Abstract
Purpose
This work aimed to evaluate genotype-phenotype associations in individuals carrying germline variants of transmembrane protein 127 gene (TMEM127), a poorly known gene that confers susceptibility to pheochromocytoma (PHEO) and paraganglioma (PGL).
Design
Data were collected from a registry of probands with TMEM127 variants, published reports, and public databases.
Main Outcome Analysis
Clinical, genetic, and functional associations were determined.
Results
The cohort comprised 110 index patients (111 variants) with a mean age of 45 years (range, 21-84 years). Females were predominant (76 vs 34, P < .001). Most patients had PHEO (n = 94; 85.5%), although PGL (n = 10; 9%) and renal cell carcinoma (RCC, n = 6; 5.4%) were also detected, either alone or in combination with PHEO. One-third of the cases had multiple tumors, and known family history was reported in 15.4%. Metastatic PHEO/PGL was rare (2.8%). Epinephrine alone, or combined with norepinephrine, accounted for 82% of the catecholamine profiles of PHEO/PGLs. Most variants (n = 63) occurred only once and 13 were recurrent (2-12 times). Although nontruncating variants were less frequent than truncating changes overall, they were predominant in non-PHEO clinical presentations (36% PHEO-only vs 69% other, P < .001) and clustered disproportionately within transmembrane regions (P < .01), underscoring the relevance of these domains for TMEM127 function. Integration of clinical and previous experimental data supported classification of variants into 4 groups based on mutation type, localization, and predicted disruption.
Conclusions
Patients with TMEM127 variants often resemble sporadic nonmetastatic PHEOs. PGL and RCC may also co-occur, although their causal link requires further evaluation. We propose a new classification to predict variant pathogenicity and assist with carrier surveillance.
Keywords: pheochromocytoma, paraganglioma, TMEM127, tumor suppressor gene, genotype-phenotype association
Pheochromocytomas (PHEOs) and paragangliomas (PGLs) are neuroendocrine tumors that arise in the adrenal gland or paraganglia, respectively, and often produce catecholamines (1). It is estimated that approximately 1000 cases are diagnosed each year in the United States (2). Currently, approximately 40% of cases are estimated to result from a recognizable germline mutation, making PHEOs/PGLs the most frequently heritable neuroendocrine tumor (3). More than 20 distinct genes have been associated with PHEO/PGL predisposition, including the transmembrane protein 127 gene (TMEM127). The TMEM127 gene encodes a multispanner, transmembrane protein conserved among vertebrates (4). Its biological role is not fully understood. TMEM127 functions as a classic tumor suppressor gene, with loss of the wild-type (WT) allele in tumor tissue (4), and it is included in diagnostic genetic panels of PHEO/PGL susceptibility (5). We have previously reported that TMEM127 colocalizes to the plasma membrane, early endosomes, and lysosomes and that it associates with a nutrient-sensing, lysosome-based protein complex (6-8), and handling of glucose and insulin in vivo (9). These earlier studies suggest that TMEM127 signals through the mTOR (mammalian target of rapamycin) pathway, a central hub of cellular homeostasis that is frequently disrupted in human malignancies (10).
Despite these advances, the precise cellular function of TMEM127 is unknown, and the clinical spectrum, disease severity, and penetrance of TMEM127-associated disease remain poorly defined. As a result, guidelines for surveillance in individuals carrying TMEM127 germline variants have not been precisely established. The first comprehensive series of TMEM127 mutations, published by Yao et al in 2010 (11), revealed that TMEM127 mutations were associated exclusively with PHEOs and presented at an age similar to that of sporadic tumors. Despite the mutation being germline in all cases, family history was reported in only a quarter of the patients, raising the question of a relatively low disease penetrance (11, 12). Long-term follow-up of a large, 6-generation family in which diagnosis of carriers was made before or post-TMEM127 identification as the predisposition gene provided support to the concept that penetrance of the driver mutation, approximately 30% at age 65 years, was not as high as some other PHEO/PGL susceptibility genes such as RET or VHL (13).
Since then, other clinical features were detected in association with TMEM127 variants, including PGL (14-16) and renal cell carcinoma (RCC) (7, 8, 17, 18). These data suggest that TMEM127 dysfunction may lead to a wider clinical spectrum. Moreover, accumulating evidence has revealed additional properties of TMEM127, which may be useful to supplement the analysis of variant pathogenicity. Our previous (4, 7, 8, 11) and recent (19, 20) work evaluating subcellular distribution and/or expression level of patient-derived TMEM127 variants has identified features consistent with pathogenicity. In brief, truncating mutations invariably lead to unstable protein products that are rapidly degraded, a profile shared by other tumor suppressor genes. Likewise, nontruncating variants (ie, missense mutations or in-frame insertions or deletions, not expected to alter the protein length) located within transmembrane domains (TMs) also lead to mislocalization and/or decreased expression that likely reflect functional disruption.
Here we examined a large series of individuals with TMEM127 gene variants, published and unpublished, with the aim of delineating genotype-phenotype associations of relevance for clinical and follow-up surveillance.
Materials and Methods
Patient data collection
Data were collected from 2 distinct sources. First, we used our registry of probands with TMEM127 variants collected through an institutional review board–approved sample repository (NCT03160274, https://clinicaltrials.gov/ct2/show/NCT03160274), through which patients were enrolled after providing signed informed consent. The data collected included sex; age at diagnosis; family history; tumor type, site and number; malignancy status (as defined by the detection of metastases at nonparaganglial tissue, as established by the World Health Organization classification [21]); and catecholamine profile. Information on catecholamine levels varied; some had only epinephrine or norepinephrine, others had only total, but not fractionated, metanephrines, others had only vanillylmandelic acid, and a few had dopamine or 3-methoxytyramine measurements. For those with detailed information, we adopted the rule of a relative increase of plasma metanephrines above 5% of the total increase of the metabolites to define an epinephrine (EPI) phenotype (22). For cases with only catecholamines (plasma or urine available), we considered an EPI phenotype when only epinephrine was increased, or combined (“NE_Epi”) to indicate those in whom norepinephrine (NE) and EPI both were elevated, either in plasma or urine. In addition to these clinical indicators, type and location of the TMEM127 variant were recorded. Anonymized samples were also obtained through this repository, collected through institutional review board–approved protocols at their respective institutions. The length of follow-up time since diagnosis was not available for most patients and was therefore not included in this study.
A second method of sample collection was performed through a comprehensive PubMed search using the terms TMEM127; pheochromocytoma; paraganglioma; hereditary cancer; mutation; variant; genetic screening; and genetic testing, spanning the period from January 2010 to February 2020, with subsequent manual review of all the references to curate relevant cases, collect pertinent information, and remove potential redundancies. In some instances, direct communication was established with the contact author for clarifications. Samples with incomplete, discrepant, or conflicting data, or those perceived as duplicated were removed.
Finally, our search extended to existing databases of germline variants, the Leiden Open Variation Database (LOVD) databases (https://databases.lovd.nl/shared/genes/TMEM127), for which we (G.A.P., S.F.K., and P.L.M.D.) serve as curators, and the ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) public archive for TMEM127 variants from other submissions. Because these sources are redundant with individual reports or publications, or may lack clinical information, they were used only as reference and were not included in the final statistical analysis (see the following sections). Variants detected in unaffected populations (Gnomad/EXAC public databases gnomad.broadinstitute.org) at a frequency of greater than 0.01% or annotated as “benign” (LOVD, ClinVar) were not included. Somatic variants identified in large data sets/cohorts of cancers were not included in the present study.
Genetic analysis
DNA from blood, saliva, and/or tumor, either fresh-frozen or from formalin-fixed, paraffin-embedded samples from our repository, were obtained using standard methods. TMEM127 gene sequencing spanning its coding region and approximately 20 bp of exon-intron boundaries was carried out as previously reported (11). Loss of heterozygosity (LOH) analysis was performed when germline and tumor samples were both available (21). In addition, TMEM127 sequencing data were also obtained from next-generation sequencing data from exomes, or PHEO/PGL gene panels, from academic centers or commercial laboratories. Finally, TMEM127 sequencing information was obtained from reported cases as originally described by the authors (Table 1; (4, 7, 8, 11, 12, 14, 16-18, 23-36)).
Table 1.
Clinical and genetic features of the patients with TMEM127 germline variants
| Sample ID | Variant(nucleotide) | Variant (protein) | Sex | Age, y | Clinical presentation | Multicentricitya | Catecholamine profile | Metastatasis | Family history | LOH | Prediction scorese | Source of data | Proposed classification | This study’s score | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.? (g.(?_96919536)_(96931129_?)del | p.? | M | 43 | HNPGL | Y | NS | N | N | 1 | i | 3j | t.s. | ||
| 2 | c.-18C > T | p.? | M | 44 | Pheo | N | N | N | 2,3 | i | 4 | (11) | |||
| 3 | c.2T > A | p.? | M | 42 | Pheo | N | Epi_NE | N | N | 0/0 | 1 | iii | 3 | t.s. | |
| 4 | c.3G > A | p.? | F | 35 | Pheo | N | Y | 0/0/3 | 2,3 | iii | 3 | (15) | |||
| 5 | c.3G > A | p.? | M | 36 | Pheo | N | Y | 0/0/3 | 2,3 | iii | 3 | (15) | |||
| 6 | c.3G > A | p.? | F | 58 | Pheo | N | N | 0/0/3 | 2,3 | iii | 3 | (15) | |||
| 7 | c.3Gv> A | p.? | F | 52 | Pheo | N | Y | 0/0/3 | 2,3 | iii | 3 | (15) | |||
| 8 | c.3G > A | p.? | M | 26 | Pheo | Y | NE | N | N | 0/0/3 | 2,3 | iii | 3 | (25) | |
| 9 | c.3G > T | p.? | F | 61 | Pheo | N | Epi | N | N | Y | 0/0/5 | 2,3 | iii | 3 | (11) |
| 10 | c.4T > G | p.Tyr2Asp | F | 68 | HNPGL | N | NS | N | N | 0.6/0 | 1 | iii | 3 | t.s. | |
| 11 | c.6C > A | p.Tyr2Ter | M | 40 | Pheo | N | N | 2 | i | 5 | (29) | ||||
| 12 | c.30del | p.Gly11Alafs*70 | F | 29 | Pheo | N | N | N | 2 | i | 5 | (12) | |||
| 13 | c.31G > T | p.Gly11Cys | M | 54 | Pheo | Y | NE | N | N | N | 0.4/0/3 | 2,3 | iii | 3f,j | (19) |
| 14 | c.50G > T | p.Ser17Ile | M | 67 | Pheob | N | Epi_NE | N | N | N | 0/0.1 | 2 | iii | 3 | (19) |
| 15 | c.53C > T | p.Pro18Leu | F | 57 | Pheo | N | Epi | 0/0.4/2 | 1,3 | iii | 3 | t.s. | |||
| 16 | c.73A > T | p.Lys25Ter | M | 68 | Pheo | N | N | 2,3 | i | 5 | (15) | ||||
| 17 | c.74dup | p.Gln26Alafs*5 | F | 41 | Pheo | N | Epi | N | Y | Y | 2 | i | 5 | (24) | |
| 18 | c.74dup | p.Gln26Alafs*5 | F | 65 | Pheo | N | Epi_NE | N | N | 1 | i | 5 | t.s. | ||
| 19 | c.74dup | p.Gln26Alafs*5 | F | 23 | Pheo | N | N | N | 1 | i | 5 | t.s. | |||
| 20 | c.76C > T | p.Gln26Ter | F | 37 | Pheo | N | N | N | NA | 2,3 | i | 5 | (11) | ||
| 21 | c.86delG | p.Arg29Leufs*52 | F | 33 | Pheo | Y | Epi_NE | N | N | 2 | i | 5 | (27) | ||
| 22 | c.109G > C | p.Gly37Arg | M | 46 | Pheo | N | Epi_NE | N | N | Y | 0.9/0.1 | 2 | ii | 4g | (19) |
| 23 | c.117_120delGTCT | p.Ile41Argfs*39 | F | 52 | Pheo | Y | Epi | N | N | Y | 1 | i | 5 | t.s. | |
| 24 | c.117_120delGTCT | p.Ile41Argfs*39 | F | 53 | Pheo | Y | Epi_NE | N | Y | 1 | i | 5 | t.s. | ||
| 25 | c.117_120delGTCT | p.Ile41Argfs*39 | F | 66 | Pheo | Y | N | Y | 2 | i | 5 | (4) | |||
| 26 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 34 | Pheo | N | N | N | NA | 2,3 | i | 5 | (11) | ||
| 27 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 45 | Pheo | N | N | N | 1 | i | 5 | t.s. | |||
| 28 | c.117_120delTGTC | p.Ile41Argfs*39 | M | 40 | Pheo | Y | Epi | N | N | 2,3 | i | 5 | (32) | ||
| 29 | c.117_120delTGTC | p.Ile41Argfs*39 | M | 48 | Pheo | Y | Epi | N | N | 2,3 | i | 5 | (32) | ||
| 30 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 26 | Pheo | N? | N | 2 | i | 5 | (30) | ||||
| 31 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 62 | Pheo | N? | N | 2 | i | 5 | (30) | ||||
| 32 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 23 | Pheo | N | N | N | 1 | i | 5 | t.s. | |||
| 33 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 49 | Pheo | N | Epi | N | N | 1 | i | 5 | t.s. | ||
| 34 | c.117_120delTGTC | p.Ile41Argfs*39 | F | 84 | Pheo | N | Epi_NE | N | N | 1 | i | 5 | t.s. | ||
| 35 | c.131T > G | p.Leu44Arg | F | 43 | Pheo | Y | N | 0.4/0 | 2 | ii | 4 | (15) | |||
| 36 | c.139G > C | p.Ala47Pro | F | 48 | Pheo | N | Epi_NE | N | N | Y | 0.6/0 | 1 | ii | 4 | t.s. |
| 37 | c.140C > A | p.Ala47Asp | F | 45 | Pheo | Y | Epi | N | Y | Y | 0.4/0/3 | 2 | ii | 4 | (12) |
| 38 | c.150insA | p.Pro51Thrfs*57 | F | 25 | Pheo | N | N | Y | 2,3 | i | 5 | (4) | |||
| 39 | c.154G > T | p.Ala52Ser | M | 35 | RPPGL | N | Epi_NE | Y | N | 0.2/0.3/3 | 2,3 | ii | 4 | (16) | |
| 40 | c.158G > C | p.Trp53Ser | M | 21 | Pheo, BMF | N | N | N | 2,3 | i | 4 | (11) | |||
| 41 | c.158G > A | p.Trp53Ter | F | 57 | Pheo | N | Epi_NE | N | Y | NA | 0.3/0/3 | 2,3 | ii | 4 | (19) |
| 42 | c.181T > C | p.Cys61Arg | F | 21 | RCCc | N | NA | N | N | 0.9/0 | 1 | iii | 2 or 3f,j | t.s. | |
| 43 | c.185C > G | p.Ser62Trp | M | 46 | Pheo | N | Epi_NE | N | N | Y | 0.4/0.1 | 2,3 | iii | 3g | (19) |
| 44 | c.190_191dup | p.Gln64Hisfs*18 | F | 28 | Pheo | N | Epi_NE | N | N | 2 | i | 5 | (12) | ||
| 45 | c.190_191dup | p.Gln64Hisfs*18 | F | 56 | Pheo | Y | Epi_NE | N | N | 2 | i | 5 | (35) | ||
| 46 | c.202del | p.Val68Serfs*13 | F | 42 | Pheo | N | N | N | 1 | i | 5 | t.s. | |||
| 47 | c.202del | p.Val68Serfs*13 | M | 32 | Pheo | Y | Epi | N | N | 1 | i | 5 | t.s. | ||
| 48 | c.202del | p.Val68Serfs*13 | F | 56 | Pheo | N | N | N | 2 | i | 5 | (12) | |||
| 49 | c.208G > A | p.Asp70Asn | M | 67 | RCC | N | NA | N | N | 0.2/0.4/2 | 2,3 | iii | 3 | (8) | |
| 50 | c.208G > A | p.Asp70Asn | F | 50 | Pheo | N | N | N | NA | 0.2/0.4/2 | 2,3 | iii | 3 | (11) | |
| 51 | c.215T > A | p.Leu72Ter | F | 58 | Pheo | N | N | N | 2 | i | 5 | (15) | |||
| 52 | c.217G > C | p.Gly73Arg | M | 44 | Pheo | N | N | N | NA | 0.8/0.1/3 | 2,3 | iii | 3 | (11) | |
| 53 | c.221A > C | p.Tyr74Ser | M | 35 | Pheo | Nd | N | 0/0.3/3 | 2,3 | iii | 3 | (30) | |||
| 54 | c.222T > A | p.Tyr74Ter | M | 58 | Pheo | Nd | Dopa | N | N | 2 | i | 5 | (18) | ||
| 55 | c.248delT | p.Phe83fs*2 | M | 54 | Pheo | Y | Epi_NE | N | N | NA | 1,3 | i | 5 | t.s. | |
| 56 | c.248delT | p.Phe83fs*2 | M | 46 | Pheo | N | Epi_NE | N | N | 2,3 | i | 5 | (28) | ||
| 57 | c.264_267delCAGA | p.Thr89fs*35 | M | 46 | Pheo | Y | N | N | Y | 2,3 | i | 5 | (4) | ||
| 58 | c.268G > A | p.Val90Met | F | 32 | Pheo | N | N | N | NA | 0.5/0/2 | 2,3 | iii | 2 | (4) | |
| 59 | c.268G > A | p.Val90Met | M | 44 | Pheo | N | Epi_NE | N | N | Y | 0.5/0/2 | 2,3 | iii | 2 | (12) |
| 60 | c.280C > T | p.Arg94Trp | F | 43 | Pheo | N | N | N | NA | 0.6/0/3 | 2,3 | iii | 4 | (11) | |
| 61 | c.292G > A | p.Ala98Thr | F | 44 | HNPGL | N | NS | N | N | N | 0.1/0.4/c | 1,3 | ii | 3f,j | t.s. |
| 62 | c.308delG | p.Gly103Alafs*20 | F | 47 | Pheo_RCC | Y | Epi | N | N | NA | 2 | i | 5 | (17) | |
| 63 | c.319_321del AGT | p.Ser107del | F | 37 | Pheo | Nd | N | 2 | ii | 4 | (30) | ||||
| 64 | c.325T > C | p.Ser109Pro | F | 34 | HNPGL | Y | NS | N | N | 0.1/0.1/3 | 2,3 | ii | 4 | (14) | |
| 65 | c.348T > C | p.Phe116Phe | M | 39 | Pheo | N | Epi_NE | N | N | n.s.a.p. | 2 | ii | 3 | (24) | |
| 66 | c.353C > T | p.Pro118Leu | M | 60 | RCC | N | NA | Y | NA | 0.0 | 2,3 | iii | 3 | (8) | |
| 67 | c.377C > T | p.Thr126Ile | F | 47 | RCC | N | NA | N | N | 0.9 | 2,3 | iii | 3 | (8) | |
| 68 | c.388G > T | p.Ala130Ser | F | 67 | Pheo | Y | N | N | 0.9 | 1 | ii | 3 | t.s. | ||
| 69 | c.398A > G | p.His133Arg | M | 46 | Pheo | Y | Epi_NE | N | N | 0.9 | 1,3 | ii | 3 | t.s. | |
| 70 | c.408G > A | p.Thr136Thr | F | 45 | Pheo | N | Epi | N | N | p.s.a./3 | 1,3 | ii | 3 | t.s. | |
| 71 | c.409 + 1G > T r.245_409del | p.Asp82_Thr136del | F | 38 | Pheo, BrCa | N | N | Y | Yi | p.s.a./5 | 2,3 | i | 5 | (11) | |
| 72 | c.409 + 22A > G | p.? | M | 65 | Pheo | N | Epi_NE | N | N | n.s.a.p. | 1 | i | 3 | t.s. | |
| 73 | c.410-1G > C | p.? | M | 45 | Pheo | Y | N | p.s.a. /5 | 2 | i | 5 | (15) | |||
| 74 | c.410-2A > C r.410_417del | p.Leu138Cysfs*12 | F | 34 | Pheo | Y | Epi_NE | N | Y | Y | p.s.a. /5 | 2,3 | i | 5 | (4) |
| 75 | c.410-2A > C r.410_417del | p.Leu138Cysfs*12 | F | 37 | Pheo | Y | N | Y | Y | p.s.a. /5 | 2,3 | i | 5 | (4) | |
| 76 | c.410-2A > C r.410_417del | p.Leu138Cysfs*12 | F | 53 | Pheo | N | Epi | N | p.s.a. /5 | 2,3 | i | 4 | (36) | ||
| 77 | c.410-2A > G r.410_417del | p.Leu138Cysfs*12 | M | 31 | Pheo | Y | Epi | N | N | Y | p.s.a. /4 | 2,3 | i | 4 | (25) |
| 78 | c.411T > A | p.Val137Val | M | 60 | Pheo | N | Epi_NE | N | N | p.s.a. /3 | 1,3 | ii | 3 | t.s. | |
| 79 | c.413T > G | p.Leu138Pro | M | 66 | RPPGL | N | N | 0.8/0 | 2 | ii | 5 | (15) | |||
| 80 | c.415C > T | p.Gln139Ter | M | 33 | Pheo | Nd | Epi | N | N | 2 | ii | 5 | (26) | ||
| 81 | c.418T > C | p.Cys140Arg | F | 47 | Pheo | N | N | N | NA | 0.9/0/4 | 2,3 | ii | 5 | (11) | |
| 82 | c.419G > A | p.Cys140Tyr | F | 59 | Pheo | Y | Y | N | 0.9/0/4 | 2,3 | ii | 5 | (11) | ||
| 83 | c.419G > A | p.Cys140Tyr | F | 22 | Pheo | Y | N | N | NA | 0.9/0/4 | 2 | ii | 5 | (31) | |
| 84 | c.424_426dup | p.Thr142dup | F | 32 | Pheo | N | N | Y | Y | 1,3 | ii | 3 | t.s. | ||
| 85 | c.424_426delACC | p.Thr142del | F | 52 | Pheo | N | Epi_NE | N | N | N | 1 | ii | 3 | t.s. | |
| 86 | c.443_445del | p.Tyr148del | F | 31 | Pheo | N | Epi | N | N | 1 | ii | 3 | t.s. | ||
| 87 | c.446G > A | p.Trp149Ter | F | 30 | Pheo | Y | Epi | N | N | 1 | i | 5 | t.s. | ||
| 88 | c.447G > A | p.Trp149Ter | F | 40 | Pheo | Y | N | N | NA | 2,3 | i | 5 | (11) | ||
| 89 | c.462C > G | p.Ile154Met | M | 76 | Pheo | Nd | N | 0.3/0.2/3 | 2,3 | iii | 3 | (15) | |||
| 90 | c.464T > A | p.Leu155Ter | F | 51 | Pheo_HNPGL | Y | Y | N | 2,3 | i | 5 | (15) | |||
| 91 | c.469C > T | p.Gln157Ter | M | 42 | Pheo | N | Epi | N | N | 2,3 | i | 5 | (12) | ||
| 92 | c.475C > T | p.Gln159Ter | F | 72 | Pheo | Y | N | Y | Y | 2,3 | i | 5 | (4) | ||
| 93 | c.492C > G | p.Tyr164Ter | F | 35 | Pheo | N | Epi_NE | N | N | 2 | i | 5 | (12) | ||
| 94 | c.518T > C | p.Phe173Ser | F | 26 | Pheo | N | N | 0.9/0 | 2 | ii | 4 | (15) | |||
| 95 | c.523G > A | p.Val175Ile | F | 27 | Pheo_HNPGL | Y | NE | N | N | N | 0/0.5 | 2 | ii | 3d,j | (24) |
| 96 | c.532_533insT | p.Tyr178Leufs*48 | F | 47 | Pheo_RCC | Y | Epi | N | Y | Yi | 2 | i | 5 | (7) | |
| 97 | c.532_533insT | p.Tyr178Leufs*48 | F | 25 | Pheo | N | N | 2 | i | 5 | (15) | ||||
| 98 | c.532-533insTCGCCGTTAGCTTCT | p.Tyr178Phefs*67 | F | 37 | Pheo | Y | Epi_NE | N | N | Y | 2 | i | 5 | (19) | |
| 99 | c.536T > C | p.Leu179Pro | F | 33 | Pheo | N | Epi | N | N | Inconclusive | 0.9/0 | 2 | ii | 3 | (24) |
| 100 | c.540_553delGGGCAGGAGCTGGT | p.Ala181Aspfs63Ter | F | 66 | Pheo | N | N | N | NA | 1 | i | 5 | t.s. | ||
| 101 | c.543_555dup | p.Ala186Argfs*44 | F | 33 | Pheo | Y | Y | 2 | i | 5 | (15) | ||||
| 102 | c.553G > A | p.Gly185Arg | F | 51 | Pheo_RPPGL | Y | Epi | N | N | 0.9/0/3 | 2,3 | ii | 4 | (14) | |
| 103 | c.556G > C | p.Ala186Pro | F | 49 | Pheo | N | N | Y | Y | 0.8/0/3 | 2,3 | ii | 3 | (24) | |
| 104 | c.556G > C | p.Ala186Pro | F | 40 | Pheo | N | Epi | N | N | Y | 0.8/0/3 | 1,3 | ii | 3 | t.s. |
| 105 | c.568G > A | p.Ala190Tyr | F | 50 | HNPGL | N | NS | N | 0.9/0/3 | 2,3 | ii | 4 | (15) | ||
| 106 | c.570delC | p.Thr191Argfs116Ter | F | 64 | Pheo | N | Nh | N | 2,3 | i | 5 | (34) | |||
| 107 | c.572delC | p.Thr191Argfs76Ter | F | 47 | Pheo | Y | N | N | 2 | i | 5 | (31) | |||
| 108 | c.572delC | p.Thr191Argfs*116 | F | 26 | Pheo | N | N | 2 | i | 5 | (15) | ||||
| 109 | c.627_640dup | p.Met214fs | F | 26 | Pheo, PTC | Y | N | N | NA | 2,3 | iv | 5 | (11) | ||
| 110 | c.640del | p.Met214fs | F | 40 | Pheo | Y | Epi_NE | N | N | 1 | i | 4 | t.s. | ||
| 111 | c.665C > T | p.Ala222Val | F | 55 | Pheo | N | NE | N | N | 0.1/0/3 | 2,3 | iii | 3 | (33) |
Source of data: 1, this study; 2, reported in the literature; 3, reported in ClinVar.
Abbreviations: BMF, bone marrow failure; BrCa, breast carcinoma; del, deletion; Epi, epinephrine; F, female; HNPGL, head and neck paraganglioma; IF, in-frame; ins, insertion; LOH, loss of heterozygosity; M, male; N, no; NA, not applicable; NE, norepinephrine; NE_Epi, elevated norepinephrine and epinephrine; N.S, non-secreting; PGL, paraganglioma; PHEO, pheochromocytoma; PTC, papillary thyroid carcinoma; RCC, renal cell carcinoma; RPPGL, retroperitoneal paraganglioma; TMEM127, transmembrane protein 127 gene; t.s., this study; WT, wild-type; Y, yes.
a Multicentricity defined as more than one tumor in the same patient; multifocality has been addressed in only 2 cases, which coincidentally were also multicentric.
b Composite tumor PHEO/ganglioneuroma.
c Papillary histology.
d Unspecified, likely single tumor.
e Prediction scores list the Polyphen2 score/SIFT score, when available. The last element, when available, corresponds to the ClinVar score (1-5 or c = conflicting); some variants (synonymous or splice site) show the splice predictor interpretation from Human Splice Finder (p.s.a., predicted splicing alteration; n.s.a.p., nonsplice altering prediction).
f Variants that are located in TM domains, but the type of substitution may not impact function, as predicted by pathogenicity algorithms—these samples have not yet been verified experimentally.
g These 2 variants were detected in the same patient, in cis, with LOH of the WT allele in this tumor. Variant Gly37Arg is disrupted by in vitro assay; variant Cys62Trp is partially stable.
h Pheochromocytomatosis, but no description of metastases in nonadjacent tissues.
i LOH in Pheo only.
j Other potential genetic driver event detected in patient’s sample: 1 = SDHD germline pathogenic variant; 13 = MAX c.296-14T > A, MAX LOH, and negative MAX immunohistochemistry; 42 = TSC2 somatic frameshift mutation, no TSC2 LOH; 61 = SDHD somatic pathogenic variant and negative SDHB immunohistochemistry; 95 = SDHD pathogenic variant, with SDHD LOH and negative SDHB immunohistochemistry.
Germline screening for other PHEO/PGL susceptibility genes was performed in all patients; however, the number of genes sequenced varied; at a minimum, RET, VHL, SDHB, or SDHD sequencing was performed, and clinical features of neurofibromatosis 1 (NF1) were also collected. In cases identified by PHEO/PGL panel (either commercial or from academic research labs), additional susceptibility genes were included (EGLN1, EGLN2, EPAS1, FH, HRAS, KIF1B, MAX, NF1, SDHA, SDHAF2, and SDHC). All patients with RCC were screened for VHL and, in some cases, other RCC susceptibility genes (FH, MET, FLCN, SDHB, and SDHD).
In silico analysis
All collected TMEM127 variants were annotated using the database dbNSFP v4.0a (https://vatlab.github.io/vat-docs/) (37, 38) and the program SnpSift (http://snpeff.sourceforge.net/SnpSift.html) (39) and uploaded onto The Human Splicing Finder program (HSF; www.umd.be/HSF/) to predict the potential effect of an amino acid substitution on the structure and function of the TMEM127 protein (PolyPhen2, SIFT [sorting intolerant from tolerant]) and putative splicing abnormalities caused by variants located at nonconsensus splice sites, including coding variants. HSF contains its own programs and other prediction platforms, including the exonic splicing enhancer (ESE) finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home), RESCUE-ESE (http://hollywood.mit.edu/burgelab/rescue-ese/), FAS-ESS (http://hollywood.mit.edu/fas-ess/), Putative Exonic Splicing Enhancers/Silencers, and splicing silencer motifs (40). The score interpretation results were extracted into Table 1. Frameshift and nonsense variants typically do not produce a score in this analysis and were considered to be “damaging.”
Variant classification
Classification of variants (“proposed classification” in Table 1) followed the original principles of 5-class grouping (41) and also incorporated the recommendations for variant interpretation specifically designed for PHEO/PGL (5). This recommendation is based on the frequency of the variant in the general population and disease databases, the variant type, co-segregation with the disease in families, its previous reporting in the literature in a context that establishes link with the disease, in silico predictions, and the results of functional or other supporting information (eg, LOH analysis, activity assays, immunohistochemistry).
A third component of the variant interpretation involved the results of in vitro assays performed in our laboratory in 28 distinct variants, corresponding to 49 samples: 26 truncating (ie, nonsense, frameshift insertions or deletions, variants involving start or stop codons) variants, and 23 nontruncating (missense and in-frame insertions or deletions) changes (7, 8, 11, 19). These analyses are based on subcellular distribution of expressed GFP-TMEM127 variant constructs in cells lacking TMEM127 by CRISPR-Cas9 modification, and time course of GFP-TMEM127 or untagged TMEM127 construct expression analysis, both relative to WT-TMEM127 or GFP-TMEM127 (7, 8, 11, 19). The possible outcomes of the confocal analysis are 1) membrane/punctate appearance, similar to WT, 2) diffuse/cytoplasmic, or 3) predominant plasma membrane. The outcomes of the time course analysis are 1) expression similar to WT, 2) reduced expression, relative to WT, or 3) markedly low or undetectable expression. For either test, outcomes 2 and 3 are considered to disrupt TMEM127 function (equivalent to a pathogenicity score of 4 or 5), while pattern 1 is currently unclear (equivalent to scores 1-3, or benign/likely benign or variant of uncertain significance).
Statistical analysis
We evaluated clinical and genetic variables related to probands with a TMEM127 mutation to determine potential correlations: age at diagnosis, tumor type and/or site, malignancy status, biochemical profile, family history, variant type (truncating vs nontruncating) and location of the variant relative to the gene sequence (exon number, and TM vs non–TM-encoding amino acids). Data were analyzed based on clinical and genotypic information. For categorical variables, Fisher exact test was used to compare proportions. For continuous variables, mean values were compared by a 2-tailed paired t test. Binomial test was used for assessing the significance of the difference between 2 proportions. All P values are 2-tailed. A P value of less than .05 was considered statistically significant.
Results
Clinical features
We collected information from 134 patients carrying 135 TMEM127 germline variants for this study. Twenty-four samples were excluded because of incomplete data or the possibility that they may have been duplicated. The remaining 110 cases, including 111 variants, were kept for further analysis. Of these, 30 have not been previously reported (see Table 1). There was a preference for women (76 vs 34 men, P < .001). The mean age of presentation was 45 years, with the youngest case reported at age 21 years, and the oldest at 84 years. The age of presentation did not vary by sex (44 years vs 47 years, women and men, respectively, P = .27).
PHEO was the sole clinical presentation in 94 patients (85.5%). Ten individuals (9%) had PGL, 5 of which were located at the head and neck (HNPGL) and 2 were retroperitoneal (RPPGL). In addition, 6 patients (5.4%) presented with RCC; histologically, 5 of these were clear cell and 1 was papillary type. Other cancer types had only a single occurrence (see Table 1) and were not further evaluated. Most patients presented with a single tumor (65.5%, n = 73) but 37 (33%; see Table 1) had multiple tumors: 30 bilateral PHEOs; 2 bilateral HNPGLs, and 5 others had PHEO in combination with HNPHL (n = 2), RPPGL (1), or RCC (n = 2). Known family history of PHEO/PGL or RCC was observed in 15.4% of those with reported data (17/104), but only half (n = 8) of these were bilateral tumors. Conversely, only one patient presenting with multiple tumors had a family history of PHEO/PGL/RCC (see Table 1). We found no clinical difference in sex, age, or biochemical profile of patients with multiple vs single tumor presentation (see Table 1). Overall, metastatic PHEO/PGL was a rare occurrence (n = 3, 2.8%, RCC cases excluded), and one patient was reported with pheochromocytomatosis (34). None of the RCC cases had reported metastases. However, although follow-up was longer than 20 years in a few patients, this information was limited and/or may not have been sufficiently long to evaluate multicentric or metastatic disease more accurately in most cases.
The catecholamine profile was reported in 56 cases: co-secretion of EPI and NE was the most frequent pattern, present in 25 cases, followed by EPI secretion in 21 cases. Increased NE alone was documented in only 4 cases, and dopamine was elevated in 1 patient. The 5 cases with HNPGL only were reported as nonsecretors, as expected (see Table 1).
Genetic findings
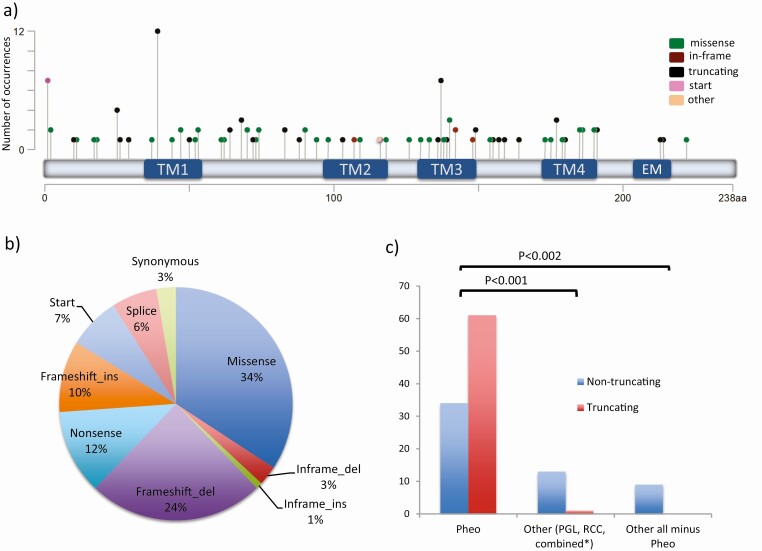
The 111 TMEM127 variants were distributed along the entire coding sequence of the gene (Fig. 1A). Overall, more variants led to predicted truncating products (nonsense, splice site, frameshift indels, start or stop codon disruptions, n = 66) than nontruncating changes (missense or in-frame indels, n = 42, P = .03, Fig. 1B). Three synonymous variants were not included in this calculation because they have not been experimentally tested. However, 2 of them are expected to alter splice and may give rise to a truncated product (see Table 1). Missense mutations represented more than one-third of all the genetic changes (see Fig. 1B). Most variants were detected only once (62%, n = 68), but there were 13 recurrent variants, including c.117_120del, which was reported in 12 distinct probands, unknown to be related (see Table 1 and Table 2).
Figure 1.
Distribution of transmembrane protein 127 gene (TMEM127) variants. A, Diagram of the 111 germline TMEM127 variants along the amino acid sequence, displayed as lollipop symbols designed using the Mutation Mapper tool of the cBioPortal website (45, 46) and color coded based on the mutation class. The Y axis represents the number of occurrences of each variant; TM, transmembrane domains; EM, endocytic motif (19). B, Distribution of the 111 TMEM127 variants based on the mutation class (% shown). C, Distribution of TMEM127 variants (truncating vs nontruncating) based on clinical presentation into 3 groups: pheochromocytoma (Pheo) only; paraganglioma (PGL), renal cell carcinoma (RCC), either alone or combined with Pheo; PGL, RCC without Pheo; truncating variants include: nonsense, frameshift indels, splice site, and start site variants; nontruncating include missense and in-frame insertions or deletions.
Table 2.
Clinical features of patients carrying recurrent TMEM127 variants
| Variant | No. of occurrences | Age at onset, y (individual cases) | Age range, y | Age median, y | Sex | Presentation (n) | Biochemical presentation (n) | Exon | Type of mutation |
|---|---|---|---|---|---|---|---|---|---|
| c.190_191dup | 2 | 28, 56 | 28-56 | 42 | 2 F | PHEO (2) | Epi_NE (2) | 2 | T |
| c.208G > A | 2 | 50, 67 | 50-67 | 58 | 1 M, 1 F | RCC (1) | NApp (1) | 2 | NT |
| PHEO (1) | NA (1) | ||||||||
| c.248delT | 2 | 46, 54 | 46-54 | 50 | 2 M | PHEO (2) | Epi_NE (2) | 3 | T |
| c.268G > A | 2 | 32, 44 | 32-44 | 38 | 1 M, 1 F | PHEO (2) | Epi_NE (1), NA (1) | 3 | NT |
| c.419G > A | 2 | 22, 59 | 22-59 | 40.5 | 1 M, 1 F | PHEO (2) | NA (2) | 4 | NT |
| c.532_533insT | 2 | 25, 47 | 25-47 | 36 | 2 F | PHEO/RCC (1) | Epi (1) | 4 | T |
| PHEO (1) | NA (1) | ||||||||
| c.556G > C | 2 | 40, 49 | 40-49 | 44 | 2 F | PHEO (2) | EPI (1), NA (1) | 4 | NT |
| c.572delC | 2 | 26, 47 | 26-47 | 36 | 2 F | PHEO (2) | NA (2) | 4 | T |
| c.410-2A > C r.410_417del | 3 | 34, 37, 53 | 34-53 | 37 | 3 F | PHEO (3) | Epi (1), Epi_NE (1), NA (1) | 4 | T |
| c.74dup | 3 | 23, 41, 65 | 23-65 | 43 | 3 F | PHEO (3) | Epi (1) | 2 | T |
| Epi_NE (1), NA (1) | |||||||||
| c.202del | 3 | 32, 42, 56 | 32-56 | 43 | 2 F | PHEO (3) | Epi (1) | 2 | T |
| 1 M | NA (2) | ||||||||
| c.3G > A | 5 | 26, 35, 36, 52, 58 | 26-58 | 41 | 3 F | PHEO (5) | NE (1) | 2 | T |
| 2 M | NA (4) | ||||||||
| c.117_120delGTCT | 12 | 23, 26, 34, 40, 45, 48, 49, 52, 53, 62, 66, 84 | 23-84 | 48 | 10 F, 2 M | PHEO (12) | Epi (4), Epi_NE (2), NA (6) | 2 | T |
Abbreviations: Epi, epinephrine; F, female; M, male; NE, norepinephrine; NE_Epi, elevated norepinephrine and epinephrine; NA, not available; NApp, not applicable; PHEO, pheochromocytoma; RCC, renal cell carcinoma; TMEM127, transmembrane protein 127 gene.
Interestingly, patients with PGLs or RCC but without PHEO were more likely to have nontruncating variants than patients presenting with PHEO (93% vs 35.8%, respectively, P < .001, Fig. 1C). It is important to note that, in addition to the germline TMEM127 variant, SDHD pathogenic variants were found in 2 HNPGLs and a somatic TSC2 frameshift variant was detected in 1 RCC (see Table 1). Two other cases, 1 with combined HNPGL and PHEO (24), and 1 patient with PHEO only, also had another attributable pathogenic variant (SDHD and MAX, respectively). In 4 of these cases no LOH was detected at the TMEM127 locus (see Table 1), supporting the alternative variants as the main driver mutation of these tumors. Therefore, we removed these ambiguous cases and reanalyzed the variant distribution. The predominance of nontruncating variants in patients with non-PHEO clinical presentations remained (100% vs 35.8%, P < .0012; see Fig. 1C), suggesting that truncating TMEM127 variants may not be compatible with transformation of nonadrenomedullary cells.
Although TMs represent approximately one-third of the TMEM127 amino acid sequence, most nontruncating variants clustered in this area (61%, P = .02; see Fig. 1A, Table 3). In independent studies, we have shown that variants located within TM domains lose their membrane binding ability and are unstable (11, 19), supporting their functional relevance.
Table 3.
Distribution of nontruncatingaTMEM127 variants relative to transmembrane domains
| Location of nontruncating variants | Amino acids involvedb | No. of nontruncating variants | Nontruncating variants, % | TMEM127 amino acid sequence, % | P, Fisher exact test |
|---|---|---|---|---|---|
| TM1 | 30-53 | 7 | 15.2 | 10.1 | |
| TM2 | 96-116 | 3 | 6.5 | 8.8 | |
| TM3 | 130-150 | 11 | 23.9 | 8.8 | |
| TM4 | 169-190 | 7 | 15.2 | 9.2 | |
| No. of amino acids (all TM domains) | 88 | 28 | 52.8 | 37.0 | |
| No. of amino acids (non-TM domains) | 150 | 18 | 34.0 | 63.0 | |
| Total nontruncating variants | NA | 46 | .0117 |
Abbreviations: NA, not available; TM, transmembrane domain; TMEM127, transmembrane protein 127 gene.
a Missense or in-frame insertions or deletions.
b The limits of each transmembrane domain are estimated based on consensus in silico and experimental predictions; precise limits will require crystal structure models, which are currently unavailable for TMEM127.
Classification of transmembrane protein 127 gene variants
We annotated the variants based on a multipronged approach, involving standard general International Agency for Research on Cancer guidelines for variant classification (41), the recommendations for variant interpretation developed specifically for PHEO/PGL (5), and our in-house in vitro experiments, to predict TMEM127 disruption. Experimental pathogenicity of TMEM127 variants involved subcellular distribution by confocal microscopy and steady-state levels of the expressed product on a cell lacking endogenous TMEM127 (19, 20). Using the parameters described in “Materials and Methods,” we were able to classify the variants into 4 groups (Table 1): i) truncating variants; ii) nontruncating, transmembrane-spanning variants; iii) nontruncating, nontransmembrane variants, and iv) others, which includes a novel group that has an internalization defect and leads to predominant plasma membrane localization (19). Groups i, ii, and iv are considered pathogenic or likely pathogenic because they result in an unstable product, often with aberrant localization and/or undetectable. Group iii remains undefined. Variants in this group were not overtly distinct from TMEM127, although more subtle disruptions may be present that were not clearly detected by our assays.
Based on this analysis, only one-third of the variants remained undefined (see Table 1). Interestingly, 1 patient had 2 simultaneous TMEM127 variants in cis: 1 located in the novel, proximal TM (Gly37Arg) and a second, Cys62Trp, located outside TMs. Although we cannot fully define the consequences of the latter, the former variant is functionally deficient and is likely the pathogenic driver of this patient’s phenotype.
Overall, comparisons between prediction algorithms and our curated prediction were highly concordant. However, a few discordances were noted. For example, some missense variants outside TMs were considered to be disease related based on clinical features and the absence of other genetic findings that might explain disease susceptibility, and yet these variants did not conclusively show disruption by either of the in vitro assays that we adopted (those indicated as group iii in Table 1). Although those variants may be functionally neutral polymorphisms, it is conceivable that they may impair other, yet undetermined motifs that impair TMEM127 activity. Furthermore, because the TM boundaries have not been precisely defined, amino acids located in the proximity of TMs might be relevant for appropriate insertion, positioning, and/or folding of the protein into the membranes (see Table 3). Last, the type of amino acid has an impact on the pathogenicity, even when it resides within a predicted TM, indicating that experimental verification is required to further validate the prediction at a TM site (19, 20). Two variants within TMs may fall into this category (Ala98 and Val175; see Table 1). Despite these limitations, combining clinical evidence with our experimental verification allowed us to classify variants as likely pathogenic that had not been called by other prediction models (p.Leu44Arg, p.Ala47Asp, p.Trp53Ser, p.Arg94Trp, p.Ser109Pro, and p.Gly185Arg).
Discussion
This is the largest analysis of TMEM127 variants to date. We found a preference for women among the cases evaluated, which was not previously observed in the TMEM127 setting. The mean age of diagnosis was 45 years, similar to the age reported in sporadic cases, and older than other hereditary PHEO/PGL syndromes, such as those related to VHL and RET, in agreement with our earlier observations (11). Family history was present in a minority of patients, including only half of patients with bilateral disease, suggesting low penetrance of TMEM127 variants as previously suspected (11, 13, 15), although a potentially high prevalence of de novo mutations cannot be excluded and should be actively investigated. This fact, along with the age at diagnosis, makes TMEM127-variant carriers easily mischaracterized as sporadic cases (11). However, despite this presentation, the potential for multiple tumors, either synchronously or metachronously, is high and should be considered for surgical planning and surveillance.
In our study, we show that the most common clinical presentation of pathogenic TMEM127 variants is PHEO, as previously reported (11, 12). However, TMEM127 variant carriers can also present with PGL or RCC, either alone or co-occurring with PHEO. In some of these cases, variants in other susceptibility genes, such as SDHD or TSC2, may in fact be the main drivers of the phenotype. However, 4 of the 7 patients presenting with HNPGL, 4 of the 6 with RCC, and all 3 with abdominal PGL had no other detectable mutations; therefore, the TMEM127 variant is presumably the primary genetic defect in these patients. Hence, associations of TMEM127 with PGLs or RCC should continue to be investigated (18). Other cancers have not been recurrently identified in patients with TMEM127 variants.
Our results also confirm earlier observations that PHEO/PGLs related to TMEM127 infrequently progress to metastasis, although some cases of metastatic and/or clinically aggressive disease have been reported (11, 15, 16, 34). However, it is important to caution that follow-up data, which were available in only a few cases, were rarely longer than 5 years; thus, the precise frequency of metastatic disease remains to be more accurately evaluated. Also, importantly, repeated occurrences of RCC in TMEM127 variant carriers should be considered when assessing broadly the risk of “malignancy” in TMEM127-related disease.
Other unique, rare circumstances worth mentioning are homozygous TMEM127 mutations and whole-gene deletion. The clinical presentation of homozygous variant carriers from 3 separate consanguineous families was not particularly distinctive compared to heterozygous individuals, though all had bilateral disease and strikingly elevated catecholamines (25, 35). However, these patients also had neurological and developmental delays. Intriguingly, TMEM127 maps to a copy number variation region on chromosome 2 that has been associated with neurodevelopmental disorders and usually spans multiple genes (42), thus the contribution of TMEM127 for the neurological phenotype remains undefined. In the single patient with HNPGL and a heterozygous deletion involving the whole TMEM127 gene, a pathogenic SDHD germline variant was also found, which likely represents a stronger candidate for the phenotype. Therefore, the role of TMEM127-gene deletion specifically in the tumor phenotype remains unclear.
Our recent studies have identified 2 novel structural domains in TMEM127: a fourth, N-terminal TM involving amino acids 30 to 53, and an endocytic domain on the C-terminus that is relevant for protein internalization (19, 20). Based on these findings, we propose that interpretation of variants should consider, in addition to clinical evidence, the type and location of the variant, especially nontruncating variants at TM regions. Importantly, this information must be evaluated considering additional evidence, such as LOH, and the additional genetic data of each patient, including co-occurring variants in other PHEO/PGL susceptibility genes, as highlighted earlier. This proposed classification should be reassessed as additional insights into TMEM127 become available and other functional assays are developed.
As our study shows, there is often minimal distinction between the most frequent presentation of PHEO/PGLs associated with TMEM127 mutations compared to sporadic cases. The present study, and ample data reported since the guidelines were published by the Endocrine Society in 2014, support recommending genetic testing to all patients with PHEOs/PGLs, and not only those with multicentric, metastatic, or early-onset disease (43). With regard to clinical surveillance of TMEM127-variant carriers, we agree with current recommendations by Favier et al (44) that suggest an initial screen involving physical exam with blood pressure measurement, baseline plasma or urine metanephrines, and imaging of the neck plus thoracic-abdomino-pelvic area with contrast magnetic resonance imaging or computed tomography, for evaluation of the adrenal glands, paraganglial tissue, and kidneys. Subsequently, yearly physical exam and metanephrines measurement is recommended with whole-body magnetic resonance imaging every 3 years. We currently lack evidence that disease penetrance is variant dependent. Therefore, for relatives of patients identified by cascade genetic screen, and the rarity of clinically detectable disease before age 21 years in probands, we suggest that surveillance, especially biochemical, of mutation carriers should start 10 years earlier, at age 11 years. As large, prospective follow-up studies of families with clearer data on penetrance and clinical spectrum become available, specific surveillance programs will be further refined.
Acknowledgments
We thank the patients and their families for contributing information and samples for this study. We are also grateful to Dr Anand Karnad, Dr Shamima Yeasmin, Dr Alberto Chavez, Dr Carolina Solis-Herrera, and UTHSA endocrinology fellows, as well as Lindsey Mette and Martha Thomas, for assistance with patient enrollment at UTHSA, Dr Mercedes Robledo, Spanish National Cancer Research Center, Madrid; Dr Picó-Alfonso, AM, Endocrinology Department, Hospital General Universitario de Alicante-ISABIAL-FISABIO, Alicante, Spain; Dr Massimo Mannelli, Department of Clinical Pathophysiology, University of Firenze, Italy, for clinical and genetic information; Dr Graeme Eisenhofer, University of Dresden, Germany, for scientific advice; and Dr Hartmut Neumann, Albert-Ludwigs-University, Freiburg, Germany, for sharing information about his published data. C.J., L.F., T.E. and P.L.M.D. are member of the A5 Adrenal Alliance.
Financial Support: P.L.M.D. was supported by the National Institute of General Medical Sciences (NIGMS GM114102), CTSA-IIMS (National Institutes of Health (NIH)/National Center for Advancing Translational Sciences Grants UL1 TR001120 and UL1 TR002645), the Mays Cancer Center (NIH-National Cancer Institute (NCI) P30 CA54174), and Alex’s Lemonade Stand Foundation for Childhood Cancer (with support from Northwest Mutual/Flashes of Hope) research grants. S.K.F. was supported by an NIGMS fellowship grant (F31GM131634) and, previously, by an NCI training grant (T32CA148724). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The Central South University Xiangya School of Medicine, Changsha, Hunan, China, provided support to X.Z. D.B., T.D., and R.C.B. were supported by grants from the National Health and Medical Research Council (APP1108032), Hillcrest Foundation (Perpetual Trustees), and PheoPara Alliance. A.P.G.R. and N.B. were supported by the Institut National du Cancer and the Direction Générale de l’Offre de Soins (PRT-K 2014, COMETE-TACTIC, INCa-DGOS_8663). L.F. was supported by the American Cancer Society Mentored Research Scholar Grant (MRSG-15-063-01-TBG).
Glossary
Abbreviations
- EPI
epinephrine
- HNPGL
head and neck paraganglioma
- LOH
loss of heterozygosity
- NE
norepinephrine
- NE_Epi
elevated norepinephrine and epinephrine
- PGL
paraganglioma
- PHEO
pheochromocytoma
- RCC
renal cell carcinoma
- RPPGL
retroperitoneal paraganglioma
- TM
transmembrane domain
- TMEM127
transmembrane protein 127 gene
- WT
wild-type
Additional Information
Disclosure Summary: O.H. reports research collaboration with Mayo Clinic and advisory board participation with Corcept Therapeutics and Pfizer outside the submitted work. The remaining authors have nothing to disclose.
Data Availability
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”
References
- 1. Dahia PL. Evolving concepts in pheochromocytoma and paraganglioma. Curr Opin Oncol. 2006;18(1):1-8. [DOI] [PubMed] [Google Scholar]
- 2. Chen H, Sippel RS, O’Dorisio MS, Vinik AI, Lloyd RV, Pacak K; North American Neuroendocrine Tumor Society (NANETS) . The North American Neuroendocrine Tumor Society consensus guideline for the diagnosis and management of neuroendocrine tumors: pheochromocytoma, paraganglioma, and medullary thyroid cancer. Pancreas. 2010;39(6):775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer. 2014;14(2):108-119. [DOI] [PubMed] [Google Scholar]
- 4. Qin Y, Yao L, King EE, et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat Genet. 2010;42(3):229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toledo RA, Burnichon N, Cascon A, et al. Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat Rev Endocrinol. 2017;13(4):233-247. [DOI] [PubMed] [Google Scholar]
- 6. Deng Y, Qin Y, Srikantan S, et al. The TMEM127 human tumor suppressor is a component of the mTORC1 lysosomal nutrient-sensing complex. Hum Mol Genet. 2018;27(10): 1794-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng Y, Flores SK, Cheng Z, et al. Molecular and phenotypic evaluation of a novel germline TMEM127 mutation with an uncommon clinical presentation. Endocr Relat Cancer. 2017;24(11):L79-L82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin Y, Deng Y, Ricketts CJ, et al. The tumor susceptibility gene TMEM127 is mutated in renal cell carcinomas and modulates endolysosomal function. Hum Mol Genet. 2014;23(9):2428-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Srikantan S, Deng Y, Cheng ZM, et al. The tumor suppressor TMEM127 regulates insulin sensitivity in a tissue-specific manner. Nat Commun. 2019;10(1):4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao L, Schiavi F, Cascon A, et al. Spectrum and prevalence of FP/TMEM127 gene mutations in pheochromocytomas and paragangliomas. JAMA. 2010;304(23):2611-2619. [DOI] [PubMed] [Google Scholar]
- 12. Abermil N, Guillaud-Bataille M, Burnichon N, et al. TMEM127 screening in a large cohort of patients with pheochromocytoma and/or paraganglioma. J Clin Endocrinol Metab. 2012;97(5):E805-E809. [DOI] [PubMed] [Google Scholar]
- 13. Toledo SP, Lourenço DM Jr, Sekiya T, et al. Penetrance and clinical features of pheochromocytoma in a six-generation family carrying a germline TMEM127 mutation. J Clin Endocrinol Metab. 2015;100(2):E308-E318. [DOI] [PubMed] [Google Scholar]
- 14. Neumann HPH, Sullivan M, Winter A, et al. Germline mutations of the TMEM127 gene in patients with paraganglioma of head and neck and extraadrenal abdominal sites. J Clin Endocrinol Metab. 2011;96(8):E1279-E1282. [DOI] [PubMed] [Google Scholar]
- 15. Bausch B, Wellner U, Bausch D, et al. Long-term prognosis of patients with pediatric pheochromocytoma. Endocr Relat Cancer. 2014;21(1):17-25. [DOI] [PubMed] [Google Scholar]
- 16. Hamidi O, Young WF Jr, Iñiguez-Ariza NM, et al. Malignant pheochromocytoma and paraganglioma: 272 patients over 55 years. J Clin Endocrinol Metab. 2017;102(9):3296-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernandez KG, Ezzat S, Morel CF, et al. Familial pheochromocytoma and renal cell carcinoma syndrome: TMEM127 as a novel candidate gene for the association. Virchows Arch. 2015;466(6):727-732. [DOI] [PubMed] [Google Scholar]
- 18. Casey RT, Warren AY, Martin JE, et al. Clinical and molecular features of renal and pheochromocytoma/paraganglioma tumor association syndrome (RAPTAS): case series and literature review. J Clin Endocrinol Metab. 2017;102(11):4013-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flores SK, Deng Y, Cheng Z, et al. Functional characterization of TMEM127 variants reveals novel insights into its membrane topology and trafficking. J Clin Endocrinol Metab. 2020;105(9):e3142-e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flores SK, Deng Y, Cheng Z, et al. Functional characterization of germline variants in the TMEM127 tumor suppressor reveals novel insights into its membrane topology and trafficking. bioRxiv. doi:10.1101/2020.04.08.031039, April 9, 2020, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lam AK. Update on adrenal tumours in 2017 World Health Organization (WHO) of endocrine tumours. Endocr Pathol. 2017;28(3):213-227. [DOI] [PubMed] [Google Scholar]
- 22. Eisenhofer G, Klink B, Richter S, Lenders JW, Robledo M. Metabologenomics of phaeochromocytoma and paraganglioma: an integrated approach for personalised biochemical and genetic testing. Clin Biochem Rev. 2017;38(2):69-100. [PMC free article] [PubMed] [Google Scholar]
- 23. Bausch B, Schiavi F, Ni Y, et al. ; European-American-Asian Pheochromocytoma-Paraganglioma Registry Study Group . Clinical characterization of the pheochromocytoma and paraganglioma susceptibility genes SDHA, TMEM127, MAX, and SDHAF2 for gene-informed prevention. JAMA Oncol. 2017;3(9):1204-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ben Aim L, Pigny P, Castro-Vega LJ, et al. Targeted next-generation sequencing detects rare genetic events in pheochromocytoma and paraganglioma. J Med Genet. 2019;56(8):513-520. [DOI] [PubMed] [Google Scholar]
- 25. Eijkelenkamp K, Olderode-Berends MJW, van der Luijt RB, et al. Homozygous TMEM127 mutations in 2 patients with bilateral pheochromocytomas. Clin Genet. 2018;93(5):1049-1056. [DOI] [PubMed] [Google Scholar]
- 26. Elston MS, Meyer-Rochow GY, Prosser D, Love DR, Conaglen JV. Novel mutation in the TMEM127 gene associated with phaeochromocytoma. Intern Med J. 2013;43(4):449-451. [DOI] [PubMed] [Google Scholar]
- 27. Fernández-Pombo A, Cameselle-Teijeiro JM, Puñal-Rodríguez JA, et al. Novel TMEM127 variant associated to bilateral phaeochromocytoma with an uncommon clinical presentation. Case Rep Endocrinol. 2019;2019:2502174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fishbein L, Leshchiner I, Walter V, et al. ; Cancer Genome Atlas Research Network . Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell. 2017;31(2):181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flynn A, Benn D, Clifton-Bligh R, et al. The genomic landscape of phaeochromocytoma. J Pathol. 2015;236(1):78-89. [DOI] [PubMed] [Google Scholar]
- 30. Papathomas TG, Oudijk L, Persu A, et al. SDHB/SDHA immunohistochemistry in pheochromocytomas and paragangliomas: a multicenter interobserver variation analysis using virtual microscopy: a multinational study of the European Network for the Study of Adrenal Tumors (ENS@T). Mod Pathol. 2015;28(6):807-821. [DOI] [PubMed] [Google Scholar]
- 31. Patócs A, Lendvai NK, Butz H, et al. Novel SDHB and TMEM127 mutations in patients with pheochromocytoma/paraganglioma syndrome. Pathol Oncol Res. 2016;22(4):673-679. [DOI] [PubMed] [Google Scholar]
- 32. Takeichi N, Midorikawa S, Watanabe A, et al. Identical germline mutations in the TMEM127 gene in two unrelated Japanese patients with bilateral pheochromocytoma. Clin Endocrinol (Oxf). 2012;77(5):707-714. [DOI] [PubMed] [Google Scholar]
- 33. Welander J, Andreasson A, Juhlin CC, et al. Rare germline mutations identified by targeted next-generation sequencing of susceptibility genes in pheochromocytoma and paraganglioma. J Clin Endocrinol Metab. 2014;99(7):E1352-E1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu R, Sharaga D, Donner C, Palma Diaz MF, Livhits MJ, Yeh MW. Pheochromocytomatosis associated with a novel TMEM127 mutation. Endocrinol Diabetes Metab Case Rep. 2017;2017:17-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laboureau S, Guichet A, Duriez T, et al. New case of bilateral pheochromocytomas involving the homozygous TMEM127 mutation. Clin Genet. 2018;94(2):278-279. [DOI] [PubMed] [Google Scholar]
- 36. Paredes SCS, Lopes SGC, Torres IMBF, Alves MLF. Pheochromocytoma due to TMEM127 mutation—the importance of genetic testing for clinical decision. Eur Endocrinol. 2020;16(1):72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37(3):235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cingolani P, Patel VM, Coon M, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plon SE, Eccles DM, Easton D, et al. ; IARC Unclassified Genetic Variants Working Group . Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kushima I, Aleksic B, Nakatochi M, et al. Comparative analyses of copy-number variation in autism spectrum disorder and schizophrenia reveal etiological overlap and biological insights. Cell Rep. 2018;24(11):2838-2856. [DOI] [PubMed] [Google Scholar]
- 43. Lenders JW, Duh QY, Eisenhofer G, et al. ; Endocrine Society . Pheochromocytoma and paraganglioma: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2014;99(6):1915-1942. [DOI] [PubMed] [Google Scholar]
- 44. Favier J, Amar L, Gimenez-Roqueplo AP. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2015;11(2):101-111. [DOI] [PubMed] [Google Scholar]
- 45. Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or in the data repositories listed in “References.”