Abstract
In 2010, the genetic testing criteria was changed to allow women diagnosed ≤ 60 years old with triple negative breast cancer (TNBC) to undergo germline testing. In the same year, estrogen receptor (ER) positivity was defined as having ≥1% ER staining cells. While tumors with 1–10% ER staining cells and HER2 negative (HER2-) status share characteristics with TNBC, the utility of germline testing in women with ER low positive/HER2- (ERLP/HER2-) tumors is not well-understood. To this end, all patients with hormone receptor positive staining cells ≤ 10% and negative HER2 status were identified. Clinical genetic test results were extracted for patients who underwent testing. Panel testing was performed for those women who had genomic DNA available for research purposes. ERLP/HER2-tumors constituted 2.7% of all tumors in the database. Patients did not differ significantly from those with TNBC by age at diagnosis, ethnicity, family history or tumor size, stage or grade (p > 0.05). Mutation frequency did not differ significantly (p = 0.757) between groups (ERLP/HER2- 16.1%; TNBC 16.7%). Hereditary forms of breast cancer were similar in both ERLP/HER2- and TNBC, thus current guidelines may result in the under testing of women with low ER tumors, resulting in missed opportunities to improve patient management.
Keywords: genetic testing, breast cancer, ER low positive, ASCO/CAP guidelines
1. Introduction
Triple negative breast cancers (TNBC), do not express estrogen receptors (ERs), progesterone receptors (PRs), or HER2, and are associated with a poor prognosis [1]. Germline mutations in the BRCA1 and BRCA2 genes have been detected in 15% of patients with TNBC [2]. Women with germline mutations in the BRCA genes and TNBC may be sensitive to platinum agents [3]. In addition, Poly ADP Ribose Polymerase (PARP) inhibitors have been approved by the FDA for the treatment of TNBC in patients with germline BRCA mutations [4].
In 2010, two events altered genetic testing eligibility for women with invasive breast cancer. First, the National Comprehensive Cancer Network (NCCN) included TNBC in their guidelines for germline testing of BRCA genes, thus expanding the number of women eligible for testing [5]. Second, the American Society of Clinical Oncology (ASCO) and College of American Pathologists (CAP) recommended that a cutoff of 1% positive cells should be used to define ER and PR positive status [6]. Using these criteria, only HER2 negative (HER2-) tumors with <1% hormone receptor staining cells would be considered TNBC, precluding testing for women with low levels of hormone receptor and HER2-tumors who do not otherwise meet test eligibility criteria. Gene expression studies, however, have demonstrated that the majority of tumors with 1–9% ER staining have ER negative intrinsic subtypes [7,8], and patients with low-staining tumors have survival rates similar to those with ER negative tumors [9]. The updated ASCO/CAP guidelines published in 2020 recommended that tumors with 1–10% ER positive cells should be classified as ER low positive (ERLP) [10].
Although ASCO/CAP has recognized that tumors with low ER staining should be recognized as a special type of tumor, the cutoff for defining ER positivity remains ≥ 1%. Under this criterion, ERLP tumors that are HER2 negative would not be classified as TNBC, potentially causing some women to miss the opportunity for clinical genetic testing. In this study, we performed panel testing in patients with ERLP and HER2 negative (ERLP/HER2-) tumors or TNBC to determine whether the germline mutation frequency and array of mutations in women with ERLP/HER2-tumors is similar to those in women with TNBC. The results suggest that ERLP/HER2-tumors should be included in the NCCN test eligibility criteria.
2. Materials and Methods
Patients from the Walter Reed National Military Medical Center (WRNMMC), Bethesda, MD, USA; the Anne Arundel Medical Center, Annapolis, MD, USA; or the Joyce Murtha Breast Cancer Center, Windber, PA, USA, were enrolled in the Clinical Breast Care Project (CBCP) starting in 2001. As both NCCN and ASCO/CAP guidelines were issued in 2010, only patients diagnosed 2010 through 2019 were included in this study. Patients were eligible for enrollment if they were ≥18 years of age and were mentally competent and willing to provide informed consent. This work was performed with approval from the WRNMMC Human Use Committee and Institutional Review Board (protocol WRNMMC IRB #20704).
Pathological characterization was performed as previously described [7]. ER and PR status were determined by IHC analysis, and the percentage of stained cells were recorded in the CBCP database. HER2 status was defined using ASCO/CAP guidelines [11]. Family history was classified by the number of first- or second-degree family members with breast, ovarian, or pancreatic cancer. Test eligibility was assigned using the NCCN Genetic/Familial High-risk Assessment: Breast, Ovarian, and Pancreatic guidelines version 1.2020 (genetics_bop.pdf (nccn.org)). The test results were extracted from the CBCP database for the 161 patients who underwent clinical genetic testing.
Blood samples were available from 117 women who did not undergo genetic testing. Genomic DNA was isolated from blood clots using the Gentra Clotspin and Puregene DNA purification kits (Qiagen, Valencia, CA, USA). Isolated DNA was quantitated using the Qubit® Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). To generate the sequencing libraries, 50 ng of DNA was processed using the Illumina DNA prep with enrichment kit and TruSight Cancer panel (Illumina, Inc, San Diego, CA, USA). The resulting libraries were sequenced on a MiSeq (Illumina, Inc, San Diego, CA, USA). Variant Interpreter (Illumina, Inc, San Diego, CA, USA) was used for the data analysis; only variants with a minor allele frequency of >25% were included [12]. Variants were classified using the ClinVar database (http://www.clinvar.com/). Statistical analyses were performed using Fisher’s exact tests or Chi-square analyses, with significance defined by p < 0.05.
3. Results
3.1. Patient Characteristics
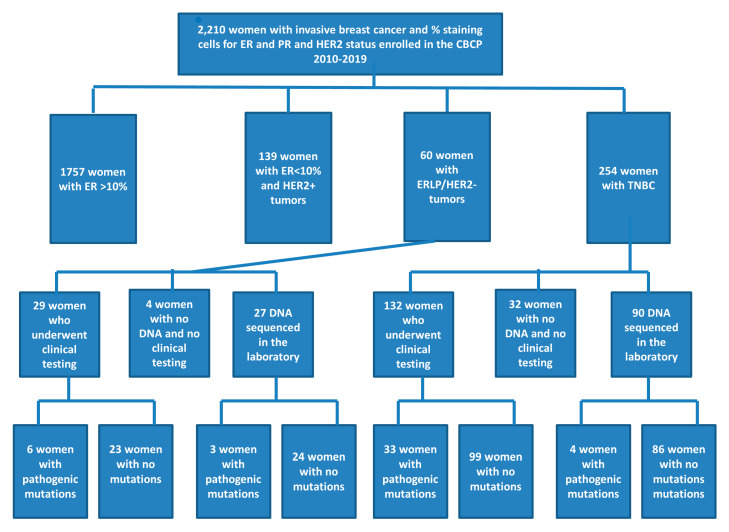
Of the 2210 women with invasive breast cancer who enrolled in the CBCP between 2010 and 2019, 60 (2.7%) had ERLP/HER2-tumors with PR staining ≤ 10% tumors and 254 (11.5%) had TNBC (Figure 1). The average age at diagnosis was 57.3 years old in women with ERLP/HER2-tumors and 56.5 years in those with TNBC (p = 0.700). Ethnicity; family history; and tumor size, stage, and grade did not differ significantly between groups (Table 1).
Figure 1.
Flow-chart describing biomarker status, test uptake, and sequencing results.
Table 1.
Demographic and pathologic characteristics of women with estrogen receptor low positive (ERLP)/ HER2 negative (HER2-) and triple negative breast cancers (TNBC) tumors.
| Characteristic | ERLPHN (n = 60) | TNBC (n = 254) | p-Value |
|---|---|---|---|
| Age | 0.443 | ||
| ≤45 years | 11 (18.3%) | 55 (21.7%) | |
| 46–60 years | 30 (50.0%) | 104 (40.9%) | |
| >60 years | 19 (31.7%) | 95 (37.4%) | |
| Ethnicity | 0.178 | ||
| African American | 13 (21.6%) | 85 (33.4) | |
| Asian | 2 (3.3%) | 5 (2.0%) | |
| Hispanic | 1 (1.7%) | 6 (2.4%) | |
| Other | 1 (1.7%) | 3 (1.2%) | |
| European American | 42 (70.0%) | 155 (61.0%) | |
| Unknown | 1 (1.7%) | 0 (0.0%) | |
| Family history | 0.197 | ||
| 0 | 18 (30.0%) | 108 (42.5%) | |
| 1 | 25 (41.7%) | 78 (30.7%) | |
| 2 | 9 (15.0%) | 46 (18.1%) | |
| ≥3 | 8 (13.3%) | 20 (7.9%) | |
| Unknown | 0 (0.0%) | 2 (0.8%) | |
| Tumor size | 0.189 | ||
| T1 | 22 (36.7%) | 131 (51.6%) | |
| T2 | 30 (50.0%) | 97 (38.2%) | |
| T3/4 | 6 (10.0%) | 22 (8.6%) | |
| Unknown | 2 (3.3%) | 4 (1.6%) | |
| Tumor grade | 0.488 | ||
| Well-differentiated | 2 (3.3%) | 4 (1.6%) | |
| Moderately-differentiated | 12 (20.0%) | 35 (13.8%) | |
| Poorly-differentiated | 45 (75.0%) | 211 (83.0%) | |
| Unknown | 1 (1.7%) | 4 (1.6%) | |
| Tumor stage | 0.092 | ||
| I | 18 (30.0%) | 109 (42.9%) | |
| II | 30 (50.0%) | 111 (43.7%) | |
| III | 9 (15.0%) | 31 (12.2%) | |
| IV | 3 (5.0%) | 3 (1.2%) |
3.2. Test Eligibility and Uptake
Significantly fewer (p = 0.015) women with ERLP/HER2- (67.7%) were eligible for genetic testing compared with those with TNBC (81.1%). The test uptake did not differ significantly (p = 0.305) in the two groups with 29/40 (67.5%) women with ERLP/HER2- and 132/206 (61.2%) women with TNBC pursuing clinical genetic testing.
3.3. Genetic Results
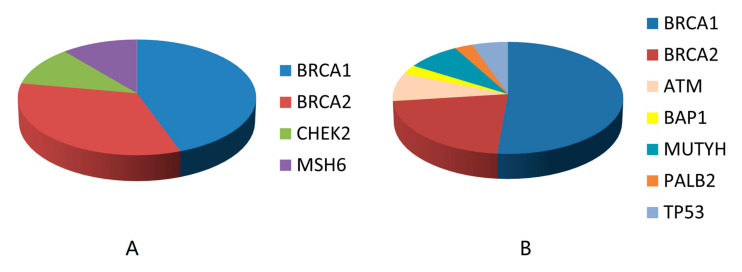
Germline data were available for 56/60 women with ERLP/HER2-tumors and 222/254 women with TNBC. The mutation frequency did not differ significantly (p = 0.757) between groups (ERLP/HER2- 16.1%; TNBC 16.7%). Mutations were detected in four genes, BRCA1 (n = 4), BRCA2 (n = 3), CHEK2 (n = 1), and MSH6 (n = 1) in women with ERLP/HER2-, and in seven genes, ATM (n = 3), BAP1 (n = 1), BRCA1 (n = 20), BRCA2 (n = 8), MUTYH (n = 2), PALB2 (n = 1), and TP53 (n = 2), in women with TNBC (Table 2). Mutations in BRCA1 were the most common in both groups (44.4% ERLP/HER2- and 54.1% TNBC; Figure 2).
Table 2.
Mutations within women with ERLP/HER2-tumors (n = 9) or TNBC (n = 37).
| Patient | Gene | Mutation | ≤45 Years at Diagnosis | <60 Years at Diagnosis | Family History a |
|---|---|---|---|---|---|
| ERLP/HER2- | |||||
| 38 | BRCA1 | Glu1250Ter | √ | √ | √ |
| 185 | BRCA1 | Mutation not provided b | √ | √ | |
| 191 | BRCA1 | Glu1250Ter | √ | √ | √ |
| 226 | BRCA1 | Gln1756Profs | √ | √ | √ |
| 78 | BRCA2 | Leu1466Phefs | √ | ||
| 117 | BRCA2 | Leu2510Pro | √ | ||
| 125 | BRCA2 | Ile1859Lysfs | √ | √ | |
| 171 | CHEK2 | Thr367Metfs | √ | ||
| 213 | MSH6 | Phe1088Leufs | √ | √ | |
| TNBC | |||||
| 280 | ATM | Glu2236Ter | |||
| 230 | ATM | 901 + 1G > A | √ | ||
| 289 | ATM | Tyr1124Ter | √ | ||
| 303 | BAP1 | Leu573fs | √ | ||
| 5 | BRCA1 | 5193 + 2del | √ | ||
| 48 | BRCA1 | Gln1408Ter | √ | √ | |
| 54 | BRCA1 | Met1775Arg | √ | √ | |
| 73 | BRCA1 | 5467 + 1G > A | √ | √ | |
| 80 | BRCA1 | Glu908Ter | √ | ||
| 110 | BRCA1 | Gln1756Profs | √ | ||
| 112 | BRCA1 | Ser1655fs | √ | √ | √ |
| 123 | BRCA1 | 4986 + 6T > G | √ | √ | √ |
| 150 | BRCA1 | Val1234fs | √ | √ | √ |
| 159 | BRCA1 | Val1688del | √ | √ | |
| 172 | BRCA1 | 4986 + 3G > C | √ | √ | |
| 182 | BRCA1 | Glu23fs | √ | √ | |
| 201 | BRCA1 | Mutation not provided b | √ | √ | √ |
| 219 | BRCA1 | Val1838Glu | √ | √ | √ |
| 237 | BRCA1 | Glu23fs | √ | √ | |
| 239 | BRCA1 | Mutation not provided b | √ | ||
| 280 | BRCA1 | Met1775Arg | |||
| 283 | BRCA1 | 135-1G > T | √ | √ | √ |
| 288 | BRCA1 | Thr276fs | √ | √ | |
| 310 | BRCA1 | Lys894fs | √ | √ | √ |
| 62 | BRCA2 | Asp1199_Cys1200insTer | |||
| 109 | BRCA2 | Tyr1894Terfs | √ | √ | |
| 139 | BRCA2 | Asn1784fs | √ | ||
| 236 | BRCA2 | Ile2315fs | |||
| 238 | BRCA2 | Asn1626Serfs | |||
| 268 | BRCA2 | Asn986fs | √ | ||
| 277 | BRCA2 | Leu2805fs | √ | √ | |
| 308 | BRCA2 | Cys1200Terfs | √ | ||
| 133 | MUTYH | Arg233Ter | √ | ||
| 221 | MUTYH | 850-2A > G | √ | √ | |
| 241 | PALB2 | Tyr1183Ter | √ | √ | |
| 220 | TP53 | Arg175Gly | √ | √ | |
| 300 | TP53 | Arg273Cys | √ | √ | |
a Family history defined using National Comprehensive Cancer Network (NCCN) 1.2020 criteria for determining hereditary testing eligibility. b for three patients, the exact mutation from clinical testing was not provided in the database.
Figure 2.
Frequency of mutations by genes in ERLP/HER2-tumors (A) and TNBC (B).
3.4. Mutation Frequency in Women Not Eligible for Genetic Testing
Eleven (18.3%) women diagnosed between 45 and 60 years of age with ERLP/HER2- did not have a significant family history of breast cancer. One (9.1%) of these test-ineligible women harbored a germline mutation in BRCA2, with tumor biomarkers reported as 10% ER, 1% PR, and 0+ HER2. Within the group of 46 women diagnosed with TNBC between 45 and 60 years of age but without a significant family history of breast cancer, three (6.5%) harbored mutations in ATM (n = 1), BAP1 (n = 1), or BRCA2 (n = 1). The mutation frequency was not significantly different (p = 0.825) between these two groups of women.
4. Discussion
Since the identification of the BRCA1 and BRCA2 genes over 25 years ago [13,14], the benefits of germline testing have expanded to include not only personal and familial risk assessment, but also treatment choice. For women with BRCA germline mutations, patients may elect contralateral prophylactic mastectomy and oophorectomy [15]. In patients with TNBC and BRCA mutations, platinum salts and PARP inhibitors, which exploit the genomic instability of cells with defective DNA repair, may be beneficial [16]. Surveillance and surgical strategies have also been developed for women with germline mutations in non-BRCA genes in order to prevent or facilitate in the early detection of additional cancers [17]. The opportunity to utilize these precision medicine approaches may not be available for women who harbor germline mutations but that are ineligible for germline testing using the current criteria.
Within our dataset, the mutation frequency and array of mutated genes did not differ significantly between women with TNBC and ERLP/HER2-tumors, suggesting that germline testing would be beneficial in women with ER low-staining tumors. Of note, under the current ASCO/CAP guidelines, which define ER tumor staining of 1–10% as ER positive, 18.3% of women in the ERLP/HER2- group diagnosed at 46–60 years of age without a family history, were not eligible for genetic testing, while the 18.1% of women with TNBC with similar characteristics would be eligible. Moreover, the mutation frequency within this test-ineligible group did not differ significantly from their counterparts with TNBC. Nine percent of these test-ineligible women may have benefited from precision medicine approaches afforded by the determination of the mutational background.
To our knowledge, one other publication examined germline mutations in low ER staining/HER2-breast tumors. A study from The University of Texas MD Anderson Cancer Center evaluated mutation frequencies in 314 patients with HER2-ER < 10% tumors with BRCA1/2 genetic test results. That study found that 238 women had TNBC and 76 women had hormone receptor (HR) 1–9%/HER2-tumors [18]. As in our study, mutation frequencies did not differ significantly between patients with TNBC and those with HR-low positive tumors. The BRCA mutation frequency in the MD Anderson study (36.9%) was, however, significantly higher (p < 0.001) than that detected in our study (16.9%). This difference is likely attributable to the following inclusion criteria: all of the patients in the MD Anderson study were referred for genetic counseling between 2004 and 2014, while our study included all of the patients diagnosed between 2010–2020 with HER2-/ER ≤ 10% tumors, 21.6% of whom were not eligible for genetic testing using the NCCN 1.2020 criteria. In addition, our study evaluated a more extensive (n = 94) set of cancer predisposition genes, and detected mutations in genes such as PALB2, which has been associated with heritable forms of TNBC [19]. Despite these differences, both data sets suggest that the ER ≥ 1% threshold defined by ASCO/CAP may lead to under testing.
This study has several limitations. While the frequency of ERLP/HER2-tumors was within the 2–3% range reported by ASCO/CAP [10], only 60 cases were reported in the past decade. Of these, four women had no germline testing and no DNA available for research purposes, which may have influenced the mutation frequency. In addition, data do not yet exist as to whether women with ERLP/HER2-tumors, especially those diagnosed at 46–60 years old without a significant family history, would pursue genetic testing or, for those with BRCA mutations, would benefit from PARP inhibitors or platinum agents.
5. Conclusions
These data demonstrate that women with ERLP/HER2-tumors have germline mutation frequencies similar to those detected in women with TNBC, with BRCA1 and BRCA2 being the most frequently mutated genes in this population. These data suggest that while ASCO/CAP guidelines classify tumors with ER staining 1–10% as ER positive, women with ERLP/HER2-tumors may benefit from germline genetic testing to improve patient management and provide the opportunity for family planning and risk assessment.
Acknowledgments
The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of the Uniformed Services University of the Health Sciences (USUHS); The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.; the Department of Defense (DoD); or the Departments of the Army, Navy, or Air Force. The mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
Author Contributions
Study concept and design, R.E.E.; Acquisition of data, L.A.L., J.M.W. and C.D.S.; Analysis and interpretation of data, R.E.E. and C.E.T.; Drafting of the manuscript R.E.E.; Critical revision of the manuscript for important intellectual content, L.A.L., J.M.W., C.E.T. and C.D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a cooperative agreement from the Uniformed Services University of the Health Sciences HU0001-16-2-0004 through the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lin N.U., Claus E., Sohl J., Razzak A.R., Amaout A., Winer E.P. Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: High incidence of central nervous system metastases. Cancer. 2008;113:2638–2645. doi: 10.1002/cncr.23930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P., Klemp J.R., Kimler B.F., Mahnken J.D., Geier L.J., Khan Q.J., Elia M., Connor C.S., McGinness M.K., Mammen J.M., et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: Implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res. Treat. 2014;145:707–714. doi: 10.1007/s10549-014-2980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garutti M., Pelizzari G., Bartoletti M., Malfatti M.C., Gerratana L., Tell G., Puglisi F. Platinum salts in patients with breast cancer: A focus on predictive factors. Int. J. Mol. Sci. 2019;20:3390. doi: 10.3390/ijms20143390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmer A.S., Gillard M., Lipkowitz S., Lee J.M. Update on PARP Inhibitors in breast cancer. Curr. Treat. Opt. Oncol. 2018;19:21. doi: 10.1007/s11864-018-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network Genetic/familial high-risk assessment: Breast and ovarian. JNCCN. 2010;8:562–594. doi: 10.6004/jnccn.2010.0043. [DOI] [PubMed] [Google Scholar]
- 6.Hammond M.E.H., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deyarmin B., Kane J.L., Valente A.L., van Laar R., Gallagher C., Shriver C.D., Ellsworth R.E. Effect of ASCO/CAP guidelines for determining ER status on molecular subtype. Ann. Surg. Oncol. 2013;20:87–93. doi: 10.1245/s10434-012-2588-8. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto T., Booser D., Valero V., Murray J.L., Koenig K., Esteva F.J., Ueno N.T., Zhang J., Shi W., Qi Y., et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J. Clin. Oncol. 2012;30:729–734. doi: 10.1200/JCO.2011.36.2574. [DOI] [PubMed] [Google Scholar]
- 9.Yi M., Huo L., Koenig K.B., Mittendorf E.A., Meric-Bernstam F., Kuerer H.M., Bedrosian I., Buzdar A.U., Symmans W.F., Crow J.R., et al. Which threshold for ER positivity? a retrospective study based on 9639 patients. Ann. Oncol. 2014;25:1004–1011. doi: 10.1093/annonc/mdu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison K.H., Hammond M.E.H., Dowsett M., McKernin S.E., Carey L.A., Fitzgibbons P.L., Hayes D.F., Lakhani S.R., Chavez-MacGregor M., Perlmutter J., et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J. Clin. Oncol. 2020;38:1346–1366. doi: 10.1200/JCO.19.02309. [DOI] [PubMed] [Google Scholar]
- 11.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. J. Clin. Oncol. 2018;36:2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 12.Rummel S.K., Lovejoy L.A., Turner C.E., Shriver C.D., Ellsworth R.E. Should genetic testing for cancer predisposition be standard-of-care for women with invasive breast cancer? The Murtha Cancer Center experience. Cancers. 2020;12:234. doi: 10.3390/cancers12010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 14.Wooster R., Bignell G., Lancaseter J., Swift S., Seal S., Mangion J., Collins N., Gregory S., Gumbs C., Micklem G., et al. Identification of the breast cancer susceptibility gene BRCA. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 15.Song C.V., Teo S.H., Taib N.A., Yip C.H. Surgery for BRCA, TP53 and PALB2: A literature review. Ecancermedicalscience. 2018;12:863. doi: 10.3332/ecancer.2018.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolas E., Bertucci F., Sabatier R., Goncalves A. Targeting BRCA deficiency in breast cancer: What are the clinical evidences and the next perspectives? Cancers. 2018;10:506. doi: 10.3390/cancers10120506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piombino C., Cortesi L., Lambertini M., Punie K., Grandi G., Toss A. Secondary prevention in hereditary breast and/or ovarian cancer syndromes other than BRCA. J. Oncol. 2020;2020:6384190. doi: 10.1155/2020/6384190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanford R.A., Song J., Gutierrez-Barrera A.M., Profato J., Woodson A., Litton J.K., Bedrosian I., Albarracin C.T., Valero V., Arun B. High incidence of germline BRCA mutation in patients with ER low-positive/PR low-positive/HER-2 neu negative tumors. Cancer. 2015;121:3422–3427. doi: 10.1002/cncr.29572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buys S.S., Sandbach J.F., Gammon A., Patel G., Kidd J., Brown K.L., Sharma L., Saam J., Lancaster J., Daly M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]