Abstract
Uveal melanoma (UM) is the most common primary intraocular malignancy of the eye. It has a high metastatic potential and mainly spreads to the liver. Genetics play a vital role in tumor classification and prognostication of UM metastatic disease. One of the driver genes mutated in metastasized UM is subunit 1 of splicing factor 3b (SF3B1), a component of the spliceosome complex. Recurrent mutations in components of the spliceosome complex are observed in UM and other malignancies, suggesting an important role in tumorigenesis. SF3B1 is the most common mutated spliceosome gene and in UM it is associated with late-onset metastasis. This review summarizes the genetic and epigenetic insights of spliceosome mutations in UM. They form a distinct subgroup of UM and have similarities with other spliceosome mutated malignancies.
Keywords: genetic, DNA repair, epigenetic, mutational analysis, chromosomes, prognosis, metastatic disease, therapy
1. Introduction
Uveal melanoma (UM) is the most common primary ocular malignancy in adults [1]. The European incidence varies between approximately 2 to 8 per million with the southern parts of Europe having the lowest incidence and the northern parts having the highest [1]. It arises from uveal melanocytes and it is located in either the choroid (90.3%), ciliary body (6.1%) or iris (3.6%) [2]. UM usually presents with symptoms such as blurred vision, photopsia, floaters and visual field loss [3]. Other patients are asymptomatic and their UM are detected during routine ophthalmic examination. Diagnosis is made by an ophthalmologist using fundoscopy with additional techniques such as ultrasonography (US) and fluorescein angiography (FAG). UM diagnosis is confirmed to be histopathological if tissue is available from biopsy or enucleation. Metastatic disease is very rare at presentation [4] and in our cohort only 10 out of 808 (1.2%) UM patients presented with detectable metastatic disease at the time of diagnosis with ultrasonographic screening of the abdomen. This is most likely an underestimation, since not all metastasis can be detected with ultrasonography of the abdomen. UM can be a highly aggressive disease, with up to 25% of the UM patients developing metastases within 10 years after diagnosis [2]. Metastasis usually spreads via the hematogenous route, with the liver as the most common site [5]. However, patients also develop metastases after enucleation, suggesting the presence of micro metastases at an early stage during tumorigenesis [6]. Current treatments aim to conserve the eye and preserve vision, which varies from stereotactic radiotherapy, proton therapy and brachytherapy to enucleation [7].
Prognostication of the tumor can be staged using The American Joint Committee on Cancer (AJCC) Cancer Staging Manual, which uses the tumor-node-metastasis (TNM) classification [8]. This includes tumor size, basal diameter, tumor thickness, ciliary body involvement and extraocular tumor extension (EOE). Large tumors, presence of EOE and ciliary body involvement are associated with poor prognosis [2,9]. Histopathological parameters include tumor cell type, mitotic activity and closed extravascular matrix patterns. Epithelioid cell type, high number of mitoses and closed vascular loops are associated with poor survival rates [10,11].
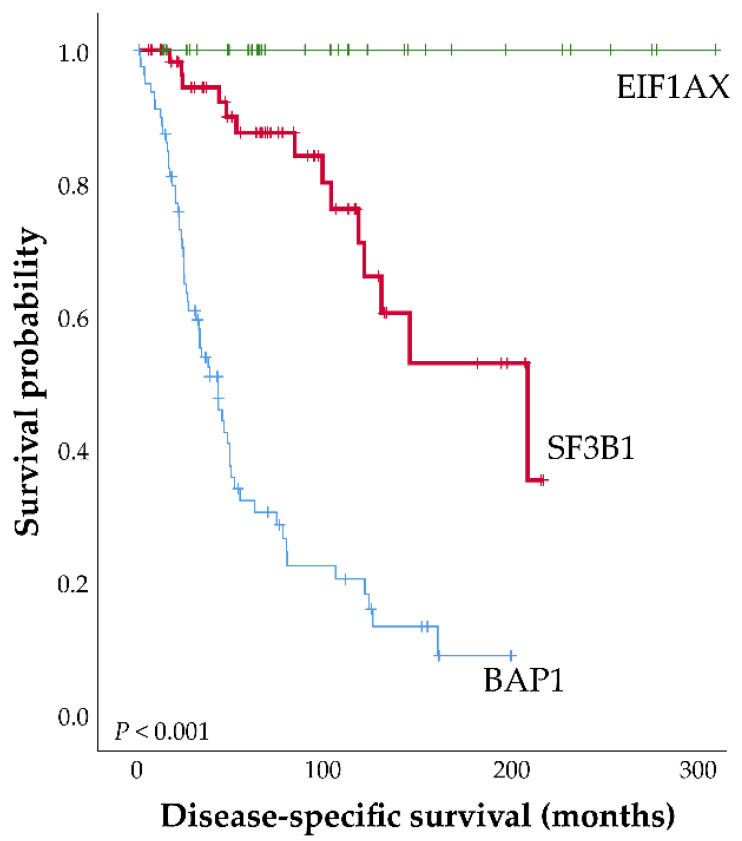
Gene mutations play an important role in prognosis. Activating mutations in GNAQ or GNA11 are seen as primary driver oncogenes in UM and mutations in these genes are usually mutually exclusive [12]. GNAQ and GNA11 encode for guanine nucleotide-binding protein alpha Q and guanine nucleotide-binding protein alpha-11 respectively, which are alpha subunits of the heterotrimeric G protein. These proteins transduce signals from G protein-coupled receptors (GPCR) to cellular signaling pathways [13]. Mutations in these genes occur in nearly all UM but are not associated with prognosis, because they are found not only in UM but also in benign blue nevi [14,15,16]. Recurrent mutations in CYSLTR2 (Cysteinyl Leukotriene Receptor 2) and PLCB4 (Phospholipase C Beta 4) were found in some of the cases lacking mutations in GNAQ or GNA11 [17,18]. Secondary driver genes are strongly associated with prognosis and include BAP1 [19], EIF1AX [20] and SF3B1 [21]. BAP1 is located on chromosome 3 (3p21) encoding the BRCA associated protein-1. BAP1 has many functions including protein de-ubiquitination, DNA damage repair, chromatin regulation and cell cycle regulation [22]. Loss of function mutations in BAP1 are often concurrent with monosomy 3, often resulting in loss of BAP1 protein expression and are associated with poor prognosis [19,23]. In our cohort of 808 patients, BAP1 mutations are seen in up to 82% of all metastasized UM patients and are correlated with monosomy 3 (87%). EIF1AX encodes the eukaryotic translation initiation factor 1A, an X-chromosomal protein and mutations in the N terminal region of this gene are found in approximately 20% of UM patients [20]. EIF1AX is a component of the 43S pre-initiation complex, which mediates the recruitment of the small 40S ribosomal subunit to the 5′ cap of mRNA to initiate translation of proteins [24]. The EIF1AX mutations are change-of-function mutations and they are associated with a more favorable prognosis in UM [20]. SF3B1 encodes splicing factor 3B subunit 1 and mutations in the SF3B1 gene occur in approximately 20% of UM cases [20,25,26]. Although mutations in SF3B1 were initially supposedly associated with a better prognosis compared to SF3B1 wild-type patients [25], our group has been able to demonstrate a biphasic development in SF3B1 mutated UM [21]. A favorable prognosis was observed in the first few years after diagnosis. However, with longer follow-up, a decline in survival was observed due to late-onset metastasis (Figure 1).
Figure 1.
Disease-specific survival plot of Uveal melanoma (UM) patients, defined as the first date of treatment until the date of metastasis or until last follow-up. Patients were censored when they were lost to follow-up or when death from a cause other than UM occurred. Updated Kaplan-Meier curve of SF3B1-mutated UM adapted and updated from Yavuzyigitoglu et al. [21]. Three groups of patients with secondary driver gene mutations: n = 185; EIF1AX, n = 33; SF3B1, n = 63; BAP1, n = 89).
2. SF3B1 and Other Spliceosome Mutations in Uveal Melanoma
This review focuses on elucidating the pathogenic features of this distinct subgroup of spliceosome gene mutated UM. RNA splicing is a fundamental process in eukaryotic species and accurate RNA splicing is essential for cell survival. Recurrent mutations in components of the spliceosome suggests these mutations are expressed and that a change in splicing accuracy that is caused by these mutations plays an important role in cancer [27].
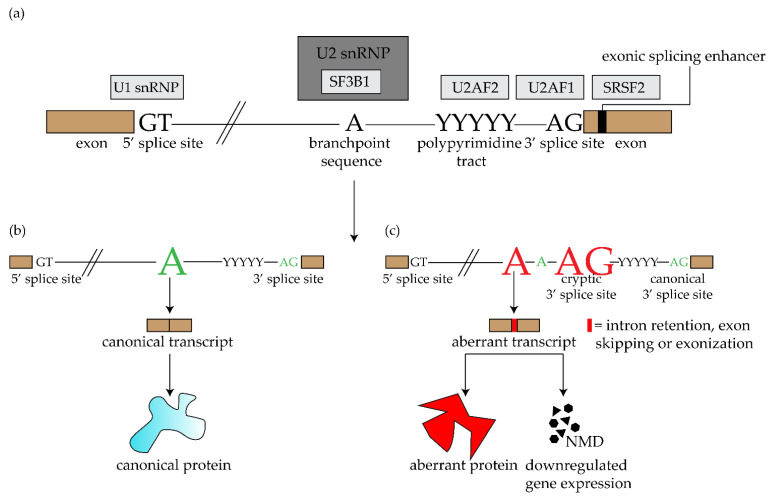
Mature mRNA is produced by removing the noncoding intronic sequences and ligating the coding sequences of pre-mRNA. This process—splicing—is regulated by the spliceosome. There are two types of this RNA/protein structure: the major spliceosome complex—involved in “general” or canonical splicing—and the minor spliceosome complex involved in non-canonical splicing (often resulting in tissue specific transcripts) [28,29]. Each of these spliceosome complexes include five crucial small nuclear ribonucleoprotein particles (snRNP): U1, U2, U4, U5 and U6 in the major spliceosome and U11, U12, U4atac, U5 and U6atac in the minor spliceosome. These particles recognize specific intronic consensus sequences: U2-type introns are recognized by the major spliceosome complex and U12 type sequences by the minor spliceosome. Canonical splicing occurs when consensus sequences in the intron: the 5′ donor splice site (5′ss), the 3′ acceptor splice site (3′ss), and the branchpoint sequence (BPS), are recognized by U1 snRNP, U2AF (U2 small nuclear RNA auxiliary factor) and U2 snRNP protein, respectively. U2AF is a heterodimer and together with the U2AF2 (U2 small nuclear RNA auciliary factor 2), it recognizes the AG dinucleotide sequence and the polypyrimidine (Py) tract of the 3′ss and coordinates the binding of the U2 snRNP at the BPS. Interaction of U1 and U2 snRNP generates junction of the 5′ end of the intron to the BPS (Figure 2). The 5′ss and 3′ss ligate together, which results in a mature mRNA product [30]. The U2 snRNP includes the SF3b1 subunit encoded by the SF3B1 gene.
Figure 2.
Splicing of pre-mRNA into mature mRNA. (a) Overview of different complexes in the spliceosome. (b) Splicing in wild-type SF3B1 (SF3B1WT) cells: SF3B1WT binds to the branchpoint of the pre-mRNA, usually an adenosine. When this process is executed correctly, mature mRNA is formed which results in a canonical protein. (c) Splicing in mutated SF3B1 (SF3B1MUT) cells: SF3B1MUT recognizes an alternative branchpoint, which leads to mis-splicing of the mRNA (e.g., intron retention, exon skipping and intronic extension of exon). The mis-spliced mRNA can be translated into aberrant proteins or be degraded by nonsense-mediated decay (NMD), resulting in downregulation of gene expression.
The SF3B1 gene, hence an essential protein in the U2 snRNP complex, is the most frequently mutated spliceosome gene and mutations are present in various malignant diseases such as UM [26], myelodysplastic syndromes (MDS) [31], chronic lymphocytic leukemia (CLL) [32], breast cancer [33], pancreatic cancer [34] and mucosal melanoma (especially in anorectal and vulvovaginal melanomas) [35]. SF3B1 mutations have an incidence of approximately 20% in UM [20,25,26], 42% in mucosal melanoma (60% in anorectal melanoma and 44% in vulvovaginal melanoma) [36], 7% in MDS (80% in refractory anemia with ring sideroblasts (RARS)-MDS, a subset of MDS) [31], 10% in CLL [32] and approximately 1.8% in breast cancer [37]. Mutated SF3B1 leads to deregulation of the U2 snRNP splicing due to an alternative usage of the 3′ss AG [38]. Mutated SF3B1 recognizes an alternative BPS located upstream of original BPS and this alternative BPS has a higher affinity to U2/SF3B1mut snRNA compared to the canonical BPS. This alternate BPS usage results in aberrant splicing. The resulting aberrant spliced transcripts could be broken down due to nonsense-mediated decay (NMD) and result in downregulated gene expression or result in aberrant proteins with changed function (Figure 2) [39,40].
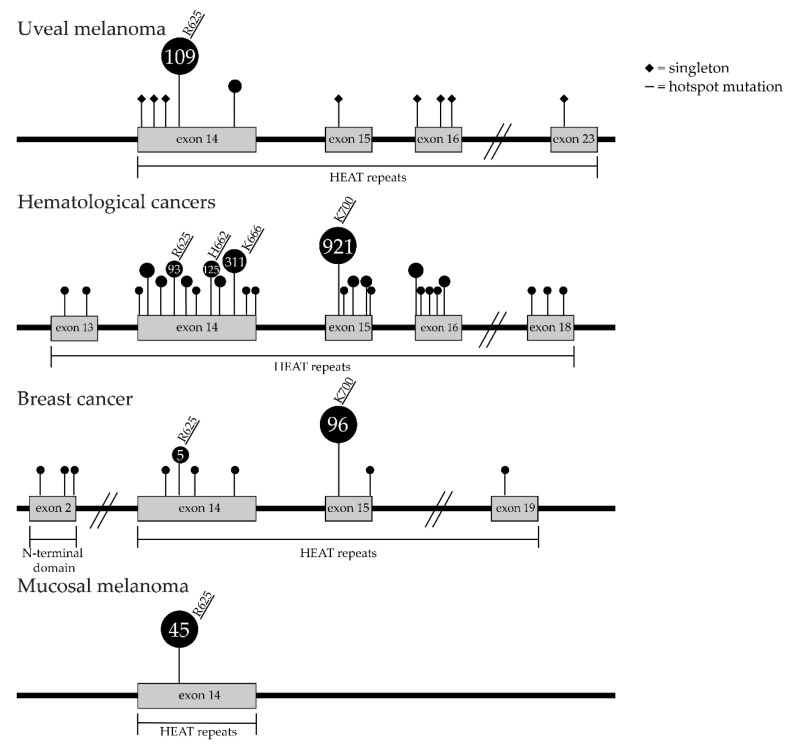
Mutations in SF3B1 are almost exclusively clustered in a specific repeat sequence motif: the 22 non-identical HEAT (Huntington, Elongation factor 3, protein phosphatase 2A, Targets of rapamycin 1) repeats domain region. Mutations in this motif have been shown to affect the interaction with the DDX46 (DEAD-Box Helicase 46) protein (prp5 in yeast) necessary for stable association of U2 snRNP to the pre-mRNA [41]. In UM, hotspot mutations are mostly in the fifth HEAT repeat at codon position arginine (R) 625, although occasionally other mutations have also been reported (Table 1). In contrast, hotspot mutations in the sixth and seventh HEAT repeat codon position lysine (K) 666 and K700, respectively, are seen in the other cancers such as hematologic cancer and breast cancer (Figure 3) [31,33,42]. The few mutations outside the HEAT repeat domains do not display aberrant splicing, suggesting they have different effects on cancer cells or have no effect and are simply passenger mutations [39]. The outcome of a large study of 533 patients with MDS indicated that patients with SF3B1 mutations were associated with a better overall survival and a lower risk of progression to acute myeloid leukemia compared to patients without SF3B1 mutations [43]. Moreover, SF3B1 mutations in MDS is an independent predictor of favorable outcome, although other studies indicated no significant effect on overall survival and were associated with shorter leukemia-free survival [44]. In breast cancer, SF3B1 mutations are associated with poor prognosis in PR-negative and luminal B subgroups [45].
Table 1.
Table of SF3B1 missense mutation frequency of uveal melanoma (UM) patients. Reference sequence: NM_012433.3(SF3B1); Build: hg38 (data from: Catalogue of Somatic Mutations in cancer v92, 27-08-2020, [46]).
| Position | Exon | AA Mutation | CDS Mutation | Effect | Frequency (n = 122, %) |
|---|---|---|---|---|---|
| 2: 197402759 | 14 | p.R625H | c.1874G > A | Missense | 61 (50%) |
| 2: 197402760 | 14 | p.R625C | c.1873C > T | Missense | 38 (31%) |
| 2: 197402759 | 14 | p.R625L | c.1874G > T | Missense | 7 (5.7%) |
| 2: 197402760 | 14 | p.R625G | c.1873C > G | Missense | 1 (0.8%) |
| 2: 197402759 | 14 | p.R625P | c.1874G > C | Missense | 1 (0.8%) |
| 2: 197402760 | 14 | p.R625S | c.1873C > A | Missense | 1 (0.8%) |
| 2: 197402775 | 14 | p.M620V | c.1858A > G | Missense | 1 (0.8%) |
| 2: 197402767 | 14 | p.E622D | c.1866G > C | Missense | 1 (0.8%) |
| 2: 197402766 | 14 | p.Y623H | c.1867T > C | Missense | 1 (0.8%) |
| 2: 197402636 | 14 | p.K666T | c.1997A > C | Missense | 2 (1.6%) |
| 2: 197402110 | 15 | p.K700E | c.2098A > G | Missense | 1 (0.8%) |
| 2: 197401765 | 16 | p.E783K | c.2347G > A | Missense | 1 (0.8%) |
| 2: 197401771 | 16 | p.D781N | c.2341G > A | Missense | 1 (0.8%) |
| 2: 197401887 | 16 | p.G742D | c.2225G > A | Missense | 1 (0.8%) |
| 2:197396227 | 23 | p.C1123Y | c.3368G > A | Missense | 1 (0.8%) |
Figure 3.
Overview of SF3B1 mutations in UM, hematological cancers, breast cancer and mucosal melanoma. Squares represent singleton mutations. Dots represent mutations seen at least two times. Underline represents hotspot mutations. Singleton mutations are only displayed for the UM cohort. For the hematological cancers and breast cancer cohorts only mutations with frequency of ≥5% are reported (see supplementary Table S1 for other data (Catalogue of Somatic Mutations in cancer v92, 27-08-2020, [46]).
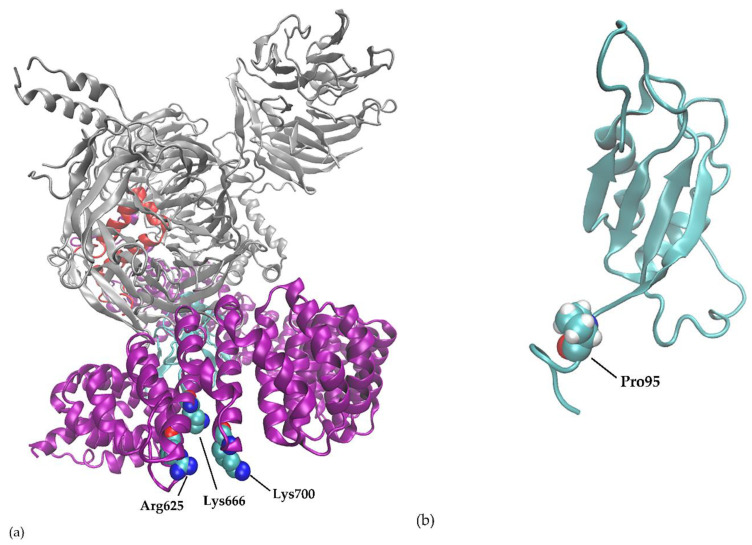
A variant of the 625R residue of SF3B1 is the most common spliceosome mutation seen in UM, but other mutations in another component of the spliceosome have also been reported [47]. Molecular modeling shows location of hotspot mutations in secondary structures of splicing genes (Figure 4).
Figure 4.
Three-dimensional view of the proteins. (a) SF3B1 protein with hotspot residue R625, K666 and K700. (b) SRSF2 protein with hotspot P95.
A second, less frequently mutated spliceosome gene is in UM is the SRSF2 (serine- and arginine-rich splicing factor 2). SRFSF2 is part of the SR protein family, which promotes spliceosome assembly by binding to exonic splicing enhancer (ESE) sequences. Van Poppelen et al. assessed SRSF2 mutations in UM patients and found five patients with SRSF2 mutations [48]. They all had in-frame deletions, but at 3 different locations. Two patients harbored in-frame deletions at protein residues 92–99, two at protein residues 92–100 and one patient harbored in-frame deletions at protein residues 173–179 [27,48]. Mutations in SRSF2 occur in approximately 12% of MDS patients [31], however not as in-frame deletions but the more common missense mutations at hotspot P95 [31]. As in case of the SF3B1 mutations, these in-frame deletions are typical for UM and are not observed in other malignancies where nucleotide change are more prevalent.
3. Chromosomal Anomalies and Epigenetic Changes Unique for SF3B1 UM
Chromosomal instability is characteristic for most cancers. In comparison to other cancers UM has relatively simple chromosomal anomalies [49]. In general, these chromosomal anomalies differ between secondary driver mutation types. BAP1 mutated UM is strongly associated with loss of chromosome 3. This in contrast to UM without a BAP1 anomaly, which retain both copies [47]. BAP1 mutated UM are characterized by gains or losses of entire chromosomes or chromosome arms. SF3B1 mutated UM karyotypes are more complex and characterized by multiple chromosomal structural variants (CSV) at the distal ends of chromosomes: loss of chromosomal region 6q (52%) and gain of chromosomal region 6p (85%) and 8q (73%) (Figure 5) [47,50]. Furthermore, SF3B1 mutated UM are most likely to have more than 3 chromosomal structural variants (CSV) per tumor, with 70% of UM with >3 CSV harboring SF3B1 mutations [50]. Between 20–36% of SF3B1 mutated CLL tumors have deletions in chromosome 11q resulting in a worse prognosis [42,51]. For UM, we have updated the data used by Yavuzyigitoglu et al. [50], and found that 30 out of 63 (47.6%) of all SF3B1 mutated UM with CSV analysis had aberrations in chromosome 11. Eight patients died due to UM, with a mean disease-specific survival of 46.5 months.
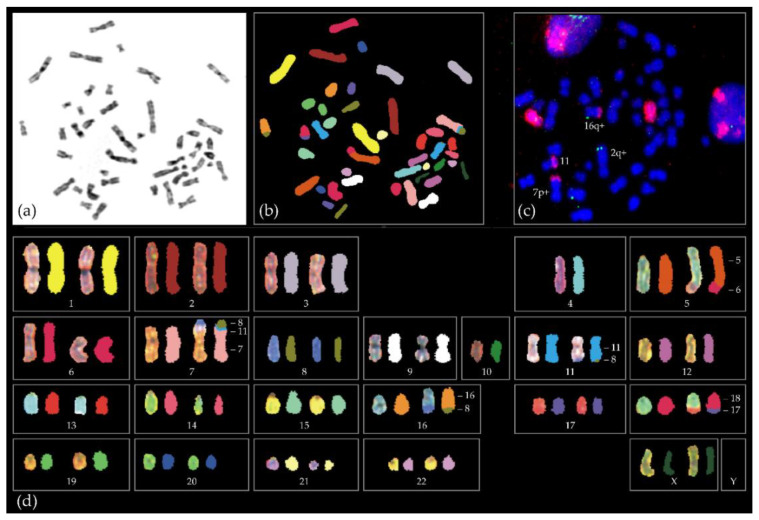
Figure 5.
Example of the complex chromosome aberrations of a SF3B1-mutated tumour of a 38 year old female. Shown is (a) metaphase spread with DAPI staining, (b) spectral karyotyping in which each chromosome has a specific color, (c) red chromosome 8 paint and green telomeric 6q probe and (d) spectral karyotype showing exchanges between different chromosomes. For instance, part of chr6 (red) is attached to the bottom part (q-arm) of chromosome 5 (orange). The upper arm (p-arm) of chromosome 7 has material of chromosome 11 (blue) and 8 (green). Full karyotype in Naus et al. [53].
Aberrant methylation can cause aberrant gene regulation, which plays an important role in tumorigenesis. We and others have assessed aberrant methylation in metastatic uveal melanoma [47,52] and the results indicate a distinct hypomethylation pattern in SF3B1 mutated patients compared to the BAP1 mutated patients. Loss of expression of genes such as tumorsupressor genes or enhanced or ectopic expression of oncogenes could play a crucial role in spliceosome induced tumorigenesis, including UM.
4. Telomere Length in SF3B1 Mutated UM
Aberrant transcripts in SF3B1 mutations affect multiple cellular pathways, including the telomere maintenance pathway [54]. Telomeres are specific gDNA domains formed by repetitive DNA sequences (TTAGGG repeats) at the end of each chromosome and protect the ends of chromosomes from degradation and are essential for maintaining chromosomal stability [55]. Growth arrest and cellular senescence occur in cells when the TTAGGG repeats reach a critical length. Maintaining the telomerase length is regulated by enzyme telomerase (TERT gene) which is activated and upregulated in cancer cells for survival. Telomerase is detected in approximately 90% of all cancers [56]. Barthel et al. assessed telomere lengths in multiple cancer types including UM, and found shorter telomeres in tumors, including UM, compared to normal tissues [57]. TERT was expressed in 73% of all tumors and a third of these had TERT abnormalities, including TERT promotor mutations and amplification or chromosomal rearrangement. However, mutations in TERT promotor occur at extremely low frequency (1%) in UM compared to other types of ocular lesions [58]. Interestingly, the TERT promotor mutated UM had normal BAP1 staining and had wild type SF3B1 as well as EIF1AX.
Our group has studied the length of telomeres in UM tumors harboring SF3B1 and BAP1 mutations. The total telomere length was significantly shorter in the BAP1MUT group compared to the control group (p < 0.001). However, no significant difference in total telomere length was observed between the SF3B1MUT group and control group (p = 0.054) or between the SF3B1MUT and BAP1MUT groups (Figure S1). We previously observed more chromosomal structural variants with recurrent distal breakpoints on chromosomes 6 and 8 in SF3B1MUT UM, suggesting a different tumorigenesis in this group compared to BAP1MUT UM [50]. Interestingly, we found that SF3B1 mutated tumors had longer telomeres in chromosome 6 compared to controls and the CCCCAA repeats were significantly more expressed in chromosomes 6 p-arm and 8 p-arm in SF3B1 mutated UM compared to BAP1 mutated UM (p < 0.05). Previously, Doherty et al. [59] observed increased levels of DNA-PK (DNA-dependent protein kinase) in UM and this activity could indeed be instrumental for the observed increase in structural changes in SF3B1-mutated UM. However, in contrast to the M3G8q (monosomy 3, gain of 8q) subgroup of UM patients described in the Doherty paper where this gene is included in the amplified region, most of the 8q gains in tumors of SF3B1 patients are gains of telomeric region of chromosome 8q and do not include the 8q11 region where DNA-PK is located. However, it would certainly be interesting to assess NHEJ (non-homologous end joining) expression upon having taken the mutational status in UM into account.
5. Therapeutic Opportunities
The prominent role of altered splicing in tumorigenesis makes it an alluring target for novel therapeutic approaches. Splicing inhibitors target specific components of the spliceosome which block spliceosome assembly [60]. Splicing inhibitors that interrupt U2 snRNP in joining the branch point sequence have been shown to target SF3B1. These include FR901464, pladienolide B and herboxidiene (Table 2) [60], Spliceostatin A, family of FR901464 and sudemycin. These splicing inhibitors have been evaluated in SF3B1 mutant breast cancer cell lines and cells of MDS patients. Mutant SF3B1 cells showed selective growth inhibition, altered splicing and increased cell death [37,61]. The effect of herboxidiene has been shown to modulate alternative pre-mRNA splicing in various human cancer cell lines [62]. H3B-8800, a pladienolide B derivative was developed and interacts directly with the SF3B1 complex [63]. It potently and preferentially kills spliceosome-mutant tumor cells. E7107, an analog of pladienolide B only affects wildtype SF3B1 and treating SF3B1 mutated colorectal cancer cells with E7107 will result in cell death [64]. Our hypothesis is that the same results will also be seen in UM cells: E7107 exposure affecting wild type SF3B1, resulting in exclusively altered splicing by mutated SF3B1 and subsequent cell death. In vivo treatment of E7107 in SRSF2 mutated murine myeloid leukemia cells resulted in preferential cell death of cells harboring mutant SRSF2 [65,66,67]. This should be replicated in SF3B1 mutated UM cell line to assess if the same results will be observed. E7107 has since been studied in two separate phase I clinical studies in patients with solid cancers [66,67]. A dose-dependent reversible inhibition of pre-mRNA was assessed in these patients. Unfortunately, unexpected toxicity of bilateral optic neuritis was reported in both studies with suspension of both trials [66,67].
Table 2.
Overview of different groups of splicing inhibitors.
| Splicing Inhibitors | Features | Trials |
|---|---|---|
|
FR901464 spliceostatins A-G, meamycins |
Targets the U2 snRNP of SF3B1; isolated from fermentation broth of bacterium Pseudomonas sp. No. 2663 [68]. | Preclinical trial in cancer cell lines [68] |
| Herboxidiene | Targets the U2 snRNP of SF3B1; isolated from Streptomyces chromofuscus ATCC 49,982 [69]. | Preclinical trial in cancer cell lines [62] |
| Pladienolide B pladienolides A-G, H3B-8800, E7107 |
Targets the U2 snRNP of SF3B1; isolated from bacterium Streptomyces platensis [70]. | Two phase I trials of E7107 on solid tumors, suspended due to side effects [66,67] |
| Sudemycin | Targets SF3B1; pharmacophore between FR901464 and pladienolide B [71]. | Preclinical trial in cancer cell lines and tumor samples [71] |
Experiments with splicing inhibitors has resulted in potential target genes and therapies in splicing mutated malignancies. For example, Aird et al. has assessed the sensitivity of splicing inhibitors in cancer cell lines [72]. Cancer cells elude apoptosis through various pathways including the upregulation of anti-apoptotic BCL2 (B-cell lymphoma 2) family genes. In this study, BCL2A1 (BCL2 related protein A1) and MCL1 (MCL1 apoptosis regulator), members of the BCL2 family genes, were sensitive to E7107, whereas BCL2L1 (BCL2 like 1), which encodes BCLxL (B-cell lymphoma-extra large), was highly resistant to E7107-induced splicing modulation. Furthermore, when E7107 was combined with BCLxL inhibitors, this enhanced the cytotoxicity in numerous cancer cell lines. These results indicate an increased cytotoxicity can be achieved with splicing inhibitors in combination with targeting MCL1 and BCLxL. A second candidate gene is the MYC Proto-Oncogene, BHLH Transcription Factor (MYC). This transcription factor is a principle regulator of the snRNP, ensuring proper splicing of mRNA [73]. Overexpression of MYC require cells to rely on upregulation of snRNP and PRMT5 (Protein Arginine Methyltransferase 5) to sustain splicing fidelity [73]. The MYC family consists of three genes: c-MYC, l-MYC and n-MYC [74]. One of these genes, the c-MYC gene, is located on chromosome 8q24 [75]. Intriguingly, SF3B1 mutated UM are associated with gain of 8q, and proto-oncogene c-MYC is located in the 8q. Targeting BUD31 (BUD31 homolog) in MYC driven tumors results in reduced cell viability by affecting the splicing machinery [76]. Third example is the Bromodomain Containing 9 (BRD9) gene. Mutated SF3B1 recognizes aberrant deep intronic branchpoint in BRD9, a core component of the non-canonical BAF (ncBAF) chromatin remodeling complex [77]. This results into inclusion of an alternate exon with a premature termination codon and therefore depletion of BRD9. Loss of BRD9 promotes melanoma tumorigenesis through perturbation of ncBAF. When mis-splicing of BRD9 in SF3B1-mutated cells was corrected, tumor growth was suppressed. Therefore, BRD9 is a tumor suppressor in UM. Targeting mis-splicing of BRD9 could therefore be a potential cancer therapy in SF3B1 mutated malignancies.
CRISPR-Cas genome editing may prove a viable treatment option. The CRISPR-Cas uses guide RNAs to direct the Cas9 nuclease to the complementary target site where it cuts the double-stranded DNA, creating a break. The break will be repaired through homology recombination (HR) or non-homologous end joining (NHEJ), the latter results in the formation of indels [78]. SF3B1 mutated UM does not lead to loss of splicing but aberrant splicing. Therefore, correcting the R625 hotspot with the CRISPR-Cas would result to normal splicing. The CRISPR-Cas technique has been studied in various cancer cell lines [79,80]. Moreover, results of a clinical trial using the CRISPR-Cas technique has recently been published [81]. However, delivery of the CRISPR gene therapy in vivo (off-target effects, targeted delivery, delivery efficiency and editing efficiency) remains challenging, and should be studied extensively before eventually applying this potential therapeutic method to patients with metastasized SF3B1 mutated UM.
To conclude, spliceosome mutated UM has been shown to differ from other gene mutated UM in prognosis, pathogenesis, genetics and epigenetics. Furthermore, SF3B1 mutated UM show similarities to other spliceosome mutated malignancies. This suggests that they should be considered as a different subgroup, most likely with different therapeutic options.
Acknowledgments
We would like to thank Tom de Vries Lentsch for his help with the figures. We would also like to thank Youri Dircken and Frank Magielsen for their contribution to the data analysis.
Abbreviations
| AJCC | American Joint Committee on Cancer |
| BAP1 | BRCA1 associated protein 1 |
| BCL2 | B-cell lymphoma 2 |
| BCL2A1 | BCL2 related protein A1 |
| BCL2L1 | BCL2 like 1 |
| BCLxL | B-cell lymphoma-extra large |
| BPS | Branchpoint sequence |
| BRD9 | Bromodomain containing 9 |
| BUD31 | BUD31 homolog |
| CLL | Chronic Lymphocytic Leukemia |
| CSV | Chromosomal structural variants |
| CYSLTR2 | Cysteinyl Leukotriene Receptor 2 |
| DDX46 | DEAD-Box Helicase 46 |
| DNA-PK | DNA-dependent protein kinase |
| EIF1AX | Eukaryotic Translation Initiation Factor 1A X-Linked |
| EOE | Extraocular tumor extension |
| ESE | Exonic splicing enhancer |
| FAG | Fluorescein Angiography |
| GNAQ | G protein Subunit Alpha Q |
| GNA11 | G protein Subunit Alpha 11 |
| GPCR | G protein-coupled receptors |
| HEAT | Huntington, Elongation factor 3, protein phosphatase 2A, targets of rapamycin 1 |
| HR | Homology recombination |
| K | Lysine |
| M3G8q | Monosomy 3, gain of 8q |
| MCL1 | MCL1 apoptosis regulator |
| MDS | Myelodysplastic syndromes |
| MUT | Mutated |
| MYC | MYC proto-oncogene, BHLH transcription factor |
| ncBAF | Non-canonical BAF |
| NHEJ | Non-homologous end joining |
| NMD | Nonsense mediated decay |
| PLCB4 | Phospholipase C Beta 4 |
| PRMT5 | Protein Arginine Methyltransferase 5 |
| Py | Polypyrimidine |
| R | Arginine |
| RARS | Refractory anemia with ring sideroblasts |
| SF3B1 | Splicing Factor 3b Subunit 1 |
| SRSF2 | Serine- and arginine-rich splicing factor 2 |
| snRNP | Small nuclear ribonucleoprotein particles |
| TERT | Telomerase reverse transcriptase |
| TNM | Tumor-node-metastasis |
| U2AF | U2 small nuclear RNA auxiliary factor |
| U2AF2 | U2 small nuclear RNA auxiliary factor 2 |
| UM | Uveal melanoma |
| US | Ultrasonography |
| WT | Wild type |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/24/9546/s1, Figure S1: Multiple comparison using TelomereHunter data, Table S1: Missense mutation frequency of SF3B1 mutation in patients with hematological cancers, breast cancer and mucosal melanoma.
Author Contributions
Conceptualization, A.d.K., E.B. and E.K.; writing—original draft preparation, J.Q.N.N.; writing—review and editing, W.D., S.Y., E.M.S., R.M.V., N.C.N., A.d.K., E.K. and E.B.; supervision, E.B. and E.K.; funding acquisition, A.d.K. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Combined Ophthalmic Research Rotterdam (CORR 5.2.0) and Uitzicht UZ-2018-4.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Virgili G., Gatta G., Ciccolallo L., Capocaccia R., Biggeri A., Crocetti E., Lutz J.M., Paci E., Group E.W. Incidence of uveal melanoma in Europe. Ophthalmology. 2007;114:2309–2315. doi: 10.1016/j.ophtha.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 2.Shields C.L., Furuta M., Thangappan A., Nagori S., Mashayekhi A., Lally D.R., Kelly C.C., Rudich D.S., Nagori A.V., Wakade O.A., et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch. Ophthalmol. 2009;127:989–998. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- 3.Damato E.M., Damato B.E. Detection and time to treatment of uveal melanoma in the United Kingdom: An evaluation of 2384 patients. Ophthalmology. 2012;119:1582–1589. doi: 10.1016/j.ophtha.2012.01.048. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman L.E., McLean I.W., Foster W.D. Does enucleation of the eye containing a malignant melanoma prevent or accelerate the dissemination of tumour cells. Br. J. Ophthalmol. 1978;62:420–425. doi: 10.1136/bjo.62.6.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diener-West M., Reynolds S.M., Agugliaro D.J., Caldwell R., Cumming K., Earle J.D., Hawkins B.S., Hayman J.A., Jaiyesimi I., Jampol L.M., et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005;123:1639–1643. doi: 10.1001/archopht.123.12.1639. [DOI] [PubMed] [Google Scholar]
- 6.Eskelin S., Pyrhönen S., Summanen P., Hahka-Kemppinen M., Kivelä T. Tumor doubling times in metastatic malignant melanoma of the uvea: Tumor progression before and after treatment. Ophthalmology. 2000;107:1443–1449. doi: 10.1016/S0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 7.Damato B. Ocular treatment of choroidal melanoma in relation to the prevention of metastatic death—A personal view. Prog. Retin. Eye Res. 2018;66:187–199. doi: 10.1016/j.preteyeres.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Amin M.B., Edge S., Greene F., Byrd D.R., Brookland R.K., Washington M.K., Gershenwald J.E., Compton C.C., Hess K.R., Sullivan D.C., et al. AJCC Cancer Staging Manual. 8th ed. Springer International Publishing; Berlin/Heidelberg, Germany: 2017. [Google Scholar]
- 9.Kujala E., Makitie T., Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003;44:4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 10.Coupland S.E., Campbell I., Damato B. Routes of extraocular extension of uveal melanoma: Risk factors and influence on survival probability. Ophthalmology. 2008;115:1778–1785. doi: 10.1016/j.ophtha.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 11.Drabarek W., Yavuzyigitoglu S., Obulkasim A., van Riet J., Smit K.N., van Poppelen N.M., Vaarwater J., Brands T., Eussen B., Verdijk R.M., et al. Multi-Modality Analysis Improves Survival Prediction in Enucleated Uveal Melanoma Patients. Investig. Ophthalmol. Vis. Sci. 2019;60:3595–3605. doi: 10.1167/iovs.18-24818. [DOI] [PubMed] [Google Scholar]
- 12.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N., et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon M.I., Strathmann M.P., Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 14.Koopmans A.E., Vaarwater J., Paridaens D., Naus N.C., Kilic E., de Klein A., Rotterdam Ocular Melanoma Study group Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br. J. Cancer. 2013;109:493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer J., Kilic E., Vaarwater J., Bastian B.C., Garbe C., de Klein A. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br. J. Cancer. 2009;101:813–815. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore A.R., Ceraudo E., Sher J.J., Guan Y., Shoushtari A.N., Chang M.T., Zhang J.Q., Walczak E.G., Kazmi M.A., Taylor B.S., et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson P., Aoude L.G., Wadt K., Glasson W.J., Warrier S.K., Hewitt A.W., Kiilgaard J.F., Heegaard S., Isaacs T., Franchina M., et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koopmans A.E., Verdijk R.M., Brouwer R.W., van den Bosch T.P., van den Berg M.M., Vaarwater J., Kockx C.E., Paridaens D., Naus N.C., Nellist M., et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod. Pathol. 2014;27:1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 20.Martin M., Masshofer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B., et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yavuzyigitoglu S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., van Bodegom A., Paridaens D., Kilic E., de Klein A., Rotterdam Ocular Melanoma Study Group Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Jensen D.E., Proctor M., Marquis S.T., Gardner H.P., Ha S.I., Chodosh L.A., Ishov A.M., Tommerup N., Vissing H., Sekido Y., et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 23.Harbour J.W., Onken M.D., Roberson E.D., Duan S., Cao L., Worley L.A., Council M.L., Matatall K.A., Helms C., Bowcock A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhuri J., Si K., Maitra U. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J. Biol. Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 25.Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S., et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harbour J.W., Roberson E.D., Anbunathan H., Onken M.D., Worley L.A., Bowcock A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiler M., Peng S., Agrawal A.A., Palacino J., Teng T., Zhu P., Smith P.G., Caesar-Johnson S.J., Demchok J.A., Felau I., et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep. 2018;23:282–296.e4. doi: 10.1016/j.celrep.2018.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Conti L., Baralle M., Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 29.Sibley C.R., Blazquez L., Ule J. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016;17:407–421. doi: 10.1038/nrg.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 32.Quesada V., Conde L., Villamor N., Ordonez G.R., Jares P., Bassaganyas L., Ramsay A.J., Bea S., Pinyol M., Martinez-Trillos A., et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 33.Ellis M.J., Ding L., Shen D., Luo J., Suman V.J., Wallis J.W., Van Tine B.A., Hoog J., Goiffon R.J., Goldstein T.C., et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biankin A.V., Waddell N., Kassahn K.S., Gingras M.C., Muthuswamy L.B., Johns A.L., Miller D.K., Wilson P.J., Patch A.M., Wu J., et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayward N.K., Wilmott J.S., Waddell N., Johansson P.A., Field M.A., Nones K., Patch A.M., Kakavand H., Alexandrov L.B., Burke H., et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 36.Hintzsche J.D., Gorden N.T., Amato C.M., Kim J., Wuensch K.E., Robinson S.E., Applegate A.J., Couts K.L., Medina T.M., Wells K.R., et al. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res. 2017;27:189–199. doi: 10.1097/CMR.0000000000000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maguire S.L., Leonidou A., Wai P., Marchio C., Ng C.K., Sapino A., Salomon A.V., Reis-Filho J.S., Weigelt B., Natrajan R.C. SF3B1 mutations constitute a novel therapeutic target in breast cancer. J. Pathol. 2015;235:571–580. doi: 10.1002/path.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alsafadi S., Houy A., Battistella A., Popova T., Wassef M., Henry E., Tirode F., Constantinou A., Piperno-Neumann S., Roman-Roman S., et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016;7:10615. doi: 10.1038/ncomms10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D., Bailey S.L., Bhavsar E.B., Chan B., Colla S., et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015;13:1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 40.Paolella B.R., Gibson W.J., Urbanski L.M., Alberta J.A., Zack T.I., Bandopadhayay P., Nichols C.A., Agarwalla P.K., Brown M.S., Lamothe R., et al. Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. Elife. 2017;6:e23268. doi: 10.7554/eLife.23268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Q., Rodriguez-Santiago S., Wang J., Pu J., Yuste A., Gupta V., Moldon A., Xu Y.Z., Query C.C. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 2016;30:2710–2723. doi: 10.1101/gad.291872.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Lawrence M.S., Wan Y., Stojanov P., Sougnez C., Stevenson K., Werner L., Sivachenko A., DeLuca D.S., Zhang L., et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malcovati L., Karimi M., Papaemmanuil E., Ambaglio I., Jadersten M., Jansson M., Elena C., Galli A., Walldin G., Della Porta M.G., et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126:233–241. doi: 10.1182/blood-2015-03-633537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y., Miao M., Han S., Qi J., Wang H., Ruan C., Wu D., Han Y. Prognostic value and clinical feature of SF3B1 mutations in myelodysplastic syndromes: A meta-analysis. Crit. Rev. Oncol. Hematol. 2019;133:74–83. doi: 10.1016/j.critrevonc.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Fu X., Tian M., Gu J., Cheng T., Ma D., Feng L., Xin X. SF3B1 mutation is a poor prognostic indicator in luminal B and progesterone receptor-negative breast cancer patients. Oncotarget. 2017;8:115018–115027. doi: 10.18632/oncotarget.22983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L., Hess J.M., Uzunangelov V., Walter V., Danilova L., et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2018;33:151. doi: 10.1016/j.ccell.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 48.van Poppelen N.M., Drabarek W., Smit K.N., Vaarwater J., Brands T., Paridaens D., Kilic E., de Klein A. SRSF2 Mutations in Uveal Melanoma: A Preference for In-Frame Deletions? Cancers. 2019;11:1200. doi: 10.3390/cancers11081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dogrusoz M., Jager M.J. Genetic prognostication in uveal melanoma. Acta Ophthalmol. 2018;96:331–347. doi: 10.1111/aos.13580. [DOI] [PubMed] [Google Scholar]
- 50.Yavuzyigitoglu S., Drabarek W., Smit K.N., van Poppelen N., Koopmans A.E., Vaarwater J., Brands T., Eussen B., Dubbink H.J., van Riet J., et al. Correlation of Gene Mutation Status with Copy Number Profile in Uveal Melanoma. Ophthalmology. 2017;124:573–575. doi: 10.1016/j.ophtha.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 51.Jeromin S., Weissmann S., Haferlach C., Dicker F., Bayer K., Grossmann V., Alpermann T., Roller A., Kohlmann A., Haferlach T., et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–117. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 52.Smit K.N., Boers R., Vaarwater J., Boers J., Brands T., Mensink H., Verdijk R.M., Van Ijcken W.F.J., Gribnau J., De Klein A., et al. Novel DNA-methylation silenced tumor suppressor genes identified for BAP1-mediated uveal melanoma metastasis. 2020 in press. [Google Scholar]
- 53.Naus N.C., van Drunen E., de Klein A., Luyten G.P., Paridaens D.A., Alers J.C., Ksander B.R., Beverloo H.B., Slater R.M. Characterization of complex chromosomal abnormalities in uveal melanoma by fluorescence in situ hybridization, spectral karyotyping, and comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30:267–273. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1088>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Brooks A.N., Fan J., Wan Y., Gambe R., Li S., Hergert S., Yin S., Freeman S.S., Levin J.Z., et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell. 2016;30:750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maciejowski J., de Lange T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 57.Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q., et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koopmans A.E., Ober K., Dubbink H.J., Paridaens D., Naus N.C., Belunek S., Krist B., Post E., Zwarthoff E.C., de Klein A., et al. Prevalence and implications of TERT promoter mutation in uveal and conjunctival melanoma and in benign and premalignant conjunctival melanocytic lesions. Investig. Ophthalmol. Vis. Sci. 2014;55:6024–6030. doi: 10.1167/iovs.14-14901. [DOI] [PubMed] [Google Scholar]
- 59.Doherty R.E., Bryant H.E., Valluru M.K., Rennie I.G., Sisley K. Increased Non-Homologous End Joining Makes DNA-PK a Promising Target for Therapeutic Intervention in Uveal Melanoma. Cancers. 2019;11:1278. doi: 10.3390/cancers11091278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S.C., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016;22:976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirai C.L., White B.S., Tripathi M., Tapia R., Ley J.N., Ndonwi M., Kim S., Shao J., Carver A., Saez B., et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun. 2017;8:14060. doi: 10.1038/ncomms14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagisetti C., Yermolina M.V., Sharma L.K., Palacios G., Prigaro B.J., Webb T.R. Pre-mRNA splicing-modulatory pharmacophores: The total synthesis of herboxidiene, a pladienolide-herboxidiene hybrid analog and related derivatives. ACS Chem. Biol. 2014;9:643–648. doi: 10.1021/cb400695j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seiler M., Yoshimi A., Darman R., Chan B., Keaney G., Thomas M., Agrawal A.A., Caleb B., Csibi A., Sean E., et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med. 2018;24:497–504. doi: 10.1038/nm.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokoi A., Kotake Y., Takahashi K., Kadowaki T., Matsumoto Y., Minoshima Y., Sugi N.H., Sagane K., Hamaguchi M., Iwata M., et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 65.Lee S.C., Dvinge H., Kim E., Cho H., Micol J.B., Chung Y.R., Durham B.H., Yoshimi A., Kim Y.J., Thomas M., et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med. 2016;22:672–678. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong D.S., Kurzrock R., Naing A., Wheler J.J., Falchook G.S., Schiffman J.S., Faulkner N., Pilat M.J., O’Brien J., LoRusso P. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Investig. New Drugs. 2014;32:436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 67.Eskens F.A., Ramos F.J., Burger H., O’Brien J.P., Piera A., de Jonge M.J., Mizui Y., Wiemer E.A., Carreras M.J., Baselga J., et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res. 2013;19:6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 68.Nakajima H., Hori Y., Terano H., Okuhara M., Manda T., Matsumoto S., Shimomura K. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. 1996;49:1204–1211. doi: 10.7164/antibiotics.49.1204. [DOI] [PubMed] [Google Scholar]
- 69.Sakai Y., Yoshida T., Ochiai K., Uosaki Y., Saitoh Y., Tanaka F., Akiyama T., Akinaga S., Mizukami T. GEX1 compounds, novel antitumor antibiotics related to herboxidiene, produced by Streptomyces sp. I. Taxonomy, production, isolation, physicochemical properties and biological activities. J. Antibiot. 2002;55:855–862. doi: 10.7164/antibiotics.55.855. [DOI] [PubMed] [Google Scholar]
- 70.Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 71.Fan L., Lagisetti C., Edwards C.C., Webb T.R., Potter P.M. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing. ACS Chem. Biol. 2011;6:582–589. doi: 10.1021/cb100356k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aird D., Teng T., Huang C.L., Pazolli E., Banka D., Cheung-Ong K., Eifert C., Furman C., Wu Z.J., Seiler M., et al. Sensitivity to splicing modulation of BCL2 family genes defines cancer therapeutic strategies for splicing modulators. Nat. Commun. 2019;10:137. doi: 10.1038/s41467-018-08150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koh C.M., Bezzi M., Low D.H., Ang W.X., Teo S.X., Gay F.P., Al-Haddawi M., Tan S.Y., Osato M., Sabo A., et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature. 2015;523:96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- 74.DePinho R., Mitsock L., Hatton K., Ferrier P., Zimmerman K., Legouy E., Tesfaye A., Collum R., Yancopoulos G., Nisen P., et al. Myc family of cellular oncogenes. J. Cell Biochem. 1987;33:257–266. doi: 10.1002/jcb.240330404. [DOI] [PubMed] [Google Scholar]
- 75.Neel B.G., Jhanwar S.C., Chaganti R.S., Hayward W.S. Two human c-onc genes are located on the long arm of chromosome 8. Proc. Natl. Acad. Sci. USA. 1982;79:7842–7846. doi: 10.1073/pnas.79.24.7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anczukow O., Krainer A.R. The spliceosome, a potential Achilles heel of MYC-driven tumors. Genome Med. 2015;7:107. doi: 10.1186/s13073-015-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inoue D., Chew G.L., Liu B., Michel B.C., Pangallo J., D’Avino A.R., Hitchman T., North K., Lee S.C., Bitner L., et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574:432–436. doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komor A.C., Badran A.H., Liu D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lentsch E., Li L., Pfeffer S., Ekici A.B., Taher L., Pilarsky C., Grutzmann R. CRISPR/Cas9-Mediated Knock-Out of Kras(G12D) Mutated Pancreatic Cancer Cell Lines. Int. J. Mol. Sci. 2019;20:5706. doi: 10.3390/ijms20225706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park M.Y., Jung M.H., Eo E.Y., Kim S., Lee S.H., Lee Y.J., Park J.S., Cho Y.J., Chung J.H., Kim C.H., et al. Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing. Oncotarget. 2017;8:36331–36338. doi: 10.18632/oncotarget.16752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.