Abstract
The recent outbreak of Covid-19 is posing a severe threat to public health globally. Coronaviruses (CoVs) are the largest known group of positive-sense RNA viruses surviving on an extensive number of natural hosts. CoVs are enveloped and non-segmented viruses with a size between 80 and 120 nm. CoV attachment to the surface receptor and its subsequent entrance into cells is mediated by Spike glycoprotein (S). For enhanced CoV entry and successful pathogenesis of CoV, proteolytic processing and receptor-binding act synergistically for induction of large-scale S conformational changes. The shape, size and orientation of receptor-binding domains in viral attachment proteins are well conserved among viruses of different classes that utilize the same receptor. Therefore, investigations unraveling the distribution of cellular receptors with respect to CoV entry, structural aspects of glycoproteins and related conformational changes are highly significant for understanding virus invasion and infection spread. We present the characteristic features of CoV S-Proteins, their significance for CoVs and related receptor binding activities for pathogenesis and viral survival. We are analyzing the novel role of S-protein of CoVs along with their interactive receptors for improving host immunity and decreasing infection spread. This is hoped that presented information will open new ways in tackling coronavirus, especially for the ongoing epidemic.
Keywords: Covid-19, Infection control, Receptors binding complex, Viral replication
1. Introduction
The end of the year 2019 marked the beginning of a deadly viral outbreak in China. The devastating virus was termed as coronavirus disease (COVID-19) (previously known as 2019-nCoV). The causal agent belongs to the family Coronaviridae and is a close relative of a SARS-related CoV (severe acute respiratory syndrome (SARS)-related CoV) [1,2] and designated as SARS-CoV-2. It is officially declared as the causal agent of disease spread among residents of Wuhan city in China and the rest of the world [3,4].
In 1960s, human coronaviruses were characterized for the very first time and observed as causative agents of many respiratory infections [5]. Coronaviruses are regarded as enveloped and non-segmented viruses with a size between 80 and 120 nm having positive-sense RNA genomes of size between 27 and 36 kb [6]. The genomic RNA and phosphorylated nucleocapsid (N) protein constitute the nucleocapsid of the virion. The nucleocapsid is buried inside phospholipid bilayers and possess three proteins, i.e. spike glycoprotein (S) trimmer and, the membrane (M) and the envelope protein (E) [7]. The genotypic and serological basis divided coronavirus subfamily is categorized into four genera, namely the alpha, beta, gamma and delta coronaviruses.
In the past, infection with different types of coronaviruses was proved to be fatal with the highest death rate. For example, SARS-CoV infection in China was reported among 8000 people with 774 deaths [8]. Similarly, the mortality rate due to MERS-CoV (Middle East respiratory syndrome coronavirus) was recorded up to 35% [9]. Many Coronaviridae members continuously flow in the human populations and can be held accountable for typical mild respiratory ailments [10]. Contrarily, SARS-CoV and MERS-CoV can spread in humans from animals and results in severe respiratory disorders in patients [10,11]. Chinese horseshoe bats have been reported as a natural reservoir of SARS-CoV-2 and it is transmitted to humans by means of intermediate hosts [12].
Recent records confirm many common symptoms for this disease like fever, cough, fatigue, dyspnea, leucopenia and lymphopenia [13]. Some less common symptoms include headache, production of sputum, haemoptysis. All patients were diagnosed with pneumonia and in some cases acute cardiac injury had also been observed. So far, collective evidence points out the human-to-human transmission for Covid-19 [13]. Symptoms such as dyspnoea, dry cough, fever and abnormalities in the chest are common between, SARS-CoV, MERS-CoV and Covid-19 [2,14]. As of December 2020, more than 76.332 million confirmed cases and 1.687 million deaths, and infection has been reported throughout the world. Unfortunately, no authentic vaccine or antiviral drug has yet been developed. Leading companies around the globe are working hard to complete the development of the vaccine to fight this epidemic.
Epithelial cells are considered as the main fence to microbial infection entering host via body cavities, i.e. the respiratory or gastric tract. Along with the polarity of epithelial cells affecting both the early as well as late infection stages, i.e. viruses entry and exit, presence of suitable cell surface receptors are a key determinant in the attachment to and movement across the cell membrane [15]. For example, ACE2 (angiotensin I-converting enzyme-2) a SARS-CoV receptor is a plasma membrane-localized in apical epithelial cells [16]. CoV attachment to the surface receptor and its subsequent entrance into cells is mediated by Spike glycoprotein [17]. Common among coronaviruses, different members reflect variations in relation to the size, shape, and distribution of the S protein on the virion surface [18]. S-proteins consist of S1 and S2 domains and belong to class 1 fusion protein. Therefore, investigations unraveling the distribution of cellular receptors with respect to CoV entry, structural aspects of glycoproteins and related conformational changes are highly significant for understanding virus invasion and infection spread.
Many missing links in our knowledge and understanding regarding the source of virus, disease epidemiology, transmission duration, treatment as well as drug discovery require extensive studies. By this article, we aim to describe a novel role of coronavirus spike glycoproteins and their interactive partners/receptors in enhancing host resistance for decreasing infection spread. We are pretty hopeful that our presented information will open new ways of tackling coronavirus, especially for the ongoing epidemic Covid-19.
2. What are spike glycoproteins and their significance for CoV and its spread
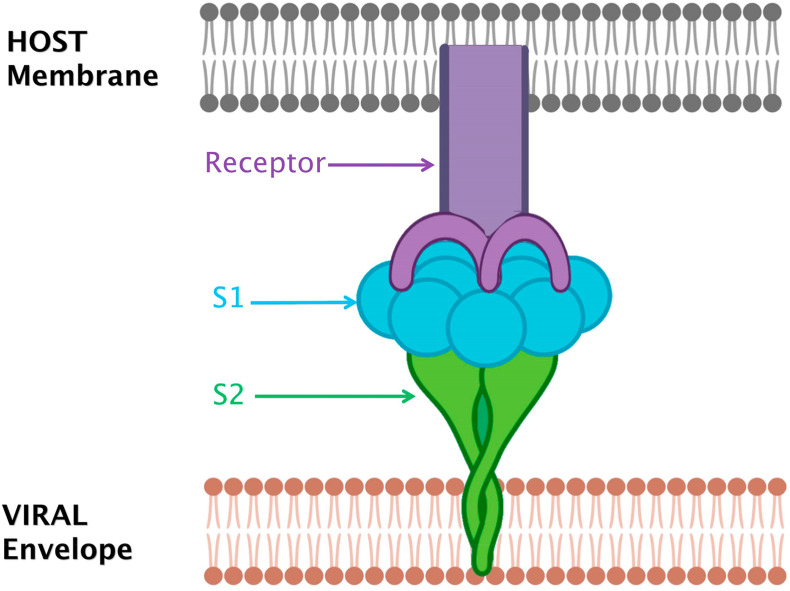
Viral penetration and infection spread depend upon the interaction between the virion and the cell surface proteins. The virion envelope comprises of three main viral proteins, namely the spike glycoprotein (S), the membrane (M) and the envelope protein (E) respectively. Assembly of CoVs takes place intra-cellularly and the M, E, and some S proteins of CoV envelope can also be retained intra-cellularly [19]. Numerous spike glycoproteins garnish the CoV virions surfaces and confer the characteristic corona shape to a virus. S use particular cell receptors and act as a chief mediator of attachment with the host cell and viral entry (Fig. 1 ) [20,21]. In coronaviruses, S-proteins (size around 200 kDa) are key players in virus virulence, tissue tropism and host range. S-proteins of SARS and 2019-nCoV share approximately 76% identity in amino acid sequence [22]. Structural analysis of S trimer in Covid-19 by cryo-EM reveals a predominant state of the trimer with one of the three RBDs rotated in an up conformation for enhancing access to a receptor. It was also confirmed that for binding with ACE2, S-protein of Covid-19 shows much higher affinity as compared to S-protein of SARS-CoV. The spike glycoprotein structures of Covid-19 and SARS-CoV display much homology (root mean square deviation (RMSD) of 3.8 Å over 959 Cα atoms). However conformational differences are also present between S in both types of CoVs. A major structural difference between these two is the RBDs (receptor binding domain) position in their respective down conformations. RBD in SARS-CoV in its down conformation reportedly packs tightly against the NTD (N-terminal domain) of the neighboring protomer while the RBD of S of Covid-19 in its down conformation is found close to the central cavity of the trimer [23].
Fig. 1.
Spike glycoproteins (S) use specialized cell receptors and act as a chief mediator of attachment with the host cell and viral entry. Successful completion of Spike glycoprotein-receptor complex results in infection. Spike glycoproteins furnish surface of CoVs and give characteristic Corona shape to these viruses. Conformational differences are also present between S among different types of CoVs. A major structural difference between S of these CoV types is the position of RBDs (Receptor Binding Domain) in their respective conformations.
On coronavirus envelope, S-proteins trimers make the distinguishing big spikes that bind to receptors for mediating virus entry, membrane fusion and elicit antibodies [24]. Present in the S1 domain (amino-terminal) of S, the conserved RBD is a key factor in defining host range [25]. S2 (carboxy-terminal) domain manages viral entry and cell fusion. Like HIV envelope (env) or influenza hemagglutinin (HA), S proteins of CoVs are Class I viral fusion protein, characteristically involving protease cleavage during assembly and exocytosis of virions. In diverse cell types and tissues as well, a broad range of host protease such as HAT, TMPRSS-2 (transmembrane protease serine 2), trypsin etc. cleave CoV S proteins. The cleavage between the S1 and S2 domains permit conformational alterations in S2, activated by receptor binding along with/without low pH. These factors mediate membrane fusion, causing virus entry as well as syncytia formation [26]. CoV S proteins possess two heptad repeats (HR) in their S2 domain that is reckoned as a typical character of class I viral fusion proteins. HR consists of abcdefg that is a repetitive heptapeptide with hydrophobic residues (a and d) essential for the formation of coiled-coil taking part in the fusion process [27].
Spike proteins are considerable actors in cell tropism activities. The changes in tropism straightly correspond with S protein [28] although the avirulence is also the result of attenuated mutations. In chimeric viruses such as MHV (mouse hepatitis virus) and FCoV (feline CoV) data corroborates direct linkage of S proteins with pathogenesis and tropism [29]. Such changes or differences in S protein-mediated tropism are linked with prime functions of proteins, i.e. fusion and receptor binding. A fusion of membranes, i.e. viral and host is mediated by changes in S-protein conformation. Such fusion is activated by different triggers. Although conformational changes get started after binding with receptor yet additional triggers like pH changes or proteolytic cleavage sometimes become essential for fusion activity [27]. Any mutation in both domains of S may lead to viral dysfunction, including replication [30] and infection spread. Therefore, avirulence can be aptly attenuated by mutation(s) in S. Besides, we have an option that mutations, deletions or exchange of genes may affect recombination as well as give rise to more virulence e. g. FIPV (feline infectious peritonitis virus) from FECV (feline enteric coronavirus) [[31], [32], [33]]. We consider this a crucial tip during infection control and management procedures. Two main functions are generally attributed to S proteins, i.e. Binding with receptor and Fusion [27]. CoVs of all four groups use one or the other receptors for binding and entry into host cells. For instance, CoVs in serogroup 1, i.e. HCoV-229E, FCoV, TGEV and CCoV use APN as receptor. HCoV-229E may use hAPN or fAPN as a receptor. It is possible that viruses of one group can bind to a common site in host APN receptor [34,35]. The dissemination and pathogenesis of Covid-19 have been attributed to receptor ACE2, the one used by SARS-CoV for entrance [22]. Some CoVs such as BCoV and HCoV-OC43 show binding activity with sialic acid [36]. Betacorona viruses are well able to bind carbohydrates with the help of a galectin fold like structure in S1 of NTD (N-terminal domain) [37]. Type 1 FCoV spike bind with heparan sulfate [27]. Likewise, neutralizing antibodies for S proteins provide protection in different animals. For example, one key neutralizing epitope has been detected in the ACE2-binding region of the S protein of SARS. Detailed mapping of neutralizing epitope-containing regions highlight some overlapping regions, particularly a neutralizing epitope 80R of a human monoclonal antibody [38]. This point out the significance of overlaps in biological functions. The most important aspect comes on the front is neutralization based vaccine against S. Such S-protein based vaccination may induce the function of neutralizing antibodies for preventing binding and entry of virus into cells. For all types of CoVs in general and Covid-19 in particular, functional analysis and characterization of S glycoprotein is immensely required for identification of susceptible hosts and cells.
3. Spike glycoproteins necessarily interact with different host receptors and regulate viral entry
For CoV entry into target cells, special receptors are bound by S-glycoproteins [21,27]. Overall, the binding between viral attachment proteins (VAPs) and their particular receptors is the first step in host-CoV interaction. Post-binding changes in VAP conformation ends in virus entry into a host cell. Such protein-receptor complex(s) are the real point of order in unwinding the host-virus relationships [39]. The shape, size and orientation of receptor-binding domains in VAP are well conserved among viruses of different classes that utilize the same receptor. Frequently, the VAP bind to a site possessing one or more hydrophobic residues in receptor protein, e.g. HA glycoprotein of influenza A virus binds to sialic acid receptors [40]. Similarly, the RGD domain of VP1 that is VAP of foot and mouth disease virus binds to integrin αvβ3 [41]. Any mutation in amino acid of the VAPs can perturb the capacity of a virus to use different integrins as receptors in vivo [41]. In 1991, a multifunctional protein CEACAM1 (Carcinoembryonic antigen-cell adhesion molecule) was identified as premier CoV receptor bound by MHV spike glycoproteins. CEACAM1 belongs to the immunoglobulin superfamily and is a type I transmembrane protein [21,27]. In both of its allelic forms, i.e. a and b, CEACAM1 act as a receptor. N-terminal domain N in ectodomain of CEACAM1 takes part in binding with S in MHV. Interesting is the fact that infection spread can be independent of CEACAM1. After the establishment of primary infection, the virus use cell-to-cell fusion and rapidly propagate independently of the receptor. This stance gets support from the fact that neurovirulence of MHV-JHM strain is linked to the quick spread of viral infection in the brain without the involvement of CEACAM1. In vitro requirement of CEACAM1 as receptor for JHM and in vivo independence of this receptor [42] advocates our opinion that JHM may use any other alternative receptor. Additionally, this neurovirulence is directly proportional to the length of the hypervariable region in S1.
The length of the hypervariable region is inversely proportional to dependence upon CEACAM1 based cell-cell fusion and the infection spread [43]. This use of receptors also provide clues about ancestors. Aminopeptidase N (APN) is a Zn2+ dependent protease of type II transmembrane protein expressed on the apical domain of respiratory and enteric epithelial cells. This is used as a common receptor by different CoVs such as CCoV (Canine coronavirus), HCoV-229E (Human coronavirus 229E), serotype 2 FCoV (Feline Coronavirus) and TGEV (Transmissible gastroenteritis virus). All these virus types have homologous S-proteins. Interestingly, besides their host-specific APN, these viruses also bind to feline APN. This proposes the origin of mentioned virus types from a common ancestor CoV infecting felines that used APN as a receptor [44]. S in MERS-CoV targets DPP4 receptor (dipeptidyl peptidase 4). Sub-domain in RBD of MERS-CoV interacts with β-propeller of DPP4. RBD in MERS-CoV and SARS-CoV present high structural resemblance in their core subdomains, but are notably divergent in the receptor-binding subdomain [45]. A type I integral membrane protein ACE2 (angiotensin-converting enzyme 2) is abundantly expressed in lung tissue. Human SARS-CoV can bind to human as well as palm civet ACE2 but the palm civet virus is unable to bind human ACE2 (hACE2). The mentioned adaptation of the virus to humans is attributed to point mutations in RBD of S protein [46]. Identification of more mutations will be very helpful in recognizing the further interactions of receptors with S proteins. In bats, SARS-CoV like viruses have been isolated [47,48]. It is noteworthy that in this case, viral entry is independent of ACE2 and receptor(s) involved still need to be explored and investigated in depth. The dissemination and pathogenesis of Covid-19 has been attributed to receptor ACE2, the one used for entrance by SARS-CoV [22]. This is a very important implication for understanding the Covid-19 infection. This highlights a very clear point common between SARS-CoV and Covid-19. We anticipate the use of the same cells by both CoVs and the earlier known diffident expression of ACE2 in the upper respiratory tract [48] might lower Covid-19 transmission. Furthermore, it is worth mentioning that the expression of ACE2 is not restricted only to the lungs and SARS-CoV may extend to extra-pulmonary regions in ACE2+ tissues [49]. Expecting the same for Covid-19 is not so irrelevant and its affinity with SARS-CoV must be matched. The S cleavage is the main determinant of effective MHV neutralization by means of the soluble receptor [50]. Perhaps the S in CoVs has sufficient flexibility to present a membrane fusion peptide without requiring protease activation for receptor binding. Here, we suggest the elaboration of receptor-mediated neutralization of Covid-19 and its entry into a host for better prevention strategies and treatment.
In SARS spike protein-ACE2 interaction, many of the residues are located on the surface of the beta-sheets as well as inter-connecting loops influence the S-ACE2 bonding. The different types of residues including charged, hydrophobic and polar depict that all such differential interactions can be directly or indirectly involved in the S RBD-ACE2 association [51]. As discussed earlier, a mutation in the form of amino acid sequence replacement, i.e. found between residues 323 and 505 with the corresponding sequence of the SARS-CoV RBD is adequate to permit use of hACE2 [52,53]. Moreover, in SARS-CoV and MERS-CoV, structures of RBD-receptor complex have been proposed by the superimposition of S-protein trimmers. It is on record that S trimmers are cross-linked by binding of CD26 to standing RBD in S of MERS-CoV. Very importantly, MERS-CoV believed to depict more eagerness in binding a receptor as compared to SARS-CoV hosted by the same surface [54]. It means that flexible RBD in S proteins is an essential component in the establishment of pathogenesis. This binding is a guarantee of the entrance of the virus into a host cell. Our view is advocated by studies of Yuan et al. [54] describing that top of S1 is found in the open state due to the presence of flexible RBD. This open state makes central stem in S trimmer accessible along with heptad repeat 1 (HR1) and central helix to inhibitors of antiviral proteins. Such accessibility of flexible RBD explains the efficiency of RBD-directed neutralization of antibodies. Any perturbance in the establishment of this binding complex would hamper the viral life cycle. Therefore, we suggest developing some antibodies that can target N-terminal domain in RBD and may interrupt the binding of S protein to a receptor. This will be very feasible for designing future epidemic control programs and disease prevention. Additionally, RBD in S proteins of Covid-19must be targeted to discover its corresponding receptor. Our view provides a very important and pragmatic framework to unravel infection control strategies and drug development for any future epidemic outbreak.
The recent investigations on Covid-19 have revealed pangolin as intermediate host and bats as the first host for this virus. Despite 96% sequence similarity between Covid-19and BatCoV, It was revealed by Wong et al. [55] that RBM of Covid-19and BatCoV show divergence in amino acid similarity (435–510 AA residues). Such divergence is usually representative of the alternative source of Covid-19 RBM coding sequence. The metagenomic investigation of pangolin genome depicts a complex origin of Covid-19 and high sequence similarity between both CoV genomes. This provides foot marks of some recombinant events in host receptor-protein binding taken place many years ago and contributed in viral evolution and involvement of intermediate hosts. The genome of RaTG13 (BatCoV) shares only one out of five key amino acids involved in RBM, while the pangolin coronavirus shares five out of five key amino acid residues [55]. Hence, it seems that due to the recombination event between CoV strains of pangolin and RaTG13 RBM was introduced in Covid-19. But more and more pangolin derived virus data sets are required for multiple genome alignments with focus on other aspects rather than S. Keeping in view the significance of host receptor-S protein binding, we can get comprehensive information about commonalities in infection progression and ultimate survival strategies.
4. Where are receptors expressed?
Expression of different receptors for a diverse range of proteins strongly correlates with infection sites in the body. Such infections are directly dependent upon the stage of tissue differentiation as well as expression of mRNA [52,56]. Irrespective of apical or basal polarity, receptors are generously expressed in well-differentiated cells and facilitate viral entry and replication. But most of the time, viral entry occurs through the apical surface. Harmer et al. [57] described the expression of ACE2 in 72 tissues of the human body. The relative expressional levels were obviously different among tissue types and positively correlated within related tissue types, e.g. duodenum, jejunum, ileum. The highest expression was recorded in cardiovascular and renal tissues. A comparatively high expression was also noticed in gastrointestinal tissues. The tissue-specific expression was altogether advocated by expression of one type ACE2 in only that tissue as compared to the other. For instance, testicular ACE2 was expressed in testes but the same receptor was not expressed/poorly expressed in jejunum and pancrease. Very limited expression of ACE2 was observed in lymphoid tissue [57]. ACE2 expression was also observed in endothelial cells of smooth muscles, lung alveolar, skin, bone marrow and cardiovascular tissues [56]. This multi-tissue immuno-localization of ACE2 and its binding by S proteins of SARS as well as newly reported Covid-19 shows that both infections can harm different body organs. This resemblance with SARS must be a focal point in future investigations for control and treatment of 2019-nCoV.Since CoVs also use receptors for entry and virulence, therefore, such information seems essential for studying infection biology of CoVs.
Taking ACE2 into consideration, apical distribution of this receptor in lungs proposes enzyme-dependent cleavage of peptides at the mucosal surface or respiratory tract [58]. For viral entry and infection, mucosa of pulmonary tract and eyes seem most suitable facilitators, e.g. SARS-CoV. Detailed tissue biopsy of persons with acute SARS symptoms revealed this CoV in proximal as well as distal epithelia of air passage [59]. This infection not only imbalance tissue metabolism but also, destruct tissue, cause type II cell hyperplasia and sometimes responsible for multinucleated syncytial cells [60]. Such deviations from normalcy reveal the collective impacts of infection, host defense reactions and therapeutic interferences as well. This specifically reflects that receptor-expressing cells have differentially high infection susceptibility as compared to receptor non expressing cells. A negative correlation of ACE2 in poorly differentiated epithelia with low infection rate as well as positive complementation of ACE2 with enhanced transduction in the same tissue with S protein-pseudotyped virions in a dose-dependent fashion [58] advocates our stance.
In the context of infection spread and viral release in close vicinity to the capillaries of pulmonary tissues, we may infer a probable general virus transmission to distant organs in the human body. For PEDV (Porcine epidemic diarrhea virus) and HCoV-229E, APN protein (aminopeptidase N) is a functional receptor. But it was confirmed by Li et al. [61] that this is not an essential receptor for PEDV entry. The overexpressing APN reduces the susceptibility of MDCK cells to TGEV contrary to PEDV. The APN knockout human and porcine cell lines revealed S1-APN binding in both cell types showing positive trait against viral spread [61]. This represents differential ability and responses of receptors to different viral spike glycoproteins. Receptor responsive to one type of viral spike glycoprotein type should not very necessarily be responsive to another type.
This has been confirmed that human CoV dissemination is regulated by the S-proteins interacting with a receptor present on the human cell surface. Protein-protein docking in binding between S and human receptor revealed that S of 2019-nCoV binds with ACE2 in humans. The 73% binding affinity between Covid-19 S and ACE2 is almost similar to SARS-CoV that depicts a similar mode of transmission in humans for both types of CoVs [62,63]. RBD in S of Covid-19, inclusive of its RBM, directly contacts ACE2 [63]. This also proves that 2019-nCoV uses ACE2 as a receptor. ACE2 from different animal species, such as cats, pigs and ferrets are also recognized by 2019-nCoV [63]. This may provide clues of intermediate hosts playing their roles in transmission. In ACE2 expressing cells, it had been recorded that DC/L-SIGN (dendritic-cell-specific ICAM3-grabbing nonintegrin) augmented SARS-CoV. But without ACE2, DC/L-SIGN are not adequate in spreading infection. Besides ACE2, L/DC-SIGN has also been reported for SARS-CoV receptors, but no receptor other than ACE2 seems to have been discussed for Covid-19. Literature supports both Covid-19 and SARS infections taking place by means of ACE, although SARS-CoV S protein binds ACE2 more strongly. The difference in the infection pathway between the two is still unclear. So we hypothesize the essential involvement of other receptor(s). This reflects that route of infection between SARS-CoV and Covid-19should be different. Identification/differentiation of route of infection has critical implications for disease management and cure strategies. The detailed and targeted studies may also offer in depth analyses of viral entry, receptor use, infectivity and origin of CoV. Because we are still lacking in detection of appropriate receptors for S protein of Covid-19, it is highly recommended to reinvigorate the role of already known and unknown receptors/co-receptors/attachment factors by means of molecular and histo-pathological data for disease control. The analogy for receptors between different types of viruses must be focused avenue and data must be kept updated.
5. Can receptors be blocked for controlling CoV infection spread?
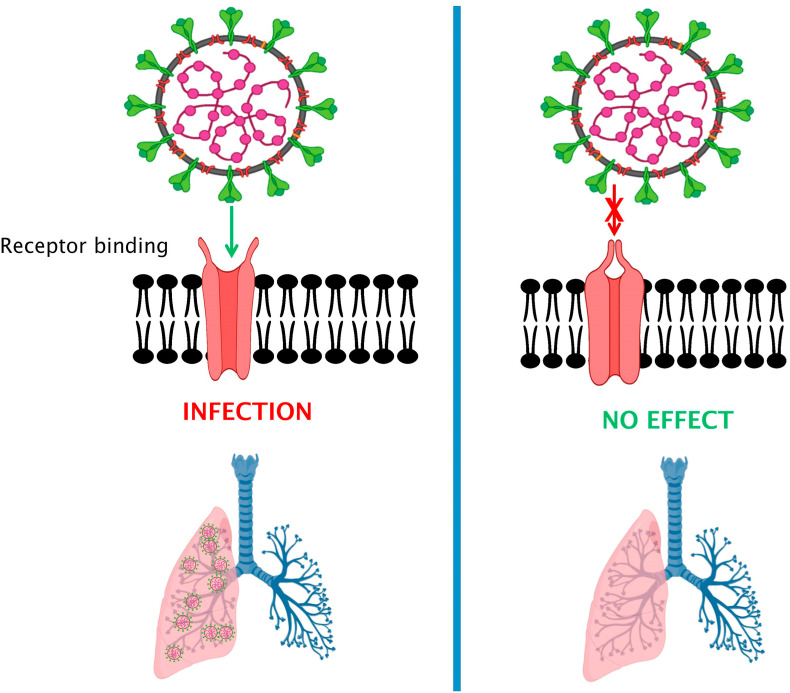
Virus entry into the host is directly linked with its early-stage propagation. All steps during the virus entry process are likely objectives for understanding unique antiviral agents called as viral entry inhibitors. Blockage at any level restricting viral entry into the host definitely decreases the chances of viruses to evolve as well as become resistant against antiviral drugs. Hence, for addressing the prime need for designing vaccines or approval of anti-Covid-19 therapies, appraisal of interactions between CoVs and their cellular receptors as well as entry blockage activities would offer new understandings into the CoV pathogenesis and related treatment (Fig. 1, Fig. 2 ). So far entry limiting inhibitors for several viruses are on record, e.g. enfuvirtide for HIV, RFI 641 for RSV [[64], [65], [66]] while many others are undergoing the developmental process. We anticipate, in the light of a growing body of evidence, entry inhibitors would be very promising besides reverse-transcriptase or protease inhibitors being used for the treatment of viral infections. Since 2000, research advances have elaborated many molecules of different characteristics and origin for controlling disease progression and management. For example, HCV (hepatitis C viruses) receptors have been searched with the help of pseudotyped HCVs [67]. Use of pseudo-typed viruses, i.e. SARS, HCV offer in-depth assay of neutralizing antibodies, cell tropism as well as recognition of drugs that inhibit the virus entry into host cell [[68], [69], [70]]. Small molecules as inhibitors can be designed chemically and synthesized e.g. TAK-779 for HIV-1 [71] or they can be isolated from natural products e.g. glycirhizin and luteolin against SARS-CoV [70]. These inhibitors differ in their potency in regulating entry of different viruses. Likewise, tryptophan dendrimers exhibit exclusive in vitro potency as antivirals against enterovirus (EV-A71). Sun et al. [72] described MADAL385 (a tryptophan dendrimer) accountable for preventing bonding and internalization of enterovirus. The 5-fold vertex of EV-A71 is targeted by MADAL385 and in turn blocks receptor. In our opinion, such findings open novel research arenas and support in anti-CoV drug development. Receptors may also induce the membrane fusion reaction and point to be noted is presence of alternative fusion triggers i.e. pH [73]. Recovering patients of SARS had shown a neutralizing antibody reaction against S protein. The resembling neutralizing antibody associated response was noticed in Covid-19 patients however its efficiency was not as high as reported in SARS-CoV [22]. This response actually reflects reduction in S protein mediated viral entry into host cell like in SARS-CoV has been noticed.
Fig. 2.
Virus entry into the host is directly linked with its early-stage propagation. Expression of different receptors for a diverse range of proteins strongly correlates with infection sites in the body. In the context of infection spread and viral release in close vicinity to the capillaries of pulmonary tissues, we may infer a probable general virus transmission to distant organs in the human body. Blockage at any level restricting viral entry into the host definitely decreases the chances of viruses to evolve. Appraisal of interactions between CoVs and their cellular receptors as well as entry blockage activities would offer new understandings into the CoV pathogenesis and related treatment.
Very possibly, the isolates of natural origin or synthetic inhibitors perform the antiviral activity by interfering CoV-host cell fusion process. Actually, the energy of receptor binding allows exposure of S2 fusion peptide which introduces this peptide into a target membrane. In Ebola Virus (EBoV), glycoproteins also manage viral entry into cells. Additionally, these glycoproteins counter the antiviral activity of host proteins, e.g. tetherin. This protein is appropriately antagonized by glycoprotein of EBoV. It was observed in mice that an antibody directed against EBoV may block glycoprotein dependent antagonism to tetherin [74]. So very simply, we can infer that virus neutralizes the antiviral protein(s) by means of glycoprotein. Normally, alteration of glycoproteins with N-glycans is attuned with effective expression and proteins dependent viral entry into cell [74]. Absence or inappropriate glycosylation can categorically disturb protein expression leading to impeded viral actions. The glycan shield of S proteins makes them stable during biogenesis [75,76]. Among viral genotypes, Glycosylation sites are found highly conserved [77]. N-terminal part of the RBD is furnished with glycosylation sites and found close to each other not only in the sequence but in structure also. We argue that glycosylation sites must be functional for S-receptor interaction(s). Dysfunctional glycosylation sites i.e. mutation in residues hampered protein expression but do not affect binding to receptor. This has been proved in case of SARS-S and ACE2 interaction. Mutagenesis of the fragment S319–518 supported the notion that atleast a single glycosylation site is essential for appropriate expression of CoV protein [78]. Inhibition of glycosylation by some compounds like tunicamycin lead to the production of virions without S glycoproteins [79]. These aspects must be taken into consideration during investigations dealing with both glycoproteins and receptors for managing CoV spread and control. Similarly, the use of nano-carriers and the associated process of protein deglycosylation must be kept in mind to modulate disease symptoms and related treatment [80]. In this regard, macrophage receptors can also be focused for checking uptake and recognizing viral ligands, protein(s) and related events. Therefore, we think that inhibitors for blocking CoV entry change this energy as well as triggers of binding process. This provides a mechanistic challenge to be accepted for designing drugs to control Covid-19. Amalgamation of in hand information with somewhat expected outcomes will certainly gear the process of drug discovery.
6. Can changes in S conformation and blocked receptors reduce infection spread?
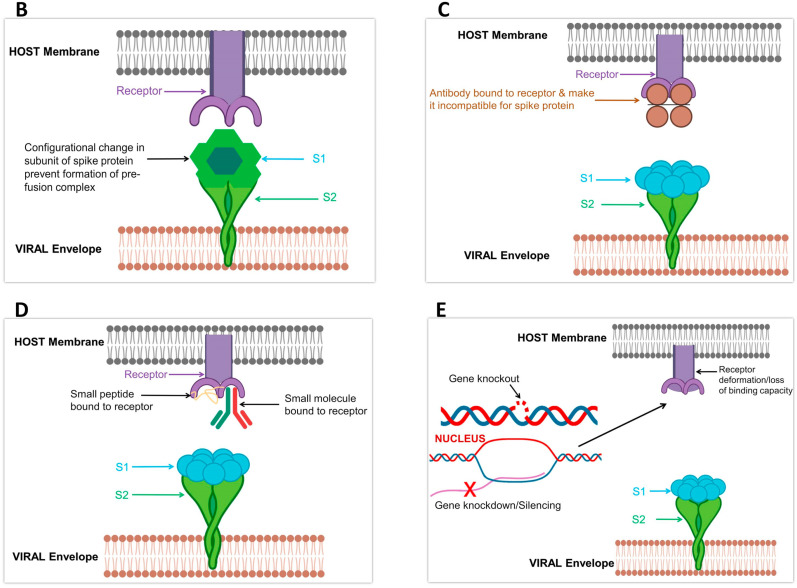
. For enhanced CoV entry and successful pathogenesis of CoV, proteolytic processing and receptor-binding act synergistically for induction of large-scale S conformational changes [75]. Viruses require different receptors for penetrating hosts. Two distinguished conformations of S-protein on a structural basis are on record, i.e. prefusion and postfusion. The transition in conformation from prefusion to postfusion of S must be triggered that leads to membrane fusion (Fig. 1, Fig. 3 B) [51]. The fusion peptide in S2 subunit is thought to the most significant in the activation of membrane fusion. The RBD in S of SARS-CoV and MERS-CoV S displays diverse conformational positions that control the capacity to work together with host cells by means of available receptor-binding motifs (RBMs) [81,82]. Previously it was considered that binding with receptor may start membrane fusion [83]. Contrarily, the S in other CoVs assumes a closed conformation. Such diversity in conformation renders it incompatible for engaging a receptor and highlights structural rearrangements preceding receptor binding as pre-requisite for entry [76,84]. Studies focusing upon structural, biochemical and functional aspects have elaborated main steps of glycoprotein induced membrane fusion and unveiled the complexity of this process in HIV [85]. Due to unusual mechanism and concealing RBM conformation, our knowledge of activation of CoV membrane fusion is limited.
Fig. 3.
Future drug design and mechanism; A) With reference to Fig. 1, the formation of the pre-fusion complex and post-fusion complex is pre-requisite for infection. Failure to form these complexes can block infection spread. B) Any conformational change in S can be beneficial in infection control. Such changes will prevent the formation of a fusion complex and can be very helpful in infection control. C) Bonding of antibodies to a receptor can also protect from infection spread. D) In case of binding between peptides or small molecules, the binding site will no more be available for S. This can be a good option for designing future drugs to control Covid-19 infection. E) By using molecular techniques, receptors can be Knockout or knockdown. This may hamper their activation or deform their binding site. By such drug design and discovery, the disease can be managed more efficiently.
Receptors are highly selective in their choice of ligand and respective performance. Many receptors function at diverse levels ranging from replication of the virus, ionic flux to in vivo spread of infection [86]. Receptor blockage by antibody molecule such as Siglec-1 by anti-Siglec-1 monoclonal antibodies can cease viral uptake, i.e. EBoV. Blocking entry into cytoplasm offers cross-protection against viruses, including HIV [87]. Moreover, some studies support knockout and knockdown of receptors (Fig. 3C and D) and reduced expression of related proteins restrict attachment and entry of some viruses such as HCV. For instance, SDC-2 gene knockout decreased HCV infection establishment by 70%. On the other side, the overexpressing SMAD6 incremented expression of SDC-1 and SDC-2 and, ultimately, increased HCV infection [88,89]. Therefore, a positive correlation is evident in the mentioned cases. In the light of previous studies focusing on viral entry and cell-to-cell dissemination, we agree that infection spread can be blocked by using neutralizing antibodies. The restricted HCV infection by E1/E2 antibodies [90] advocates our stance. In the same way, monoclonal antibodies (mAbs) recovered from survivors of the disease are also a good option to be used against viral infection in general [91]. A recent report by Wrapp et al. [23] has revealed that RBD of Covid-19 did not present much binding affiliation for some mAbs, i.e. S230, m396 and 80R but contrary to SARS-CoV. Besides the relatively small surface area of the mAbs epitopes interacting with Covid-19 RBD, the absence of observed binding proposes that SARS-directed mAbs would not essentially be cross-reactive and that isolation of antibody in future and drug design efforts would be benefited by using S proteins of Covid-19 as probes [23].
Other than antibodies, some peptides and small molecules can also block receptors and function in developing immunity against viral invasion and other disease-causing agents (Fig. 3D and E). LDLR (low-density lipoprotein receptor) and ApoER2 are bound by Proprotein convertase subtilisin/kexin type 9 (PCSK9) for induction of its internalization as well as degradation. This binding is dependent upon EGF-A peptide (epidermal growth factor-like repeat A). Contrastingly, EGF-A constrains bonding between PCSK9 and VLDLR in the mouse. Although it is apparent that different receptors can be approached by the same molecule but interaction involves other crucial factors that can be beneficial for one but infeasible for the other [92]. Adding to previous evidence, lipidated peptides such as pepducins do not activate intracellular receptors, i.e. G-proteins by the same molecular mechanism as done by other agonists in routine. This is because of lipid conjugate added to the peptide that particularly needs C-tail of a receptor for interaction [93]. Such distinguished interfaces between effector-receptor offer a baseline for the development of new targeted immunogenic strategies. Equally, synthetic peptides can be useful in making antibodies well able to elicit a special immune response and restrict viral entry. Such synthetic peptides with biologically active sequences have raised antibodies and successfully tested against S proteins in SARS-CoV [94]. For designing such synthetic peptides to control viral infection, some key points must be borne in mind. Choy et al. stated that [94] sequences in such peptides should be able to target the surface of S-proteins. The designed peptide should be of a rigid structure by adopting a helix motif. This will increase its antigenicity. Some linkers can be added for cycling the linear peptides or disulfide bonds to stabilize peptide. The loop structures in the peptides can be additionally accustomed and reinforced to improve the conforming antigenicity.
Keeping in view all of these data, with particular reference of Covid-19, some very important issues come to the front. The first is a determination of infection spread mode, i.e. whether Covid-19 is spread cell-to-cell or this CoV infect uninfected cells using its extra-cellular progeny. Besides, the most important issue in tackling Covid-19 is the molecular basis of the underlying mechanism(s) of surface receptors and related antibodies, small molecules or peptides for controlling transmission. The role of attachment and post-attachment events at receptor levels in Covid-19 infection and cell-to-cell spread is elusive. Like HCV, these attachment and post attachment receptors [88] should be thoroughly investigated. It must be unraveled that whether the neighboring cells are preferred over random cells for infection spread or not?
7. Conclusion and future perspectives
Since 2000, two outbreaks of super pathogenic CoVs, i.e. SARS and MERS have revealed that CoVs would remain circulating among humans due to facilitated interaction between animals and humans. The advanced molecular biology tools have identified CoV strains among different animals with resembling pathogenic characteristics. This shows a constant risk of emergence of highly hazardous CoVs outbreak. These emerging CoVs can be similar symptomatically or antigenically to previously recorded CoVs with disastrous implications. Therefore, strict public health measures must be implemented to check CoV infection spread along with disease surveillance.
This emerging COVID-19 has offered a daunting challenge to researchers in all fields of natural, physical and social sciences. The imperative measures are linked together for formulating any model to control the epidemic. In the light of current prevention strategies, social scientists must engage public by devising and launching public awareness strategies about infection spread and prevention. Some surveys regarding losses done due to non-compliance of prevention strategies must be conducted and information should be disseminated through media among the public.
It is imperative to conduct strategic research programs to target control of CoVs for saving global public health from such emerging threats. Like many other viruses or diseases, seasonal recurrence of COVID-19 is not out of the question. For sustained control of CoV infection, the development of a vaccine is essential. So far, we lack any absolute vaccine for control of this disease. Therefore, we recommend the development of vaccines, antivirals as well as a synthetic peptide-based strategy as a feasible measure to prevent infection. Synthetic peptides can be a basic step in probing COVID-19 related research involving spike proteins as well as diverse receptors. RBD in S proteins of COVID-19 must be targeted to discover its corresponding receptor(s). Structural data and functional information of spike glycoproteins with emphasis on binding sites must be focused to find out their receptors and for related blockage efforts. Investigations must be carried out to mimic conformation of epitope regions for the production of recombinant proteins analogous to proteins working for immunogenic purposes in hosts. The antigenic sites in viral proteins should also be brought into focus for generating new recombinant proteins with high antiviral efficacy. Data banks having DNA, RNA, protein sequences of Covid-19 should be designed to monitor polymorphism. For all types of proteins related to antiviral as well as viral activity binding kinetics and biophysical attributes must be studied for unveiling structural and functional relations between different CoVs. With the best efforts being carried out for control of Covid-19, the increasing evidence has suggested bats as potential host and reservoir like other CoV epidemics in the past. With a theme alarmingly indicative of the co-infection or re-assortment mechanisms used by other viruses, the animals inhabiting the similar habitat like bats or adjoining area must be phylogenetically evaluated to assess the relationship of CoVs on the basis of reservoirs and hosts for harboring infections. This may address any future epidemic outbreak due to pathogenic CoVs in humans Inter-species spread ought to be systematically inspected together by processes of mutation-based evolution as well as RNA recombination. For researchers in all domains of biology, this is the real-time to accept the challenge of developing vaccines/antivirals to fight a genetically improved virus capable of quick emergence. Absolutely, ecology and epidemiology of current outbreak reiterate the extreme necessity for developing proficient therapeutics against viruses existing heterogeneously and in highly irregular reservoirs. Inter-species spread ought to be systematically inspected together by processes of mutation-based evolution as well as RNA recombination. This Covid-19 epidemic is an indication of the utter requirement of efforts and strategies from a united platform to thwart emerging disease extortions before their occurrence.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
We are thankful to Biorender for providing us platform for image drawing and visualization.
References
- 1.King A.M., Adams M.J., Carstens E.B., Lefkowitz E.J. 2012. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; pp. 486–487. [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. 10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020:1–5. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. J. Am. Med. Assoc. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 6.Walls A., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F., et al. Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo‐electron microscopy. Protein Sci. 2017;26:113–121. doi: 10.1002/pro.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Fan Y, Lai Y, Han T, Li Z, Zhou P, et al. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahn J.S., McIntosh K. History and recent advances in coronavirus discovery. Pediatr. Infect. Dis. J. 2005;24:S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 9.Organization WH. Middle East Respiratory Syndrome Coronaviurs (MERS-CoV) Fact Sheet [cited 2017 Oct 30].
- 10.Corman V., Lienau J., Witzenrath M. Der Internist; 2019. Coronaviruses as the cause of respiratory infections; pp. 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr A.R., Channappanavar R., Perlman S. Middle East respiratory syndrome: emergence of a pathogenic human coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.-W., Wong B.H., et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. Unit. States Am. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. 10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du B., Zhang W., Liu B., Hu J., Wei Z., Shi Z., et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren X., Glende J., Al-Falah M., de Vries V., Schwegmann-Wessels C., Qu X., et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J. Gen. Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 16.Tseng C.-T.K., Tseng J., Perrone L., Worthy M., Popov V., Peters C.J. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 2005;79:9470–9479. doi: 10.1128/JVI.79.15.9470-9479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siddell S.G. Springer; 1995. The Coronaviridae. The Coronaviridae; pp. 1–10. [Google Scholar]
- 19.Lontok E., Corse E., Machamer C.E. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Krueger N., Mueller M.A., Drosten C., Poehlmann S. he novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv; 2020. T. [Google Scholar]
- 23.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PloS One. 2013;8:e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., et al. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105:19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgson T., Casais R., Dove B., Britton P., Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J. Virol. 2004;78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navas S., Seo S.-H., Chua M.M., Sarma J.D., Lavi E., Hingley S.T., et al. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 2001;75:2452–2457. doi: 10.1128/JVI.75.5.2452-2457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leparc-Goffart I., Hingley S.T., Chua M.M., Phillips J., Lavi E., Weiss S.R. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J. Virol. 1998;72:9628–9636. doi: 10.1128/jvi.72.12.9628-9636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen N.C. Springer; Coronaviruses: 1987. Virologic and Immunologic Aspects of Feline Infectious Peritonitis Virus Infection; pp. 529–550. [DOI] [PubMed] [Google Scholar]
- 32.Vennema H., Poland A., Foley J., Pedersen N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology. 1998;243:150–157. doi: 10.1006/viro.1998.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benbacer L., Kut E., Besnardeau L., Laude H., Delmas B. Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J. Virol. 1997;71:734–737. doi: 10.1128/jvi.71.1.734-737.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hegyi A., Kolb A.F. Characterization of determinants involved in the feline infectious peritonitis virus receptor function of feline aminopeptidase N. J. Gen. Virol. 1998;79:1387–1391. doi: 10.1099/0022-1317-79-6-1387. [DOI] [PubMed] [Google Scholar]
- 36.Schwegmann-Weßels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K., et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan K., Zelus B.D., Meijers R., Liu Jh, Bergelson J.M., Duke N., et al. Crystal structure of murine sCEACAM1a [1, 4]: a coronavirus receptor in the CEA family. EMBO J. 2002;21:2076–2086. doi: 10.1093/emboj/21.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiley D.C., Skehel J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 41.Jackson T., Sharma A., Ghazaleh R.A., Blakemore W.E., Ellard F.M., Simmons D.L., et al. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin alpha (v) beta3 in vitro. J. Virol. 1997;71:8357–8361. doi: 10.1128/jvi.71.11.8357-8361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura T.A., Travanty E.A., Oko L., Bielefeldt-Ohmann H., Weiss S.R., Beauchemin N., et al. The spike glycoprotein of murine coronavirus MHV-JHM mediates receptor-independent infection and spread in the central nervous systems of Ceacam1a−/− mice. J. Virol. 2008;82:755–763. doi: 10.1128/JVI.01851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips J.J., Chua M., Seo S-h, Weiss S.R. Multiple regions of the murine coronavirus spike glycoprotein influence neurovirulence. J. Neurovirol. 2001;7:421–431. doi: 10.1080/135502801753170273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss S.R., Navas-Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005;69:635–664. doi: 10.1128/MMBR.69.4.635-664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren W., Qu X., Li W., Han Z., Yu M., Zhou P., et al. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;82:1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., et al. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS One. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zelus B.D., Schickli J.H., Blau D.M., Weiss S.R., Holmes K.V. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37 C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 2003;77:830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F. Structure, function, and evolution of coronavirus spike proteins. Annual review of virology. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., et al. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong M.C., Cregeen S.J.J., Ajami N.J., Petrosino J.F. bioRxiv; 2020. Evidence of Recombination in Coronaviruses Implicating Pangolin Origins of nCoV-2019. [Google Scholar]
- 56.Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 58.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.To K., Tong J.H., Chan P.K., Au F.W., Chim S.S., Allen Chan K., et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in‐situ hybridization study of fatal cases. J. Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeffers S.A., Tusell S.M., Gillim-Ross L., Hemmila E.M., Achenbach J.E., Babcock G.J., et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W., Luo R., He Q., van Kuppeveld F.J.M., Rottier P.J.M., Bosch B.-J. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res. 2017;235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Q., Herrmann A. vol. 2020. bioRxiv; 2020. (Fast Assessment of Human Receptor-Binding Capability of 2019 Novel Coronavirus (2019-nCoV)). 02.01.930537. [Google Scholar]
- 63.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020 doi: 10.1128/JVI.00127-20. JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esté J.A. Virus entry as a target for anti-HIV intervention. Curr. Med. Chem. 2003;10:1617–1632. doi: 10.2174/0929867033457098. [DOI] [PubMed] [Google Scholar]
- 65.Moore J.P., Doms R.W. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:10598–10602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Razinkov V., Gazumyan A., Nikitenko A., Ellestad G., Krishnamurthy G. RFI-641 inhibits entry of respiratory syncytial virus via interactions with fusion protein. Chem. Biol. 2001;8:645–659. doi: 10.1016/s1074-5521(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 67.Hsu M., Zhang J., Flint M., Logvinoff C., Cheng-Mayer C., Rice C.M., et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartosch B., Bukh J., Meunier J.-C., Granier C., Engle R.E., Blackwelder W.C., et al. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. Unit. States Am. 2003;100:14199–14204. doi: 10.1073/pnas.2335981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wool-Lewis R.J., Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yi L., Li Z., Yuan K., Qu X., Chen J., Wang G., et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strizki J.M., Xu S., Wagner N.E., Wojcik L., Liu J., Hou Y., et al. SCH-C (SCH 351125) An orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. Unit. States Am. 2001;98:12718–12723. doi: 10.1073/pnas.221375398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun L., Lee H., Thibaut H.J., Lanko K., Rivero-Buceta E., Bator C., et al. Viral engagement with host receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallagher T.M., Buchmeier M.J. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brinkmann C., Nehlmeier I., Walendy-Gnirß K., Nehls J., González Hernández M., Hoffmann M., et al. The tetherin antagonism of the ebola virus glycoprotein requires an intact receptor-binding domain and can Be blocked by GP1-specific antibodies. J. Virol. 2016;90:11075–11086. doi: 10.1128/JVI.01563-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039. doi: 10.1016/j.cell.2018.12.028. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A., et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parsons L.M., Bouwman K.M., Azurmendi H., de Vries R.P., Cipollo J.F., Verheije M.H. Glycosylation of the viral attachment protein of avian coronavirus is essential for host cell and receptor binding. J. Biol. Chem. 2019;294:7797–7809. doi: 10.1074/jbc.RA119.007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chakraborti S., Prabakaran P., Xiao X., Dimitrov D.S. The SARS coronavirus S glycoprotein receptor binding domain: fine mapping and functional characterization. Virol. J. 2005;2:73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossen J., De Beer R., Godeke G.-J., Raamsman M., Horzinek M., Vennema H., et al. The viral spike protein is not involved in the polarized sorting of coronaviruses in epithelial cells. J. Virol. 1998;72:497–503. doi: 10.1128/jvi.72.1.497-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghazaryan A., Landfester K., Mailänder V. Protein deglycosylation can drastically affect the cellular uptake. Nanoscale. 2019;11:10727–10737. doi: 10.1039/c8nr08305c. [DOI] [PubMed] [Google Scholar]
- 81.Park J.-E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Luo C., et al. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melikyan G.B. In: Current Topics in Membranes. Chernomordik L.V., Kozlov M.M., editors. Academic Press; 2011. Chapter 4 - membrane fusion mediated by human immunodeficiency virus envelope glycoprotein; pp. 81–106. [DOI] [PubMed] [Google Scholar]
- 86.Chen S., Shenk T., Nogalski M.T. P2Y2 purinergic receptor modulates virus yield, calcium homeostasis, and cell motility in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:18971–18982. doi: 10.1073/pnas.1907562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez-Zsolt D., Erkizia I., Pino M., García-Gallo M., Martin M.T., Benet S., et al. Anti-Siglec-1 antibodies block Ebola viral uptake and decrease cytoplasmic viral entry. Nature Microbiology. 2019;4:1558–1570. doi: 10.1038/s41564-019-0453-2. [DOI] [PubMed] [Google Scholar]
- 88.Fan H., Qiao L., Kang K.-D., Fan J., Wei W., Luo G. Attachment and postattachment receptors important for hepatitis C virus infection and cell-to-cell transmission. J. Virol. 2017;91:e00280. doi: 10.1128/JVI.00280-17. -17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang F., Sodroski C., Cha H., Li Q., Liang T.J. Infection of hepatocytes with HCV increases cell surface levels of heparan sulfate proteoglycans, uptake of cholesterol and lipoprotein, and virus entry by up-regulating SMAD6 and SMAD7. Gastroenterology. 2017;152:257–270. doi: 10.1053/j.gastro.2016.09.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Keck Z.-Y., Xia J., Cai Z., Li T.-K., Owsianka A.M., Patel A.H., et al. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 2007;81:1043–1047. doi: 10.1128/JVI.01710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mire C.E., Geisbert J.B., Borisevich V., Fenton K.A., Agans K.N., Flyak A.I., et al. Therapeutic treatment of Marburg and Ravn virus infection in nonhuman primates with a human monoclonal antibody. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shan L., Pang L., Zhang R., Murgolo N.J., Lan H., Hedrick J.A. PCSK9 binds to multiple receptors and can be functionally inhibited by an EGF-A peptide. Biochem. Biophys. Res. Commun. 2008;375:69–73. doi: 10.1016/j.bbrc.2008.07.106. [DOI] [PubMed] [Google Scholar]
- 93.Covic L., Gresser A.L., Talavera J., Swift S., Kuliopulos A. Activation and inhibition of G protein-coupled receptors by cell-penetrating membrane-tethered peptides. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:643–648. doi: 10.1073/pnas.022460899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choy W.-Y., Lin S.-G., Chan P.K.-S., Tam J.S.-L., Lo Y.M.D., Chu I.M.-T., et al. Synthetic peptide studies on the severe acute respiratory syndrome (SARS) coronavirus spike glycoprotein: perspective for SARS vaccine development. Clin. Chem. 2020;50:1036–1042. doi: 10.1373/clinchem.2003.029801. [DOI] [PMC free article] [PubMed] [Google Scholar]