Abstract
Glutathione peroxidase 3 (GPx3) belongs to the glutathione peroxidase family of selenoproteins and is a key antioxidant enzyme in multicellular organisms against oxidative damage. Downregulation of GPx3 affects tumor progression and metastasis and is associated with liver and heart disease. However, the physiological significance of GPx3 in vertebrate embryonic development remains poorly understood. The current study aimed to investigate the functional roles of gpx3 during embryogenesis. To this end, we determined gpx3’s spatiotemporal expression using Xenopus laevis as a model organism. Using reverse transcription polymerase chain reaction (RT-PCR), we demonstrated the zygotic nature of this gene. Interestingly, the expression of gpx3 enhanced during the tailbud stage of development, and whole mount in situ hybridization (WISH) analysis revealed gpx3 localization in prospective tail region of developing embryo. gpx3 knockdown using antisense morpholino oligonucleotides (MOs) resulted in short post-anal tails, and these malformed tails were significantly rescued by glutathione peroxidase mimic ebselen. The gene expression analysis indicated that gpx3 knockdown significantly altered the expression of genes associated with Wnt, Notch, and bone morphogenetic protein (BMP) signaling pathways involved in tailbud development. Moreover, RNA sequencing identified that gpx3 plays a role in regulation of cell death in the developing embryo. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and phospho-histone 3 (PH3) staining confirmed the association of gpx3 knockdown with increased cell death and decreased cell proliferation in tail region of developing embryos, establishing the involvement of gpx3 in tailbud development by regulating the cell death. Furthermore, these findings are inter-related with increased reactive oxygen species (ROS) levels in gpx3 knockdown embryos, as measured by using a redox-sensitive fluorescent probe HyPer. Taken together, our results suggest that gpx3 plays a critical role in posterior embryonic development by regulating cell death and proliferation during vertebrate embryogenesis.
Keywords: gpx3, tailbud, apoptosis, posterior development, embryogenesis
1. Introduction
Reactive oxygen species (ROS) are highly reactive chemical species that contain oxygen in the form of reactive superoxide anions, hydrogen peroxides, or hydroxyl radicals [1]. In recent years, ROS have been recognized as important signaling molecules that play a role in early embryonic development and regeneration [2]. Maintaining high levels of hydrogen peroxide is required for Wnt/β-catenin signaling and fibroblast growth factor (FGF) signaling activation during Xenopus tail regeneration [3]. An ROS burst is observed after fertilization in sea-urchins and Xenopus [4]. ROS play essential roles during egg cleavage and mesoderm formation during vertebrate embryogenesis [5]. However, excessive ROS damage can affect various cells and biological processes [1,6]. Therefore, ROS homeostasis is required for steady-state biological and cellular processing [1,6,7].
Cells use different enzymatic and non-enzymatic antioxidants to maintain ROS homeostasis [6,7]. Glutathione peroxidase (GPx) is a multiple isozyme group that is involved in converting hydrogen peroxide (H2O2) or organic hydrogen peroxide to water using reduced glutathione as an electron donor [8]. Some GPxs have selenium-dependent glutathione peroxidase activity [9]. In mammals, four selenium dependent GPxs are broadly distributed in different mammalian tissues: GPx1, GPx2, Gpx3, and GPx4, [10].
Glutathione peroxidase 3 (GPx3), also known as plasma GPx, is present in various organs, including kidney, lung, heart, muscles, epididymis, vas deferens, placenta, and seminal vesicles [11,12,13]. GPx3 forms a homotetramer and is a secreted protein that is also present in the cytoplasm [14]. GPx3 plays a crucial role in ROS detoxification. Studies have shown that GPx3 silencing promotes tumor metastasis in human gastric and thyroid cancer [15]. Recent reports suggest that GPx3 ablation enhances tumor inflammation, injury, proliferation, and DNA damage in mice subjected to inflammatory carcinogenesis and chronic colitis [16]. Hyper-methylation-mediated downregulation of GPx3 was also observed in breast, ovarian, gastric, esophagus, and prostate cancer [17,18,19,20,21]. In addition to its anticancer role, high GPx3 levels alleviate cardiovascular disease-associated mortality [22]. GPx3 downregulation enhances liver aging, and an inadequate GPx3 level is a significant factor in human heart disease [23,24]. However, GPx3’s role during early embryonic development remains elusive.
In this study, we investigated gpx3 developmental functions during Xenopus embryogenesis. Analyzing gpx3’s spatiotemporal expression pattern revealed that it is expressed in the tailbud region. gpx3 knockdown resulted in a shortened post-anal tail and increased ROS levels. Whole mount in situ hybridization (WISH) analysis indicated that gpx3 is not involved in the tailbud structure. However, gpx3 depletion induced cell death and decreased cell proliferation in the tailbud region. These findings suggest that gpx3 plays a significant role in the tailbud extension during Xenopus embryonic development.
2. Materials and Methods
2.1. Xenopus Growth Conditions and In Vitro Fertilization
Adult X. laevis were obtained from the Korean Xenopus Resource Center for Research and were housed at 18 °C with a 12 h photoperiod in containers recommended by the Ulsan National Institute of Science and Technology Institutional Review Board. To induce ovulation, Xenopus females were injected with 1000 IU of human chorionic gonadotropins into the dorsal lymph sac the evening before the experiment. The next morning, females were transferred to a 1x high salt solution for egg harvesting. For fertilization, male Xenopus were kept in a 1x benzocaine solution for 5–15 min and were then sacrificed to isolate the testes. The isolated testes were kept in 1x modified Barth’s saline (MBS) at 4 °C. The eggs were washed with 0.1x MBS three times and were then fertilized with a sperm suspension solution derived from the isolated testes. After fertilization, the eggs were de-jellied by swirling them in 2% L-cysteine and were then washed five times with 0.5x MBS. The unfertilized and dead eggs were removed. The fertilized eggs were cultured at 15–18 °C in 0.5x MBS containing 2% Ficoll 400 (GE Healthcare, Little Chalfont, UK) after injections.
2.2. Plasmids and mRNA Synthesis
As gpx3 has a UGA opal codon within the coding sequence, to avoid translation termination and confirm morpholino oligonucleotides (MO) specificity, we cloned a mutant construct that has the selenocysteine changed to cysteine. DNA was isolated from tailbud stage Xenopus. gpx3 primers were designed based on the gpx3 sequences listed in Xenbase and NCBI. Flag-tagged U73C gpx3 mRNA was synthesized using PCR, and the plasmid was constructed using the pCS107 vector with ClaI and XhoI restriction sites.
The HyPer-cyto gene was obtained from pHyPer-cyto (Evrogen, Moscow, Russia) and subcloned into the pCS2+ vector. HyPer-pCS2+ was linearized using the NotI restriction enzyme. SP6 mMessage mMachine kit was used to synthesize mRNA (Ambion, Woodward Street, Austin, TX, USA).
2.3. MO Design and Xenopus Embryo Microinjection
A 25-nucleotide gpx3 MO (Gene Tools) was designed to bind the gpx3 mRNA translation initiation region. The Xenopus gpx3 translation-blocking MO sequence was 5′-atgggggtcaaatttagggggctct-3′, and the splicing-blocking MO sequence was 5′-acgcacatacctggcagtcaaaaac-3′. MOs were injected into the embryo’s ventral blastomere at the 4-cell stage, and green fluorescent protein mRNA was co-injected as a lineage tracer. Ebselen, a GPx chemical mimic, was used to perform the rescue experiments.
2.4. Whole-Mount In Situ Hybridization
The gpx3 MO was injected into the Xenopus embryos ventral region at the 4-cell stage. The MO-injected embryos were collected at the desired stage and were fixed in fixative MEMFA (4% paraformaldehyde, 0.1 M MOPS buffer (pH 7.4), 1 mM MgSO4, 2 Mm ethylene glycol-bis(β-amino ethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) overnight at 4 °C. The embryos were dehydrated before storage in 100% methanol at −20 °C. To prepare the antisense digoxigenin (DIG)-labeling probes, DNA templates were linearized using appropriate restriction enzymes. Probes were synthesized using SP6 or T7 RNA polymerase (Ambion) and were detected using an alkaline phosphatase-labeled antidigoxigenin antibody (1:1000, Roche, Basel, Switzerland) or nitro blue tetrazolium/5-bromo-4-chloro-3indolyl phosphate (NBT/BCIP) [25].
2.5. Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from Xenopus embryos using Isol-RNA lysis reagent (5 Prime GmbH, Hilden, Germany). cDNA was synthesized from total RNA of Xenopus embryos using the PrimeScript first-strand cDNA synthesis kit (Takara, Kusatsu, Japan). PCR was performed using customized primers, (Table 1) and the products were loaded on 1% agarose gels. Images were captured using WiseCapture I-1000 (Daihan Scientific, Wonju, South Korea).
Table 1.
List of Primers for RT-PCR
| Gene | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| odc | 5′-CAGCTAGCTGTGGTGTGG-3′ | 5′-CAACATGGAAACTCACACC-3′ |
| wnt5b | 5′-AAGCAGAGAGGCGGCATTT-3′ | 5′-CCGAGACACCATGGCACTT-3′ |
| notch1 | 5′-ACCTACAAATGCTCCTGCCC-3′ | 5′-AACAAGGGTGGGAAGCACAA-3′ |
| tbx6 | 5′-TATCCGGGGGAAGAGGAAGG-3′ | 5′-CCCTTGACTGTGGCTGATGT-3′ |
| Xnot2 | 5′-TAATCTCCTGCACCCCCAGA-3′ | 5′-TTGATGCGTCGGTTCTGGAA-3′ |
| fgf8 | 5′-CCAACTGGCAACTGAGCAAC-3′ | 5′-ACCGTGTCCTACCGAGAACT-3′ |
| Xbra | 5′-CGTGCAGTACCGGGTAGATC-3′ | 5′-TGGCAAATGGGTTGTGCTTG-3′ |
| wnt3a | 5′-TCCTCTGTGGGCTACACCAA-3′ | 5′-CGCCAATCACCCTGAAGTCT-3′ |
| wnt5a | 5′-GGCAGTGCAATGGTCTCTCA-3′ | 5′-GCGACATCAGCCAAGGTACT-3′ |
2.6. Transcriptomic Analysis
An RNA sequencing (RNA-seq) library was constructed from total RNA extracted from each sample with polyA enrichment, according to the manufacturer’s instructions (Illumina). To estimate the mRNA abundance, X. laevis cDNA sequence reads were mapped from the genome project consortium [26] using bwa (version 0.7.15). EdgeR (version 3.20.7) was used to analyze differentially expressed genes (DEGs). Significantly differentially expressed genes had a >2-fold change with a false discovery rate (FDR) <0.01 in an exact test. Over-represented biological processes in the DEGs were tested using Fisher’s test and PANTHER database (released 20171205). Human orthologous genes were determined based on best hits using the BLASTP search. Raw data for the RNA-seq is available at the NCBI GEO database (PRJEB39362) [27].
2.7. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) and Phospho-Histone 3 (PH3) Staining
Xenopus embryos were fixed for 24 h in MEMFA at 4 °C and bleached (3% H2O2, 5% formamide, and 5xsaline sodium citrate (SSC)) after washing with phosphate buffer saline (PBS). The embryos were processed for TUNEL and PH3 staining, as described previously [28].
2.8. In Vivo Imaging of ROS
HyPer cyto (HyPer) was used to detect H2O2 related ROS. HyPer mRNAs (10 ng) were injected into the 1-cell staged embryos. Images were taken using live anesthetized embryos using 1:1000 diluted benzocaine and an Olympus FV1200 confocal microscope. Images were analyzed by ImageJ [Cite Schneider, C. A.; Rasband, W. S. & Eliceiri, K. W. (2012), “NIH Image to ImageJ: 25 years of image analysis”, Nature methods 9(7): 671-675, PMID 22930834 ].
2.9. Statistical Analysis
WISH and RT-PCR data were analyzed using ImageJ. Results are presented as the mean ± standard error from three independent experiments. Results were interpreted by unpaired t-test (two-tailed) using GraphPad Prism 7 software (GraphPad Software Inc., La Jolla, CA, USA). P-values of less than 0.05 were considered statistically significant. P-values were represented by asterisk (*).
3. Results
3.1. Gpx3 Spatiotemporal Expression Pattern during Xenopus Embryogenesis
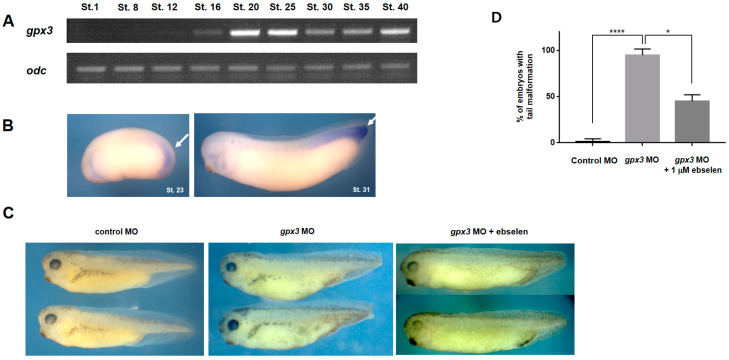
To determine the specific gpx3 roles during Xenopus embryonic development, we first analyzed the gpx3 expression pattern in Xenopus. RT-PCR was conducted to assess the temporal gpx3 expression. RT-PCR indicated that gpx3 is first expressed at the gastrula stage (Nieuwkoop and Faber; St.12) of Xenopus embryos (Figure 1A). The highest expression levels were observed at the tailbud stage (NF. St.20-25). Expression was observed until later development stages, such as the late tailbud stage (St.40; Figure 1A). RT-PCR revealed the zygotic nature of gpx3, with the highest expression level at the early tailbud stage in Xenopus embryos.
Figure 1.
gpx3 is required for tailbud development during Xenopus embryonic development. (A) RT-PCR analysis indicated the zygotic gpx3 gene expression. gpx3 expression started at the gastrula stage of development (St.12) and continued until later stages (St.40). The highest expression levels were observed at the tailbud stage (St.20-25). Ornithine decarboxylase (odc) was used as an internal control. (B) Whole mount in situ hybridization (WISH) analysis demonstrated gpx3 expression at the prospective tailbud region (St.23) and exhibited its localization in the developing embryo tail region (St.31). (C) gpx3 morpholino oligonucleotides (MO) microinjection into the 4-cell stage embryo ventral side resulted in short post-anal tails in gpx3 morphant embryos compared to control MO-injected embryos. Malformed phenotypes induced by gpx3 knockdown were effectively rescued by ebselen, a GPx mimic. (D) Embryo phenotype quantification revealed that more than 90% of the injected embryos with gpx3 MO developed short post-anal tails compared to control embryos. The short tail phenotypes were 50% recovered in ebselen-treated embryos. * p < 0.05, **** p < 0.0001.
WISH analysis determined the gpx3 spatial expression during Xenopus embryonic development. Our results showed the gpx3 expression in the developing embryo prospective tailbud region (NF. St.23) and in the tail region of later stage embryos (NF. St. 31; Figure 1B). These results suggest that gpx3 may be involved in tailbud development during Xenopus embryogenesis.
3.2. Gpx3 Knockdown Leads to a Reduced Post-Anal Tail during Embryonic Development
To evaluate our hypothesis that gpx3 is involved in tailbud development, loss of function experiments were performed using antisense MO against gpx3. To check the specificity and the efficiency of MO against gpx3, we used translational blocking MO and splicing blocking MO. These gpx3 MOs were microinjected into the ventral region of Xenopus embryos at the 4-cell stage to repress gpx3 expression. Microinjection of the gpx3 MOs resulted in a shortened post-anal tail in the morphant embryos compared to control embryos (Figure 1C). It was revealed that more than 90% of the embryos injected with the gpx3 MO exhibited reduced tail development (Figure 1D).
To further determine the gpx3 role in tailbud development and to confirm the gpx3 specificity, we conducted rescue experiments using ebselen, which mimics glutathione peroxidase [29]. Ebselen has free-radical and singlet oxygen quenching properties and catalyzes the hydrogen peroxide reduction similar to glutathione peroxidase at the expense of thiol [29]. The reason for performing rescue experiments using ebselen was the non-availability of gpx3 specific antibodies in Xenopus. Thus, to rescue the gpx3 MO-induced malformations, we injected gpx3 MO into the Xenopus embryos at the 4-cell stage and transferred them to a 1 μΜ ebselen solution. Approximately 50% of the gpx3 MO-injected embryos grown in the ebselen media recovered from the malformations that were observed in gpx3 morphant embryos (Figure 1C,D). Taken together, these findings demonstrate the gpx3 MO specificity, which is responsible for a shortened post-anal tail in gpx3 morphant embryos.
3.3. Gpx3 Mediates Tailbud Development through Wnt, Notch, and FGF Signaling Pathway Regulation
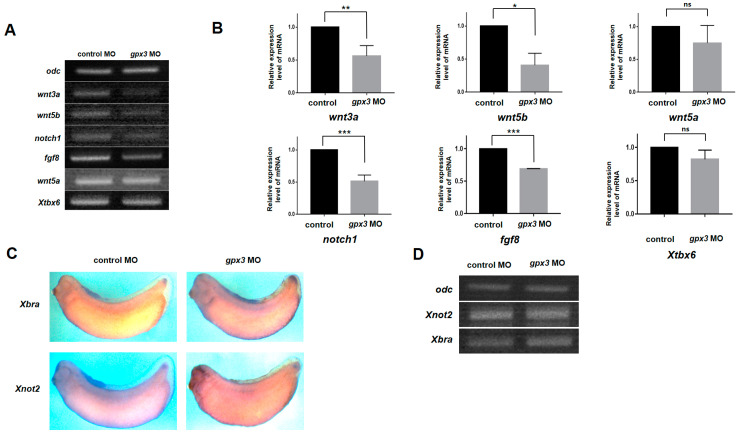
In Xenopus, tailbud development is regulated by different signaling pathways. Active Wnt/β-catenin signaling is required for posterior development, and many Wnt signaling genes, such as wnt3a, wnt5, and wnt8, are expressed in the posterior portion of the developing embryos [30,31]. In addition to Wnt, Notch and bone morphogenetic protein (BMP) signaling also play critical roles in tailbud outgrowth [32,33]. FGF proteins are also involved in posterior patterning, and Xtbx6 mediates posterior development through Wnt and FGF signaling regulation [34]. To evaluate gpx3’s role in posterior development signaling pathway regulation, we microinjected Xenopus embryos with gpx3 MO and assessed Wnt gene expression via RT-PCR. We assessed wnt3a, wnt5a, wnt5b, notch1, fgf8, and Xtbx6 expression in the excised posterior part of morphant embryos. The expressions of wnt3a, wnt5b, notch1, and fgf8 were significantly reduced in gpx3-depleted embryos compared to control embryos (Figure 2A,B). Overall, gpx3 is involved in posterior development by influencing Wnt, Notch, and BMP signaling pathways to regulate tailbud development.
Figure 2.
gpx3 knockdown perturbed Wnt, Notch, and FGF signaling gene expression but did not affect the early tailbud gene expression. (A,B) gpx3 MO was injected into the ventral regions of 4-cell stage embryos, and RT-PCR was used to determine wnt3a, wnt5a, wnt5b, notch1, fgf8, and Xtbx6 gene expression after removing the anterior part of embryos. wnt3a, wnt5b, notch1, and fgf8 expressions were significantly reduced in gpx3 MO-injected embryos. * p < 0.05; ** p < 0.001; *** p < 0.0001; ns, not significant. (C) WISH analysis of gpx3 morphant embryos using Xbra and Xnot2 (chordoneural hinge and posterior wall markers). gpx3 knockdown did not affect the early tailbud gene expression. (D) RT-PCR of Xbra and Xnot2 relative expression revealed no significant differences between the control and gpx3 MO-injected embryos.
3.4. Gpx3 Knockdown Does Not Perturb Early Tailbud Gene Expression
Tailbud outgrowth is regulated by the expression of two gene sets, which are categorized as early and late genes [35,36]. Early genes (X-delta-1, Xlim1, Xbra, Xnot2, and Xcad3) are expressed in the future tailbud regions, while late genes (lunatic fringe, LFNG) are expressed at the time of tailbud extension, forming the axial tail tissues [37].
Because gpx3 knockdown disrupted normal tail development in Xenopus embryos, we speculated that gpx3 may be involved in the early tailbud gene regulation. To investigate gpx3’s functional role in tailbud development, we performed a loss of function experiment using the gpx3 MO and analyzed its effect on the expression of two tailbud early genes, Xbra and Xnot2. The T-box factor, Xbra, is expressed in the chordoneural hinge and the posterior wall, whereas the homeobox factor, Xnot2, is expressed in the ventral neural tube and the chordoneural hinge. Embryos at the 4-cell stage were ventrally injected with a gpx3 MO, and WISH was used to evaluate Xbra and Xnot2 tailbud expression. Surprisingly, the WISH analysis showed that gpx3 knockdown did not affect Xbra and Xnot2 tailbud expression. The expression of both tailbud markers was the same in control embryos and gpx3 morphant embryos, and no significant physical alterations were observed (Figure 2C). To confirm the WISH data, we performed RT-PCR using primers specific for Xbra and Xnot2. The RT-PCR results demonstrated that there were no appreciable expression differences for Xbra and Xnot2 in gpx3 MO-injected embryos (Figure 2D), consistent with WISH data. Taken together, these findings suggest that gpx3 depletion had no functional significance in regulating early tailbud gene expression.
3.5. Gpx3 Regulates ROS Level in the Tailbud Region
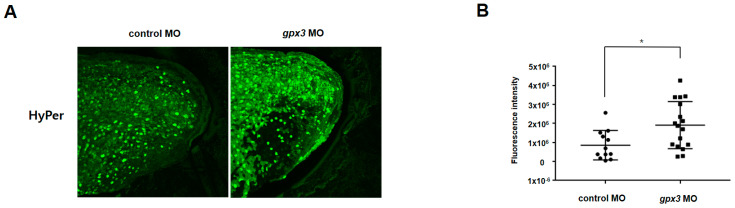
To assess gpx3’s antioxidant function, we microinjected HyPer mRNA into 1-cell staged embryos and subsequently microinjected gpx3 MO into the embryos once at the 4-cell stage. Compared to control embryos, gpx3 knockdown embryos showed increased HyPer fluorescent intensity, representing increased H2O2 levels related to ROS (Figure 3A,B). These results demonstrate that gpx3 has an antioxidant function, and gpx3 disruption in the tailbud region leads to ROS accumulation with post-anal tail defects.
Figure 3.
Perturbed gpx3 leads to increased reactive oxygen species (ROS) levels. Embryos were injected with 10 ng of HyPer mRNA at the 1-cell stage. MOs were injected into the ventral regions at the 4-cell stage. (A) gpx3 MO-injected embryos show increased HyPer fluorescent compared to the control. (B) HyPer fluorescence intensity quantification using ImageJ. * p < 0.05.
3.6. Gpx3 Is Required for Regulating Programmed Cell Death during Xenopus Embryogenesis
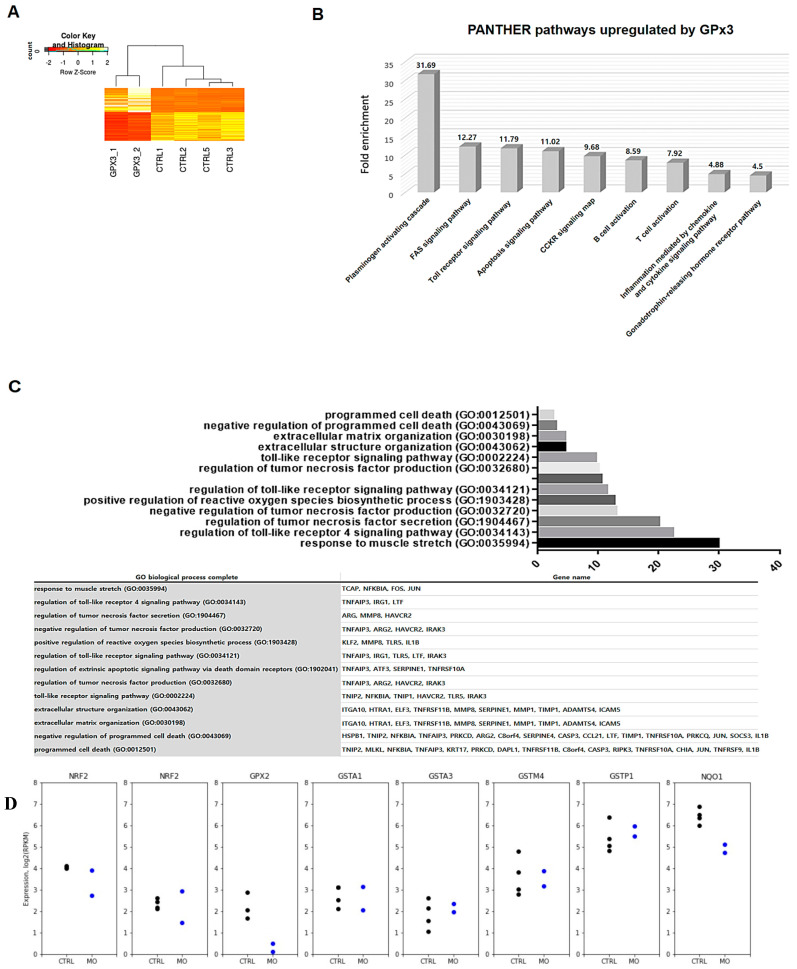
We performed transcriptome analysis of gpx3-depleted embryos to identify specific genes that are regulated by gpx3 during Xenopus embryonic development. Total RNA from gpx3 morphants was extracted and processed for transcriptome analysis. Our transcriptome analysis revealed that there were significant differences in the overall transcript expression between the control and the gpx3 MO-injected embryos (Figure 4A). gpx3 knockdown led to upregulation in genes related to apoptosis signaling, toll receptor signaling, and Fas signaling pathways (Figure 4B,C). Thus, our transcriptome analysis suggests that the shortened tail in gpx3 morphants is the result of cell death in the posterior regions.
Figure 4.
Effects of gpx3 knockdown on the Xenopus embryo transcriptome. (A) Heat map showing significant differences between control and gpx3 MO-injected embryos. (B) Transcriptome enrichment analysis using PANTHER indicated that cell death processes are significantly upregulated in gpx3-depleted embryos. (C) Gene ontology (GO-terms) and genes also indicated that genes associated with apoptosis and cell-death-related pathways were significantly affected by gpx3 depletion. (D) The expression of Nrf2 and its putative targets were not affected by gpx3 knockdown.
Since the knockdown of gpx3 is associated with increased level of ROS in morphant embryos, and enhanced ROS levels can lead to activation of Nrf2. Thus, we checked our RNA-seq data to see whether Nrf2 (Nfe2l2) and their putative targets were induced by gpx3 knockdown. However, expression levels of two duplicated genes (homologs) of nfe2l2 (nfe2l2.L and nfe2l2.S) were not changed significantly (Figure 4D). Further, we also checked the target of Nrf2 related to ROS, and they also did not show any change by depleting gpx3 (Figure 4D). Although Nrf2 regulates many ROS detoxifying genes, our transcriptomic data showed that the gpx3 knockdown might not activate Nrf2 regulatory network.
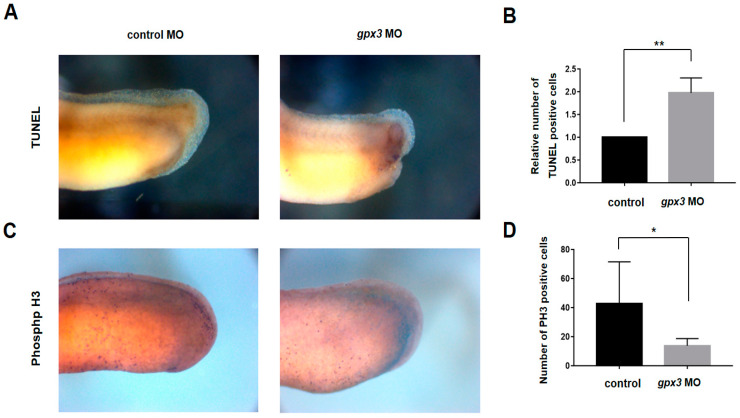
To verify gpx3’s role in cell death gene expression, a TUNEL assay and phospho-H3 staining were performed. In the gpx3 knockdown embryos, TUNEL-positive cells were significantly increased in the tailbud regions compared to control embryos (Figure 5A,B). In contrast, cell proliferation in the tailbud region was significantly decreased in gpx3 morphant embryos (Figure 5C,D). Together, our data suggest that gpx3 regulates tailbud development by regulating cell death and proliferation.
Figure 5.
gpx3 suppression activates apoptosis and inhibits cell proliferation in gpx3 morphants. (A) The expression and number of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells were higher in gpx3 knockdown embryos compared to control embryos. (B) Relative TUNEL expression, indicating a significant increase in apoptotic cells in gpx3 morphants. ** p < 0.001. (C) gpx3 loss resulted in reduced phospho-histone 3 (PH3)-positive cells in the tail region of gpx3 knockdown embryos. PH3-positive cells are indicated by the color purple, whereas β-gal staining is indicated by the color blue. (D) The relative PH3 expression showed cell proliferation was significantly reduced by gpx3 knockdown. * p < 0.05.
4. Discussion
GPx3 belongs to an important class of selenoproteins [8] and is among the strongest antioxidant enzymes by catalyzing peroxide reduction and free radicals through reduced glutathione to oxidized glutathione [17]. GPx3 is well known for its roles in tumor suppression, and increased GPx3 expression may also play a protective role in cardiomyocytes by ROS detoxification [15,17,22,38]. Studies have shown that reduced GPx3 expression is associated with liver and heart disease in humans [24,39]. However, GPx3’s role in embryonic development and ontogenesis has not yet been elucidated.
In this study, we systematically assessed gpx3’s spatiotemporal expression pattern during embryonic development in Xenopus. RT-PCR analysis revealed that gpx3 is expressed at the late blastula stage, with the highest expression at the early tailbud stage (Figure 1A). The spatial expression pattern showed that gpx3 is localized at the tail region during Xenopus embryogenesis (Figure 1B). gpx3’s involvement in tailbud development was verified by gpx3 MO microinjection into the embryo ventral blastomere at the 4-cell stage. gpx3 depletion led to a shortened post-anal tail compared to control embryos (Figure 1C,D). To our surprise, gpx3 depletion did not affect early tailbud gene expression required for tailbud development (Figure 2C,D). Instead, gpx3 ablation perturbed gene expression associated with signaling pathways involved in tailbud outgrowth during Xenopus embryonic development (Figure 2A,B). These data suggest that gpx3 is associated with signaling pathways that are required for proper embryogenesis.
Cell death and renewal are indispensable processes in multicellular organisms and are required for proper tissue and organ development [40]. Apoptosis occurs in multicellular organisms and is required for removing abnormal cells and aids in internal environment stability and multiple system development [41]. GPx3 induces apoptosis in cardiomyocytes, and excessive ROS levels can lead to apoptotic induction through caspase-3 activation [38,41]. In this study, we found that gpx3 suppression results in increased ROS levels (Figure 3) and gene expression associated with apoptosis signaling, Fas signaling, and toll receptor signaling. These findings suggest that gpx3 regulates embryonic development by controlling ROS levels and cell death processes (Figure 4). To further verify these findings, we performed TUNEL and PH3 staining to detect the gpx3 effect on cell death and cell proliferation. The cell death rate was significantly increased, and cell proliferation was decreased in gpx3 knockdown embryos (Figure 5).
5. Conclusions
In conclusion, apoptosis and cell proliferation are fundamental physiological processes for embryonic development. gpx3 knockdown perturbs proper tailbud development by activating cell death and inhibiting cell proliferation. However, further work is needed to determine how gpx3 regulates Wnt, BMP, and Notch signaling pathways. Additionally, further investigation is required to identify the regulatory mechanisms involved in gpx3-associated cell death during vertebrate embryonic development, as our data confirmed that gpx3 knockdown is not associated with activation of Nrf2 regulatory network.
Author Contributions
Conceptualization, H.-S.L. and T.K.; methodology, H.-Y.R. and D.-S.L.; formal analysis, H.L., T.I., and Y.K.; investigation, T.K.K., H.-S.L.; writing—original draft preparation, T.I., H.L., and S.C.; writing—review and editing, H.-S.L., T.J.P.; visualization, H.L.,T.K.K., and H.-S.L.; supervision, H.-S.L., T.K., and H.-Y.R.; funding acquisition, T.K., and H.-S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (Grant No. 2020R1A4A1018280 & Grant No. 2020R1F1A1048235), and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2018R1A6A1A03025810) The Republic of Korea.
Conflicts of Interest
The authors declared no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ismail T., Kim Y., Lee H., Lee D.S., Lee H.S. Interplay between mitochondrial peroxiredoxins and ROS in cancer development and progression. Int. J. Mol. Sci. 2019;20:4407. doi: 10.3390/ijms20184407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brieger K., Schiavone S., Miller F.J., Jr., Krause K.H. Reactive oxygen species: From health to disease. Swiss Med. Wkly. 2012;142:w13659. doi: 10.4414/smw.2012.13659. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covarrubias L., Hernández-García D., Schnabel D., Salas-Vidal E., Castro-Obregón S. Function of reactive oxygen species during animal development: Passive or active? Dev. Biol. 2008;320:1–11. doi: 10.1016/j.ydbio.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 5.Hernández-García D., Wood C.D., Castro-Obregón S., Covarrubias L. Reactive oxygen species: A radical role in development? Free Radic. Biol. Med. 2010;49:130–143. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Sies H. Oxidative stress: Oxidants and antioxidants. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 7.Pisoschi A.M., Pop A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015;97:55–74. doi: 10.1016/j.ejmech.2015.04.040. [DOI] [PubMed] [Google Scholar]
- 8.Flohé L. The fairytale of the GSSG/GSH redox potential. Biochim. Biophys. Acta. 2013;1830:3139–3142. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Flohe L., Günzler W.A., Schock H.H. Glutathione peroxidase: A selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 10.Margis R., Dunand C., Teixeira F.K., Margis-Pinheiro M. Glutathione peroxidase family–an evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 11.Wendel A. Glutathione peroxidase. Methods Enzymol. 1981;77:325–333. doi: 10.1016/s0076-6879(81)77046-0. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., Leng J.Y., Gao F., Zhao Z.A., Deng W.B., Liang X.H., Zhang Y.J., Zhang Z.R., Li M., Sha A.G., et al. Differential expression and anti-oxidant function of glutathione peroxidase 3 in mouse uterus during decidualization. FEBS Lett. 2014;588:1580–1589. doi: 10.1016/j.febslet.2014.02.043. [DOI] [PubMed] [Google Scholar]
- 13.Schwaab V., Faure J., Dufaure J.P., Drevet J.R. GPx3: The plasma-type glutathione peroxidase is expressed under androgenic control in the mouse epididymis and vas deferens. Mol. Reprod. Dev. 1998;51:362–372. doi: 10.1002/(SICI)1098-2795(199812)51:4<362::AID-MRD2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Kho C.W., Lee P.Y., Bae K.H., Kang S., Cho S., Lee D.H., Sun C.H., Yi G.S., Park B.C., Park S.G. Gpx3-dependent responses against oxidative stress in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 2008;18:270–282. [PubMed] [Google Scholar]
- 15.Barrett C.W., Ning W., Chen X., Smith J.J., Washington M.K., Hill K.E., Coburn L.A., Peek R.M., Chaturvedi R., Wilson K.T., et al. Tumor suppressor function of the plasma glutathione peroxidase gpx3 in colitis-associated carcinoma. Cancer Res. 2013;73:1245–1255. doi: 10.1158/0008-5472.CAN-12-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang S.-N., Lee J.M., Oh H., Park J.H. Glutathione peroxidase 3 inhibits prostate tumorigenesis in TRAMP mice. Prostate. 2016;76:1387–1398. doi: 10.1002/pros.23223. [DOI] [PubMed] [Google Scholar]
- 17.An B.C., Choi Y.D., Oh I.J., Kim J.H., Park J.I., Lee S.W. GPx3-mediated redox signaling arrests the cell cycle and acts as a tumor suppressor in lung cancer cell lines. PLoS ONE. 2018;13:e0204170. doi: 10.1371/journal.pone.0204170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai M., Sikong Y., Wang Q., Zhu S., Pang F., Cui X. Gpx3 prevents migration and invasion in gastric cancer by targeting NFKB/Wnt5a/JNK signaling. Int. J. Clin. Exp. Pathol. 2019;12:1194–1203. [PMC free article] [PubMed] [Google Scholar]
- 19.Worley B.L., Kim Y.S., Mardini J., Zaman R., Leon K.E., Vallur P.G., Nduwumwami A., Warrick J.I., Timmins P.F., Kesterson J.P., et al. GPx3 supports ovarian cancer progression by manipulating the extracellular redox environment. Redox Biol. 2019;25:101051. doi: 10.1016/j.redox.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H., Li J., Li X., Han C., Zhang Y., Zheng L., Guo M. Silencing GPX3 Expression promotes tumor metastasis in human thyroid cancer. Curr. Protein Pept. Sci. 2015;16:316–321. doi: 10.2174/138920371604150429154840. [DOI] [PubMed] [Google Scholar]
- 21.Zhou C., Pan R., Li B., Huang T., Zhao J., Ying J., Duan S. GPX3 hypermethylation in gastric cancer and its prognostic value in patients aged over 60. Future Oncol. 2019;15:1279–1289. doi: 10.2217/fon-2018-0674. [DOI] [PubMed] [Google Scholar]
- 22.Buijsse B., Lee D.H., Steffen L., Erickson R.R., Luepker R.V., Jacobs D.R., Jr., Holtzman J.L. Low serum glutathione peroxidase activity is associated with increased cardiovascular mortality in individuals with low HDLc’s. PLoS ONE. 2012;7:e38901. doi: 10.1371/journal.pone.0038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao S., Yan B., Lu Y., Zhang G., Li J., Zhai W., Guo W., Zhang S. Methylation of promoter and expression silencing of GPX3 gene in hepatocellular carcinoma tissue. Clin. Res. Hepatol. Gastroenterol. 2015;39:198–204. doi: 10.1016/j.clinre.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Kanno S.I., Tomizawa A., Yomogida S., Hara A. Glutathione peroxidase 3 is a protective factor against acetaminophen-induced hepatotoxicity in vivo and in vitro. Int. J. Mol. Med. 2017;40:748–754. doi: 10.3892/ijmm.2017.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chae S., Lee H.K., Kim Y.K., Jung Sim H., Ji Y., Kim C., Ismail T., Park J.W., Kwon O.S., Kang B.S., et al. Peroxiredoxin1, a novel regulator of pronephros development, influences retinoic acid and Wnt signaling by controlling ROS levels. Sci. Rep. 2017;7:8874. doi: 10.1038/s41598-017-09262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y., Jeong Y., Kwon K., Ismail T., Lee H.K., Kim C., Park J.W., Kwon O.S., Kang B.S., Lee D.S., et al. Physiological effects of KDM5C on neural crest migration and eye formation during vertebrate development. Epigenetics Chromatin. 2018;11:72. doi: 10.1186/s13072-018-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura Y., Feng Q., Kumagai T., Torikai K., Ohigashi H., Osawa T., Noguchi N., Niki E., Uchida K. Ebselen, a glutathione peroxidase mimetic seleno-organic compound, as a multifunctional antioxidant. Implication for inflammation-associated carcinogenesis. J. Biol. Chem. 2002;277:2687–2694. doi: 10.1074/jbc.M109641200. [DOI] [PubMed] [Google Scholar]
- 30.Beck C.W. Development of the vertebrate tailbud. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:33–44. doi: 10.1002/wdev.163. [DOI] [PubMed] [Google Scholar]
- 31.Beck C.W., Slack J.M. A developmental pathway controlling outgrowth of the Xenopus tail bud. Development. 1999;126:1611–1620. doi: 10.1242/dev.126.8.1611. [DOI] [PubMed] [Google Scholar]
- 32.Beck C.W., Slack J.M. Notch is required for outgrowth of the Xenopus tail bud. Int. J. Dev. Biol. 2002;46:255–258. doi: 10.1387/ijdb.011489. [DOI] [PubMed] [Google Scholar]
- 33.Beck C.W., Whitman M., Slack J.M.W. The role of BMP signaling in outgrowth and patterning of the Xenopus tail bud. Dev. Biol. 2001;238:303–314. doi: 10.1006/dbio.2001.0407. [DOI] [PubMed] [Google Scholar]
- 34.Lou X., Fang P., Li S., Hu R.-Y., Kuerner K.M., Steinbeisser H., Ding X. Xenopus Tbx6 mediates posterior patterning via activation of Wnt and FGF signalling. Cell Res. 2006;16:771–779. doi: 10.1038/sj.cr.7310093. [DOI] [PubMed] [Google Scholar]
- 35.Davis R.L., Kirschner M.W. The fate of cells in the tailbud of Xenopus laevis. Development. 2000;127:255–267. doi: 10.1242/dev.127.2.255. [DOI] [PubMed] [Google Scholar]
- 36.Gont L.K., Steinbeisser H., Blumberg B., de Robertis E.M. Tail formation as a continuation of gastrulation: The multiple cell populations of the Xenopus tailbud derive from the late blastopore lip. Development. 1993;119:991–1004. doi: 10.1242/dev.119.4.991. [DOI] [PubMed] [Google Scholar]
- 37.Ho D.M., Yeo C.Y., Whitman M. The role and regulation of GDF11 in Smad2 activation during tailbud formation in the Xenopus embryo. Mech. Dev. 2010;127:485–495. doi: 10.1016/j.mod.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong Y., Yang J., Cai J., Liu Q., Zhang J.m., Zhang Z. Effect of Gpx3 gene silencing by siRNA on apoptosis and autophagy in chicken cardiomyocytes. J. Cell. Physiol. 2019;234:7828–7838. doi: 10.1002/jcp.27842. [DOI] [PubMed] [Google Scholar]
- 39.Pastori D., Pignatelli P., Farcomeni A., Menichelli D., Nocella C., Carnevale R., Violi F. Aging-related decline of glutathione peroxidase 3 and risk of cardiovascular events in patients with atrial fibrillation. J. Am. Heart Assoc. 2016;5 doi: 10.1161/JAHA.116.003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmore S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi M., Honda T., Proske R.J., Yeh E.T. Regulation of reactive oxygen species-induced apoptosis and necrosis by caspase 3-like proteases. Oncogene. 1998;17:2753–2760. doi: 10.1038/sj.onc.1202211. [DOI] [PubMed] [Google Scholar]