γ heterotrimeric enzyme is well-suited to replicate through regions of the genome that may have adopted secondary structures.
Strand displacement
Because the mitochondrial genome is circular, each round of mtDNA replication can be expected to result in the replication machinery encountering the 5′-end of either primer RNA or DNA. Based on the in vitro findings here, p140 is able to displace a downstream RNA strand hybridized to the DNA template but does not appreciably displace an adjacent DNA strand hybridized distal to this RNA. The Pol γ heterotrimer, on the other hand, facilitated displacement of both downstream RNA and DNA oligos, although both forms of the polymerase exhibited a greater than 10-fold reduction in the rate constant kcat during strand displacement synthesis. Flap endonuclease 1 (Fen1) localizes to the mitochondria (48, 49) and has been proposed to process the 5′-RNA primers at the end of DNA replication (50, 51). Furthermore, Kazak et al. (52) showed that Fen1 displays a preference for ssRNA and has an affinity for substrates containing a 5′-RNA flap. We suggest that, in the absence of efficient removal of the RNA primer by RNaseH1 and ExoG (53, 54), the strand displacement activity of Pol γ may generate a substrate for Fen1 (48, 50). Alternatively, in the event of more extensive strand displacement, DNA2 can digest the 5′-portion of a displaced primer to produce a substrate for Fen1 (51). A flap structure must be generated in either case, and we show here that the Pol γ complex is sufficient to generate such 5′-flaps.
G-quadruplexes
The mtDNA heavy strand has a propensity for G-quadruplex formation (31). Additionally, G4-forming regions have been shown to exist in close proximity to known mtDNA deletion breakpoints (20, 21), and they may interfere with mitochondrial gene transcription (55). Recently, Butler et al. (56) analyzed two Italian cohorts and found that overlap between G-quadruplex–forming regions in mtDNA and increased rates of mutagenesis. Although activity of Pol γ was measured in the above-mentioned body of work, the in vitro experiments focused on DNA helicase activity and not on the kinetics of Pol γ. Therefore, we utilized steady-state kinetics to assess polymerase activity and stalling on a larger set of G4-forming substrates. A small library of eight quadruplex-forming sequences were chosen from the predicted G4 sites. To our surprise, most of these substrates did not pose an absolute barrier to Pol γ primer extension (Table 3). Substrate G4_1 in our study is modeled on the same G-quadruplex (MitoG4-29) used by Butler et al. (56). Consistent with results from that study, G4_1 posed a moderate block to synthesis of full-length product by p140 in our hands. However, we found the enzyme capable of extending primers to form both intermediate- and full-length products in the presence of the accessory subunit p55. Only one quadruplex-forming oligo that we tested posed a strong block to primer extension by the Pol γ heterotrimer, and we decided to investigate this G4_5 substrate more carefully.
Substrate G4_5 mimics a quadruplex in the heavy strand of mtDNA that spans nucleotides 581–525 (Fig. 6), which not only includes the HSP1 but also houses one end of a 3,895-bp deletion first identified by Moraes et al. in 1991 (41). More than a decade later in 2004, Krishnan et al. (43, 57) discovered that incidence of this large deletion was strongly correlated with UV exposure in human patients. Additionally, the formation of this deletion could be triggered in cell culture through exposure to UV radiation. Ensuing studies have found that this deletion is also present in human corneal stroma and macular regions of the retina (58).
Along these lines, it is known that UV irradiation induces thymine dimers and 6–4 cyclobutane pyrimidine photoproducts in DNA. Although UV photoproducts are excised and repaired by nucleotide excision repair in the nucleus, nucleotide excision repair is absent in mitochondria (59). Persistent UV photoproducts are a barrier to DNA replication, and we have previously demonstrated that UV photoproducts essentially block DNA replication by human Pol γ, although a low level of lesion bypass was detectable in vitro (13). Such roadblocks can act as initiation sites for deletions, and others have correlated UV damage in mtDNA with an increase in mtDNA deletions (60, 61).
Intrigued by these findings, we sought to evaluate whether UV exposure could affect primer extension by Pol γ across the G-quadruplex structure coinciding with the 3,895-bp deletion breakpoint. Indeed, we found that exposure to UV radiation resulted in a dose-dependent increase in intermediate formation with a concomitant decrease in formation of full-length products.
Taken together, the data presented here suggest that the quadruplex formed at the HSP1 marks a notable exception from the overall finding that many of the structured regions tested are unlikely to pose a strong barrier to DNA synthesis by Pol γ and are therefore unlikely to be major contributors to the generation of mtDNA deletions. We recently identified millions of mtDNA deletions in a large scale, ultrasensitive sequencing analysis of human muscle tissues (62). Although we did detect, albeit infrequently, deletions with one end-point matching the mtDNA location of our G4_5 substrate, a general correlation between predicted quadruplex sites overall and mtDNA deletions observed in vivo could not be established (62). Substrate G4_5 generated a strong block to primer extension by Pol γ over a range of conditions in vitro. The 3,895-bp deletion is flanked by two 13-bp direct repeat sequences, and the first repeat encompasses the first two strands and the first loop of the G4_5-quadruplex (shown in orange in Fig. 6B). Stalling or disassociation of the replication machinery upon encountering this structure may lead to polymerase slippage and/or primer relocation to the corresponding distal repeat sequence to facilitate generation of this deletion.
We show here that DNA synthesis catalyzed by Pol γ through this quadruplex, in vitro, is UV-sensitive, a finding that supports current in vivo data showing that the 3,895-bp deletion correlates with UV exposure. These findings, combined with the current literature, present the exciting possibility that the quadruplex structure formed at HSP1 plays an important regulatory role in mtDNA replication, mtDNA transcription, or both. For example, transcription of 12S and 16S rRNA genes requires HSP1 to be intact. As a structural barrier to Pol γ DNA synthesis, the HSP1 quadruplex could function as a natural pause site for L-strand synthesis pending the completion of transcription. This may be similar to observations by Jemt et al. (63) of sequence-specific structures near the 5′- and 3′-ends of the NCR that correlate transcription termination and aborted DNA replication. A deeper understanding of the crossover between mtDNA replication and transcription may prove helpful in explaining the biological implications of these findings.
Experimental procedures
Recombinant enzyme preparation
Polymerase γ catalytic subunit p140 was overexpressed in baculovirus-infected Spodoptera frugiperda (Sf9) cells. The cells were grown in suspension with shaking at 27 °C. His6 tagged Pol γ was purified according to Kasiviswanathan et al. (64) with the following exception. Phosphocellulose P11 resin was replaced with a GE HiTrap Heparin HP column. The column was pre-equilibrated with 85% Heparin Buffer A (HBA) (25 mm KPO4, pH 7.5, 10% glycerol, and 25 mm NaCl) and 15% Heparin Buffer B (HBB) (25 mm KPO4, pH 7.5, 10% glycerol, and 1,000 mm NaCl). After loading lysate, the column was washed with 5 column volumes (CV) of 85% HBA and 15% HBB. Protein was eluted in a stepwise gradient of 28% HBB for 5 CV and 75% HBB for 5 CV. Fractions containing p140 (75% HBB elution) were pooled, and purification continued as previously described. Accessory subunit p55 was expressed in Escherichia coli and purified as previously described by Kasiviswanathan et al. (64).
Oligonucleotide substrates
All oligos were prepared by Integrated DNA Technologies. Unless otherwise stated, fluorophore-labeled oligos were labeled at the 5′ terminus. Oligonucleotides were resuspended in TE buffer (10 mm Tris-HCl, pH 7.5, and 0.1 mm EDTA) and combined in annealing buffer (5 mm Tris-HCl, pH 7.5, 0.05 mm EDTA, and 100 mm NaCl). When annealing template oligos and fluorophore-labeled primers, templates were used at a 1.2× concentration to ensure that all labeled oligo was annealed. During annealing, oligos were heated to 90 °C and allowed to cool slowly to room temperature. Annealing was verified by native-polyacrylamide gel electrophoresis. Oligonucleotide sequences are shown in Table S1.
G-quadruplex mapping
Prediction of G-quadruplex sites within mtDNA (NC_012920) was carried out using both G4-Hunter (34) and QGRS (35) software. G4-Hunter was fed the heavy-strand mtDNA sequence and predicts structure on both strands from this information. QGRS was run separately with both heavy- and light-strand sequences. Prediction results and parameters are shown in File SA. Quadruplex-forming regions of interest were plotted on mtDNA using SnapGene.
CD
Oligos containing G-quadruplex–forming sequences were diluted into TE buffer with 100 mm NaCl. Oligos were then heated to 90 °C for 5 min and allowed to slowly cool to room temperature. Oligo solutions (10 µm) were pipetted into a 1-mm path quartz cuvette and analyzed under N2 gas in a Jasco J-1500 Spectrophotometer. CD spectral data were collected at 25 °C from 220–320 nm with a 1-nm bandwidth and a 0.1-nm pitch.
Primer extension assays
Pol γ enzymatic assays were all prepared on ice. Aliquots of catalytic subunit p140 and accessory subunit p55 were thawed on ice, and p140 was first diluted to a working stock concentration in 25 mm HEPES, pH 8.0, 50 mm NaCl, 1 mm tris(2-carboxyethyl)phosphine, 50 µm EDTA, and 0.5 µm BSA. p140 and p55 were pre-incubated in reaction tubes with 1 µm BSA and the corresponding oligo substrates in assay buffer containing 25 mm HEPES, pH 8.0, 1 mm tris(2-carboxyethyl)phosphine, and 100 µm EDTA. Unless stated otherwise, reactions with p140 alone had 100 mm NaCl present and reactions with p140 and p55 in complex had 200 mm NaCl present. Reactions utilizing the G-quadruplex substrate library contained 200 nm oligonucleotide substrate (G4_1–G4_8), 10–30 nm Pol γ p140, and when present, 30–60 nm p55. p55 was used at a minimum 2:1 stoichiometric ratio with p140. After being mixed on ice, reactions were moved to 37 °C for 1 min. Reactions were then initiated by addition of 20 µm dNTPs and 2 mm MgCl2. Time points were removed and quenched into 95% formamide, 15 mm EDTA, and 0.01% Bromphenol Blue at 4 °C. Substrate and product were then separated on 20-cm urea-PAGE denaturing gels. Gels were run in 1× Tris borate-EDTA buffer. Using an Amersham Biosciences Typhoon 9000, total fluorescence was measured. Fluorescence intensity was quantified with ImageQuant software and used to determine product formation following primer extension reactions.
UV irradiation
Irradiation of oligonucleotide substrates with 254-nm UV irradiation was accomplished with a Stratalinker 1800 UV source. Oligonucleotides were heated to 90 °C in annealing buffer and slowly cooled to room temperature to facilitate formation of secondary structure. Samples of oligonucleotide were then spotted on a glass surface, placed in the Stratalinker UV box, and dosed with either 10,000 J/m2 or 30,000 J/m2 of UV irradiation, as indicated. Identical samples were placed in an opaque box inside the UV Stratalinker and treated concurrently as unexposed control samples.
Data availability
All data are contained within this article and in the supporting information.
Supplementary Material
Acknowledgments
We thank Drs. Bradley Klemm, Margaret Gustafson, and Amanda Riccio at the NIEHS, National Institutes of Health for critical evaluation of the manuscript. We also thank Dr. Geoff Mueller of the Nuclear Magnetic Resonance group for analysis of the DNA substrates and Dr. Robert Petrovich of the Structural Biology core for access to the circular dichroism spectrophotometer.
This article contains supporting information.
Author contributions—E. D. S. data curation; E. D. S. formal analysis; E. D. S. investigation; E. D. S. methodology; E. D. S. and W. C. C. writing-original draft; E. D. S., M. J. L., and W. C. C. writing-review and editing; M. J. L. and W. C. C. supervision; W. C. C. conceptualization; W. C. C. resources; W. C. C. funding acquisition; W. C. C. project administration.
Funding and additional information—Funding was provided the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences Grant ES065078 (to W. C. C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- mtDNA
- mitochondrial DNA
- Pol γ
- DNA polymerase γ
- mtSSB
- mitochondrial ssDNA binding protein
- Ori
- origin
- H
- heavy
- L
- light
- NCR
- noncoding span of the mtDNA
- G4
- G-quadruplex
- nt
- nucleotide
- CV
- column volumes
- HSP
- heavy-strand promoter.
References
- 1. Wallace D. C. (1999) Mitochondrial diseases in man and mouse. Science 283, 1482–1488 10.1126/science.283.5407.1482 [DOI] [PubMed] [Google Scholar]
- 2. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., and Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- 3. Korhonen J. A., Pham X. H., Pellegrini M., and Falkenberg M. (2004) Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 10.1038/sj.emboj.7600257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Graziewicz M. A., Longley M. J., and Copeland W. C. (2006) DNA polymerase γ in mitochondrial DNA replication and repair. Chem. Rev. 106, 383–405 10.1021/cr040463d [DOI] [PubMed] [Google Scholar]
- 5. Kaguni L. S. (2004) DNA polymerase γ, the mitochondrial replicase. Annu. Rev. Biochem. 73, 293–320 10.1146/annurev.biochem.72.121801.161455 [DOI] [PubMed] [Google Scholar]
- 6. Lim S. E., Longley M. J., and Copeland W. C. (1999) The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 274, 38197–38203 10.1074/jbc.274.53.38197 [DOI] [PubMed] [Google Scholar]
- 7. Fan L., Sanschagrin P. C., Kaguni L. S., and Kuhn L. A. (1999) The accessory subunit of mtDNA polymerase shares structural homology with aminoacyl-tRNA synthetases: Implications for a dual role as a primer recognition factor and processivity clamp. Proc. Natl. Acad. Sci. U. S. A. 96, 9527–9532 10.1073/pnas.96.17.9527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson A. A., Tsai Y., Graves S. W., and Johnson K. A. (2000) Human mitochondrial DNA polymerase holoenzyme: reconstitution and characterization. Biochemistry 39, 1702–1708 10.1021/bi992104w [DOI] [PubMed] [Google Scholar]
- 9. Falkenberg M., Larsson N. G., and Gustafsson C. M. (2007) DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699 10.1146/annurev.biochem.76.060305.152028 [DOI] [PubMed] [Google Scholar]
- 10. Miralles Fusté J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson O., Sabouri N., Gustafsson C. M., and Falkenberg M. (2014) In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 10, e1004832 10.1371/journal.pgen.1004832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaur P., Longley M. J., Pan H., Wang H., and Copeland W. C. (2018) Single-molecule DREEM imaging reveals DNA wrapping around human mitochondrial single-stranded DNA binding protein. Nucleic Acids Res. 46, 11287–11302 10.1093/nar/gky875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cline S. D. (2012) Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim, Biophys, Acta 1819, 979–991 10.1016/j.bbagrm.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasiviswanathan R., Gustafson M. A., Copeland W. C., and Meyer J. N. (2012) Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 287, 9222–9229 10.1074/jbc.M111.306852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kasiviswanathan R., Minko I. G., Lloyd R. S., and Copeland W. C. (2013) Translesion synthesis past acrolein-derived DNA adducts by human mitochondrial DNA polymerase γ. J. Biol. Chem. 288, 14247–14255 10.1074/jbc.M113.458802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graziewicz M. A., Sayer J. M., Jerina D. M., and Copeland W. C. (2004) Nucleotide incorporation by human DNA polymerase γ opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 32, 397–405 10.1093/nar/gkh213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graziewicz M. A., Bienstock R. J., and Copeland W. C. (2007) The DNA polymerase γ Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2′-deoxyguanosine. Hum. Mol. Genet. 16, 2729–2739 10.1093/hmg/ddm227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rahman S., and Copeland W. C. (2019) POLG-related disorders and their neurological manifestations. Nat. Rev. Neurol. 15, 40–52 10.1038/s41582-018-0101-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Copeland W. C. (2008) Inherited mitochondrial diseases of DNA replication. Ann. Rev. Med. 59, 131–146 10.1146/annurev.med.59.053006.104646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Persson O., Muthukumar Y., Basu S., Jenninger L., Uhler J. P., Berglund A. K., McFarland R., Taylor R. W., Gustafsson C. M., Larsson E., and Falkenberg M. (2019) Copy-choice recombination during mitochondrial L-strand synthesis causes DNA deletions. Nat. Commun. 10, 759 10.1038/s41467-019-08673-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong D. W., Pereira F., Barrett S. P., Kolesar J. E., Cao K., Damas J., Yatsunyk L. A., Johnson F. B., and Kaufman B. A. (2014) Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints. BMC Genomics 15, 677 10.1186/1471-2164-15-677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bharti S. K., Sommers J. A., Zhou J., Kaplan D. L., Spelbrink J. N., Mergny J. L., and Brosh R. M. Jr (2014) DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 289, 29975–29993 10.1074/jbc.M114.567073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wanrooij S., Fusté J. M., Farge G., Shi Y., Gustafsson C. M., and Falkenberg M. (2008) Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. U. S. A. 105, 11122–11127 10.1073/pnas.0805399105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong T. W., and Clayton D. A. (1985) In vitro replication of human mitochondrial DNA: accurate initiation at the origin of light-strand synthesis. Cell 42, 951–958 10.1016/0092-8674(85)90291-0 [DOI] [PubMed] [Google Scholar]
- 24. Fusté J. M., Wanrooij S., Jemt E., Granycome C. E., Cluett T. J., Shi Y., Atanassova N., Holt I. J., Gustafsson C. M., and Falkenberg M. (2010) Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell 37, 67–78 10.1016/j.molcel.2009.12.021 [DOI] [PubMed] [Google Scholar]
- 25. Shadel G. S., and Clayton D. A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66, 409–435 10.1146/annurev.biochem.66.1.409 [DOI] [PubMed] [Google Scholar]
- 26. Kasiviswanathan R., and Copeland W. C. (2011) Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem. 286, 31490–31500 10.1074/jbc.M111.252460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murakami E., Feng J. Y., Lee H., Hanes J., Johnson K. A., and Anderson K. S. (2003) Characterization of novel reverse transcriptase and other RNA-associated catalytic activities by human DNA polymerase γ: importance in mitochondrial DNA replication. J. Biol. Chem. 278, 36403–36409 10.1074/jbc.M306236200 [DOI] [PubMed] [Google Scholar]
- 28. Kaguni L. S., and Clayton D. A. (1982) Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc. Natl. Acad. Sci. U. S. A. 79, 983–987 10.1073/pnas.79.4.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eddy S., Tillman M., Maddukuri L., Ketkar A., Zafar M. K., and Eoff R. L. (2016) Human translesion polymerase κ exhibits enhanced activity and reduced fidelity two nucleotides from G-Quadruplex DNA. Biochemistry 55, 5218–5229 10.1021/acs.biochem.6b00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamath-Loeb A. S., Loeb L. A., Johansson E., Burgers P. M., and Fry M. (2001) Interactions between the Werner syndrome helicase and DNA polymerase Δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 276, 16439–16446 10.1074/jbc.M100253200 [DOI] [PubMed] [Google Scholar]
- 31. Bedrat A., Lacroix L., and Mergny J. L. (2016) Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 44, 1746–1759 10.1093/nar/gkw006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huppert J. L., and Balasubramanian S. (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 10.1093/nar/gki609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Todd A. K., Johnston M., and Neidle S. (2005) Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 33, 2901–2907 10.1093/nar/gki553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brázda V., Kolomazník J., Lýsek J., Bartas M., Fojta M., Šťastný J., and Mergny J.-L. (2019) G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics 35, 3493–3495 10.1093/bioinformatics/btz087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kikin O., D'Antonio L., and Bagga P. S. (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–682 10.1093/nar/gkl253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhattacharyya D., Mirihana Arachchilage G., and Basu S. (2016) Metal cations in G-Quadruplex folding and stability. Front Chem 4, 38 10.3389/fchem.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Longley M. J., Ropp P. A., Lim S. E., and Copeland W. C. (1998) Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry 37, 10529–10539 10.1021/bi980772w [DOI] [PubMed] [Google Scholar]
- 38. DeBalsi K. L., Longley M. J., Hoff K. E., and Copeland W. C. (2017) Synergistic effects of the in cis T251I and P587L mitochondrial DNA polymerase γ disease Mutations. J. Biol. Chem. 292, 4198–4209 10.1074/jbc.M116.773341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang D. D., and Clayton D. A. (1984) Precise identification of individual promoters for transcription of each strand of human mitochondrial-DNA. Cell 36, 635–643 10.1016/0092-8674(84)90343-X [DOI] [PubMed] [Google Scholar]
- 40. Zollo O., Tiranti V., and Sondheimer N. (2012) Transcriptional requirements of the distal heavy-strand promoter of mtDNA. Proc. Natl. Acad. Sci. U. S. A. 109, 6508–6512 10.1073/pnas.1118594109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moraes C. T., Andreetta F., Bonilla E., Shanske S., DiMauro S., and Schon E. A. (1991) Replication-competent human mitochondrial DNA lacking the heavy-strand promoter region. Mol. Cell Biol. 11, 1631–1637 10.1128/mcb.11.3.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moraes C. T., Ricci E., Petruzzella V., Shanske S., DiMauro S., Schon E. A., and Bonilla E. (1992) Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nat. Genet. 1, 359–367 10.1038/ng0892-359 [DOI] [PubMed] [Google Scholar]
- 43. Krishnan K. J., Harbottle A., and Birch-Machin M. A. (2004) The use of a 3895 bp mitochondrial DNA deletion as a marker for sunlight exposure in human skin. J. Invest. Dermatol. 123, 1020–1024 10.1111/j.0022-202X.2004.23457.x [DOI] [PubMed] [Google Scholar]
- 44. Smith-Carpenter J. E., and Taylor J. S. (2019) Photocrosslinking of G-quadruplex-forming sequences found in human promoters. Photochem. Photobiol. 95, 252–266 10.1111/php.12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su D. G., Fang H., Gross M. L., and Taylor J. S. (2009) Photocrosslinking of human telomeric G-quadruplex loops by anti cyclobutane thymine dimer formation. Proc. Natl. Acad. Sci. U. S. A. 106, 12861–12866 10.1073/pnas.0902386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith J. E., Lu C., and Taylor J. S. (2014) Effect of sequence and metal ions on UVB-induced anti cyclobutane pyrimidine dimer formation in human telomeric DNA sequences. Nucleic Acids Res. 42, 5007–5019 10.1093/nar/gku163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gustafsson C. M., Falkenberg M., and Larsson N. G. (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 10.1146/annurev-biochem-060815-014402 [DOI] [PubMed] [Google Scholar]
- 48. Liu P., Qian L., Sung J. S., de Souza-Pinto N. C., Zheng L., Bogenhagen D. F., Bohr V. A., Wilson D. M. 3rd, Shen B., and Demple B. (2008) Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell Biol. 28, 4975–4987 10.1128/MCB.00457-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Szczesny B., Tann A. W., Longley M. J., Copeland W. C., and Mitra S. (2008) Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 283, 26349–26356 10.1074/jbc.M803491200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Copeland W. C., and Longley M. J. (2008) DNA2 resolves expanding flap in mitochondrial base excision repair. Mol. Cell 32, 457–458 10.1016/j.molcel.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Copeland W. C., and Longley M. J. (2014) Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 19, 190–198 10.1016/j.dnarep.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kazak L., Reyes A., He J., Wood S. R., Brea-Calvo G., Holen T. T., and Holt I. J. (2013) A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS ONE 8, e62340 10.1371/journal.pone.0062340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cerritelli S. M., Frolova E. G., Feng C., Grinberg A., Love P. E., and Crouch R. J. (2003) Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell 11, 807–815 10.1016/S1097-2765(03)00088-1 [DOI] [PubMed] [Google Scholar]
- 54. Wu C. C., Lin J. L. J., Yang-Yen H. F., and Yuan H. S. (2019) A unique exonuclease ExoG cleaves between RNA and DNA in mitochondrial DNA replication. Nucleic Acids Res. 47, 5405–5419 10.1093/nar/gkz241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Falabella M., Kolesar J. E., Wallace C., de Jesus D., Sun L., Taguchi Y. V., Wang C., Wang T., Xiang I. M., Alder J. K., Maheshan R., Horne W., Turek-Herman J., Pagano P. J., St Croix C. M., et al. (2019) G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci. Rep. 9, 5605 10.1038/s41598-019-41464-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Butler T. J., Estep K. N., Sommers J. A., Maul R. W., Moore A. Z., Bandinelli S., Cucca F., Tuke M. A., Wood A. R., Bharti S. K., Bogenhagen D. F., Yakubovskaya E., Garcia-Diaz M., Guilliam T. A., Byrd A. K., et al. (2020) Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro. Hum. Mol. Genet. 29, 1292–1309 10.1093/hmg/ddaa043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harbottle A., Maki J., Reguly B., Wittock R., Robinson K., Parr R., and Birch-Machin M. A. (2010) Real-time polymerase chain reaction analysis of a 3895-bp mitochondrial DNA deletion in epithelial swabs and its use as a quantitative marker for sunlight exposure in human skin. Br. J. Dermatol. 163, 1291–1295 10.1111/j.1365-2133.2010.10001.x [DOI] [PubMed] [Google Scholar]
- 58. Gendron S. P., Bastien N., Mallet J. D., and Rochette P. J. (2013) The 3895-bp mitochondrial DNA deletion in the human eye: a potential involvement in corneal ageing and macular degeneration. Mutagenesis 28, 197–204 10.1093/mutage/ges071 [DOI] [PubMed] [Google Scholar]
- 59. Clayton D. A., Doda J. N., and Friedberg E. C. (1974) The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl. Acad. Sci. U. S. A. 71, 2777–2781 10.1073/pnas.71.7.2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ray A. J., Turner R., Nikaido O., Rees J. L., and Birch-Machin M. A. (2000) The spectrum of mitochondrial DNA deletions is a ubiquitous marker of ultraviolet radiation exposure in human skin. J. Invest. Dermatol. 115, 674–679 10.1046/j.1523-1747.2000.00092.x [DOI] [PubMed] [Google Scholar]
- 61. Berneburg M., Plettenberg H., Medve-König K., Pfahlberg A., Gers-Barlag H., Gefeller O., and Krutmann J. (2004) Induction of the photoaging-associated mitochondrial common deletion in vivo in normal human skin. J. Invest. Dermatol. 122, 1277–1283 10.1111/j.0022-202X.2004.22502.x [DOI] [PubMed] [Google Scholar]
- 62. Lujan S. A., Longley M. J., Humble M. H., Lavender C. A., Burkholder A., Blakely E. L., Alston C. L., Gorman G. S., Turnbull D. M., McFarland R., Taylor R. W., Kunkel T. A., and Copeland W. C. (2020) Ultrasensitive deletion detection links mitochondrial DNA replication, disease, and aging. Genome Biol. 21, 248 10.1186/s13059-020-02138-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jemt E., Persson O., Shi Y., Mehmedovic M., Uhler J. P., Dávila López M., Freyer C., Gustafsson C. M., Samuelsson T., and Falkenberg M. (2015) Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 43, 9262–9275 10.1093/nar/gkv804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kasiviswanathan R., Longley M. J., Young M. J., and Copeland W. C. (2010) Purification and functional characterization of human mitochondrial DNA polymerase gamma harboring disease mutations. Methods 51, 379–384 10.1016/j.ymeth.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within this article and in the supporting information.