Abstract
A methanogenic enrichment growing on a medium with methanol was obtained from a petroleum reservoir (Republic of Azerbaijan) and stored for 33 years without transfers to fresh medium. High-throughput sequencing of the V4 region of the 16S rRNA gene revealed members of the genera Desulfovibrio, Soehngenia, Thermovirga, Petrimonas, Methanosarcina, and Methanomethylovorans. A novel gram-positive, rod-shaped, anaerobic fermentative bacterium, strain 1933PT, was isolated from this enrichment and characterized. The strain grew at 13–55 °C (optimum 35 °C), with 0–3.0% (w/v) NaCl (optimum 0–2.0%) and in the pH range of 6.7–8.0 (optimum pH 7.0). The 16S rRNA gene sequence similarity, the average nucleotide identity (ANI) and in silico DNA–DNA hybridization (dDDH) values between strain 1933PT and the type strain of the most closely related species Soehngenia saccharolytica DSM 12858T were 98.5%, 70.5%, and 22.6%, respectively, and were below the threshold accepted for species demarcation. Genome-based phylogenomic analysis and physiological and biochemical characterization of the strain 1933PT (VKM B-3382T = KCTC 15984T) confirmed its affiliation to a novel species of the genus Soehngenia, for which the name Soehngenia longivitae sp. nov. is proposed. Genome analysis suggests that the new strain has potential in the degradation of proteinaceous components.
Keywords: Soehngenia longivitae, new species, genome analysis, polyphasic taxonomy, oilfield
1. Introduction
The genus Soehngenia belongs to the family Tissierellaceae [1] of the order Tissierellales of the class (not validly published) ‘Tissierellia’ [2] within the phylum Firmicutes. At the time of writing, the genus Soehngenia was represented by the type strain BOR-YT (=DSM 12858T = ATCC BAA-502T) of the only published species Soehngenia saccharolytica isolated from an anaerobic sludge bed reactor treating potato starch waste [3]. Strain DSM 12858T is a gram-positive, rod-shaped, motile, mesophilic, neutrophilic anaerobic spore-former, which ferments a wide range of carbohydrates and other carbon sources, including yeast extract, cysteine, and serine. Sulfite and thiosulfate may be used as electron acceptors (reduced to H2S). The strain performs dismutation of benzaldehyde to benzoate and benzyl alcohol [4]. Using 16S rRNA gene sequencing, members of the genus Soehngenia were detected in petroleum reservoirs [5,6,7,8] and in water droplets dispersed in heavy oil [9], in methanogenic crude oil- and propionate-degrading enrichments [10,11], in marine sediments contaminated with heavy metals and polycyclic aromatic hydrocarbons [12], and in cyanide-degrading enrichments [13].
In 1977, in the course of ecological investigation of microbial communities of the Binagady petroleum reservoir (Republic of Azerbaijan) [14], methanogenic enrichment cultures were obtained from production water samples. This enrichment was routinely maintained in our laboratory for 8 years by means of successive 3–6 month transfers into the medium with methanol. Fermentative bacteria and methanogens of the genus Methanosarcina were identified in the enrichments using culture-based techniques [15]. Since 1985, several flasks with the methanogenic enrichment growing on the medium with methanol have been stored at room temperature (18–24 °C) for 33 years without transfers to fresh medium in order to investigate survival of oilfield communities. In 2018, the enrichment was used to inoculate fresh medium with methanol, and methane production was registered in this culture. Sarcinae were present in the environment, as well as the cells of other morphological types. Methanosarcinae and other microorganisms occurring in the enrichment were isolated by inoculating the media with various carbohydrate and proteinaceous substrates, with or without antibiotics, and incubating the media at various temperatures. Thus, several stable enrichments were obtained from the methanogenic enrichment. One of the first isolates was a pure culture of a fermenting bacterium, strain 1933PT (VKM B-3382T = KCTC-15984T), straight motile rods growing in the medium with peptone at 28 °C [16]. The 16S rRNA gene sequence of the strain 1933PT had 98.5% similarity to that of Soehngenia saccharolytica DSM 12858T. This value was below the threshold accepted for species demarcation at 98.65% 16S rRNA gene sequence similarity [17].
The present study aimed to describe the phylogenetic diversity of prokaryotes in methanogenic enrichment stored for 33 years and details of the isolation procedure, the morphological and chemotaxonomic properties of strain 1933PT, as well as the results of a comparative analysis of Tissierellaceae genomes. These data allowed us to assign strain 1933PT to a novel species of the genus Soehngenia, for which the name Soehngenia longivitae sp. nov. is proposed.
2. Materials and Methods
2.1. Characteristics of the Sampling Site
The Binagady oilfield is located near Baku city in the Republic of Azerbaijan. Exploitation of this oilfield involves flooding with the Lake Beyuk Shor water (salinity 7–10 g L−1). In 1977, a water sample was obtained from production well 1933, which operated on a sandstone oil-bearing horizon, was located at a depth 590–597 m below sea level, and has a temperature of 28 °C. The total salinity of the studied formation water was 84.8 g L−1. The oil recovered was heavy, paraffin-free, with specific density of 0.899–0.964 g/cm3 (at 20 °C). Information on the physicochemical conditions and microbial numbers in production water of the Binagady oilfield was presented earlier [14]. The sample of oil–water mixture taken directly from production wellhead was collected in a sterile 0.5 L serum bottle. The bottle was sealed with a rubber stopper and screw cap and transported at ambient temperature to the field laboratory, where it was immediately used for inoculation of the nutrient media.
2.2. Isolation of Methanogenic Enrichment and Pure Culture of a Fermenting Bacterium
In 1977, a methanogenic enrichment culture was obtained in the mineral medium (MM) [18] containing, per liter distilled water, 0.2 g KH2PO4, 0.25 g NH4Cl, 15 g NaCl, 0.4 g MgCl2 · 6H2O, 0.5 g KCl, 0.1 g CaCl2 · 2H2O, 0.5 g Na2S · 9H2O, supplemented with methanol (5%, vol/vol), and inoculated with production water (10%, vol/vol) from the Binagady oilfield. Since 1985, the culture was stored at room temperature for 33 years without transfers to fresh medium. In 2018, using this long-term storage methanogenic enrichment as inoculum, a pure culture, strain 1933PT (VKM B-3382T = KCTC 15984T), was isolated by sequential transfers from the highest dilutions of enrichment to the mineral medium MM with peptone (2 g L−1), yeast extract (0.2 g L−1), NaCl (15 g L−1), and Na2S · 9H2O (0.2 g L−1), at pH 7.0 and 30 °C. The medium was supplemented with 1 mL L−1 of the following solutions: 0.1% (w/W) Mohr’s salt (FeSO4 · (NH4)2SO4 · 6H2O), vitamins [19], and microelements [20]. The medium was prepared anaerobically under a flow of Ar, dispensed into Hungate tubes [21], and sealed with butyl rubber stoppers. Strain 1933PT was subsequently maintained in this medium. Strain 1933PT did not form colonies in solid medium. The culture purity was confirmed by microscopic studies, as well as by the 16S rRNA gene and genome sequencing.
2.3. DNA Isolation, Amplification and Sequencing of the 16S rRNA Genes from the Methanogenic Enrichment
Cell biomass from 20 mL of the methanogenic enrichment culture stored for 33 years was collected on the membrane filter, washed off with a lysing solution containing 0.15 M NaCl and 0.1 M Na2-EDTA (pH 8.0) and used for DNA extraction. Isolation of the total DNA was carried out using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA), according to the manufacturer’s recommendations. DNA was stored in a freezer at −20 °C. Total genomic DNA was amplified using the 515f/806r primer set that amplifies the V4 region of the 16S rRNA gene [22]. Sequencing was carried out on a MiSeq platform (Illumina, San Diego, CA, USA) using the MiSeq Reagent Kit v3 (600 cycles) (Illumina, United States) according to the manufacturer’s recommendations. The obtained 250-bp paired-end reads were further processed according to the workflow implementing suitable scripts from USEARCH version 10 [23]. Reads were demultiplexed (-fastx_demux), trimmed to remove the primer sequences (-fastx_truncate), and then quality filtered (-fastq_filter). UNOISE3 [24] was used to generate zero radius operational taxonomic units (zOTUs). zOTU is a term specific to analysis with UNOISE, referring to operational taxonomic units which were generated by an error correction algorithm as opposed to a sequence similarity clustering algorithm [25]. Raw merged read pairs were mapped back to zOTUs using the -otutab command. zOTUs were submitted for taxonomic analysis in the SILVA database (SINA, https://www.arb-silva.de/aligner/, October 2020, version 1.2.11 [26], SILVA reference database release 138.1) using default settings.
2.4. Morphological, Physiological and Chemotaxonomic Characterization
Strain 1933PT was characterized using a polyphasic taxonomic approach and compared with the reference strain Soehngenia saccharolytica DSM 12858T kindly provided by Dr. S.N. Parshina from the Research Center of Biotechnology of the Russian Academy of Sciences. Unless otherwise stated, both strains were cultivated in a liquid MM medium. This medium contained peptone (2 g L−1), yeast extract (0.2 g L−1), 1.5% NaCl for strain 1933PT, sucrose (2 g L−1), yeast extract (1.0 g L−1) and 0.1% (w/v) NaCl for strain DSM 12858T. Optimal temperature conditions were determined by growing the strain 1933PT in liquid medium at 8, 13, 20, 25, 30, 35, 42, 45, 50, 55, and 60 °C. Salt tolerance tests for strains 1933PT and DSM 12858T were carried out in liquid medium supplemented with 0–6 % (w/v) (0, 0.1, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, and 6.0 %) of NaCl at 35 °C. The pH range for growth of the isolate was tested in the media adjusted to pH 5.5, 6, 6.3, 6.7, 7.0, 7.5, 8.0, 8.5, and 9.0 with the addition of the appropriate buffer at optimal temperature (35 °C) and NaCl concentration (1.5% (w/v) NaCl) [27].
The cells’ sizes were measured on living cells using an Axio Imager.D1 epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) with an Axio Cam HRc digital camera and Axio Vision computer software. The gram reaction and cell ultrastructure were studied as described previously [27]. Ultrathin sections were examined under a JEM-100C transmission electron microscope (JEOL, Tokyo, Japan) at an accelerating voltage of 80 kV. Biochemical and enzyme characteristics of strains 1933PT and DSM 12858T were determined by using API 50CH, API ZYM, and API 20E kits (bioMérieux, Marcy-l’Étoile, France) according to the manufacturer’s instructions, and incubated 7 days at 35 °C; to prevent oxygen access, mineral oil was added to each well. Catalase activity was determined by the standard method involving addition of 3% (v/v) H2O2 to concentrated cell suspensions. Oxidase activity was determined using the oxidase reagent (bioMérieux, France). Growth of the strain 1933PT was additionally tested in MM medium with yeast extract (0.2 g L−1) and various substrates. The concentrations of sugars and biopolymers were 2 g L−1; those of organic acids and alcohols were 20 mM, and the H2 + CO2 mixture was tested at 80: 20 (v/v). Growth was monitored by the optical density (OD) at 660 nm. OD increases of <10, 10–50, and >50% obtained with the test substrates were scored as no utilization (−), weak utilization (W) and good utilization (+). Growth was registered for up to 14 days in three successive transfers. All tests were performed in duplicate. Strain 1933PT was tested for its ability to use sulfate (20 mM), thiosulfate (15 mM), sulfite (15 mM), sulfur (5 g L−1), and nitrate (20 mM) as electron acceptors. Sulfide was measured by the colorimetric method with N,N-dimethyl-p-phenylenediamine in the modification by Trüper and Schlegel [28]; nitrite was determined using the Griess reagent. Fermentation products were analyzed by gas chromatography as described previously [29]. Nitrogen fixation was estimated by acetylene reduction assay in mineral medium with maltose (2.0 g L−1) and yeast extract (0.1 g L−1) under the N2 gas phase amended with 10% (v/v) acetylene. Ethylene production in each vial was quantified, using a gas chromatograph equipped with a flame ionization detector and a capillary column, as recommended [30].
The fatty acid composition was analyzed using a Maestro gas chromatograph–mass spectrometer (Interlab, Moscow, Russia). The cell biomass was dried with methanol and subjected to acidic methanolysis (1.2 M HCl/MeOH, 80 °C, 45 min) as described earlier [31]. The analysis of polar lipids of strains 1933PT and S. saccharolytica DSM 12858T was performed at the All-Russian Collection of Microorganisms according to the method described by Minnikin et al. [32]. Polar lipids were extracted from freeze-dried cells. The lipids were separated by two-dimensional TLC on Silica Gel 60F TLC-plates (Merck) using the following solvent systems: chloroform/methanol/water (65: 25: 4, by vol.) in the horizontal dimension and chloroform/acetic acid/methanol/water (80: 15: 12: 4, by vol.) in the vertical dimension. Total lipids were visualized by spraying with a 5 % (w/v) solution of phosphomolybdic acid in ethanol. Phospholipids were further characterized by spraying with ninhydrin (specific for amino groups), molybdenum blue (specific for phosphates) and α-naphthol (specific for glycolipids).
2.5. 16S rRNA Gene Sequencing and Phylogenetic Analysis
DNA was extracted from 1933PT culture using the PowerSoil DNA Isolation Kit (MoBio, USA), according to the manufacturer’s recommendations. The 16S rRNA gene of the strain 1933PT was amplified with the 27F and 1492R primers [33], and purified PCR products were sequenced with an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) using the Big Dye Terminator reagent kit, version 3.1. The 16S rRNA gene sequence analysis was performed using the EzBioCloud [34]. Phylogenetic analysis of the 16S rRNA gene sequences was carried out using the maximum-likelihood, neighbour-joining, and maximum-parsimony algorithms. The sequences were first aligned by MUSCLE [35], and the maximum-likelihood tree was inferred using the GTR + F + I + G4 model recommended by ModelFinder [36] in IQ-Tree [37]. Neighbour-joining and maximum-parsimony trees were reconstructed using the MEGA7 software package [38]. Bootstrap values were calculated from 1000 alternative trees.
2.6. Genome Analysis
The genome of the strain 1933PT was sequenced and annotated as described previously [16]. Phylogenomic analysis of strain 1933PT and members of the families Tissierellaceae and Gottschalkiacecae was conducted using a concatenated alignment of 120 single-copy phylogenetic marker genes obtained using the software GTDB-Tk version 1.0.2 [39]. A maximum likelihood phylogenomic tree was calculated using IQ-Tree [37] according to the model recommended by ModelFinder [36] and branch support was estimated using UFBoot2 [40]. Maximum parsimony and neighbour-joining trees were reconstructed using MPBoot [41] and MEGA7 [38], respectively. The pair-wise average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values among 1933PT, S. saccharolytica DSM 12858T, and strains of other genera of the family Tissierellaceae based on their whole genomes were calculated using the ANI calculator (https://ani.jgi.doe.gov/html/calc.php?) [42] and the genome-to-genome distance calculator version 2.1 [43] with BLAST+ for genome alignments [44], respectively. Average amino acid identity (AAI) values were calculated using CompareM 0.0.23 (https://github.com/dparks1134/CompareM) with default blastp parameters (i.e., e-value ≤ 0.001, percent identity ≥ 30% and alignment length ≥ 70%). The pairwise percentage of conserved proteins (POCP) was calculated using the runPOCP.sh script [45,46], which was based on a previously published approach [47]. Eleven Tissierellaceae genomes were used for a pangenomic analysis. The analysis was done following the bioinformatic pipeline proposed [48] with the anvi’o program version 6.2 [49]. The genomes were organized based on the distribution of gene clusters using the MCL algorithm (distance: Euclidean; linkage: Ward). Functional genome annotations were performed using DRAM [50].
2.7. Nucleotide Sequence Accession Numbers
The GenBank/EMBL/DDBJ accession number of the 16S rRNA gene sequence of strain 1933PT is MN698738.1. The GenBank/EMBL/DDBJ accession number of the genome of strain 1933PT is SRIB00000000 (version SRIB01000000) [16]. The 16S rRNA gene library of the methanogenic enrichment was deposited to NCBI, project PRJNA673732, SRR12971726.
3. Results and Discussion
3.1. Phylogenetic Diversity of Prokaryotes in Methanogenic Enrichment
The methanogenic enrichment 1933, which was stored for 33 years at room temperature in the medium with methanol without transfer to fresh medium, was used for sequencing of the V4 hypervariable region of prokaryotic 16S rRNA genes. The resulting dataset upon filtration consisted of 35,373 reads assigned to Bacteria and 104 reads assigned to Archaea. A minority of archaea in methanogenic enrichment is likely due to death and lysis of methanogens over the 33 years period. Archaeal sequences (˂0.25% in the library) belonged to members of Euryarchaeota (genera Methanosarcina and Methanomethylovorans). Bacterial groups revealed in the enrichment were affiliated to the class Deltaproteobacteria (56.9% of sequences in the library) and to the phyla Firmicutes (26.3%), Synergistetes (9.8%), Bacteroidetes (4.7%), and Actinobacteria (1.1%). Minor components of the enrichment belonging to the phyla Thermotogae, Spirochaetes, and Chloroflexi were each responsible for ˂1% of the sequences in the library. At the species level the enrichment was dominated by a sulfate-reducing bacterium Desulfovibrio aminophilus (55.9% of sequences), fermenting bacterium Soehngenia sp. (26.1%), Thermovirga sp. (8.8%), Petrimonas sulfuriphila (3.2%), and Proteiniphilum sp. (0.8%). The fragments of their 16S rRNA genes had more than 99.6% similarity with the genes of respective bacteria.
In this enrichment only methanogenic archaea of the genera Methanomethylovorans and Methanosarcina, which were among strains able to grow on methanol, survived for 33 years [51,52]. Desulfovibrio aminophilus was detected in the enrichment, despite sulfate not being added into the medium. Sulfate may come from sulfur organic molecules, including amino acids and yeast extract. This bacterium is known to be capable of growing on amino acids, H2/CO2, formate, and ethanol as electron donors with sulfate as an electron acceptor, and fermented pyruvate, casamino acids, or peptone in the absence of sulfate in the medium [53]. A moderately thermophilic, anaerobic, amino acid-degrading bacterium Thermovirga lienii and a mesophilic fermentative sulfur-reducing bacterium P. sulfuriphila were detected in the enrichment. These bacteria were originally isolated from petroleum reservoirs and also could not use methanol for growth [54,55]. It is known that Desulfovibrio aminophilus and Thermovirga sp. are capable of growth by fermenting proteinaceous components of the biomass [53,54]. The products of fermentation of peptone, proteinaceous substrates, some amino acids, and a limited number of organic acids (but not sugars, fatty acids, or alcohols) by T. lienii include acetic and propionic acids, ethanol, H2, and CO2 [54]. The possible function of Proteiniphilum in the community is utilization of protein substrates and carbohydrates from cellular debris and production of acetate and CO2 [56,57]. Bacteria of the genus Petrimonas are capable of fermenting carbohydrates and some organic acids with production of acetate, hydrogen, and CO2 [55]. However, as a whole, metagenome analysis likely provides a picture of a part of the original methanogenic culture, without any possibility to associate a role to the detected microbes in an attempt to extrapolate a nutritional network that likely misses some components. Some of the bacteria and archaea that had a certain role in the crossfeeding relationship, are probably not present anymore, and their nucleic acids have been degraded a long time ago.
3.2. Phenotypic Characterization of Strain 1933PT
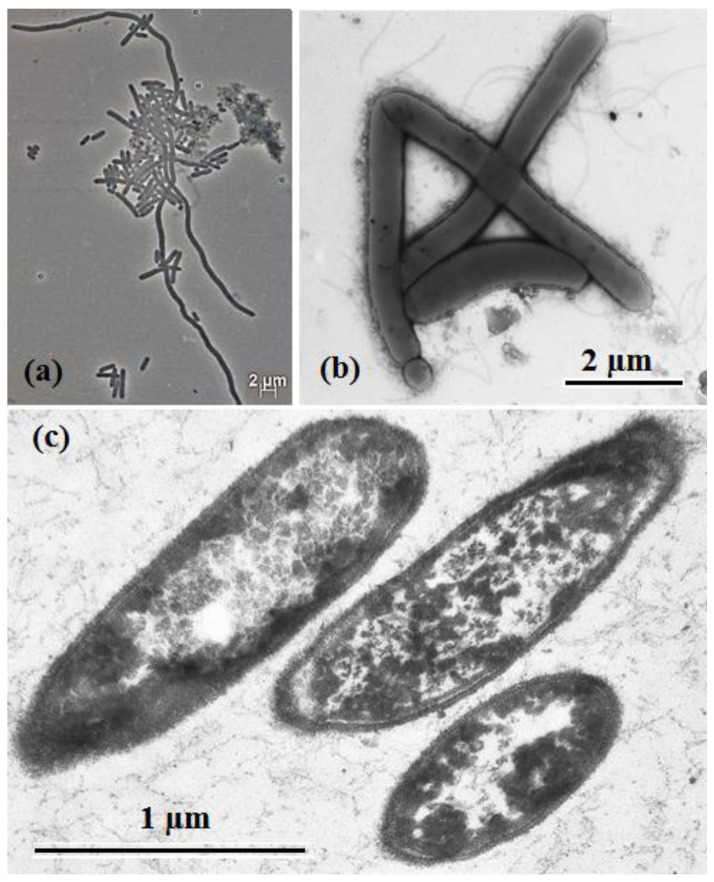
Strain 1933PT and its closely related species, S. saccharolytica DSM 12858T, were phenotypically characterized. Cells of the strain 1933PT were 0.5 μm in width and 2–5 μm in length, gram-stain-positive, motile, peritrichously flagellated rods with rounded ends. Cell division was usually symmetric, but small cells and chains up to 150 μm in length were also visible in the culture (Figure 1a,b). On ultrathin sections the cells had a gram-positive cell wall structure (Figure 1c). Endospores occurred rarely and were terminal, round, and did not distend the mother cell. Strain 1933PT grew at 13–55 °C (optimum 35 °C), at pH 6.7–8.0 (optimum pH 7.0) and with 0–3.0% (w/v) NaCl (optimum 0–2.0%) (Figure S1a–c). Strain S. saccharolytica DSM 12858T grew in the presence of 0–2.0% (w/v) NaCl (optimum, 0–0.5% NaCl) (Figure S1d). Comparative morphological, physiological, and biochemical characteristics of strain 1933PT and of phylogenetically related bacteria S. saccharolytica DSM 12858T and Gudongella oleilytica W6T [1] are summarized in Table 1.
Figure 1.
Phase contrast micrographs of cells: (a) transmission electron micrographs of negatively stained flagellated cells, (b) sections (c) of strain 1933PT grown for 120 h in mineral medium (MM) with peptone and yeast extract at 35 °C.
Table 1.
Characteristics differentiating strain 1933PT, S. saccharolytica DSM 12858T [3] *, and Gudongella oleilytica [1].
| Characteristics | 1933PT | S. saccharolytica DSM 12858T | Gudongella oleilytica W6T |
|---|---|---|---|
| Cell size, µM | 0.5 × 2–5 | 0.5–0.7 × 2–11 | 2–4 × 9–21 |
| Spore formation | + | + | – |
| Temperature range (optimum), °C | 13–55 (35) | 15–40 (30–37) | 20–45 (40) |
| pH range (optimum) | 6.7–8.0 (7.0) | 6.5–7.5 (7.0) | 6.5–9.0 (7.5) |
| NaCl range (optimum), % w/v | 0–5.0 (0–2.0) | 0–2.0 (0) * | 0–3.5 (0) |
| Genomic G + C content, % | 31.9 | 32.94 | 42.4 |
| Genome size (Mb) | 1.92 | 2.0 | 2.36 |
| Utilization of | |||
| Pyruvate | − | + | W |
| Arabinose | − | + | W |
| Cellobiose | − | + | W |
| Galactose | − | + | − |
| Glucose | − | + | − |
| Fructose | − | + | − |
| Lactose | − | + | − |
| Maltose | + | + | − |
| Mannose | W | + | − |
| Mannitol | − | + | |
| Ribose | − | + | − |
| Sucrose | − | + | − |
| Raffinose | − | − | |
| Xylose | − | + | − |
| Xylan | – | + | ND |
| Peptone | + | − | + |
| Methionine | − | − | + |
| Serine | − | + | ND |
| Hydrolysis of gelatin | W | − | ND |
| Molecular nitrogen fixation | − | – * | + |
| The main products of the yeast extract fermentation | Acetate, H2, CO2 | Acetate, H2, CO2 | ND |
| Electron acceptors: | |||
| Elemental sulfur | + | − | ND |
| Sulfite | − | + | ND |
| Major fatty acids | C14:0, C16:0, iso-C15:0 | C16:0, C18:1, C16:1 * | iso-C15:0, C14:0, C16:0, iso-C13:0 |
| Major polar lipids | ND | PGL, PL, L, DPG * | L, AL |
| Isolation source | Methanogenic enrichment from oilfield | Anaerobic-digester sludge | Oily sludge from oilfield |
* Data from this study. ND—no data. Designations: +, positive; −, negative; (W), weakly positive. All strains were rod-shaped, gram-stain-positive, utilized yeast extract, were negative for growth on formate, acetate, and propionate, and could not use sulfate as an electron acceptor.
Strain 1933PT was an anaerobic bacterium. It did not grow in the medium without reductant and in non-reduced medium under 7% (v/v) air in the gas phase. Electron acceptors including nitrate, sulfate, and sulfite were not reduced by strain 1933PT in the MM medium with yeast extract (2 g L−1), but sulfur and thiosulfate stimulated the growth of the strain and were reduced with production up to 90 mg H2S per liter. Nitrate was not reduced to nitrite. The major fermentation products of strain 1933PT growing in MM medium with yeast extract (2 g L−1) were acetic acid, H2, and CO2, while minor amounts of ethanol, propionic, n-butyric and iso-valeric acids were formed. The major products of yeast extract fermentation by strain DSM 12858T were acetic acid, ethanol, acetone, H2, and CO2, and minor amounts of propionic, iso-butyric and n-butyric acids were formed. Although the ability to fix dinitrogen was stated in the description of S. saccharolytica, acetylene assay experiments did not detect this ability in strains DSM 12858T and 1933PT. Strain 1933PT did not grow on benzaldehyde.
As determined using the API ZYM and API 20E tests (bioMérieux, France), strain 1933PT had high activities of alkaline phosphatase and β-glucosidase, as well as weak activities of esterase-lipase (C8), acid phosphatase and naphthol-AS-BI-phosphohydrolase and was negative for indole production (Table S1). S. saccharolytica strain DSM 12858T exhibited positive activity of acid phosphatase and naphthol-AS-BI-phosphohydrolase, was weakly positive on β-glucosidase and there was negative activity of alkaline phosphatase and esterase-lipase (C8). Both strains had negative activities of esterase (C4), lipase (C14), leucine arylamidase, valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, urease, tryptophan deaminase, lysine decarboxylase, ornithine decarboxylase, α-fucosidase, and citrate utilization.
In growth experiments strain 1933PT fermented yeast extract, peptone, tryptone, mannose, but could not grow on formate, acetate, propionate, pyruvate, casein hydrolysate, ethanol, methanol, propanol, leucine, glycine, alanine, and cannot produce acid from fructose, glucose, sucrose, trehalose, cellobiose, raffinose, arabinose, cellulose, starch, xylan, mannitol, or glycerol. In our experiments using the API 50CH kit, strains 1933PT and DSM 12858T were negative for acid production from d-adonitol, d-arabinose, d-arabitol, d-fucose, d-lactose, d-melezitose, d-sorbitol, dulcitol, erythritol, l-rhamnose, l-sorbose, l-xylose, methyl α-d-mannopyranoside, and xylitol. Strain 1933PT did not produce acid from amygdalin, l-arabinose, d-xylose, d-galactose, inositol, methyl α-d-glucopyranoside, d-melibiose, gentiobiose, d-tagatose, and l-arabitol; these tests were positive for strain DSM 12858T.
The major fatty acids of strain 1933PT were C14:0 (36.7%), C16:0 (29.7%), and iso-C15:0 (8.3%), while S. saccharolytica DSM 12858T was mainly composed of C16:0 (41.4%), C18:1 (21.1%), and C16:1 (20.5%) (Table S2). Because of the problems recovering enough biomass of strain 1933PT, we could not reliably determine the polar lipid profile of this strain (Figure S2a, Table 1). Strain DSM 12858T had unidentified phosphoglycolipids, phospholipids, diphosphoglycolipids, and lipids (Figure S2b), which was consistent with results of Wu et al. [1].
3.3. Phylogenetic Analysis of 16S rRNA Gene Sequences
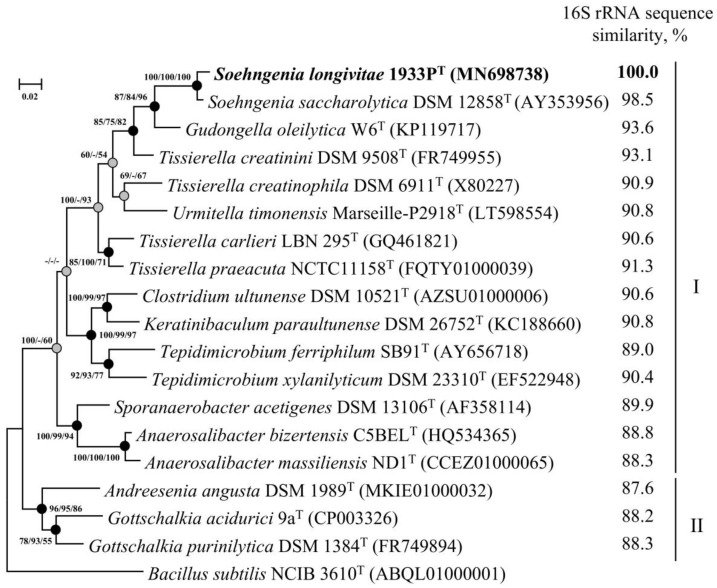
The complete 1476-bp 16S rRNA gene sequence of strain 1933PT was obtained using PCR. The V4 hypervariable region of the sequenced 16S rRNA gene was identical to zOTU of Soehngenia sp., determined from methanogenic enrichment 1933. On phylogenetic trees inferred from the maximum-likelihood, neighbour-joining and maximum-parsimony algorithms, strain 1933PT formed a distinct lineage in the clade with S. saccharolytica DSM 12858T separated from strains of the other genera of the family Tissierellaceae, indicating that strain 1933PT is a member of the genus Soehngenia (Figure 2). The 16S rRNA gene sequences of strains 1933PT and DSM 12858T had 98.5% similarity, which was below the threshold accepted for species demarcation [17] and suggested affiliation of the strain 1933PT to a new species.
Figure 2.
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences (1498 nucleotide sites) reconstructed with the evolutionary model GTR + F + I + G4, showing the position of strain 1933PT and closely related members of the families Tissierellaceae (I) and Gottschalkiaceae (II). Grey circles indicate that the corresponding nodes were recovered in the tree that was reconstructed based on the maximum parsimony algorithm; black circles indicate that the corresponding nodes were also recovered based on the neighbour-joining and maximum-parsimony algorithms. Bootstrap values (>50%) are listed as percentages at the branching points. Scale bar, 0.02 substitutions per nucleotide position. The 16S rRNA gene sequence similarity refers to strain 1933PT compared to the rest of the species. The tree was rooted using Bacillus subtilis NCIB 3610T as the outgroup. GenBank accession numbers for 16S rRNA genes are indicated in brackets.
3.4. Whole Genome Sequencing and Phylogenomic Analyses
The final assembled 1,917,091-bp-long genome of strain 1933PT was comprised of 33 scaffolds, with an N50 value of 132,646 bp, G+C content of 31.9%, and coverage of 630×. The genome contained 1853 genes, of which 1789 were protein-coding sequences, 23 were pseudogenes, and 41 coded RNAs. Functional annotation of the genome was performed with a RAST server [58,59], via the RASTtk pipeline with the default settings [60]; it revealed that 150 of the genes were associated with protein metabolism, 106 genes, with metabolism of amino acids and derivatives, 101 genes, with carbohydrate metabolism, and 40 genes, with metabolism of cofactors, vitamins, and pigments (Figure S3). The genome of strain 1933PT was compared with that of S. saccharolytica DSM 12858T. The genome of strain DSM 12858T was obtained from the JGI IMG database (accession number 2571042347) [61]. General properties of the genomes of both strains are summarized in Table S3.
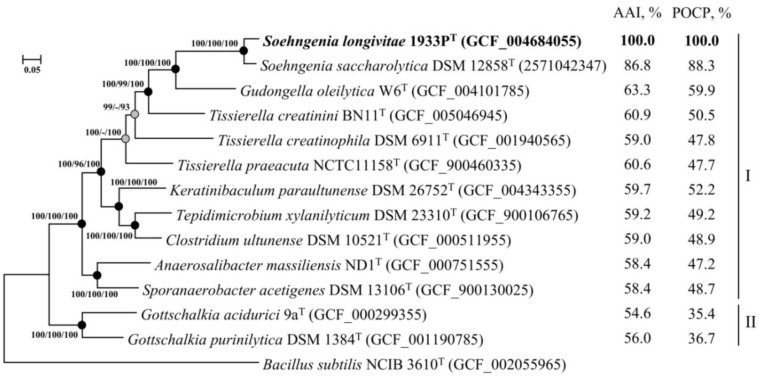
On the phylogenomic tree, strain 1933PT formed a branch with S. saccharolytica DSM 12858T (Figure 3). The ANI and dDDH values of 83.5% and 27.0%, respectively, to the genome of the closest species S. saccharolytica DSM 12858T were below the species cutoff (95–96% for ANI and 70% for dDDH) [62], which indicates that the strain 1933PT belongs to a new species (Table S4). Comparison of strain 1933PT with S. saccharolytica DSM 12858T yielded AAI and POCP values of 86.8% and 88.3%, respectively, which were higher than the proposed genus thresholds of 65% for AAI [63] and 50% for the POCP [47] values. Thus, according to the results of genome-based phylogenomic analysis, strain 1933PT may be classified as a novel species within the genus Soehngenia, for which the name Soehngenia longivitae sp. nov. is proposed.
Figure 3.
Maximum-likelihood phylogenetic tree derived from concatenated 120 single copy marker proteins showing the position of strain 1933PT in relation to taxonomically characterized members of the families Tissierellaceae (I) and Gottschalkiacecae (II). Phylogenetic analysis was performed with an LG + F + I + G4 model based on 34,747 amino acid positions. Scale bar, 0.05 amino acid substitutions per site. The tree was rooted using Bacillus subtilis NCIB 3610T as the outgroup. Accession numbers for the genomes are indicated in brackets. The average amino acid identity (AAI) and pairwise percentage of conserved proteins (POCP) values refer to strain 1933PT compared to the rest of the species.
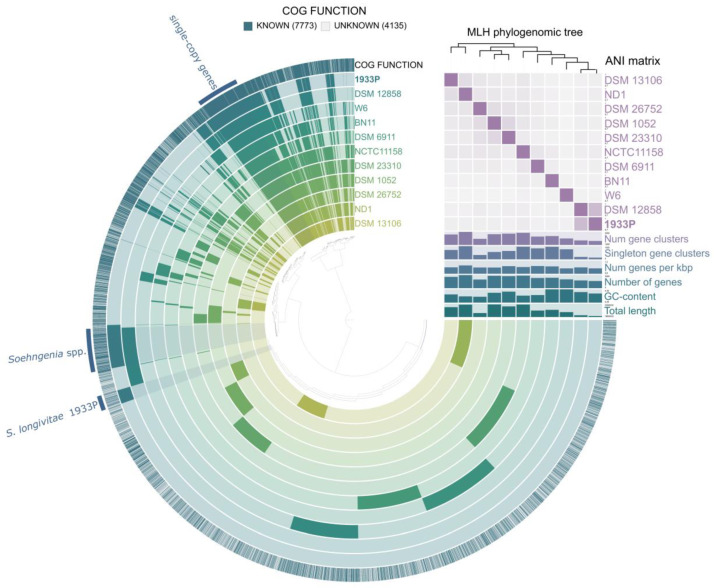
A total of nine genomes were used for the pangenomic analysis of the Tissierellaceae species. The pangenome of Tissierellaceae comprised 29,296 genes in 11,908 gene clusters, and 482 in the core genome (Figure 4). Functional analysis of the core genome proteins revealed that 222 proteins were associated with processing of the genetic information, 31 with nucleotide metabolism, and 30 with carbohydrate metabolism. All Tissierellaceae genomes contained genes for glycolysis, pyruvate oxidation, and phosphate acetyltransferase-acetate kinase pathway. All genomes lacked electron transport chain genes (Figure S4). Strain 1933PT and S. saccharolytica DSM 12858T had 417 shared gene clusters that are not present among other members of Tissierellaceae, and 222 of them were functionally annotated. Shared genes were involved in genetic (57) and environmental (30) information processing, signaling and cellular processes (21), carbohydrates (18), amino acids (12), and vitamins/cofactors (11) metabolisms. Soehngenia genomes carried 22 ABC transporters, which were responsible for iron (III) (afuABC), molybdate (modABC), raffinose/stachyose/melibiose (msnEFG), sugars (chvE, gguAB), phosphate (pstACS), oligopeptide peptides (oppAC), and branched-chain amino acids (livGHKM) acquisition. The genes responsible for fixation of molecular nitrogen were absent in both strains, which correlated with the results of our in vitro tests.
Figure 4.
Pangenome analysis of eleven genomes of Tissierellaceae was calculated with Anvi’o version 6.2. The dendrogram at the center represents the relationship between the 11,908 gene clusters (29,926 genes) found in the analyzed genomes. Dark regions in colored circles represent the genes found in that area for each genome. The average nucleotide identity (ANI) heatmap in purple squares varies between 70 and 100%. The phylogenomic tree was reconstructed using the single copy genes.
The genome of strain 1933PT contained 145 unique gene clusters, but only 36 of them had a predicted function. Among them, the genes responsible for lycopene biosynthesis (phytoene desaturase crtI, 15-cis-phytoene synthase crtB, and phytol kinase VTE5) were found. Unlike the S. saccharolytica DSM 12858T genome, the 1933PT strain lacked ribose/D-xylose transporters (rbsABC).
Genome analysis shows that the metabolism of bacteria of the genus Soehngenia in the methanol-degrading methanogenic enrichment was probably based on fermentation of proteins and amino acids. Genome sizes for Soehngenia sp. 1933PT and S. saccharolytica DSM 12858T (accession number in the JGI IMG database is 2571042347) were 1.92 and 2.00 Mb, respectively (Table S3), which indicated the low catabolic potential of these bacteria. Growth of these rarely occurring bacteria is probably dependent on activity of microorganisms of the previous trophic level, which provide them with available substrates, and is possibly based on metabiotic and syntrophic interactions. It was suggested that rare taxa may represent a reservoir of genetic diversity that actively responds to environmental change [64,65,66]. Oilfields have low levels of water and mass exchange and contain oil, gaseous hydrocarbons, and products of oil oxidation as the major sources of organic matter. Microbial populations of these habitats, including rare species, are adapted to the conditions, as was demonstrated by analysis of the methanogenic enrichment. The 1933PT strain was the only component of the community that could survive adverse conditions due to spore formation. Elucidation of the mechanisms for dormancy preservation by other members of a long-stored methanogenic enrichment requires further investigation.
3.5. Description of Soehngenia Longivitae sp. nov
Soehngenia longivitae (lon.gi.vi’tae. L. masc. adj. longus, long; L. fem. n. vita life; N.L. gen. n. longivitae of long life).
The description is based on a single strain: gram-stain-positive rods, 0.5 μm in diameter and 2–5 μm long, motile by peritrichous flagella, with gram-positive structure of the cell wall. Endospores occur rarely and are terminal, round, and do not distend the mother cell. Catalase- and oxidase-negative. They are chemoorganoheterotrophic anaerobes; fermentative growth is observed with proteinaceous substrates. They are mesophilic and neutrophilic. The temperature range is 13 to 55 °C (optimum, 35 °C), the pH range is 6.7–8.0 (optimum, pH 7.0) and the NaCl range is 0–3.0% (w/v) (optimum, 0–2.0% NaCl). Sulfate, sulfite, and nitrate are not used as electron acceptors. Yeast extract, peptone, tryptone, and mannose are fermented. Acetic acid, butyric acid, H2, and CO2 are the major products of yeast extract fermentation. It cannot grow with formate, acetate, propionate, pyruvate, casein hydrolysate, ethanol, methanol, propanol, fructose, glucose, sucrose, trehalose, cellobiose, raffinose, arabinose, cellulose, starch, xylan, mannitol, and glycerol. The main cellular fatty acids were C14:0, C16:0, and iso-C15:0.
The type strain is 1933PT (= VKM B-3382T = KCTC 15984T), isolated from a methanogenic enrichment obtained from a production water sample from the Binagady oilfield (Baku city, Republic of Azerbaijan). The G + C content of the genome of the strain is 31.9 mol%, its approximate size is 1.917 Mbp. The GenBank/EMBL/DDBJ accession numbers of the 16S rRNA gene and genome sequences of strain 1933PT are MN698738.1 and SRIB00000000 (version SRIB01000000), respectively.
Emended description of the genus Soehngenia Parshina et al. 2003.
The description of the genus Soehngenia is as that given by Parshina et al. 2003, with the following modification. They are anaerobic or facultatively anaerobic, and saccharolytic and proteolytic. The genomic G + C content is about 32–33%. The size of genomes varies by around 2.0 Mbp. Members of this genus form a monophyletic clade in phylogenetic trees based on concatenated sequences for different large datasets of proteins and also in a tree based on 16S rRNA gene sequences. The type species is Soehngenia saccharolytica BOR-YT (= DSM 12858T = ATCC BAA-502T). The genomic G + C content of S. saccharolytica DSM 12858T is 32.9%. The GenBank/EMBL/DDBJ accession number of the 16S rRNA gene and the JGI IMG accession number of the genome sequence of strain DSM 12858T are AY353956 and 2571042347, respectively.
4. Conclusions
A long-stored methanogenic enrichment obtained from a petroleum reservoir in Azerbaijan was studied by 16S rRNA gene sequence analysis and its bacterial and archaeal diversity was revealed. From this enrichment a new mesophilic fermentative bacterium, strain 1933PT, was isolated in pure culture. The taxonomic study including a phylogenetic 16S rRNA gene sequence and genome analyses, chemotaxonomic, and phenotypic studies showed that this strain represented a new species within the genus Soehngenia, for which we propose the name Soehngenia longivitae sp. nov. Genome analysis of the novel strain revealed its potential in the destruction of proteinaceous components of the biomass in the enrichment and the dominance of genes involved in protein metabolism.
Acknowledgments
We thank A. Oren from The Hebrew University of Jerusalem (Israel) for his help on the nomenclature of the new species. We are grateful to Professor Nikos Kyrpides for his kind permission to use the genome sequence of the strain DSM12858T in our analysis. We are especially grateful to the Reviewers for their comments, which resulted in improvement of the article.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/12/1967/s1. Figure S1: Growth profiles of strain 1933PT at various temperatures (a), pH (b) and NaCl concentration (w/v, %) (c), and of S. saccharolytica strain DSM 12858T (d) at various NaCl concentrations (w/v, %). Figure S2: Thin layer chromatogram of polar lipids extracts from strain 1933PT (a) and S. saccharolytica DSM 12858T (b). Designations: GL—glycolipids; PGL—phosphoglycolipids, PL—phospholipids; L—lipids; DPG—diphosphoglycolipids. Figure S3: Subsystems of strain 1933PT based on the SEED database. Figure S4: Heatmap profile showing the abundance of functional genes detected within the Tissierellaceae genomes. Heatmaps were automatically generated by DRAM. Sections of the heatmap are ordered to highlight information about pathway completion and ETC subunit completion (A). Boxes colored by presence/absence in (B) represent 1–2 genes necessary to carry out a particular process. Table S1: Comparison of enzymatic activities of strains 1933PT and S. saccharolytica DSM 12858T determined by the api®ZYM test (bioMérieux, France). Designations: +, positive; –, negative; W, weakly positive. Table S2: Fatty acid compositions of strain 1933PT and type strain of Soehngenia saccharolytica DSM 12858T. Table S3: General properties and relationship of the genomes of strain 1933PT and its closely related species S. saccharolytica DSM 12858T. Table S4: AAI and POCP values of strain 1933PT and other related members of the families Tissierellaceae and Gottschalkiacecae.
Author Contributions
Conceptualization and design of the work, project administration, obtaining and maintenance of enrichment culture, T.N.N.; formal analysis, D.S.G. and T.P.T.; investigation, S.K.B., D.S.S., S.N.P., A.N.A., A.B.P., and A.K.T.; writing—original draft preparation, and editing, S.K.B., T.N.N. and D.S.G. All authors have read and agreed to the published version of the manuscript.
Funding
The work was funded by the Russian Science Foundation (grant 16-14-00028). The Ministry of Science and Higher Education of the Russian Federation partly funded cultivation of the methanogenic enrichment. The funds had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu K., Dai L., Tu B., Zhang X., Zhang H., Deng Y., Lawson P.A., Cheng L. Gudongella oleilytica gen. nov., sp. nov., an aerotorelant bacterium isolated from Shengli oilfield and validation of family Tissierellaceae. Int. J. Syst. Evol. Microbiol. 2020;70:951–957. doi: 10.1099/ijsem.0.003854. [DOI] [PubMed] [Google Scholar]
- 2.Alauzet C., Marchandin H., Courtin P., Mory F., Lemée L., Pons J.-L., Chapot-Chartier M.-P., Lozniewski A., Jumas-Bilak E. Multilocus analysis reveals diversity in the genus Tissierella: Description of Tissierella carlieri sp. nov. in the new class Tissierellia classis nov. Syst. Appl. Microbiol. 2014;37:23–34. doi: 10.1016/j.syapm.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Parshina S.N., Kleerebezem R., Sans J.L., Lettinga G., Nozhevnikova A.N., Kostrikina N.A., Lysenko A.M., Stams A.J.M. Soehngenia saccharolytica gen. nov., sp. nov., and Clostridium amygdalinum sp. nov., two novel, anaerobic, benzaldehyde-converting bacteria. Int. J. Syst. Evol. Microbiol. 2003;53:1791–1799. doi: 10.1099/ijs.0.02668-0. [DOI] [PubMed] [Google Scholar]
- 4.Parshina S.N., Kleerebezem R., van Kempen E., Nozhevnikova A.N., Lettinga G., Stams A.J.M. Benzaldehyde conversion by two anaerobic bacteria isolated from an upflow anaerobic sludge bed reactor. Process Biochem. 2000;36:423–429. doi: 10.1016/S0032-9592(00)00230-2. [DOI] [Google Scholar]
- 5.Zhang F., She Y.-H., Chai L.-J., Banat I.M., Zhang X.-T., Shu F.-C., Wang Z.-L., Yu L.-J., Hou D.-J. Microbial diversity in long-term water-flooded oil reservoirs with different in situ temperatures in China. Sci. Rep. 2012;2:760. doi: 10.1038/srep00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavlova-Kostryukova N.K., Tourova T.P., Poltaraus A.B., Feng Q., Nazina T.N. Microbial diversity in formation water and enrichment cultures from the Gangxi bed of the Dagang terrigenous oilfield (PRC) Microbiology. 2014;83:616–633. doi: 10.1134/S0026261714050208. [DOI] [Google Scholar]
- 7.Nazina T.N., Shestakova N.M., Semenova E.M., Korshunova A.V., Kostrukova N.K., Tourova T.P., Min L., Feng Q., Poltaraus A.B. Diversity of metabolically active Bacteria in water-flooded high-temperature heavy oil reservoir. Front. Microbiol. 2017;8:707. doi: 10.3389/fmicb.2017.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokolova D.S., Semenova E.M., Grouzdev D.S., Ershov A.P., Bidzhieva S.K., Ivanova A.E., Babich T.L., Sissenbayeva M.R., Bisenova M.A., Nazina T.N. Microbial diversity and potential sulfide producers in the Karazhanbas oilfield (Kazakhstan) Microbiology. 2020;89:459–469. doi: 10.1134/S0026261720040128. [DOI] [Google Scholar]
- 9.Pannekens M., Voskuhl L., Meier A., Müller H., Haque S., Frösler J., Brauer V.S., Meckenstock R.U. Densely populated water droplets in heavy-oil seeps. Appl. Environ. Microbiol. 2020;86:e00164-20. doi: 10.1128/AEM.00164-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng L., Shi S., Li Q., Chen J., Zhang H., Lu Y. Progressive degradation of crude oil n-alkanes coupled to methane production under mesophilic and thermophilic conditions. PLoS ONE. 2014;9:e113253. doi: 10.1371/journal.pone.0113253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H.Z., Lv X.M., Yi Y., Zheng D., Gou M., Nie Y., Hu B., Nobu M.K., Narihiro T., Tang Y.Q. Using DNA-based stable isotope probing to reveal novel propionate- and acetate-oxidizing bacteria in propionate-fed mesophilic anaerobic chemostats. Sci. Rep. 2019;9:17396. doi: 10.1038/s41598-019-53849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quero G.M., Cassin D., Botter M., Perini L., Luna G.M. Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs) Front. Microbiol. 2015;6:1053. doi: 10.3389/fmicb.2015.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luque-Almagro V.M., Cabello P., Sáez L.P., Olaya-Abril A., Moreno-Vivián C., Roldán M.D. Exploring anaerobic environments for cyanide and cyano-derivatives microbial degradation. Appl. Microbiol. Biotechnol. 2018;102:1067–1074. doi: 10.1007/s00253-017-8678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nazina T.N., Rozanova E.P., Kuznetsov S.I. Microbial oil transformation processes accompanied by methane and hydrogen-sulfide formation. Geomicrobiol. J. 1985;4:103–130. doi: 10.1080/01490458509385927. [DOI] [Google Scholar]
- 15.Nazina T.N. Communities of methane-producing bacteria from Apsheron oil formations. Microbiology. 1984;53:122–127. [Google Scholar]
- 16.Grouzdev D.S., Bidzhieva S.K., Sokolova D.S., Tourova T.P., Poltaraus A.B., Nazina T.N. Draft genome sequence of a fermenting bacterium, Soehngenia sp. strain 1933P, isolated from a petroleum reservoir in Azerbaijan. Microbiol. Resour. Announc. 2019;8:e00689-19. doi: 10.1128/MRA.00689-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 18.Zeikus J.G., Weimer P.J., Nelson D.R., Daniels L. Bacterial methanogenesis: Acetate as a methane precursor in pure culture. Arch. Microbiol. 1975;104:129–134. doi: 10.1007/BF00447312. [DOI] [Google Scholar]
- 19.Wolin E.A., Wolin M.J., Wolfe R.S. Formation of methane by bacterial extracts. J. Biol. Chem. 1963;238:2882–2888. [PubMed] [Google Scholar]
- 20.Kevbrin V.V., Zavarzin G.A. The effect of sulfur compounds on growth of halophilic homoacetic bacterium Acetohalobium arabaticum. Mikrobiologiia. 1992;61:812–817. (In Russian) [Google Scholar]
- 21.Hungate R.E. Chapter IV. A roll tube method for cultivation of strict anaerobes. In: Norris J.R., Ribbons D.W., editors. Methods in Microbiology. Academic Press; London, UK: 1969. pp. 117–132. [Google Scholar]
- 22.Bergmann G.T., Bates S.T., Eilers K.G., Lauber C.L., Caparoso J.G., Walters W.A., Knight R., Fierer N. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol. Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Edgar R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv. 2016:081257. doi: 10.1101/081257. [DOI] [Google Scholar]
- 25.Edgar R.C., Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 26.Pruesse E., Peplies J., Glöckner F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenova E.M., Sokolova D.S., Grouzdev D.S., Poltaraus A.B., Vinokurova N.G., Tourova T.P., Nazina T.N. Geobacillus proteiniphilus sp. nov., a thermophilic bacterium isolated from a high-temperature heavy oil reservoir in China. Int. J. Syst. Evol. Microbiol. 2019;69:3001–3008. doi: 10.1099/ijsem.0.003486. [DOI] [PubMed] [Google Scholar]
- 28.Trüper H.G., Schlegel H.G. Sulfur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek. 1964;30:321–323. doi: 10.1007/BF02046728. [DOI] [PubMed] [Google Scholar]
- 29.Bidzhieva S.K., Sokolova D.S., Tourova T.P., Nazina T.N. Bacteria of the genus Sphaerochaeta from low-temperature heavy-oil reservoirs (Russia) Microbiology. 2018;87:757–765. doi: 10.1134/S0026261718060048. [DOI] [Google Scholar]
- 30.Holguin G., Guzman M.A., Bashan Y. Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees: Their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol. Ecol. 1992;101:207–216. doi: 10.1016/0378-1097(92)90817-8. [DOI] [Google Scholar]
- 31.Bidzhieva S.K., Sokolova D.S., Grouzdev D.S., Kostrikina N.A., Poltaraus A.B., Tourova T.P., Shcherbakova V.A., Troshina O.Y., Nazina T.N. Sphaerochaeta halotolerans sp. nov., a novel spherical halotolerant spirochete from a Russian heavy oil reservoir, emended description of the genus Sphaerochaeta, reclassification of Sphaerochaeta coccoides to a new genus Parasphaerochaeta gen. nov. as Parasphaerochaeta coccoides comb. nov. and proposal of Sphaerochaetaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2020;70:4748–4759. doi: 10.1099/ijsem.0.004340. [DOI] [PubMed] [Google Scholar]
- 32.Minnikin D.E., Collins M.D., Goodfellow M. Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J. Appl. Bacteriol. 1979;47:87–95. doi: 10.1111/j.1365-2672.1979.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 33.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons; New York, NY, USA: 1991. pp. 115–175. [Google Scholar]
- 34.Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang D.T., Vinh L.S., Flouri T., Stamatakis A., von Haeseler A., Minh B.Q. MPBoot: Fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol. Biol. 2018;18:11. doi: 10.1186/s12862-018-1131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varghese N.J., Mukherjee S., Ivanova N., Konstantinidis K.T., Mavrommatis K., Kyrpides N.C., Pati A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantiukh K., Grouzdev D. POCP-Matrix calculation for a number of genomes. Figshare. 2017 doi: 10.6084/m9.figshare.5602957.v1. [DOI] [Google Scholar]
- 46.Grouzdev D.S., Rysina M.S., Bryantseva I.A., Gorlenko V.M., Gaisin V.A. Draft genome sequences of ‘Candidatus Chloroploca asiatica’ and ‘Candidatus Viridilinea mediisalina’, candidate representatives of the Chloroflexales order: Phylogenetic and taxonomic implications. Stand. Genom. Sci. 2018;13:24. doi: 10.1186/s40793-018-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin Q.L., Xie B.B., Zhang X.Y., Chen X.L., Zhou B.C., Zhou J.H., Oren A., Zhang Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delmont T.O., Eren E.M. Linking pangenomes and metagenomes: The Prochlorococcus metapangenome. PeerJ. 2018;6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eren A.M., Esen O.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L., Delmont T.O. Anvi’o: An advanced analysis and visualization platform for ’omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaffer M., Borton M.A., McGivern B.B., Zayed A.A., La Rosa S.L., Solden L.M., Liu P.F., Narrowe A.B., Rodríguez-Ramos J., Benjamin Bolduc B., et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48:8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bryant M.P., Boone D.R. Emended description of strain MST (DSM 800T), the type strain of Methanosarcina barkeri. Int. J. Syst. Bacteriol. 1987;37:169–170. doi: 10.1099/00207713-37-2-169. [DOI] [Google Scholar]
- 52.Lomans B.P., Maas R., Luderer R., Op den Camp H.J.M., Pol A., van der Drift C., Vogels G.D. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl. Environ. Microbiol. 1999;65:3641–3650. doi: 10.1128/AEM.65.8.3641-3650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baena S., Fardeau M.L., Labat M., Ollivier B., Garcia J.L., Patel B.K. Desulfovibrio aminophilus sp. nov., a novel amino acid degrading and sulfate reducing bacterium from an anaerobic dairy wastewater lagoon. Syst. Appl. Microbiol. 1998;21:498–504. doi: 10.1016/S0723-2020(98)80061-1. [DOI] [PubMed] [Google Scholar]
- 54.Dahle H., Birkeland N.-K. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 2006;56:1539–1545. doi: 10.1099/ijs.0.63894-0. [DOI] [PubMed] [Google Scholar]
- 55.Grabowski A., Tindall B.J., Bardin V., Blanchet D., Jeanthon C. Petrimonas sulfuriphila gen. nov., sp. nov., a mesophilic fermentative bacterium isolated from a biodegraded oil reservoir. Int. J. Syst. Evol. Microbiol. 2005;55:1113–1121. doi: 10.1099/ijs.0.63426-0. [DOI] [PubMed] [Google Scholar]
- 56.Chen S., Dong X. Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 2005;55:2257–2261. doi: 10.1099/ijs.0.63807-0. [DOI] [PubMed] [Google Scholar]
- 57.Hahnke S., Langer T., Koeck D.E., Klocke M. Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 2016;66:1466–1475. doi: 10.1099/ijsem.0.000902. Erratum in 2016, 66, 2454. [DOI] [PubMed] [Google Scholar]
- 58.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kyrpides N.C., Woyke T., Eisen J.A., Garrity G., Lilburn T.G., Beck B.J., Whitman W.B., Hugenholtz P., Klenk H.P. Genomic encyclopedia of type strains, phase I: The one thousand microbial genomes (KMG-I) project. Stand. Genom. Sci. 2014;9:9031278. doi: 10.4056/sigs.5068949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., Rooney A.P., Yi H., Xu X.W., De Meyer S., et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 63.Konstantinidis K.T., Rosselló-Móra R., Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;1:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sogin M.L., Morrison H.G., Huber J.A., Welch D.M., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl. Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pedros-Alió C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 66.Jones S.E., Lennon J.T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA. 2010;107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.