Abstract
Background
Patients with glioblastoma (GBM) have a dismal prognosis, and there is an unmet need for new therapeutic options. This study aims to identify new therapeutic targets in GBM.
Methods
mRNA expression data of patient-derived GBM (n = 1279) and normal brain tissue (n = 46) samples were collected from Gene Expression Omnibus and The Cancer Genome Atlas. Functional genomic mRNA profiling was applied to capture the downstream effects of genomic alterations on gene expression levels. Next, a class comparison between GBM and normal brain tissue was performed. Significantly upregulated genes in GBM were further prioritized based on (1) known interactions with antineoplastic drugs, (2) current drug development status in humans, and (3) association with biologic pathways known to be involved in GBM. Antineoplastic agents against prioritized targets were validated in vitro and in vivo.
Results
We identified 712 significantly upregulated genes in GBM compared to normal brain tissue, of which 27 have a known interaction with antineoplastic agents. Seventeen of the 27 genes, including EGFR and VEGFA, have been clinically evaluated in GBM with limited efficacy. For the remaining 10 genes, RRM2, MAPK9 (JNK2, SAPK1a), and XIAP play a role in GBM development. We demonstrated for the MAPK9 inhibitor RGB-286638 a viability loss in multiple GBM cell culture models. Although no overall survival benefit was observed in vivo, there were indications that RGB-286638 may delay tumor growth.
Conclusions
The MAPK9 inhibitor RGB-286638 showed promising in vitro results. Furthermore, in vivo target engagement studies and combination therapies with this compound warrant further exploration.
Keywords: glioblastoma, MAPK9, RRM2, therapeutic targets, XIAP
Key Points.
There is an unmet need for novel therapeutic targets in glioblastoma (GBM).
We identified RRM2, MAPK9, and XIAP as novel potential therapeutic targets in GBM.
The MAPK9 inhibitor RGB-286638 shows promising in vitro results.
Importance of the Study.
Despite optimal first-line treatment of newly diagnosed glioblastoma (GBM), recurrence is inevitable and universally results in death in most cases. Therefore, there is a need for more effective treatment strategies and new therapeutic targets. Here, we applied functional genomic mRNA (FGmRNA) profiling, a method that corrects gene expression profiles for physiological and experimental factors irrelevant to the observed tumor phenotype, on publicly available microarray expression data of patient-derived GBM samples and normal brain tissue samples. Based on the class comparison of FGmRNA profiles, known interactions with antineoplastic drugs and the association with pathways involved in GBM carcinogenesis, we identified RRM2, MAPK9, and XIAP as potential therapeutic targets in GBM. Out of the available drugs targeting RRM2, MAPK9, and XIAP, the MAPK9 inhibitor RGB-286638 showed promising in vitro results warranting further exploration.
Glioblastoma (GBM) is the most common and aggressive primary brain tumor in adults. Currently, the standard first-line treatment for patients with newly diagnosed GBM consists of maximal surgical resection followed by postoperative radiation with concomitant and adjuvant temozolomide therapy. A randomized phase 3 trial in GBM patients, who had completed standard chemoradiotherapy, reported that adding tumor treating fields to maintenance temozolomide chemotherapy prolonged progression-free survival (7.1 months) and overall survival (OS; 20.5 months) as compared to controls (4.0 and 15.6 months, respectively).1 Unfortunately, despite optimal first-line treatment, recurrence is still almost inevitable. The prognosis of these patients remains poor with a median survival of 12–20 months.2,3
At the time of recurrence, treatment options are limited due to limitations in the use of surgery and re-irradiation, as well as the limited efficacy of systemic treatment.1,4–6 In the past decades, research focused on the molecular genetic profiles of GBM to provide insights into the pathogenesis of GBM and the tenacious resistance to conventional and targeted therapies. The Cancer Genome Atlas Research Network (TCGA) made a significant contribution and performed a comprehensive genomic and transcriptomic analysis on 206 GBM samples. They demonstrated relevant genomic alterations in the p53, retinoblastoma (Rb), and receptor tyrosine kinase (RTK)/phosphoinositide 3-kinase (PI3k) signaling pathways.7 Furthermore, unsupervised hierarchical clustering analysis of the TCGA GBM expression data linked transcriptomic alterations on the mRNA level with distinct molecular subtypes of GBM, which were confirmed on the single-cell transcriptomic level.8–10 Collectively, these data helped to identify frequently amplified genes in GBM, including EGFR, PDGFRA, MET, CDK4, and PIK3CA, and commonly deleted genes, such as PTEN and RB1.7,11,12 Genomic alterations can translate into downstream effects, such as changes in protein structures (with gain or loss of function) or changes of gene expression levels (with activation or inactivation of a gene or pathway).13 Therefore, genomic alterations (eg, somatic copy number alterations [SCNAs]) hold valuable information on the biological behavior of GBM, its resistance mechanisms to conventional therapy, and possible new therapeutic targets.
The method of functional genomic mRNA (FGmRNA) profiling demonstrated that the expression level of all genes can be affected by SCNAs. However, this effect is often subtle and obscured mainly by major, non-genetic factors (eg, physiological, metabolic, and experimental factors). FGmRNA profiling is capable of correcting gene expression data for these factors, resulting in a residual gene expression signal that highly correlates with SCNAs.14 Thus, FGmRNA profiling is capable of capturing the downstream effects of genomic alterations on gene expression levels.
We hypothesize that FGmRNA profiling of publicly available, raw microarray expression data of patient-derived GBM samples and normal brain tissue harbors valuable new insights on the downstream effects of genomic alterations in GBM. Therefore, this proof of concept study used FGmRNA profiling, followed by prioritization, to identify highly expressed genes in GBM with known drug interactions that could serve as new potential therapeutic targets. Subsequently, we investigated the preclinical antitumor activity of targeted agents directed against these potential therapeutic targets in GBM.
Materials and Methods
Detailed methods information is provided in Supplementary Methods.
Data Acquisition
We collected publicly available raw microarray expression data from the Gene Expression Omnibus (GEO). We obtained gene expression data from GEO for samples that were processed on the HG-U133A (GPL96) and HG-U133 plus 2.0 (GPL570) Affymetrix platforms. Simple Omnibus Format Text (SOFT) files were downloaded for both platforms. These SOFT files contain information on the samples as provided by the investigator who uploaded the data to GEO. To identify GBM samples, we first applied automated filtering with GBM-related keywords on the SOFT files (Supplementary Table S1). This search strategy was aimed at sensitivity to minimize the chance of missing relevant samples. Therefore, manual curation was necessary to remove all non-relevant and false-positive samples. Cell lines, cultured samples, and postmortem or animal tissues were excluded. In addition, we collected raw gene expression data from the TCGA GBM multiforme data set. These data were generated with the Affymetrix HT HG-U133A and were integrated with the GEO data set. Preprocessing and aggregation of raw data were performed according to the robust multi-array average algorithm. Quality control of the resulting expression data was executed, as previously described.15
Class Comparison
FGmRNA profiling was used to capture the downstream effect of genomic alterations at gene expression levels. For a detailed description of FGmRNA profiling, we refer to the work of Fehrmann et al.14 We used the FGmRNA profiles of healthy brain tissue and GBM tissue to perform a genome-wide class comparison analysis. A Welch’s T-test was used to identify genes with differential FGmRNA expression. To assess the degree of multiple testing, we performed our analysis within a multivariate permutation test (1.000 permutations) with a false discovery rate of 1% and a confidence level of 99%. This resulted in a list of significantly associated genes, which contains no more than 1% false positives.
Prioritization of Upregulated Genes
We manually curated the list of significantly upregulated genes to exclude duplicate results of multiple probes targeting the same gene, nonspecific probes mapping to multiple genes and probes that did not map to a known gene. Subsequently, we explored the resulting list of genes with the use of the Drug–Gene Interaction Database (DGIDb; http://www.dgidb.org/). The DGIDb integrates data of disease-relevant human genes, drugs, and proven or potential drug–gene interactions from 13 primary sources.16 This allowed us to select upregulated genes that interact with antineoplastic agents. We assessed all antineoplastic agents per gene with an additional PubMed and www.clinicaltrials.gov search to determine the mechanism of drug–gene interaction and the current drug development status in humans. Genes interacting with antineoplastic agents currently tested in various malignancies were prioritized. Furthermore, we assessed these prioritized genes with www.genecards.org and PubMed to identify their biological pathways. Ultimately, we selected the genes with a biological pathway involved with GBM carcinogenesis and a known interaction with antineoplastic agents tested in clinical cancer trials.
In Vitro Experiments: Cell Lines and Description of Proliferation Assay
The established GBM cell lines U87, U251, T98G, and U138 were acquired from the ATCC and were cultured in Dulbecco’s modified Eagle’s medium supplemented with 5% FBS. The HT29 colorectal cell line was included as a control. The GBM8 primary cell culture was kindly provided by Dr. Bakhos Tannous (Harvard/MGH). Glioblastoma sphere cultures (GSCs) were obtained from single patient surgical specimens at MD Anderson and the Amsterdam University Medical Centers, location VUmc.17 GBM8 and GSCs were cultured at 37°C in Neurobasal-A Medium (Life Technologies) and supplemented with N2 (Life Technologies), B27 without vitamin A (Life Technologies), Glutamax (Life Technologies), human EGF (Tebu Bio), human FGF basic (Tebu Bio), heparin (Leo Pharma), and penicillin/streptomycin (Sigma).18 Cells were certified mycoplasma free by regular testing http://www.microbiome.nl/. RGB-286638 (Bio-Connect) was dissolved in DMSO to prepare a 20 mM stock solution.
For the U87, U251, T98G, and U138 GBM cell lines, response to RGB-286638 was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay as described previously.19 In short, cells were seeded on a transparent flat-bottom 96-well plate in a density of 2000 cells per well and were allowed to adhere for 24 h. After 24 h of incubation, at = 0, the measurement was carried out, and cells were treated with 100 µL of drug solution according to a concentration dilution series ranging from 0 to 20 µM. Subsequently, after 72 h of treatment, 100 µL of MTT indicator dye (5 mg/mL) was added to the wells, and cells were incubated for 2 h at 37°C. After the addition of 100 µL of 10% SDS/0.01 M HCl to the wells, absorption was measured at 540 nm in a microplate reader. The reading from the wells with cells cultured in control medium-plus DMSO was used as a 100% viability value. For GBM8 primary and GSC cell cultures, response to RGB-286638 was determined through CellTiter-Glo 3D assay to measure the ATP content of viable cells. Cells were plated at an optimal density (which varied between GSC lines) in 384-well plates 24 h prior to drug treatment. Subsequently, cells were exposed to a serial dilution of RGB-286638 for 72 h in triplicate. Cell viability was determined using CellTiter-Glo 3D (Promega). Relative light units (RLUs) were measured using the Tecan’s Connect microplate stacker, and RLUs were normalized against the DMSO controls.20 All experiments were performed in triplicate and repeated at least 2 times. Levels of response (ie, complete response, incomplete response, or resistance) were based on the viability at the highest concentration of RGB-286638 and the IC50. Thresholds were below or higher than 5% viability for complete versus incomplete responses and below or higher than an IC50 of 1 μM for incomplete responses versus resistance.
Orthotopic In Vivo Mouse Model
Female athymic nude-Fox1nu mice (age 6–8 weeks; Envigo) were maintained in accordance with animal welfare guidelines and regulations of the VU University in Amsterdam, The Netherlands. GBM8 cells, stably expressing Firefly luciferase and mCherry, were intracranially injected in a volume of 5 µL (0.5 × 106 cells) into the striatum, as previously described.18 Tumor engraftment and growth were determined by measuring the Firefly luciferase (Fluc) activity with a CCD camera after an intraperitoneal injection of 150 µL of d-luciferin (Gold Biotechnology). Fluc activity was measured 5 days after intracranial injection of GBM8 cells and subsequently 1–2 times a week during treatment and follow-up for a maximum total amount of 15 times. Mice with incomplete tumor engraftment (Fluc activity <104 RLU) were excluded from the experiment. Based on the Fluc activity, mice were subsequently stratified into 3 treatment groups of 7 animals each and were all treated for 5 consecutive days with (1) Vehicle (PBS: 200 µL/day), (2) the pan-CDK inhibitor Flavopiridol (5 mg/kg/day) as a control for CDK inhibition, or (3) RGB-286638 (40 mg/kg/day). The treatment dose for RGB-286638 (40 mg/kg/day) was based on the in vivo study of Cirstea et al.21 in a multiple myeloma mice model. RGB-286638 and Flavopiridol were administered intravenously by tail vein. Experiments were performed under ethical review permission (AVD114002017841) and are reported according to the ARRIVE guidelines.
Results
Sample Identification
Following automated filtering, manual curation, removal of duplicates, and quality control, mRNA expression data of 46 normal brain tissue samples and 1279 patient-derived GBM samples were included for further analysis. In-depth information on the GEO and TCGA samples and their corresponding citations are provided in Supplementary Table S2.
Class Comparison Between Normal Brain Tissue and Clinical GBM Samples
We identified 712 significantly upregulated and unique genes using a class comparison analysis between FGmRNA profiles of normal brain tissue samples and GBM samples (false discovery rate 1%, confidence level 99%). Detailed results are provided in Supplementary Table S3.
Prioritization of Druggable Genes
Of the 712 upregulated genes, 27 genes interacted with 116 antineoplastic drugs, according to the DGIDb (Table 1). Seventeen of the 27 druggable genes, including EGFR and VEGFA, were previously tested in clinical GBM trials and demonstrated limited efficacy (Supplementary Table S4). Therefore, these genes were excluded from further review. For the 10 remaining genes, 14 of the 20 interacting antineoplastic drugs are currently being tested in clinical trials for various cancers but have not been tested in clinical trials for GBM (Table 2). Drugs interacting with S1PR5 and ADAMTS5 are currently not being evaluated in patients with cancer according to www.clinicaltrials.gov and were therefore also excluded. For the remaining 8 genes (HRH1, TYK2, RRM2, MAPK9, PDK3, XIAP, NR3C1, and NCOA1), an additional literature search in Pubmed and http://www.genecards.org/ was performed, which identified RRM2, MAPK9, and XIAP as members of biological pathways that play an important role in the development of GBM (Figure 1).
Table 1.
Significantly Upregulated Genes Interacting With Antineoplastic Drugs
| Rank | Gene Symbol | Gene Title | Drug |
|---|---|---|---|
| 2 | EGFR | Epidermal growth factor receptor | Afatinib | Crizotinib | Dacomitinib | Erlotinib | Gefitinib | Icotinib | Lapatinib | Mubritinib | Neratinib | Pelitinib | Poziotinib | Vandetanib | Brigatinib | Canertinib | Rociletinib | Carboplatin | Cisplatinum | Paclitaxel | Sirolimus | Cetuximab | Panitumumab | Geldanamycin | AEE788 | AZD8931 | BIBX 1382 | BMS-599626 | BPIQ-I | CUDC-101 | PD 158780 | PD 174265 |
| 36 | HRH1 | Histamine receptor H1 | Tesmilifene | Tranilast |
| 59 | VEGFA | Vascular endothelial growth factor A | Apatinib | Axitinib | Bevacizumab | Bevasiranib | Brivanib | Cabozantinib | Cediranib | Dovitinib | Endostatin-(84–114)-NH2 | Golvatinib | Linifanib | Lenvatinib | Lenalidomide | Motesanib | Nintedanib | Pazopanib | Pegaptanib | Ponatinib | Ranibizumab | Regorafenib | Semaxanib | Sorafenib | Sunitinib | Thalidomide | Tivozanib | Telatinib | Vandetanib | Vatalanib | Ziv-Aflibercept | 4SC-202 | ABT-510 | AEE788 | AZD2932 | BMS-794833 | CYC116 | ENMD-2076 | KI8751 | KRN633 | LY2874455 | MGCD-265 | RAF265 | SKLB1002 | SU5402 | TAK-593 | ZM306416 | ZM323881 |
| 77 | GPER | G protein-coupled estrogen receptor 1 | Fulvestrant | Tamoxifen |
| 147 | PSMA2 | Proteasome subunit, alpha 2 | Bortezomib | Carfilzomib |
| 163 | TUBA3C | Tubulin, alpha 3c | CYT997 | Epothilone B |
| 232 | TYK2 | Tyrosine kinase 2 | AT9283 |
| 319 | RRM2 | Ribonucleotide reductase M2 | Gallium nitrate |
| 369 | MAPK9 | Mitogen-activated protein kinase 9 | RGB-286638 |
| 381 | MAPK1 | Mitogen-activated protein kinase 1 | Erlotinib | Purvalanol A |
| 399 | PDK3 | Pyruvate dehydrogenase kinase, isozyme 3 | CPI-613 |
| 417 | PSMC2 | Proteasome 26S subunit, ATPase 2 | Bortezomib | Carfilzomib | MLN9708 |
| 432 | DDR1 | Discoidin domain receptor tyrosine kinase 1 | Imatinib |
| 434 | SRD5A1 | Steroid-5-alpha-reductase, alpha polypeptide 1 | Finasteride |
| 440 | WEE1 | WEE1 G2 checkpoint kinase | MK-1775 |
| 450 | IL8 | Interleukin 8 | ABT-510 |
| 458 | PSMD2 | Proteasome 26S subunit, non-ATPase 2 | Bortezomib | Carfilzomib | Oprozomib |
| 470 | S1PR5 | Sphingosine-1-phosphate receptor 5 | Fingolimod |
| 498 | ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif 5 | Batimastat |
| 507 | CFLAR | CASP8 and FADD-like apoptosis regulator | Bicalutamide | Cabozantinib | Dovitinib | Nintedanib |
| 513 | CYP3A5 | Cytochrome P450 family 3 subfamily A member 5 | Erlotinib | Lovastatin |
| 540 | MMP14 | Matrix metallopeptidase 14 | Marimastat |
| 577 | XIAP | X-linked inhibitor of apoptosis | AT-406 | AZD5582 | Birinapant | GDC-0917 | GDC-0152 | LCL161 | SM-337 |
| 639 | NOTCH1 | Notch 1 | MK-0752 | RO4929097 |
| 661 | NR3C1 | Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) | Fluoxymesterone | Megestrol | Onapristone |
| 664 | CDK6 | Cyclin-dependent kinase 6 | Flavopiridol | LY2835219 | Palbociclib | RGB-286638 | Ribociclib |
| 708 | NCOA1 | Nuclear receptor coactivator 1 | Genistein |
Table 2.
Significantly Upregulated Genes and Interacting Antineoplastic Drugs Tested in Clinical Trials for Various Cancers
| Rank | Gene | Interacting Drug | Interaction | Status | Tumor Type(s) |
|---|---|---|---|---|---|
| 36 | HRH1 | Tesmilifene | Antagonist | Phase 2 | Breast cancer |
| Tranilast | Inhibitor | Phase 2 | Prostate cancer | ||
| 232 | TYK2 | AT9283 | Inhibitor | Phase 2 | Advanced solid tumors, ALL, AML, childhood solid neoplasms, CML, MDS, multiple myeloma, non-Hodgkin lymphoma |
| 319 | RRM2 | Gallium nitrate | Inhibitor | Phase 2 | Advanced solid tumors, cervical cancer, colorectal cancer, non-Hodgkin lymphoma, NSCLC, prostate cancer, SCLC, transitional cell carcinoma |
| 369 | MAPK9 | RGB-286638 | Inhibitor | Phase 1 | Advanced solid tumors |
| 399 | PDK3 | CPI-613 | N/A | Phase 2 | AML, colorectal cancer, MDS, multiple myeloma, non-Hodgkin lymphoma, SCLC |
| 470 | S1PR5 | Fingolimod | N/A | None | None |
| 498 | ADAMTS5 | Batimastat | N/A | None | None |
| 577 | XIAP | AT-406 | Antagonist | Phase 1 | Advanced solid tumors, AML |
| AZD5582 | Antagonist | None | None | ||
| Birinapant | Antagonist | Phase 2 | Advanced solid tumors, ALL, AML, MDS, ovarian cancer | ||
| GDC-0917 | Antagonist | Phase 1 | Advanced solid tumors | ||
| GDC-0152 | Inhibitor | Phase 1 | Advanced solid tumors, non-Hodgkin lymphoma | ||
| LCL161 | Inhibitor | Phase 2 | Advanced solid tumors, breast cancer | ||
| SM-337 | N/A | None | None | ||
| 661 | NR3C1 | Fluoxymesterone | Antagonist | Phase 2 | Breast cancer |
| Megesterol | Antagonist | Phase 3 | Breast cancer | ||
| Onapristone | Antagonist | Phase 2 | Breast cancer, prostate cancer | ||
| 708 | NCOA1 | Genistein | N/A | Phase 3 | Prostate cancer |
Figure 1.
Prioritization process of druggable genes.
The Potential Therapeutic Targets RRM2, MAPK9, and XIAP
Ribonucleotide reductase regulatory subunit M2 (RRM2) contributes to the upregulation of ribonucleotide reductase activity during the S phase of the cell cycle. It plays an essential role in regulating the total rate of DNA synthesis.22,23 The gene is implicated in temozolomide therapy resistance and is transcriptionally co-activated by BRCA1, protecting cells from endogenous replication stress, DNA damage, and apoptosis.24–26 Subsequently, in vitro and in vivo studies with inhibition of RRM2 expression showed a significant decrease in tumor growth in various tumors, including GBM, and improved animal survival.27
The protein encoded by mitogen-activated protein kinase 9 (MAPK9), also known as c-Jun N-terminal kinase 2 (JNK2), is involved in regulating various cellular processes, including cell growth, transformation, and apoptosis.28 This pathway can also be activated by growth factors, such as epidermal growth factor and platelet-derived growth factor.29–31 Targeted inhibition of MAPK9 with specific antisense oligonucleotides resulted in marked growth suppression in human GBM T98 cells, suggesting that MAPK9 inhibition could have therapeutic benefit.32
The X-linked inhibitor of apoptosis protein (XIAP) is a strong caspase-binding protein and inhibits both the intrinsic and extrinsic apoptosis pathways.33 Overexpression of XIAP has been linked to tumor recurrence in prostate cancer and resistance to systemic and targeted therapy in breast cancer cells.34–36 Interestingly, the second mitochondria-derived activator of caspases (Smac) mimetics, which neutralizes XIAP, can sensitize GBM cells for temozolomide and can prime them for apoptosis.37,38
Based on the known drug–gene interactions and the current status of clinical evaluation in patients, the prioritized agents of interest are gallium nitrate as an inhibitor of RRM2, RGB-286638, as an inhibitor of MAPK9, and AT-406, Birinapant, GDC-0152, GDC-0917, and LCL161 as inhibitors of XIAP. Subsequently, the preclinical efficacy of these drugs was evaluated in GBM.
The Sensitivity of GBM Cell Lines to Antineoplastic Agents Targeting MAPK9, RRM2, and XIAP
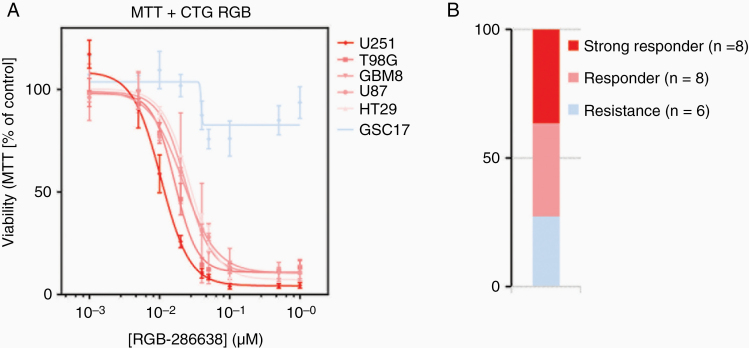
All cell lines, including the colorectal cell line HT29 as a control and the GBM8 primary cell culture, were exposed to various concentrations of the drugs of interest (ranging from 0 to 20 µM) for 72 h. Exceptionally, for gallium nitrate, higher concentrations up to 2 mM were necessary. Interestingly, low exposure to RGB-286638 demonstrated near-complete inhibition in all cell lines (IC50 ranging from 0.01 to 0.03 µM; Figure 2), compared to all the other drugs in which high concentrations resulted in no or limited inhibition (Supplementary Figure S1A–F). Additional analyses in GBM sphere culture models, which resemble GBM heterogeneity more accurate, showed that about one-third of the tumor models showed a similarly high sensitivity to RGB-286638, and around one-third showed an incomplete response resulting in 5–30% viability after exposure to 1 µM (Supplementary Figure S2). Therefore, based on its antitumor activity, RGB-286638 was selected for preclinical evaluation in an orthotopic in vivo model.
Figure 2.
(A) Viability assay of exposure to MAPK9 inhibitor RGB-286638 for 72 h and (B) the total number of GBM cell cultures responding or resistant to RGB-286638.
The Efficacy of RGB-286638 in an Orthotopic GBM8 Primary GBM In Vivo Model
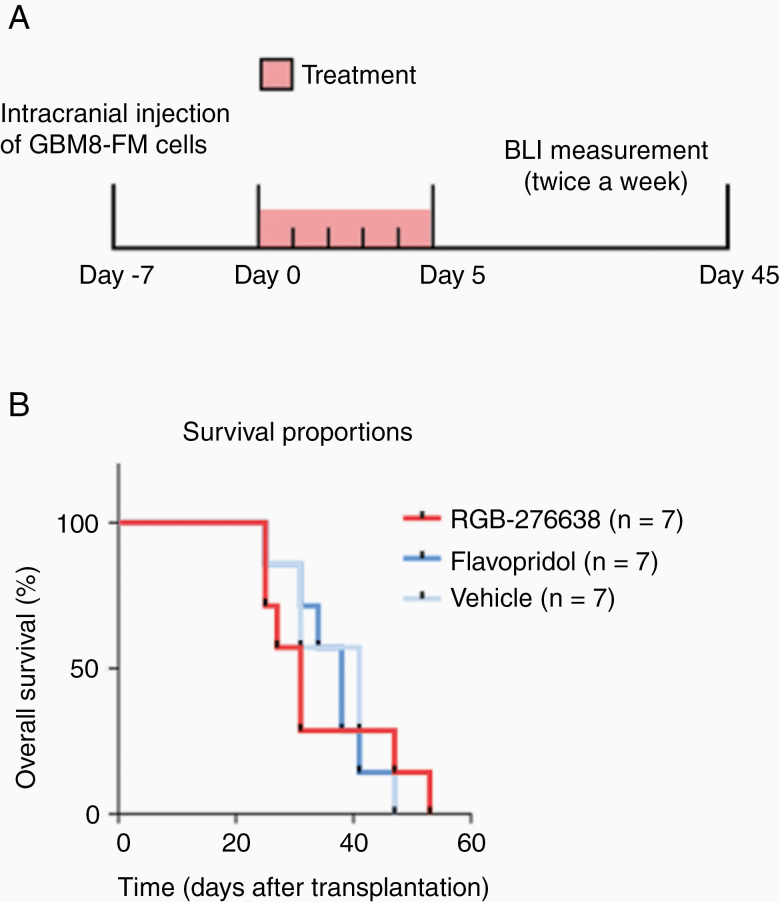
The antitumor efficacy of RGB-286638 in an orthotopic in vivo mouse model, using primary GBM8 cells (Figure 3A), was studied. As RGB-286638 is an inhibitor of MAPK9, but also inhibits multiple cyclin-dependent kinases (CDKs), the pan-CDK inhibitor Flavopiridol was used as an extra control group. Orthotopic growth was assessed with in vivo luminescence measurement, which showed a significantly lower signal in mice treated with RGB-286638 compared to the vehicle and Flavopiridol group (P = 5.5 × 10–8 and P = .01, respectively). Flavopiridol did not show significantly lower signals compared to the vehicle-treated animals. In comparison to the vehicle control, no significant difference in median OS was observed for the animals treated with RGB-286638 (47 vs 53 days, respectively, P = .93; Figure 3B). These combined results indicate that RGB-286638 may delay tumor growth, possibly by its MAPK9 and CDK inhibitory properties. Overall, all treatments were well tolerated based on the animals’ conditions and weight during the follow-up period. However, some of the animals treated with RGB-286638 developed local skin lesions on their tail as a possible reaction to the administration via tail vein injection. The skin lesions in these mice recovered successfully after the topical administration of an antibacterial ointment. Therefore, the endpoint of this animal study was not limited by toxicity.
Figure 3.
(A) Schematic overview of the in vivo experiment and (B) median overall survival in days of each treatment group.
Discussion
We applied FGmRNA profiling to correct gene expression data for the effect of non-genetic and experimental factors on gene expression levels, followed by data-driven prioritization to identify RRM2, MAPK9, and XIAP as new potential therapeutic targets in GBM. The preclinical efficacy of several clinical available compounds targeting RRM2, MAPK9, and XIAP was studied in GBM.
FGmRNA profiling is a method that corrects gene expression data for major, non-genetic factors (eg, physiological, metabolic, cell-type-specific, and experimental factors). The FGmRNA profiles enabled us to capture the downstream effect of SCNAs at gene expression levels. With this innovative method, we discovered MAPK9 as a potential therapeutic target. Subsequently, we used RGB-286638, a small molecule to target MAPK9. As with most small-molecule kinase inhibitors, this compound has multiple targets, including the family of CDKs.21,39 Interestingly, low concentrations of RGB-286638 (ie, 100 nM) showed near-complete inhibition of viability in all classical GBM cell lines and approximately one-third of all primary GSC cultures. RGB-286638 has been clinically evaluated in a phase I study in patients with advanced solid tumors.40 In this phase I study, RGB-286638 was well tolerated in a dose of 120 mg/day for 5 consecutive days every 28 days. Furthermore, prolonged disease stabilization, ranging from 2 to 14 months, was seen across the different dose levels. Unfortunately, although RGB-286638 showed a consistent decrease in tumor growth, as determined by the luciferase signal in our orthotropic GBM8 mouse model, it did not result in a better OS.
We also investigated gallium nitrate, a simple gallium salt used for the treatment of cancer-related hypercalcemia, interacts with the iron-dependent M2 subunit of ribonucleotide reductase (RRM2) and thereby inhibits DNA synthesis.41 Interestingly, gallium nitrate has demonstrated antineoplastic activity in various cancers, such as advanced bladder cancer and non-Hodgkin’s lymphoma.42–44 So far, experiments with gallium nitrate showed inconsistent results in 2 GBM cell lines.45 Here, we demonstrated that high concentrations of up to 2 mM of gallium nitrate result in a heterogeneous and poor response in all GBM cell lines, including the GBM8 primary cell culture.
Lastly, exposure to the XIAP inhibitors AT-406, birinapant, GDC-0152, GDC-0917, and LCL161 showed limited efficacy in the GBM cell lines. This is in contrast to 2 previous preclinical studies. Tchoghandjian et al.46 showed that in vitro and in vivo targeting of IAP with GDC-0152 triggered apoptosis in multiple GBM cell lines and improved the outcome in GBM-bearing mice. Furthermore, a comprehensive preclinical study of Zakaria et al.47 with birinapant in various GBM cell lines, as a single agent or combined with temozolomide, showed remarkable differences in treatment responses. However, in line with our results, the preclinical study of Houghton et al.48 with the Smac mimetic LCL161 demonstrated limited in vitro efficacy, but significant growth delay in a GBM xenograft. These conflicting results in which in vivo responses were better than the in vitro efficacy may demonstrate the critical effect of the tumor microenvironment on therapeutic responses in GBM or could indicate that insufficient target engagement was reached in the tumor.47,49
Over the past decades, progress in the improvement of the treatment and survival of patients with GBM has been frustratingly slow due to multiple factors, including the blood–brain barrier, intra- and intertumoral heterogeneity, and the tumor microenvironment. As with most drugs tested in GBM, it is possible that insufficient intracranial concentrations of the antineoplastic agents are reached due to the inadequate delivery across the blood–brain barrier.50 Unfavorable physicochemical properties for blood–brain barrier transfer are the relatively large size (molecular weight = 545.64; 8 rotational bonds) as well as the charge (3 hydrogen bond donors) of RGB-286638.51 This could explain our conflicting preclinical results with RGB-286638, in which in vitro responses were more promising than the in vivo efficacy. Therefore, RGB-286638 might be subjected to drug discovery to overcome these unfavorable characteristics.
Furthermore, exposure to these drugs may sensitize GBM cells for treatment with temozolomide or radiotherapy. For instance, Wagner et al.38 showed that combinational treatment with the Smac mimetic BV6 and temozolomide synergistically reduces cell viability and triggers apoptosis in GBM cells. Similarly, RRM2 inhibition could sensitize cells to temozolomide chemotherapy.25,26 Lastly, colleagues have recently demonstrated that the MAPK-targeting agent MEK162 was found to enhance the effect of radiotherapy on GBM cells in their in vitro and in vivo GBM model.18 Based on the results of these studies, the efficacy of the antineoplastic drugs targeting RRM2, XIAP, and MAPK9 combined with temozolomide and/or radiotherapy still warrants further investigation.
An important aspect of our work is that we used publicly available gene expression data collected from GEO and TCGA. A significant advantage of this approach is that these databases contain large amounts of data, which have yet to be fully explored in all their details. Therefore, the use and reanalysis of publicly available gene expression data may lead to novel insights and discoveries. Importantly, FGmRNA profiling was applied on mRNA expression data, which should be carefully interpreted, since the FGmRNA expression levels of genes might not always be strongly correlated with the corresponding protein expression levels. For instance, protein levels could be lower due to various cellular processes, such as a high turn-over. Furthermore, it is difficult to distinguish the effect of target gene overexpression in GBM cells from the impact of target gene overexpression in surrounding nontumor cells. To minimalize this effect caused by surrounding nontumor cells, we used sufficient normal brain tissue samples as a reference and created a threshold for overexpression.
In conclusion, with our innovative method of FGmRNA profiling followed by data-driven prioritization, we identified RRM2, MAPK9, and XIAP as potential therapeutic targets for GBM. The MAPK9 inhibitor RGB-286638 showed promising in vitro results and warrants further investigation.
Supplementary Material
Acknowledgment
Presented in the Poster Discussion Session at the ASCO Annual Meeting 2017.
Funding
This project was partially supported by the Dutch Cancer Society grants KWF-4874 and KWF-11038 and Brain Tumour Charity grant 488097.
Conflict of interest statement. A.W. received research funding (outside this work) from Novartis and Ipsen. H.V. is a member of the advisory board of Erbitux (Merck). H.V. also received honoraria from Boehringer Ingelheim and Roche for his consultancy/advisory work. H.V. received research funding (outside this work) from Amgen, Vitromics Healthcare, Immunovo BV, Roche, and Novartis. The other authors declare no disclosures.
Authorship Statement. All authors were involved in the writing of the manuscript at draft and any revision stages and have read and approved the final version. Conceptualization, C.B., M.L., H.V., A.W., and R.F.; methodology, C.B., U.A., M.H., T.L., B.W., and R.F.; formal analysis, C.B., U.A., M.H., B.W., and R.F.; resources, C.B., U.A., M.H., T.L., B.W., and R.F.; data collection, C.B., U.A., M.H., T.L., B.W., and R.F; writing—original draft preparation, C.B., U.A, M.H., H.V., B.W., A.W., and R.F.; writing—review and editing, C.B., U.A., M.H., M.L., T.L., H.V., B.W., A.W., and R.F.; supervision, H.V., B.W., A.W., and R.F.
References
- 1. Stupp R, Taillibert S, Kanner AA, et al. Maintenance therapy with tumor-treating fields plus temozolomide vs temozolomide alone for glioblastoma: a randomized clinical trial. JAMA. 2015;314(23):2535–2543. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups ; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 4. Almeida JP, Chaichana KL, Rincon-Torroella J, Quinones-Hinojosa A. The value of extent of resection of glioblastomas: clinical evidence and current approach. Curr Neurol Neurosci Rep. 2015;15(2):517. [DOI] [PubMed] [Google Scholar]
- 5. Romanelli P, Conti A, Pontoriero A, et al. Role of stereotactic radiosurgery and fractionated stereotactic radiotherapy for the treatment of recurrent glioblastoma multiforme. Neurosurg Focus. 2009;27(6):E8. [DOI] [PubMed] [Google Scholar]
- 6. De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol. 2010;12(3):304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fehrmann RS, Karjalainen JM, Krajewska M, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47(2):115–125. [DOI] [PubMed] [Google Scholar]
- 15. Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of tumorinfiltrating immune cell composition and functionality for disease outcome in breast cancer. J Natl Cancer Inst. 2017;109(1):djw192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10(12):1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Narayan RS, Gasol A, Slangen PLG, et al. Identification of MEK162 as a radiosensitizer for the treatment of glioblastoma. Mol Cancer Ther. 2018;17(2):347–354. [DOI] [PubMed] [Google Scholar]
- 19. Rovithi M, de Haas RR, Honeywell RJ, et al. Alternative scheduling of pulsatile, high dose sunitinib efficiently suppresses tumor growth. J Exp Clin Cancer Res. 2016;35(1):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narayan RS, Fedrigo CA, Brands E, et al. The allosteric AKT inhibitor MK2206 shows a synergistic interaction with chemotherapy and radiotherapy in glioblastoma spheroid cultures. BMC Cancer. 2017;17(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cirstea D, Hideshima T, Santo L, et al. Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013;27(12):2366–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Engström Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260(16):9114–9116. [PubMed] [Google Scholar]
- 23. Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63(1):22–34. [DOI] [PubMed] [Google Scholar]
- 24. Rasmussen RD, Gajjar MK, Tuckova L, et al. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun. 2016;7:13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuckerman JE, Hsueh T, Koya RC, Davis ME, Ribas A. siRNA knockdown of ribonucleotide reductase inhibits melanoma cell line proliferation alone or synergistically with temozolomide. J Invest Dermatol. 2011;131(2):453–460. [DOI] [PubMed] [Google Scholar]
- 26. Teng J, Hejazi S, Hiddingh L, et al. Recycling drug screen repurposes hydroxyurea as a sensitizer of glioblastomas to temozolomide targeting de novo DNA synthesis, irrespective of molecular subtype. Neuro Oncol. 2018;20(5):642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee Y, Vassilakos A, Feng N, et al. GTI-2040, an antisense agent targeting the small subunit component (R2) of human ribonucleotide reductase, shows potent antitumor activity against a variety of tumors. Cancer Res. 2003;63(11):2802–2811. [PubMed] [Google Scholar]
- 28. Potapova O, Gorospe M, Dougherty RH, Dean NM, Gaarde WA, Holbrook NJ. Inhibition of c-Jun N-terminal kinase 2 expression suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner. Mol Cell Biol. 2000;20(5):1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antonyak MA, Moscatello DK, Wong AJ. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273(5):2817–2822. [DOI] [PubMed] [Google Scholar]
- 30. Bost F, McKay R, Dean N, Mercola D. The JUN kinase/stress-activated protein kinase pathway is required for epidermal growth factor stimulation of growth of human A549 lung carcinoma cells. J Biol Chem. 1997;272(52):33422–33429. [DOI] [PubMed] [Google Scholar]
- 31. Lopez-Ilasaca M, Li W, Uren A, et al. Requirement of phosphatidylinositol-3 kinase for activation of JNK/SAPKs by PDGF. Biochem Biophys Res Commun. 1997;232(2):273–277. [DOI] [PubMed] [Google Scholar]
- 32. Potapova O, Gorospe M, Bost F, et al. c-Jun N-terminal kinase is essential for growth of human T98G glioblastoma cells. J Biol Chem. 2000;275(32):24767–24775. [DOI] [PubMed] [Google Scholar]
- 33. Holcik M, Korneluk RG. XIAP, the guardian angel. Nat Rev Mol Cell Biol. 2001;2(7):550–556. [DOI] [PubMed] [Google Scholar]
- 34. Seligson DB, Hongo F, Huerta-Yepez S, et al. Expression of X-linked inhibitor of apoptosis protein is a strong predictor of human prostate cancer recurrence. Clin Cancer Res. 2007;13(20):6056–6063. [DOI] [PubMed] [Google Scholar]
- 35. Xia W, Bacus S, Hegde P, et al. A model of acquired autoresistance to a potent ErbB2 tyrosine kinase inhibitor and a therapeutic strategy to prevent its onset in breast cancer. Proc Natl Acad Sci U S A. 2006;103(20):7795–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aird KM, Ghanayem RB, Peplinski S, Lyerly HK, Devi GR. X-linked inhibitor of apoptosis protein inhibits apoptosis in inflammatory breast cancer cells with acquired resistance to an ErbB1/2 tyrosine kinase inhibitor. Mol Cancer Ther. 2010;9(5):1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fulda S, Wick W, Weller M, Debatin KM. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat Med. 2002;8(8):808–815. [DOI] [PubMed] [Google Scholar]
- 38. Wagner L, Marschall V, Karl S, et al. Smac mimetic sensitizes glioblastoma cells to temozolomide-induced apoptosis in a RIP1- and NF-κB-dependent manner. Oncogene. 2013;32(8):988–997. [DOI] [PubMed] [Google Scholar]
- 39. Kanev GK, de Graaf C, de Esch IJP, et al. The landscape of atypical and eukaryotic protein kinases. Trends Pharmacol Sci. 2019;40(11):818–832. [DOI] [PubMed] [Google Scholar]
- 40. van der Biessen DA, Burger H, de Bruijn P, et al. Phase I study of RGB-286638, a novel, multitargeted cyclin-dependent kinase inhibitor in patients with solid tumors. Clin Cancer Res. 2014;20(18):4776–4783. [DOI] [PubMed] [Google Scholar]
- 41. Narasimhan J, Antholine WE, Chitambar CR. Effect of gallium on the tyrosyl radical of the iron-dependent M2 subunit of ribonucleotide reductase. Biochem Pharmacol. 1992;44(12):2403–2408. [DOI] [PubMed] [Google Scholar]
- 42. Einhorn L. Gallium nitrate in the treatment of bladder cancer. Semin Oncol. 2003;30(2 suppl 5):34–41. [DOI] [PubMed] [Google Scholar]
- 43. Straus DJ. Gallium nitrate in the treatment of lymphoma. Semin Oncol. 2003;30(2 Suppl 5):25–33. [DOI] [PubMed] [Google Scholar]
- 44. Chitambar CR. Gallium nitrate for the treatment of non-Hodgkin’s lymphoma. Expert Opin Investig Drugs. 2004;13(5):531–541. [DOI] [PubMed] [Google Scholar]
- 45. Whelan HT, Przybylski C, Chitambar CR. Differential effects of gallium nitrate on proliferation of brain tumor cells in vitro. Pediatr Neurol. 1991;7(1):23–27. [DOI] [PubMed] [Google Scholar]
- 46. Tchoghandjian A, Soubéran A, Tabouret E, et al. Inhibitor of apoptosis protein expression in glioblastomas and their in vitro and in vivo targeting by SMAC mimetic GDC-0152. Cell Death Dis. 2016;7(8):e2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zakaria Z, Tivnan A, Flanagan L, et al. Patient-derived glioblastoma cells show significant heterogeneity in treatment responses to the inhibitor-of-apoptosis-protein antagonist birinapant. Br J Cancer. 2016;114(2):188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Houghton PJ, Kang MH, Reynolds CP, et al. Initial testing (stage 1) of LCL161, a SMAC mimetic, by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2012;58(4):636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones TS, Holland EC. Standard of care therapy for malignant glioma and its effect on tumor and stromal cells. Oncogene. 2012;31(16):1995–2006. [DOI] [PubMed] [Google Scholar]
- 50. Omidi Y, Barar J. Impacts of blood-brain barrier in drug delivery and targeting of brain tumors. Bioimpacts. 2012;2(1):5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rankovic Z. CNS drug design: balancing physicochemical properties for optimal brain exposure. J Med Chem. 2015;58(6):2584–2608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.