Abstract
A growing body of evidence indicates that the levels of fucosylation correlate with breast cancer progression and contribute to metastatic disease. However, very little is known about the signaling and functional outcomes that are driven by fucosylation. We performed a global proteomic analysis of 4T1 metastatic mammary tumor cells in the presence and absence of a fucosylation inhibitor, 2-fluorofucose (2FF). Of significant interest, pathway analysis based on our results revealed a reduction in the NF-κB and TNF signaling pathways, which regulate the inflammatory response. NF-κB is a transcription factor that is pro-tumorigenic and a prime target in human cancer. We validated our results, confirming that treatment of 4T1 cells with 2FF led to a decrease in NF-κB activity through increased IκBα. Based on these observations, we conclude that fucosylation is an important post-translational modification that governs breast cancer cell signaling.
Keywords: fucosylation, N-glycosylation, breast cancer, metastasis, NF-κB
1. Introduction
Fucosylation is a type of N-linked glycosylation defined as the addition of fucose sugars in a branched structure to a carrier protein. Although it has been suggested as a putative biomarker in pancreatic and hepatocellular carcinoma [1,2,3,4,5,6,7,8], research linking breast cancer prognosis to N-glycosylation is at an early stage [9]. In mammals, two distinct metabolic pathways supply L-fucose, i.e., a de novo synthesis pathway and a salvage pathway that both rely on the transport of sugars (i.e., mannose, glucose, and fucose) into the cell. Once synthesized or transported in the cell, L-fucose is converted to GDP-L-fucose and attached to proteins within the Golgi–endoplasmic reticulum (ER).
Evidence suggests that being able to target or utilize glyco-modifications and the enzymes that regulate this process has significant clinical potential for cancer [10,11]. Global changes in glycosylation can be detected between samples from breast cancer patients, when compared to those from non-cancer populations, and are associated with metastatic progression of breast cancer [7,12,13,14,15,16,17]. Increased fucosylation is found in invasive breast cancer cell lines and patient cohorts with aggressive breast cancer [9,13,18]. Several studies show that treatment of 4T1 metastatic mammary tumor cells with a fucosylation inhibitor reduces the migratory and invasive qualities of these cells [9,19,20,21]. However, fucosylation signaling in cancer needs further investigation.
In the present study, we performed tandem-mass-tag (TMT) proteomics on 4T1 metastatic mammary tumor cells treated with a fucosylation inhibitor, 2-fluorofucose (2FF) or vehicle (DMSO) control. Over 5000 proteins were quantified, and over 400 proteins were significantly changed in the 2FF-treated samples compared to the DMSO-treated samples. While the proteins that increased in abundance were involved in spliceosome, ribosome biogenesis, and DNA replication, proteins that decreased in abundance upon treatment with 2FF highlighted the NF-κB and TNF signaling pathways, as well as membrane and vesicle-mediated transport. NF-κB is of particular interest because it is a transcription factor that is pro-tumorigenic and a prime target in human cancer. We used traditional proteomic and signaling pathway analysis—western blotting and transcript analysis—to validate our findings. This validation not only confirmed our pathway analysis but also demonstrated the sensitivity of our proteomic methodology.
2. Experimental Section
2.1. Cell Culture
(Full Details for Reagents and Global Proteomics in Supplemental File S2)
We purchased 4T1 cells from ATCC® (CRL-2539™). Cells were maintained in a humidified 5% CO2 incubator at 37 °C and grown in RPMI 1640 medium (Corning, Corning, NY, USA) supplemented with 10% heat-inactivated Fetal Bovine Serum (FBS) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), L-Glutamine (Corning, Corning, NY, USA), and a Penicillin–Streptomycin solution (Corning, Corning, NY, USA); 2-deoxy-2-fluoro-L-fucose (2FF) was purchased from Synthose Inc. (Concord, ON, Canada); 4T1 cells were treated with 500 µM 2FF for 48 h prior to analysis.
2.2. Protein Analysis
Cells were lysed in a buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl2, 1 mM EDTA, 1% Triton X-100 with the HALT protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA). Western and lectin blot analysis were performed on the Protein Simple FluorChem-R imaging system.
2.3. RNA Analysis
RNA was isolated from cells using the GeneJet RNA isolation kit (Thermo Fisher Scientific, Waltham, MA, USA), and cDNA was synthesized using BioRad iScript Supermix. Reactions were run on either a StepOne or a ViiA7 thermal cycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). mRNA expression was quantified using a standard curve or the ΔΔCt method, normalized to the expression levels of Gapdh, Hprt, or Tbp, and compared to controls.
2.4. Quantitative Global Proteomic Comparison of Protein Levels
Sample preparation, mass spectrometry analysis, bioinformatics, and data evaluation were performed in collaboration with the Proteomics Core Facility at the Indiana University School of Medicine (IUSM) [22,23,24,25]. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD021413 and 10.6019/PXD021413 [26].
3. Results
3.1. Pharmacological Inhibition of Fucosylation Alters N-Glycan Processing
To identify signaling nodes that are regulated by fucosylation, we treated 4T1 metastatic mammary tumor cells with the fucosylation inhibitor, 2FF, or DMSO (Supplementary Figure S1A) and performed TMT proteomics (Supplementary Figure S1B) [27]. This analysis identified 5750 proteins and quantified 5288 of them by MS2-based TMT. Over 400 proteins showed significant changes (abundance ratio p-value ≤ 0.05) in 2FF-treated samples compared to DMSO-treated samples (Supplementary File S1).
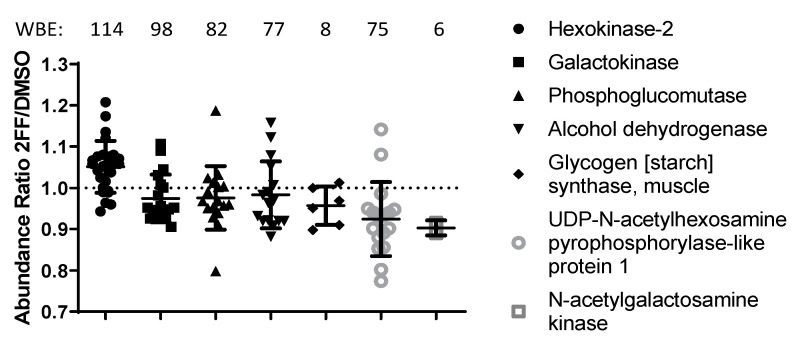
Pathway analysis using STRING.db, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome [28,29,30,31] highlighted several cancer-related pathways as downregulated in 2FF-treated cells, in contrast with DNA/RNA processing pathways which were upregulated (Table 1). In particular, we saw decreases in the abundance of proteins that participate in the calnexin/calreticulin cycle, N-glycan trimming in the ER, ER-to-Golgi anterograde and retrograde transport, transport and subsequent modification of glycans in the Golgi, and translocation of glucose transporter 4 (GLUT4) to the plasma membrane. Several key proteins within the glycan synthesis pathway were significantly altered in our 2FF-treated samples (Table 1; Figure 1, p ≤ 0.05). Our results showed that the levels of an upstream enzyme of this pathway, hexokinase-2, were significantly higher in DMSO-treated samples compared with 2FF-treated samples. This enzyme converts d-glucose to α-d-glucose-6-phosphate, an early step in glycolysis and de novo biosynthesis of monosaccharides used for N-glycan processing [32]. However, we also saw a reduction in 2FF-treated samples of 15 enzymes involved in asparagine N-linked glycosylation and of 6 major enzymes downstream of hexokinase that generate monosaccharides for N-glycan processing [32].
Table 1.
Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathway analysis. Top pathways with increased or decreased levels of proteins in 2FF- versus DMSO-treated 4T1 cells. FDR calculated through STRING-DB.org. We note that NF-κB and TNF signaling pathways contain common proteins (Supplemental File S2).
| KEGG Pathways | Reactome Pathways | ||||
|---|---|---|---|---|---|
| Decreased | |||||
| Description | Count in network | FDR | Description | Count in network | FDR |
| NF-κB signaling pathway | 9 or 93 | 0.0002 | Vesicle-mediated transport | 23 of 553 | 1.39 × 10−5 |
| RIG-1-like receptor signaling pathway | 6 of 68 | 0.0062 | Membrane trafficking | 22 of 523 | 1.39 × 10−5 |
| Tight Junction | 9 of 165 | 0.0062 | ER to Golgi Anterograde Transport | 12 of 147 | 1.45 × 10−5 |
| Protein processing in the ER | 9 of 161 | 0.0062 | Asparagine N-linked glycosylation | 15 of 269 | 3.02 × 10−5 |
| TNF signaling pathway | 7 of 108 | 0.0068 | Immune system | 38 of 1523 | 4.78 × 10−5 |
| Increased | |||||
| Spliceosome | 16 of 130 | 1.73 × 10−11 | Metabolism of RNA | 33 of 448 | 4.42 × 10−18 |
| Ribosome biogenesis in eukaryotes | 7 of 76 | 5 × 10−4 | Processing of Capped Intron-containing Pre-mRNA | 23 of 212 | 1.02 × 10−15 |
| Huntington’s disease | 8 of 187 | 0.0134 | mRNA splicing – Major Pathway | 19 of 156 | 8.94 × 10−14 |
| DNA replication | 4 of 35 | 0.0134 | Gene Expression (transcription) | 30 of 858 | 7.30 × 10−9 |
Figure 1.
Galactose catabolism and N-glycan trimming in the the ER is altered in cells treated with the fucosylation inhibitor. Graph showing the abundance ratios of individual peptides quantified for select proteins of carbohydrate pathways that significantly change in abundance. For each protein, the western blot equivalents (WBE) or total peptidespectrum matches (PSMs) is shown.
3.2. NF-κB Activity Is Reduced in 2FF-Treated 4T1 Cells
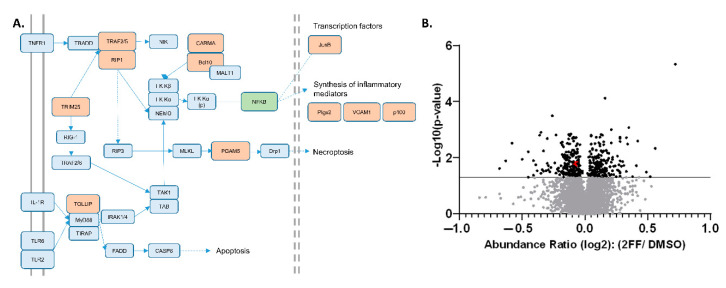
STRING-DB.org pathway analysis (Table 1) allowed us to pinpoint multiple proteins decreasing in abundance clustered around the pathways regulating nuclear factor κB (NF-κB), including tumor necrosis factor (TNF) and toll-like receptor (TLR) signaling. This finding was supported by KEGG pathway analysis, which showed that our proteomic analysis identified at least 11 different proteins which decreased significantly in abundance in response to 2FF treatment within the NF-κB and TNF signaling pathways, which have significant overlap (Figure 2A and Supplementary File S2: Supplementary Figures S2–S4).
Figure 2.
Significantly decreasing proteins associated with NF-κB and TNF pathways. (A) Pathway analysis; significantly decreased proteins are shown in orange (see Supplemental Figures S1–S3 for source KEGG pathways). (B) Volcano plot of over 5000 proteins quantified in the global proteomics experiment. Black points above the solid line indicate proteins with abundance ratio p-values ≤ 0.05; with Tollip and Bcl10 are highlighted (red dots, overlapping).
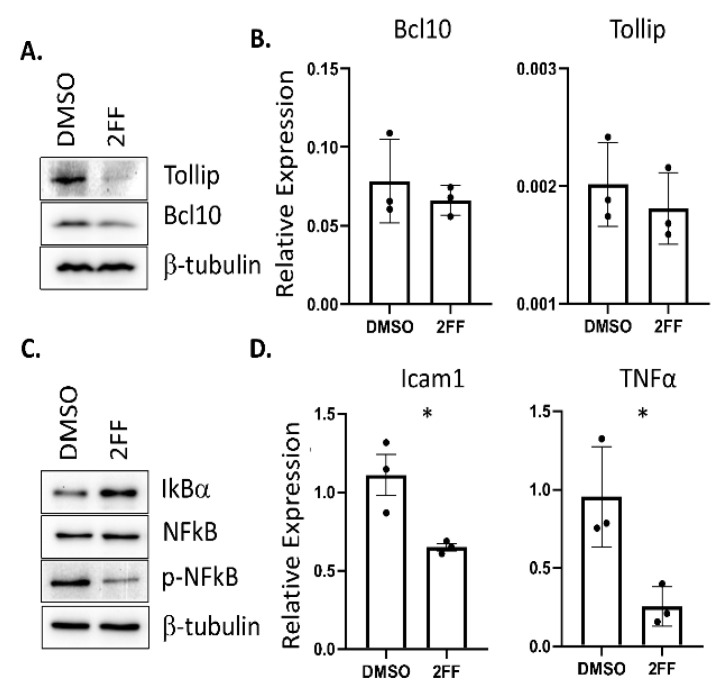
To highlight the sensitivity and precision of our semi-quantitative proteomic findings, we focused on two specific NF-κB upstream signaling effectors that showed relatively low abundance changes, Tollip and Bcl10 (Figure 2B, red dots), which were among 11 proteins across the NF-κB, TNF, and TLR pathways with NF-κB as a central mechanism in our KEGG and Reactome analyses (Figure 2A, Supplemental Figures S2–S4) [33]. We directed our analysis to these two proteins based on low abundance changes to highlight the technical sensitivity of our TMT–LC–MS technique. Functionally, Tollip and Bcl10 are linked to the regulation of pro-inflammatory responses through the activation of NF-κB, a transcription factor that is pro-tumorigenic and a prime target in human cancer [34,35,36]. Furthermore, prior studies showed that these pathways are important for the progression of cancer, including breast cancer ([37,38,39,40]. NF-κB has been implicated in the development of hormone-independent, invasive, high-grade, and late-stage breast cancer phenotypes [41]. To validate the reduction in these proteins that were identified in the TMT proteomic analysis, we used western blot to probe for protein expression levels in DMSO- or 2FF-treated 4T1 cells (Figure 3A). The western blot analysis confirmed that Tollip and Bcl10 were significantly decreased in the 2FF-treated 4T1 cells. Furthermore, the decrease in protein expression of these two factors was not due to transcriptional downregulation, as we found no difference in the transcript levels of either Tollip or Bcl10 (Figure 3B).
Figure 3.
NF-κB activity is reduced in 2FF-treated 4T1 cells. (A) Western blots showing a decrease in Tollip and Bcl10 levels after 2FF treatment. (B) Quantitative real-time PCR analysis showing no significant change in the transcripts of Bcl10 and Tollip upon treatment with 2FF. (C) Western blots showing a decrease in phospho-NF-κB and an increase in IκBα upon 2FF treatment. (D) Quantitative real-time PCR analysis showing a decrease in Icam1 and Tnfa upon treatment with 2FF; * p < 0.05 for both Icam1 and Tnfa, Student’s T-test.
To determine if the reduced abundance of Tollip and Bcl10 resulted in a decrease in NF-κB expression or activity, we performed western blotting to probe for changes in NF-κB levels and phosphorylation. We treated 4T1 cells with either DMSO or 2FF for 48 hrs. Using an antibody that detects the phosphorylation of the NF-κB subunit p65 at serine 536 (pS536), we found that the phosphorylation of NF-κB was decreased but that NF-κB levels were unaltered (Figure 3C). The regulation of NF-κB occurs through I kappa B alpha (IκBα), which inhibits NF-κB transcriptional activity. Since IκBα was not identified in our global proteomics experiment, we probed for its expression in DMSO- and 2FF-treated 4T1 cells using an antibody. We found that IκBα expression was elevated in 2FF-treated cells compared to DMSO-treated cells, consistent with our finding that NF-κB was less active in these cells (Figure 3C).
NF-κB is a transcription factor; therefore, to further validate these findings, we quantified the expression of NF-κB target genes. We found a decrease of NF-κB-regulated genes that affect angiogenesis, a process that is critical for metastasis. Specifically, we saw decreases in Icam1 and Tnfa (Figure 3D). TNF-alpha activates TNF receptor 1 (TNFR1), leading to the activation of NF-κB transcriptional regulation of Icam1 [42,43,44,45]. This finding is consistent with the pathway analysis of global proteomic changes, which showed a reduction in TNF and TNFR1-induced NF-κB signaling (Table 1 and Figure 3).
4. Discussion
Fucosylation, a post-translational modification that regulates intracellular signaling, is poorly understood. In this study, we performed TMT proteomics to identify signaling pathways affected by fucosylation. The results of the pathway analysis of our global proteomics data identified 11 members of the NF-κB and TLR pathways whose levels were significantly decreased. We chose two upstream regulators of NF-κB with a low change in abundance to highlight the innate sensitivity of this type of proteomic analysis and validate this analysis.
In consideration of our validated targets, Tollip and Bcl10, the role of Tollip in breast cancer has not been extensively explored, making our present study highly novel. Tollip is an adaptor protein that interacts with target of Myb protein 1 (TOM1), TLRs, and interleukin-1 receptor accessory protein (IL1RAP) [46,47,48]. These factors, and in particular TLRs, are readily expressed in breast cancer cells and likely serve an important function in coordinating tumor and immune cell interactions within the tumor microenvironment [49,50]. Of note, we identified one unique peptide from TLR2, a receptor upstream of Tollip (see Figure 3A), that was marginally outside the quality control cutoff for quantification and reporting. This observation indicates that a deeper proteome fractionation will likely yield further signaling insights. Although the upstream regulation of Bcl10 via receptor activation is less well understood, recent advances have demonstrated that Bcl10 is associated with the caspase recruitment domain (CARD)-and membrane-associated guanylate kinase-like domain-containing protein (CARMA-3) and mucosa-associated lymphoid tissue translocation protein 1 (MALTA) [51,52,53,54]. This complex, consisting of CARMA-3, MALTA, and Bcl10, is downstream of epidermal growth factor receptor (EGFR) and is required for EGFR-mediated activation of NF-κB [52]. EGFR was previously shown to be fucosylated, indicating a functional role for fucosylation in EGFR signaling [55,56]. Interestingly, CARMA3 (aka CARD10) was also found in our study to be decreased upon 2FF treatment (Supplementary File S1).
Taken together, these results point towards fucosylation as a regulator of the inflammatory response in metastatic breast cancer. We note limitations exist to quantitation by either Western blot or discovery proteomics—for instance, antibodies commonly utilized may lack specificity or not recognize various protein epitopes, whereas mass spectrometry is still limited by speed, ability to ionize peptides/proteins of interest, and need to separate species prior to ion detection. Future work will focus on how post-translational modifications are affected by fucose inhibition and on the identification of fucose carrier proteins and of potential druggable targets in invasive metastatic breast cancer.
Acknowledgments
The authors would like to acknowledge Tao Lu for discussion on NF-κB signaling.
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-9059/8/12/600/s1, File S1: Full list of proteins identified by global proteomics including abundances, abundance ratios, and p-values for the abundance ratios. File S2: Supplementary Methods and Figures S1–S4: 2FF treatment of 4T1 cells and schematic for TMT analysis, KEGG source pathways of NF-κB, TLR, and TNF signaling pathways used to make Figure 3A.
Author Contributions
Conceptualization, E.S.Y.; methodology, E.S.Y., A.M., E.H.D., A.L.M.; formal analysis, E.S.Y., E.H.D., T.S., M.A.; writing. E.S.Y., E.H.D.; supervision, E.S.Y., A.L.M., T.W.C.; funding acquisition, E.S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funds provided to ESY by Indiana University and the Simon Cancer Center and to TWC by NIH/NEI R01EY025641 and R01EY031939. Work in the IUSM Proteomics Core was supported, in part, by the Indiana Clinical and Translational Sciences Institute which is funded by Award Number UL1TR002529 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. Acquisition of the IUSM Proteomics core instrumentation used for this project, the Orbitrap Fusion Lumos, was provided by the Indiana University Precision Health Initiative.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang M., Mehta A., Block T.M., Marrero J., Di Bisceglie A.M., Devarajan K. A comparison of statistical methods for the detection of hepatocellular carcinoma based on serum biomarkers and clinical variables. BMC Med. Genomics. 2013;6(Suppl. 3):S9. doi: 10.1186/1755-8794-6-S3-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton P., Comunale M.A., Herrera H., Wang M., Houser J., Wimmerova M., Romano P.R., Mehta A. Development and application of a novel recombinant Aleuria aurantia lectin with enhanced core fucose binding for identification of glycoprotein biomarkers of hepatocellular carcinoma. Proteomics. 2016;16:3126–3136. doi: 10.1002/pmic.201600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comunale M.A., Wang M., Hafner J., Krakover J., Rodemich L., Kopenhaver B., Long R.E., Junaidi O., Bisceglie A.M., Block T.M., et al. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J. Proteome Res. 2009;8:595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comunale M.A., Rodemich-Betesh L., Hafner J., Wang M., Norton P., Di Bisceglie A.M., Block T., Mehta A. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: Implications for a biomarker of hepatocellular carcinoma. PLoS ONE. 2010;5:e12419. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norton P.A., Comunale M.A., Krakover J., Rodemich L., Pirog N., D’Amelio A., Philip R., Mehta A.S., Block T.M. N-linked glycosylation of the liver cancer biomarker GP73. J. Cell. Biochem. 2008;104:136–149. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyoshi E., Kamada Y. Application of glycoscience to the early detection of pancreatic cancer. Cancer Sci. 2016;107:1357–1362. doi: 10.1111/cas.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamczyk B., Tharmalingam T., Rudd P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi E., Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: Detailed analyses of oligosaccharide structures. Proteomics. 2008;8:3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 9.Herrera H., Dilday T., Uber A., Scott D., Zambrano J.N., Wang M., Angel P.M., Mehta A.S., Drake R.R., Hill E.G., et al. Core-Fucosylated Tetra-Antennary N-Glycan Containing A Single N-Acetyllactosamine Branch Is Associated with Poor Survival Outcome in Breast Cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20102528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinho S.S., Reis C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 11.Fuster M.M., Esko J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 12.Lin S.Y., Chen Y.Y., Fan Y.Y., Lin C.W., Chen S.T., Wang A.H., Khoo K.H. Precise mapping of increased sialylation pattern and the expression of acute phase proteins accompanying murine tumor progression in BALB/c mouse by integrated sera proteomics and glycomics. J. Proteome Res. 2008;7:3293–3303. doi: 10.1021/pr800093b. [DOI] [PubMed] [Google Scholar]
- 13.Kyselova Z., Mechref Y., Kang P., Goetz J.A., Dobrolecki L.E., Sledge G.W., Schnaper L., Hickey R.J., Malkas L.H., Novotny M.V. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin. Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 14.Alley W.R., Jr., Novotny M.V. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J. Proteome Res. 2010;9:3062–3072. doi: 10.1021/pr901210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christiansen M.N., Chik J., Lee L., Anugraham M., Abrahams J.L., Packer N.H. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y., Esteva F.J., Song J., Zhang H. Altered expression of sialylated glycoproteins in breast cancer using hydrazide chemistry and mass spectrometry. Mol. Cell. Proteomics MCP. 2012;11:M111-011403. doi: 10.1074/mcp.M111.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S., Kemmner W., Grigull S., Schlag P.M. Cell surface alpha 2,6 sialylation affects adhesion of breast carcinoma cells. Exp. Cell Res. 2002;276:101–110. doi: 10.1006/excr.2002.5521. [DOI] [PubMed] [Google Scholar]
- 18.Milde-Langosch K., Karn T., Schmidt M., zu Eulenburg C., Oliveira-Ferrer L., Wirtz R.M., Schumacher U., Witzel I., Schutze D., Muller V. Prognostic relevance of glycosylation-associated genes in breast cancer. Breast Cancer Res. Treat. 2014;145:295–305. doi: 10.1007/s10549-014-2949-z. [DOI] [PubMed] [Google Scholar]
- 19.Yuan K., Kucik D., Singh R.K., Listinsky C.M., Listinsky J.J., Siegal G.P. Alterations in human breast cancer adhesion-motility in response to changes in cell surface glycoproteins displaying alpha-l-fucose moieties. Int. J. Oncol. 2008;32:797–807. [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan K., Listinsky C.M., Singh R.K., Listinsky J.J., Siegal G.P. Cell surface associated alpha-L-fucose moieties modulate human breast cancer neoplastic progression. Pathol. Oncol. Res. 2008;14:145–156. doi: 10.1007/s12253-008-9036-x. [DOI] [PubMed] [Google Scholar]
- 21.Tu C.F., Wu M.Y., Lin Y.C., Kannagi R., Yang R.B. FUT8 promotes breast cancer cell invasiveness by remodeling TGF-beta receptor core fucosylation. Breast Cancer Res. 2017;19:111. doi: 10.1186/s13058-017-0904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levasseur E.M., Yamada K., Pineros A.R., Wu W., Syed F., Orr K.S., Anderson-Baucum E., Mastracci T.L., Maier B., Mosley A.L., et al. Hypusine biosynthesis in beta cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aax0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto W.R., Bone R.N., Sohn P., Syed F., Reissaus C.A., Mosley A.L., Wijeratne A.B., True J.D., Tong X., Kono T., et al. Endoplasmic reticulum stress alters ryanodine receptor function in the murine pancreatic beta cell. J. Biol. Chem. 2019;294:168–181. doi: 10.1074/jbc.RA118.005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plubell D.L., Wilmarth P.A., Zhao Y., Fenton A.M., Minnier J., Reddy A.P., Klimek J., Yang X., David L.L., Pamir N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol. Cell. Proteomics MCP. 2017;16:873–890. doi: 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peck Justice S.A., Barron M.P., Qi G.D., Wijeratne H.R.S., Victorino J.F., Simpson E.R., Vilseck J.Z., Wijeratne A.B., Mosley A.L. Mutant thermal proteome profiling for characterization of missense protein variants and their associated phenotypes within the proteome. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.014576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutsch E.W., Bandeira N., Sharma V., Perez-Riverol Y., Carver J.J., Kundu D.J., Garcia-Seisdedos D., Jarnuczak A.F., Hewapathirana S., Pullman B.S., et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020;48:D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins D.S., True J.D., Mosley A.L., Baucum A.J., 2nd Proteomic Analysis of the Spinophilin Interactome in Rodent Striatum Following Psychostimulant Sensitization. Proteomes. 2018;6 doi: 10.3390/proteomes6040053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanley P., Taniguchi N., Aebi M. N-Glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor; New York, NY, USA: 2015. pp. 99–111. [DOI] [Google Scholar]
- 33.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kowalski E., Geng S., Rathes A., Lu R., Li L. Toll-interacting protein differentially modulates HIF1alpha and STAT5-mediated genes in fibroblasts. J. Biol. Chem. 2018;293:12239–12247. doi: 10.1074/jbc.RA118.003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T., Zhang L., Joo D., Sun S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017;2 doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Multhoff G., Molls M., Radons J. Chronic inflammation in cancer development. Front. Immunol. 2011;2:98. doi: 10.3389/fimmu.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharyya S., Borthakur A., Tyagi S., Gill R., Chen M.L., Dudeja P.K., Tobacman J.K. B-cell CLL/lymphoma 10 (BCL10) is required for NF-kappaB production by both canonical and noncanonical pathways and for NF-kappaB-inducing kinase (NIK) phosphorylation. J. Biol. Chem. 2010;285:522–530. doi: 10.1074/jbc.M109.050815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez-Garcia L.A., Nava-Castro K.E., Ochoa-Mercado T.L., Palacios-Arreola M.I., Ruiz-Manzano R.A., Segovia-Mendoza M., Solleiro-Villavicencio H., Cazarez-Martinez C., Morales-Montor J. Breast Cancer Metastasis: Are Cytokines Important Players During Its Development and Progression? J. Interferon Cytokine Res. 2019;39:39–55. doi: 10.1089/jir.2018.0024. [DOI] [PubMed] [Google Scholar]
- 39.Tsao S.M., Hsu H.Y. Fucose-containing fraction of Ling-Zhi enhances lipid rafts-dependent ubiquitination of TGFbeta receptor degradation and attenuates breast cancer tumorigenesis. Sci. Rep. 2016;6:36563. doi: 10.1038/srep36563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tunset H.M., Feuerherm A.J., Selvik L.M., Johansen B., Moestue S.A. Cytosolic Phospholipase A2 Alpha Regulates TLR Signaling and Migration in Metastatic 4T1 Cells. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Nag S.A., Zhang R. Targeting the NFkappaB signaling pathways for breast cancer prevention and therapy. Curr. Med. Chem. 2015;22:264–289. doi: 10.2174/0929867321666141106124315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melotti P., Nicolis E., Tamanini A., Rolfini R., Pavirani A., Cabrini G. Activation of NF-kB mediates ICAM-1 induction in respiratory cells exposed to an adenovirus-derived vector. Gene Ther. 2001;8:1436–1442. doi: 10.1038/sj.gt.3301533. [DOI] [PubMed] [Google Scholar]
- 43.Numata T., Ito T., Maeda T., Egusa C., Tsuboi R. IL-33 promotes ICAM-1 expression via NF-kB in murine mast cells. Allergol. Int. 2016;65:158–165. doi: 10.1016/j.alit.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 44.Alhusaini A., Fadda L.M., Ali H.M., Hasan I.H., Ali R.A., Zakaria E.A. Mitigation of acetamiprid - induced renotoxicity by natural antioxidants via the regulation of ICAM, NF-kB and TLR 4 pathways. Pharmacol. Rep. 2019;71:1088–1094. doi: 10.1016/j.pharep.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Son E.W., Mo S.J., Rhee D.K., Pyo S. Vitamin C blocks TNF-alpha-induced NF-kB activation and ICAM-1 expression in human neuroblastoma cells. Arch. Pharm. Res. 2004;27:1073. doi: 10.1007/BF02975434. [DOI] [PubMed] [Google Scholar]
- 46.Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2000;2:346–351. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- 47.Yamakami M., Yoshimori T., Yokosawa H. Tom1, a VHS domain-containing protein, interacts with tollip, ubiquitin, and clathrin. J. Biol. Chem. 2003;278:52865–52872. doi: 10.1074/jbc.M306740200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang G., Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J. Biol. Chem. 2002;277:7059–7065. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- 49.Shi S., Xu C., Fang X., Zhang Y., Li H., Wen W., Yang G. Expression profile of Tolllike receptors in human breast cancer. Mol. Med. Rep. 2020;21:786–794. doi: 10.3892/mmr.2019.10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palha De Sousa C., Blum C.M., Sgroe E.P., Crespo A.M., Kurt R.A. Murine mammary carcinoma cells and CD11c(+) dendritic cells elicit distinct responses to lipopolysaccharide and exhibit differential expression of genes required for TLR4 signaling. Cell Immunol. 2010;266:67–75. doi: 10.1016/j.cellimm.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wegener E., Krappmann D. CARD-Bcl10-Malt1 signalosomes: Missing link to NF-kappaB. Sci. STKE. 2007;2007:pe21. doi: 10.1126/stke.3842007pe21. [DOI] [PubMed] [Google Scholar]
- 52.Jiang T., Grabiner B., Zhu Y., Jiang C., Li H., You Y., Lang J., Hung M.C., Lin X. CARMA3 is crucial for EGFR-Induced activation of NF-kappaB and tumor progression. Cancer Res. 2011;71:2183–2192. doi: 10.1158/0008-5472.CAN-10-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ekambaram P., Lee J.L., Hubel N.E., Hu D., Yerneni S., Campbell P.G., Pollock N., Klei L.R., Concel V.J., Delekta P.C., et al. The CARMA3-Bcl10-MALT1 Signalosome Drives NFkappaB Activation and Promotes Aggressiveness in Angiotensin II Receptor-Positive Breast Cancer. Cancer Res. 2018;78:1225–1240. doi: 10.1158/0008-5472.CAN-17-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvalho G., Le Guelte A., Demian C., Vazquez A., Gavard J., Bidere N. Interplay between BCL10, MALT1 and IkappaBalpha during T-cell-receptor-mediated NFkappaB activation. J. Cell Sci. 2010;123:2375–2380. doi: 10.1242/jcs.069476. [DOI] [PubMed] [Google Scholar]
- 55.Wang X., Gu J., Ihara H., Miyoshi E., Honke K., Taniguchi N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J. Biol. Chem. 2006;281:2572–2577. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Y., Fukuda T., Hang Q., Hou S., Isaji T., Kameyama A., Gu J. Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci. Rep. 2017;7:11563. doi: 10.1038/s41598-017-11911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.