Heart failure is an important cause of morbidity and mortality in patients with congenital heart disease (CHD). 1 , 2 It occurs in ≈25% of those aged ≥30 years and is not limited to severe cardiac defects. 3 , 4 Individuals with lower‐complexity malformations, which constitute most of the CHD population, have a higher burden of adverse cardiovascular events relative to the general population, with hazard ratios ranging from 2 for coronary artery disease to 13 for heart failure. 5 This increased risk may reflect some primitive frailty of the heart, yet the underpinning molecular determinants remain unknown.
Several factors have been acknowledged to participate in the pathogenesis of CHD‐related heart failure, such as inherited architectural disorganization of myocytes and vascular cells, cardiac damage attributable to the surgical trauma and insufficient protection during cardiopulmonary bypass, and the hemodynamic load to the heart from residual defects and failing grafts. 6 Moreover, genetic and epigenetic factors play both distinct and additive roles in maladaptive myocardial remodeling. 4 Investigation of mechanisms has mainly focused on canonical pathways, such as the renin‐angiotensin‐aldosterone system and adrenergic system. 7 , 8 Nonetheless, trials using renin‐angiotensin‐aldosterone system inhibitors in CHD‐related heart failure failed to show any benefit on ventricular function 9 , 10 ; thus, conventional therapy is unsupported by clinical evidence. 11
Should the cause be searched in the developing cardiomyocyte? The heart is the first organ formed during mammalian embryonic development. In the initial stage, the embryonic heart undergoes a series of critical migration and remodeling events instrumental to the formation of the 4 chambers. The maturation of cardiomyocytes initiates at midgestation and continues until adulthood. During this process, these cells change morphology and organize to form effcient contractile structures. An extraordinary, tightly regulated molecular program regulates these structural changes, but in babies with CHD the program is seemingly deranged toward incomplete finalization. Specific errors could lead to distinct defects in single or multiple components of the developing heart. The initial molecular handicap is amplified by the occurrence of multiple and repeated stress, related not only to the defect severity, but also to the damage from reconstructive cardiac surgery and implantation of foreign graft material (Figure 1). New holistic efforts are urgently needed to deploy effective treatments for correction of both the initial errors and the deranged compensatory mechanisms.
Figure 1. Stress‐related mechanisms leading to heart failure in congenital heart disease (CHD).
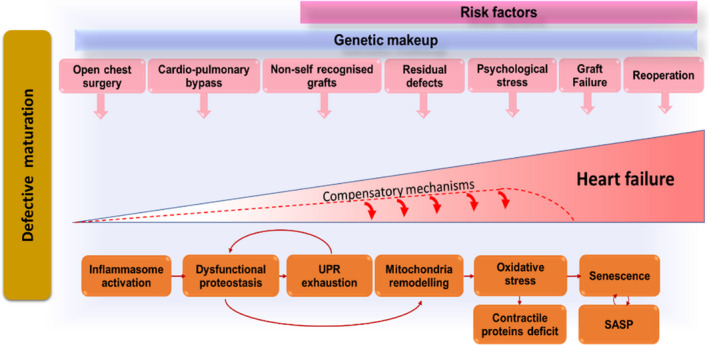
Congenitally defective cardiomyocytes are immature, fragile, and susceptible to stress‐induced damage. Cardiac cells (especially stromal cells, which have immunomodulatory roles) respond to the early and repeated stress by activating the inflammasome. Stress‐induced response pathways could attempt to reduce protein synthesis and increase the cellular capacity for protein folding and degradation. With unremitting stress, proteostasis becomes overloaded, resulting in misfolded protein accumulation in subcellular compartments, including the mitochondria, where proteotoxic stress can incite reactive oxygen species production. An increasing number of cells become senescent in the heart with CHD and transfer proteostatic stress to neighboring cells through secretion of inflammatory chemokines and misfolded peptides. This vicious cycle will lead to cardiomyocyte loss, microvascular rarefaction, and fibrocalcific interstitial remodeling of the heart, compromising myocardial perfusion and contractile function. The stress from the defect and surgical trauma is unavoidable, but drugs able to improve proteostasis and/or interfere with the inflammasome could halt cardiac deterioration in patients with CHD, thus succeeding where conventional therapy has failed. Improvement in graft biocompatibility (eg, cellularization before implantation) may reduce the inflammatory reaction to the prosthesis. SASP indicates senescence associated secretory phenotype; and UPR, unfolded protein response.
Multidisciplinary and technology‐enhanced science refers to an emerging approach where new theories are interrogated using a combination of techniques, tools, and data processing to decipher compless biological processes. The article from Lam et al, published in the current issue of the Journal of the American Heart Association (JAHA), provides a paradigmatic example of the potential from this approach. 12 The authors compared human induced pluripotent stem cells from healthy subjects and patients with pulmonary atresia but intact ventricular septum. This is a sporadic CHD characterized by the complete obstruction of the right ventricular outflow and malformation of the tricuspid valve and right ventricle–coronary artery communications (Figure 2). The authors discovered that the differentiation of human induced pluripotent stem cells to cardiomyocytes could be implemented by incorporating the provisional cardiomyocytes in bioengineered tissue constructs.
Figure 2. Cartoon showing main characteristics of pulmonary atresia with intact septum.
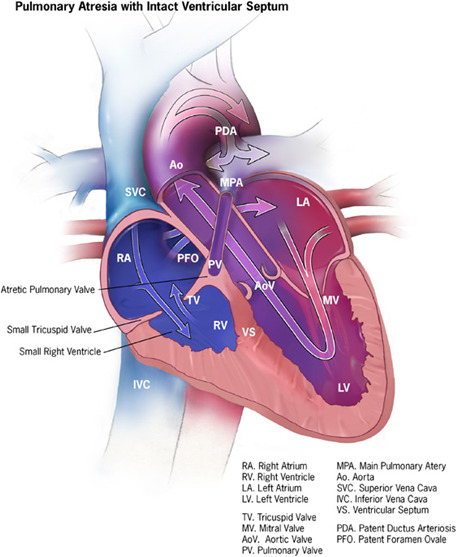
This cardiac defect involves the pulmonary valve, which does not form at all; therefore, no blood can go from the right ventricle of the heart out to the lungs. The septum between the ventricles remains complete and intact. During pregnancy when the heart is developing, little blood flows into the right ventricle, and therefore this section of the heart does not fully develop and remains small. If the right ventricle is underdeveloped, the heart can have problems pumping blood to the lungs and the body. Pulmunary atresia is considered a critical congenital heart defect: these babies may need surgery or other procedures soon after birth. New investigation suggests that the anatomical defect of the valve is associated with failure of the cardiomyocytes to reach full maturation. This discovery is based on studies in vitro, using blood cells that were induced first to become pluripotent and then forced to differentiate into cardiomyocytes. Future studies are necessary to understand if the altered program seen studying cells in culture is the same responsible for the contractility defect of the heart of babies with pulmunary atresia. The image is provided free of any copyright restrictions as a courtesy from Centers for Disease Control and Prevention, National Center on Birth Defects and Developmental Disabilities.
More important, the differentiation process followed different molecular trajectories in cells from patients compared with healthy controls, with downregulation of cardiac contractile apparatus and cardiac maturation transcripts and upregulation of their immature isoforms. These new findings may provide a molecular explanation for the persistent right ventricular systolic and diastolic dysfunction in adolescent and adult patients despite successful biventricular surgical repair of the cardiac defect. In addition, the authors report that alteration in contractile protein developments was dissociated from defects in calcium handling in the pulmonary atresia but intact ventricular septum cellular model.
Pluripotent stem cells hold great promise for drug discovery and disease modeling. Moreover, the analysis of differentiation into cardiomyocytes could provide important insights into the molecular changes responsible for proper maturation of the fetal heart. On the other hand, the information from cellular models may be valid only with reference to the early stage of development. In line with this, an algorithm generated by the analysis of hundreds of microarray data sets from early embryonic to adult hearts showed that human induced pluripotent stem cell–derived cardiomyocytes mature early in culture but are arrested at the late embryonic stage with aberrant regulation of key transcription factors. 13 Recent studies, including the one from Lam et al, have provided convincing evidence that culturing on micropatterned substrates may improve cell morphological features and enhance cardiomyocyte maturation. 12 , 14 This is an important technical advancement, increasing the value of modeling and possibly improving the validity of identified molecular trajectories beyond the early stages of cardiac development.
Single‐cell transcriptome sequencing (scRNA‐seq) can profile transcript abundance and developmental trajectories at the resolution of an individual cell. scRNA‐seq allows for separation of widely distinct cell subpopulations, which were, until recently, simply averaged together with bulk tissue transcriptome sequencing. Nonetheless, although validated methods exist for the analysis of bulk transcriptome sequencing time‐series data, the processing of gene expression and pseudotime in an scRNA‐seq environment is more complex. Moreover, pseudotimes are continuous and single cells are never at the exact identical pseudotime value. Methods such as Monocle and TSCAN, used by Lam et al, can test if gene expression is associated with pseudotime by fitting additive models of gene expression as a function of pseudotime. However, these approaches can only handle a single lineage. It would be useful to confirm the data with newly developed tools, such as tradeSeq, a method and software package that compares differential expression pattern along a lineage or between lineages and generates smooth functions for the gene expression outputs along pseudotime for each lineage. 15
Beside being finely tuned, cardiogenesis is a highly complex process involving synergistic actions of different cell lineages. 16 Using scRNA‐seq to profile the gene expression landscapes of a thousand cardiac cells from human embryos, Cui et al identified that the 4 major types of cells display distinct trajectories that are temporaly modulated. Atrial and ventricular cardiomyocytes acquired distinct features during early development. Furthermore, both cardiomyocytes and fibroblasts showed stepwise changes in gene expression. As development proceeded, valvular interstitial cells were involved. There were evident differences between humans and mice, which highlighted the challanges of using rodent models to reproduce the unique features of human heart development. In their article, Lam et al admit that more studies are warranted to determine if the altered maturation trajectories observed in pulmonary atresia but intact ventricular septum cardiomyocytes can be detected in other cardiac cells. 12 Engineering diverse cardiac cell populations in a humanized 3‐dimensional platform (heart on a chip) that reproduces the architecture of the human heart would allow to couple the direct visualization of single cell events, such as migration, survival, and biointegration, with refined understanding of molecular mechanisms implicated in cardiac maturation.
Finally, the study from Lam et al opens new exciting horizons for early diagnosis, prevention, and treatment. 12 In the past decade, the use of 3‐ and and 4‐dimensional fetal echocardiography has been introduced into clinical practice, increasing the diagnostic power of imaging for early detection of congenital heart defects. Moreover, recent advancements in computational techniques coupled with fetal magnetic resonance imaging have shown the potential to overcome some of current limitations of prenatal ultrasonography, where acoustic windows are frequently obscured by maternal or fetal anatomic structures or motion artifacts. 17 Molecular analysis of magnetic resonance imaging–guided biopsies from suspected sectors of the heart could improve diagnostic precision and inform timely corrective strategies. This exciting scenario is not devoid of challanges. Translation of initial scRNA‐seq outputs into diagnostic kits requires validation on batches collected at different times and different individuals. Deep learning is making a major breakthrough in several areas of bioinformatics. The approach could help remove complex batch effects, preserve biological variations, and reveal both discrete and pseudotemporal structure of cells across different patients. 18 , 19 In perspective, follow‐up of the congenitally defective heart from fetal‐to‐adult age using scRNA‐seq derived molecular kits may help tailoring personalized therapies with the best response and highest safety margin to ensure better patient care.
Sources of Funding
This study was supported by grants from the Medical Research Council to Dr Madeddu.
Disclosures
None.
(J Am Heart Assoc 2020;9:e019433 DOI: 10.1161/JAHA.120.019433.)
The opinions expressed in this article are not necessarily those of the editors or of the American Heart Association.
For Sources of Funding and Disclosures, see page 4.
See Article by Lam et al.
References
- 1. Rossano JW, Shaddy RE. Heart failure in children: etiology and treatment. J Pediatr. 2014;165:228–233. [DOI] [PubMed] [Google Scholar]
- 2. Massin MM, Astadicko I, Dessy H. Epidemiology of heart failure in a tertiary pediatric center. Clin Cardiol. 2008;31:388–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Konstam MA, Kiernan MS, Bernstein D, Bozkurt B, Jacob M, Kapur NK, Kociol RD, Lewis EF, Mehra MR, Pagani FD, et al; American Heart Association Council on Clinical Cardiology, Council on Cardiovascular Disease in the Young, Council on Cardiovascular Surgery and Anesthesia . Evaluation and management of right‐sided heart failure: a scientific statement from the American Heart Association. Circulation. 2018;137:e578–e622. [DOI] [PubMed] [Google Scholar]
- 4. Hinton RB, Ware SM. Heart failure in pediatric patients with congenital heart disease. Circ Res. 2017;120:978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saha P, Potiny P, Rigdon J, Morello M, Tcheandjieu C, Romfh A, Fernandes SM, McElhinney DB, Bernstein D, Lui GK, et al. Substantial cardiovascular morbidity in adults with lower‐complexity congenital heart disease. Circulation. 2019;139:1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Budts W, Roos‐Hesselink J, Radle‐Hurst T, Eicken A, McDonagh TA, Lambrinou E, Crespo‐Leiro MG, Walker F, Frogoudaki AA. Treatment of heart failure in adult congenital heart disease: a position paper of the Working Group of Grown‐Up Congenital Heart Disease and the Heart Failure Association of the European Society of Cardiology. Eur Heart J. 2016;37:1419–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andersen S, Andersen A, Nielsen‐Kudsk JE. The renin‐angiotensin‐aldosterone‐system and right heart failure in congenital heart disease. Int J Cardiol Heart Vasc. 2016;11:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyamoto SD, Stauffer BL, Nakano S, Sobus R, Nunley K, Nelson P, Sucharov CC. Beta‐adrenergic adaptation in paediatric idiopathic dilated cardiomyopathy. Eur Heart J. 2014;35:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Bom T, Winter MM, Bouma BJ, Groenink M, Vliegen HW, Pieper PG, van Dijk AP, Sieswerda GT, Roos‐Hesselink JW, Zwinderman AH, et al. Effect of valsartan on systemic right ventricular function: a double‐blind, randomized, placebo‐controlled pilot trial. Circulation. 2013;127:322–330. [DOI] [PubMed] [Google Scholar]
- 10. Hsu DT, Zak V, Mahony L, Sleeper LA, Atz AM, Levine JC, Barker PC, Ravishankar C, McCrindle BW, Williams RV, et al; Pediatric Heart Network Investigators . Enalapril in infants with single ventricle: results of a multicenter randomized trial. Circulation. 2010;122:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stout KK, Daniels CJ, Aboulhosn JA, Bozkurt B, Broberg CS, Colman JM, Crumb SR, Dearani JA, Fuller S, Gurvitz M, et al. 2018 AHA/ACC guideline for the management of adults with congenital heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e698–e800. [DOI] [PubMed] [Google Scholar]
- 12. Lam Y‐Y, Keung W, Chan C‐H, Geng L, Wong N, Breniere‐Letuffe D, Li RA, Cheung Y‐F. Single‐cell transcriptomics of engineered cardiac tissues from patient‐specific induced pluripotent stem cell‐derived cardiomyocytes reveals abnormal developmental trajectory and intrinsic contractile defects in hypoplastic right heart syndrome. J Am Heart Assoc. 2020;9:e016528 DOI: 10.1161/JAHA.120.016528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015;13:1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salick MR, Napiwocki BN, Sha J, Knight GT, Chindhy SA, Kamp TJ, Ashton RS, Crone WC. Micropattern width dependent sarcomere development in human ESC‐derived cardiomyocytes. Biomaterials. 2014;35:4454–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van den Berge K, Roux de Bezieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. Trajectory‐based differential expression analysis for single‐cell sequencing data. Nat Commun. 2020;11:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5:a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lloyd DFA, Pushparajah K, Simpson JM, van Amerom JFP, van Poppel MPM, Schulz A, Kainz B, Deprez M, Lohezic M, Allsop J, et al. Three‐dimensional visualisation of the fetal heart using prenatal MRI with motion‐corrected slice‐volume registration: a prospective, single‐centre cohort study. Lancet. 2019;393:1619–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petegrosso R, Li Z, Kuang R. Machine learning and statistical methods for clustering single‐cell RNA‐sequencing data. Brief Bioinform. 2020;21:1209–1223. [DOI] [PubMed] [Google Scholar]
- 19. Li X, Wang K, Lyu Y, Pan H, Zhang J, Stambolian D, Susztak K, Reilly MP, Hu G, Li M. Deep learning enables accurate clustering with batch effect removal in single‐cell RNA‐seq analysis. Nat Commun. 2020;11:2338. [DOI] [PMC free article] [PubMed] [Google Scholar]


