Abstract
Identification of toxicants that underlie neurological diseases is a neglected area awaiting a valid strategy to identify such toxicants. We sought biomarkers that respond to known neurotoxicants in LUHMES immortalized neurons and evaluated these biomarkers for use in screening libraries of environmental toxicants. LUHMES immortalized human dopaminergic neurons were surveyed by RNA sequencing following challenge with parkinsonian toxicants rotenone, 6-hydroxydopamine, MPP+, and ziram (zinc dimethyldithiocarbamate; Zn2+DDC2), as well as additional toxicants paraquat, MS275, and methylmercury. The metallothionein gene MT1G was the most dynamic gene expression response to all seven toxicants. Multiple toxicants also increased transcripts for SLC30A1 and SLC30A2 zinc secretion transporters, the SLC7A11 xCT cystine/glutamate antiporter important for glutathione synthesis, DNA damage inducible transcript 3 (DDIT3), and secreted growth factors FIBIN and CXCL12, whereas several toxicants decreased expression of the apelin growth factor (APLN). These biomarker genes revealed stress responses to many toxicants at sub-cytotoxic concentrations. Since several of these biomarker genes and prior neurological disease studies implicated disruption of metal distribution, we tested metal chelator thiram (dimethyldithiocarbamate, DDC), ziram, and several other metals and metal chelates for cytotoxicity and induction of MT1G expression. Metals and chelators that caused dynamic increases in MT1G expression also caused cytotoxicity, except Ni2+DDC2 induced MT1G at 5 μM, but lacked cytotoxicity up to 100 μM. These results bolster prior work suggesting that neurons are characteristically sensitive to depletion of glutathione or to disruption of cellular metal distribution and provide biomarkers to search for such neurotoxicants in chemical libraries.
Keywords: Dopaminergic neurons, LUHMES, Metallothionein, MT1G, Metal metabolism, Chelator
Introduction
Neurodegenerative diseases are characterized by the death of mature neurons. Although Alzheimer’s disease (AD) is primarily a genetic disease, other neurodegenerative diseases appear to have a large environmental component such as Parkinson’s disease (PD), amyotrophic lateral sclerosis, and frontotemporal dementia (Tanner et al. 2014). Research on AD and PD has focused on preventing accumulation of Aβ and synuclein, respectively. Drugs that inhibit Aβ accumulation have failed to ameliorate AD, raising the possibility that protein deposits may be more symptomatic than causal.
Identification of chemicals that kill mature neurons is paramount, but few toxicants have been implicated in neurodegenerative toxicity. Manganese and lead show neurodegenerative effects; however heavy metals are often regarded as interfering in neurodevelopment rather than killing mature neurons (Grandjean and Herz 2015; Tanner et al. 2014). Rotenone (ROT) and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) or its active metabolite 1-methyl-4-phenylpyridinium (MPP+, called here MPP) were identified as risk factors for PD and parkinsonism, respectively, largely by epidemiology studies, then supported by acute animal studies. Epidemiology has identified MPTP, rotenone, and other pesticides in triggering parkinsonism or PD; however, the causative pesticide(s) aside from rotenone are less clear, and high lifetime exposures to pesticides by spraying or industrial exposure result in only a 2–3-fold increased risk of PD. Exposure to the fungicide ziram is also implicated as a risk factor for PD (Wang et al. 2011); however, it has been difficult to distinguish risk factors among subjects who are administering multiple pesticides (Wang et al. 2011). Exposure of rats to lead along with thiram (dimethyldithiocarbamate, or DDC), or ziram, which is a Zn2+DDC2 complex, may facilitate lead or other heavy metal uptake into the brain (Oskarsson 1987). The distinction between parkinsonism caused by MPTP or 6-hydroxydopamine (6HD) administration and PD is that the cause(s) of PD are unknown. Since the effects and order of progression are so similar, we consider that the shared effects of rotenone, MPTP, and 6HD, all inhibitors of mitochondrial electron transport, may be useful tools to establish screens for neurodegenerative toxicity.
Neurotoxicology faces a major hurdle in identifying toxicants that act chronically to cause diseases. Because chronic exposures are impractical using cell culture models and chemical screening, chronic effects are often extrapolated from acute exposures, often using biomarkers. Recent years have seen improved neuronal cell culture models and validation of such models, including LUHMES conditionally immortalized dopaminergic neurons, neurons differentiated from stem cells, “3-D culture” using neurospheres, and co-culture of neurons with glia. Our lab has shown that LUHMES neurons are particularly sensitive to known neurotoxicants, and that low levels of glutathione and sensitivity to disruption of metal distribution underlay some of these sensitivities (Tong et al. 2017, 2018).
Using RNA sequencing to survey neurotoxicants revealed that the MT1G gene is a dynamic biomarker that responded to cell stresses in LUHMES neurons. Metallothioneins (MTs) are a family of small intracellular proteins that bind metal ions. They can bind up to 18 metals but exist mostly as Zn2+ chelates in healthy cells. MTs have a range of affinities for metal ions such that Zn2+ is preferentially displaced by metals with higher affinities, Cu+, Cd2+, Pb2+, Ag+, Hg2+, or Bi2+ (Dong et al. 2015). In addition to sequestering toxic heavy metals and participating in cellular zinc and copper homeostasis, MTs also sense oxidative stress in the following manner. If an electron acceptor oxidizes cysteine to cystine, MTs release zinc ions. Free zinc displaced from MTs by oxidants or other metals may bind to the six zinc finger domains in metal-responsive transcription factor 1 (MTF1) to activate transcription of adaptive genes such as MTs and NQO1 (Dong et al. 2015). Mice have four Mt genes, Mt1–4, whereas humans have MT2–4 orthologs, and eight MT1- genes that are expressed and inducible in various cell types. MT3 is reportedly expressed selectively in neurons and astrocytes where it does not respond to cell stresses by increasing in expression (Hozumi et al. 1998). Mt1 and Mt2 mRNAs and proteins, in contrast, are thought to increase dramatically in mouse brain following toxicant treatment or a variety of other stresses (Chung and West 2004; Miyazaki et al. 2011). This study identifies MT1G as a highly dynamic biomarker in LUHMES human immortalized neuron cell line and examines its responses to various toxicants in several cell lines.
Materials and Methods
Chemicals
Chemicals included Ferbam from TCI America, Portland, OR; ML385 and MS275 from Toris Biosciences, Minneapolis, MN; APTO235 (MedChemExpress, Princeton, NJ); and sankel (Muse Chem, Fairfield, NJ). Other chemical compounds in Table 1 were purchased from Sigma-Aldrich Chemical Co (now Merck KGaA, Darmstadt, Germany). Chemicals were dissolved in dimethylsulfoxide (DMSO, Sigma-Aldrich) at a concentration of 10 mM to make stock solutions, and stored at − 20 °C.
Table 1.
Chemicals used and abbreviations
| Abbreviation | Name | CAS number |
|---|---|---|
| Ziram | Zn2+DDC2 | 137-30-4 |
| ROT | Rotenone | 83-79-4 |
| PQ | Paraquat | 11089-65-9 |
| 6HD | 6-Hydroxydopamine | 28094-15-7 |
| MS5 | MS275 | 209783-80-2 |
| MPP | 1-Methyl-4-phenylpyridinium | 911295-24-4 |
| CAP | Captan | 133-06-2 |
| MHG | Methyl mercury (II) chloride | 115-09-3 |
| HCP | Hexachlorophene | 70-30-4 |
| MAP | 4-(Methylamino)phenol hemisulfate salt | 55-55-0 |
| CR2 | Sodium dichromate | 7789-12-0 |
| CLM | Chlorambucil | 305-03-3 |
| STA | Staurosporine | 62996-74-1 |
| MNC | Manganese chloride | 64333-01-3 |
| TER | Terfenadine | 50679-08-8 |
| Thiram | DDC; dimethyldithiocarbamate | 137-26-8 |
| Ferbam | Fe3+DDC3 | 14484-64-1 |
| AC1LAOJX | Fe2+DDC2 | 15339-38-5 |
| Sankel | Ni2+DDC2 | 15521-65-0 |
| CuCl2 | Cu2+Cl2 | 7447-39-4 |
| APTO253 | APTO-253 | 916151-99-0 |
| ML385 | ML385 | 846557-71-9 |
Cell Culture and Differentiation
The five cell lines were cultured until nearly confluent, trypsinized, and then seeded at 1/4th the previous density. When cell lines reached 9 passages, they were discarded and replaced from frozen stocks. Cell line identities were verified by short tandem repeat profiling (WiCell Research Inst. Madison, Wisconsin, USA), and checked for mycoplasma contamination using the MycoAlert™ Mycoplasma Detection Kit (Lonza). LUHMES cells were provided by Professor Marcel Leist, University of Konstanz, Germany. LUHMES cells were grown in culture vessels coated with 50 μg/ml each poly-L-ornithine and human fibronectin in Advanced DMEM/F12 medium containing 2 mM L-gluta-mine, 1X N2-supplement and 40 ng/ml bFGF. LUHMES differentiation was induced by 1 μg/ml tetracycline and 1 mM cAMP contained in advanced DMEM/F12 medium with 2 mM L-glutamine, 1X N2-supplement, and 2 ng/ml GDNF (Schildknecht et al. 2009), and proceeded for 7 days prior to toxicant treatments. SHSY-5Y cells were obtained from ATCC (American Type Culture Collection, Manassas, VA), and cultured in EMEM/F12, a 1:1 mixture of Eagle’s Minimum Essential (EMEM, ATCC) and F12 (Invitrogen) including 100 μg/mL penicillin and 100 μg/mL streptomycin, as well as 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA). SH-SY5Y cell differentiation was induced in culture medium with 3% FBS, by adding 10 μM all trans-retinoic acid. StemPro®Human Neural Stem Cells (NSCs) were obtained from Invitrogen (now Thermo-Fisher, Waltham, MA, USA). Per the manufacturer’s manual, NSCs were cultured adherently in vessels coated with Geltrex and grown in StemPro® NSC SFM defined complete medium consisting of KnockOut® D-MED/F-12 with 2% StemPro Neural Supplement, 2 mM GlutaMax-I Supplement, and 20 ng/ml each human recombinant bFGF and EGF. NSCs were treated to induce neuronal differentiation in StemPro® NSC SFM medium without bFGF and EGF. Cell differentiation was started when the cells were at 60% confluence in flasks and continued for 7 days (Tong et al. 2017), followed immediately by toxicant treatments. Non-neural cell lines were cultured using the media and protocols indicated by ATCC (Gaithersburg, MD). HEK293(ATCC CRL1573), HepG2 (ATCC HB-8065), and SA7K (Sigma-Aldrich, St. Louis, MO, USA) were cultured in ATCC-formulated EMEM medium (ATCC #30–2003) containing penicillin and streptomycin 100 μg/mL, and 10% fetal bovine sera (Hyclone, Thermo-Fisher).
Cytotoxicity Assays
Cells were distributed in 384 well plates (~ 10,000 cells per well in 30 μL medium) for cytotoxicity assays. Toxicant stock solutions in DMSO were diluted with culture medium to make 10X working solutions for each concentration and added to the cells at 10% of the volume in each well. Control cells were treated with 1% DMSO to match treatments. Cells in duplicate 384 well plates were assayed for intracellular [ATP] using the CellTitreGlo® luminescence assay (Promega, Madison, WI, USA) 24 h after toxicant treatment. Luminescence was read in an EnVision plate reader (Perkin Elmer, Waltham Massachusetts, USA). Luminescence readings were scaled to DMSO treated control wells representing 100% viability.
qRT-PCR
For gene expression assays, 3 × 105 cells were cultured per well in 12 well plates with 2 mL medium to enable RNA extraction and quantitative reverse transcriptase PCR (qRT-PCR). Cells were treated with each toxicant for 6 h in 2 mL of medium in 12-well microplates. A single toxicant concentration was used for undifferentiated and differentiated cells from each cell line at approximately the threshold of toxicity determined in prior experiments (Tong et al. 2017). Total RNA was isolated from three biological replicate treatments using Qiagen RNeasy Plus Mini Kits (Qiagen, Hilden, Germany; Cat#:74106) including DNAse treatment to remove genomic DNA. Total RNA was quantitated in a NanoDrop-8000 and stored at − 80 °C. cDNA synthesis was carried out from 0.5 μg total RNA per sample using RT2 HT First Strand Kit (Qiagen). TaqMan® assays (Thermo-Fisher, Waltham MA, USA) used were: MT1G, Hs04401199_s1; APLN, Hs00175572_m1, CNN2, Hs00854264_s1; DDIT3, Hs00358796_g1; FIBIN, Hs00831591_s1; PDK4, Hs01037712_m1; and GAPDH, Hs02786624_g1. Resulting qRT-PCR data were first subjected to thresholding: Values > 36 and “undetermined” values were changed to 36. Sample data were normalized to the CT value for GAPDH. Standard deviations were calculated from the 3 replicates for each gene. Standard deviations > 2 were flagged and outlier data points were excluded from further analysis. The median was used to represent each set of replicates to minimize variance from sporadic failed or outlier reactions. Gene expression that was undetectable was assigned a value of 1.0, so that a lower-limit fold-increase could be estimated as expression level for treated cells divided by expression level for control cells. Ratios: The Treated/Control Ratio of expression was calculated from the normalized median Ct values as follows: 2(Control Ct - Treated Ct).
RNA Sequencing
RNA-Seq was performed using solely dLUHMES cells after 7 days of differentiation followed by 6 hours of treatment. Three biological replicates were generated for each of eight treatments: 0.5% DMSO vehicle, 6-hydroxydopamine (6HD) 1 μM, 1-methyl-4-phenylpyridinium (MPP) 5 μM, rotenone (ROT) 1 μM, paraquat (PQ) 5 μM, ziram 2 μM, methylmercury (MHG) 0.5 μM, and MS275 (Vashishta and Hetman 2014) 5 μM. Total RNA samples were prepared from cultured cells using Qiaquick kit (Qiagen, Hilden Germany) and selected for polyA+ transcripts using the Qiagen Oligotex midi kit. Resulting samples were depleted of ribosomal sequences using two rounds of RiboZero Gold™, Illumina Inc. RNAseq sequencing was performed using the TruSeq® kit and Illumina HiSeq 2000 instrument by the NHLBI-NIH DNA Sequencing and Genomics Core. RNA-Seq data were mapped to human genes according to Genome Reference Consortium Human genome build 38 (GRCh38) of the human genome using STAR Aligner software. HTSeq-count was used to count the aligned reads in genomic features to produce count data for each annotated gene. RNA-Seq for the eight treatments yielded an average of 72.0 million mapped reads per treatment and a minimum of 46.7 million mapped reads for ROT. Gene expression data are available in the Gene Expression Omnibus accession # GSE150005 and sample information are summarized in Supporting Information, SI, Table S1. Gene count data were normalized between samples and statistical analysis was performed using DESeq. Two group statistical tests were performed in DESeq using the nbinomTest method which applies a Benjamini-Hochberg correction to provide an estimate of False Discovery Rate (FDR). Significant gene responses were defined as genes that showed ≥ 25 counts in at least one treatment group, exceeding 2-fold change, and < 0.1 FDR. According to these criteria, 2797 genes responded significantly to one or more treatments.
Results and Discussion
LUHMES Neurons’ Transcriptional Responses to Neurotoxicants by Deep RNA Sequencing
We began by searching for biomarkers for neurodegenerative toxicity by performing genome-wide RNA sequencing (RNA-Seq) after treating differentiated LUHMES neurons (dLUHMES; Tong et al. 2017) with vehicle, parkinsonian toxicants ROT, 6HD, or MPP; suspected parkinsonian toxicants ziram, paraquat and MS275 (Vashishta and Hetman 2014), or developmental neurotoxicant MHG.
LUHMES neurons responded to treatments using seven toxicants with transcriptional changes that shared many genes in common. The MT1G gene responded statistically significantly, and most dynamically, to all seven toxicants. The MT1E and MT2A metallothionein genes were the second and third most dynamic responses, verifying the co-regulation of MT1-subfamily and MT2A genes that has been reported in other cells and tissues (Hardyman et al. 2016). Figure 1b summarizes the normalized counts for MT1G, MT1E, and MT2A that are masked by the ratios depicted in the Fig. 1a heatmap. MT1G was represented in control samples by only 2 counts among 92 million mapped reads, hence it is difficult to precisely quantify this transcript in control dLUHMES cells. Figure 1b also shows that MT1G, MT1E and MT2A genes were robustly and similarly increased by all seven toxicants, with ziram provoking the greatest increases. MT1X, MT1H, MT1L, and MT1F transcripts were also increased markedly by ziram, and to a lesser extent by MHG in this study, whereas MT1B was detected at an average of seven copies per replicate exclusively in the ziram-treated samples. The quantitative agreement between the three MT genes in relative responses to seven toxicants (Fig. 1b) is consistent with the known co-regulation of MT1- and MT2A genes in other cell types. This agreement among MT-gene responses also adds confidence to the veracity of each individual observation. MT3 transcripts were not found in control samples and yielded only seven copies in total distributed among all 24 samples from this study (Table 2). The MT3 gene is selectively expressed in neurons and astrocytes in vivo, however the MT3 protein is known to block growth of neurons (Hozumi et al. 1998), so we speculate the LUHMES cell line has been selected against MT3 expression lest it block cell growth.
Fig. 1.
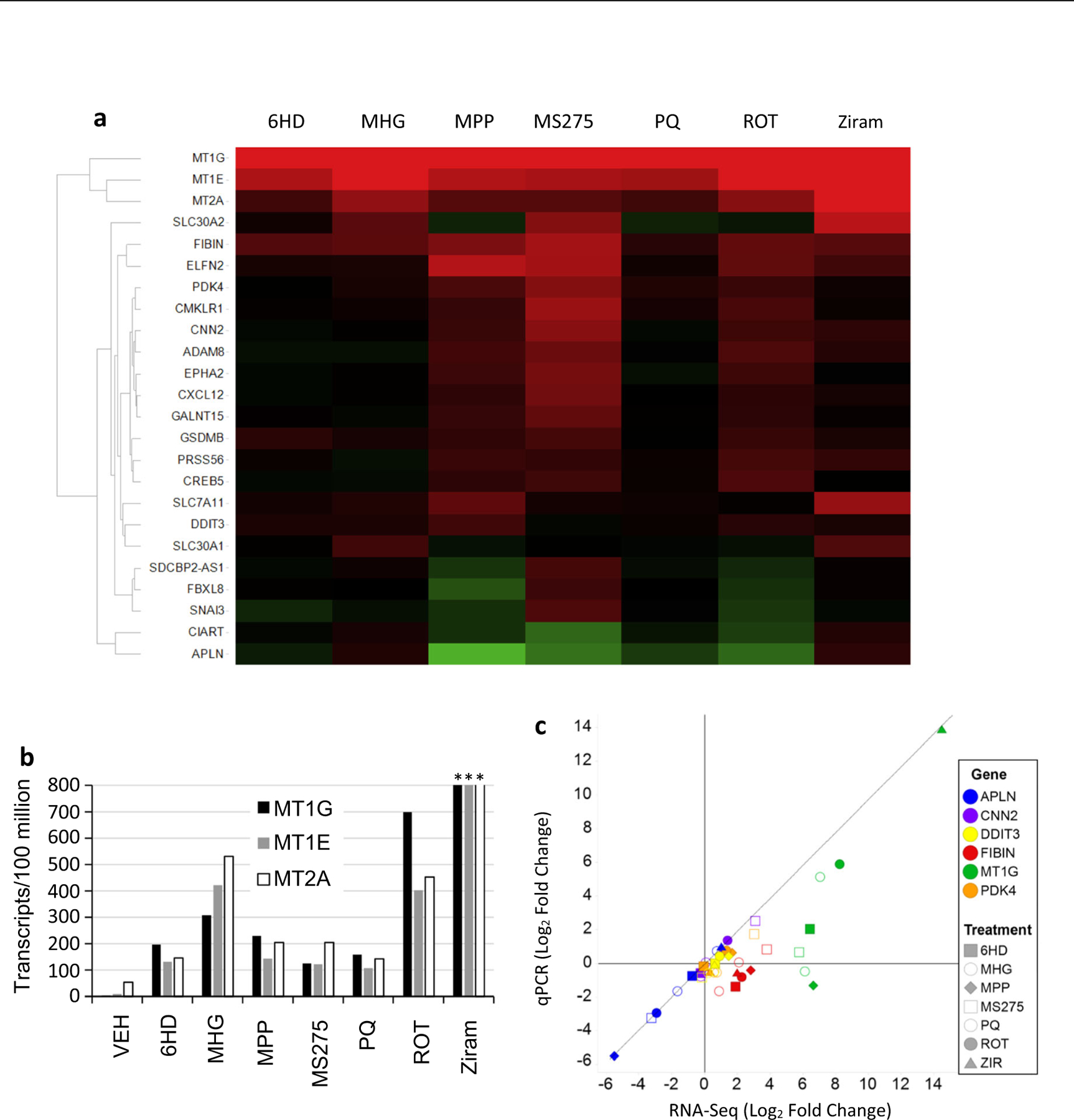
RNA sequencing responses of LUHMES neurons to neurotoxicants. dLUHMES neurons were treated with parkinsonian toxicants 6HD, 6-hydroxydopamine 1 μM; MPP, 1-methyl-4-phenylpyridinium 5 μM, ROT, rotenone 1 μM; and additional neurotoxicants PQ 5 μM, ziram (ZIR) 2 μM, MHG 0.5 μM, and possible neurotoxicant MS275 5 μM (Vashishta and Hetman 2014). a Heatmap shows genes as rows and chemical treatments as columns. Colors indicate increased (red) or decreased (green) mRNA relative to vehicle control with color saturation at 32-fold. No cytotoxicity was observed after 24 h at these concentrations. b Bar graph shows mRNA reads counted per 100 million normalized mRNA transcripts for three metallothionein genes. Among 83 million mRNA reads for three replicate vehicle-treated samples, MT1G, MT1E, and MT2A reads numbered 2, 8, and 42 reads, respectively. ***Ziram-treated dLUHMES neurons yielded 53,000, 31,000 and 31,000 transcripts per 100 million for MT1G, MT1E, and MT2A, respectively. c Dot plot comparing RNA-Seq and qRT-PCR quantitation of responses by six biomarker genes using the samples from “a” above
Table 2.
Expression of MT genes in astrocytes or medium spiny neurons from mouse striatum compared with dLUHMES neurons. Expression is shown as reads per million using mouse striatum scRNA-seq (Gokce et al. 2016) and RNA-seq analysis of human dLUHMES in this study
| Mouse gene | Astrocytes | Neurons | Human gene | dLUHMES + vehicle | dLUHMES + ziram |
|---|---|---|---|---|---|
| Mt1 | 630 | 170 | MT1G | 0.024 | 530 |
| Mt2 | 400 | 13 | MT2A | 0.53 | 310 |
| Mt3 | 470 | 190 | MT3 | <0.1 | 0.028 |
| Mt4 | <2 | <2 | MT4 | <0.1 | <0.1 |
Several toxicants caused increased expression of SLC30A1 and SLC30A2 genes in dLUHMES neurons, particularly methylmercury chloride (MHG) and the pesticide ziram. Both of these transporter genes play roles in secretion of excess Zn2+ ions from cells (Schweigel-Rontgen 2014). The SLC30A1 and SLC30A2 genes responded to the two toxicants that provoked the greatest MT responses, ziram and MHG, and SLC30A2 also responded to MS275. The increased expression of MT genes by all seven neurotoxicants, and the increased expression of these SLC30A1 and SLC30A2 zinc secretion genes in response to the toxicants that provoked the greatest MT responses collectively suggest a common response mechanism. Given the roles of MTs and SLC30- transporters in regulating metal ion distribution in cells, these observations suggest that these neurotoxicants are dysregulating metal distribution and that LUHMES neurons are susceptible to disruption of metal homeostasis.
Several additional genes that responded to multiple neurotoxicants play roles relevant to brain responses to stress, hence are possible biomarkers. For example, the SLC7A11 gene was dynamically upregulated by neurotoxicants ziram, MHG, and MPP. SLC7A11 encodes xCT, the light chain of the cystine/glutamate antiporter xC−. xC− plays a vital role in uptake of cystine for synthesis of glutathione, suggesting that these three toxicants may deplete glutathione (Ottestad-Hansen et al. 2018). It is notable that neurons in vivo are known to contain low levels of glutathione (Sun et al. 2006) and that dLUHMES neurons are particularly sensitive to toxicants that deplete glutathione (Tong et al. 2018). The CNN2 gene was also upregulated by MPP, MS275, ROT and ziram. Transgenic and knockdown studies in PC12 cells indicate that the CNN2 gene positively regulates neurite length (Pape et al. 2008), possibly by organizing actin filaments. Since CNN2 responded to four neurotoxicants and can facilitate neurite growth, CNN2 may be considered a stress response gene in neurons.
Secreted peptides are of special interest as candidate biomarkers. Three peptide hormone genes responded to at least three toxicants in this study, FIBIN, APLN, and CXCL12 (Fig. 1). FIBIN mRNA was increased significantly by MPP, MS275, and ROT, and showed increases that did not reach significance for the other four toxicants. The FIBIN gene encodes a secreted peptide hormone that plays a poorly defined role in development. Glucocorticoid stress hormones have been found to upregulate expression of FIBIN (Rashidi-Nezhad et al. 2014). Chemokine CXCL12 was similarly increased by MPP, MS275, and ROT treatments. The CXCL12 chemokine is made by stromal cell types and promotes angiogenesis and inflammation (Janssens et al. 2018). In contrast, the APLN gene was downregulated by MPP, MS275 and ROT. APLN encodes another peptide hormone, apelin, which plays neuroprotective functions by inhibiting neuronal apoptosis and promoting angiogenesis (Ishimaru et al. 2017; Wu et al. 2017).
Master regulatory genes that control metabolism or cell cycle arrest in response to cell stress were also examined. The PDK4 gene encodes Pyruvate Dehydrogenase Kinase 4, a regulatory enzyme that preferentially converts pyruvate into lactate rather than Acetyl-CoA, thereby increasing glycolysis and limiting aerobic metabolism. PDK4 was increased significantly in response to MPP, MS275, and rotenone. Another example was the DDIT3 gene for “DNA damage inducible transcript 3.” DDIT3 was statistically significantly increased only by MPP but showed a trend toward increase for all the seven toxicants in Fig. 1 except MS275 (Supplementary data, Gene Expression Omnibus, Accession # GSE150005). DDIT3 encodes a transcription factor that responds to DNA damage and to oxidative stress by regulating a series of other genes to halt the cell cycle and limit cell movement (Jauhiainen et al. 2012).
Further analyses were performed using qRT-PCR to analyze selected biomarker genes MT1G, APLN, CNN2, DDIT3, FIBIN, and PDK4. Therefore, these qRT-PCR assays were also qualified by analyzing the RNA samples used for the RNAseq experiment shown in Fig. 1a, b. Figure 1c compares the mRNA Log2 fold change values from RNA-Seq and qRT-PCR for these six biomarker genes and GAPDH to normalize data. Figure 1c shows that APLN, CNN2, DDIT3, and PDK4 showed excellent agreement between the two techniques. MT1G and FIBIN show a systematic under-representation of fold change values by qRT-PCR relative to RNA-Seq. This under-representation by qRT-PCR appears to stem from the low-level expression of these two genes in the control samples, requiring 30.6 and 26.3 cycles for detection of MT1G and FIBIN, respectively. This comparison also suggested that deep RNA sequencing of 30 million or more matched reads per sample showed greater sensitivity and dynamic range than qRT-PCR.
Expression of Metallothionein Family Genes in Neurons
The dynamic increases in mRNAs for MT2A and at least four of the eight MT1 genes in the LUHMES neuron’s responses to neurotoxicants were surprising in light of prior reports that indicated expression and increases of MT protein or mRNA in astrocytes but not in neurons of mice in vitro or in vivo (Gokce et al. 2016; Hozumi et al. 1998; Miyazaki et al. 2011). This led us to reexamine expression of mRNA for the four mouse Mt genes in neurons and astrocytes. Single cell RNA sequencing data were used from Gokce et al. who isolated single cells from mouse striatum and used RNA sequencing to gather approximately 2000–12,000 mRNA reads from each cell (Gokce et al. 2016). Table 2 shows a comparison of Mt1–4 abundances from 107 astrocytes and 837 medium spiny neurons from mouse striatum, as well as a summary of four MT genes from human. These results confirmed the lack of Mt4 expression and abundance of Mt3 expression in mouse neurons and astrocytes. These results do reveal relatively abundant expression of Mt1 (170 reads per million) and weak expression of Mt2 (13 reads per million) in mouse neurons relative to mouse astrocytes. The scRNA-seq cells were derived from healthy mice, however it is possible that the preparation of these cells caused stress that may have increased expression of Mt1 and Mt2. In any case, these results do indicate expression of Mt1 and Mt2 in mouse neurons, albeit at lower mRNA levels than in astrocytes. Although these results do not directly contradict the protein immunostaining and in situ hybridization results from prior studies, they do provide quantitative evidence that neurons in vivo can express Mt1 and Mt2 genes at significant levels.
Surveying Three Biomarker Genes and Cytotoxicity Responses to 15 Toxicants in Six Cell Lines
We next asked whether biomarker genes MT1G, PDK4, and DDIT3 responded similarly to toxicants in other neural and non-neural cell types. These three candidate stress response biomarker genes were surveyed for responses in six varied cell types, to a panel of fifteen toxicants (Fig. 2). In these experiments, qRT-PCR was used to assay MT1G, PDK4, and DDIT3 expression by comparison with housekeeping gene GAPDH. Responses were tested in human cell lines: dLUHMES immortalized neurons(Lotharius et al. 2005), HEK293 immortalized neuronal lineage cells derived from embryonic kidney tissue(Shaw et al. 2002), HepG2 hepatocarcinoma cells(Donato et al. 2015), HUVEC primary umbilical endothelial cells(Chu et al. 2020), SA7K immortalized proximal tubule epithelial cells(Li et al. 2017), and SHSY-5Y neuroblastoma cells(Odelstad et al. 1981). A panel of fifteen known or suspected neurotoxicants (Tong et al. 2017) was used to challenge these six cell lines at concentrations that were threshold-cytotoxic in dLUHMES and SH-SY5Y cells. Figure 2 shows that these toxicants exhibited cytotoxicity that varied considerably by cell type. Notably, dLUHMES cells were most sensitive to killing by 6HD, MAP, CLM, ROT, MHG, and ziram; consistent with prior comparisons between dLUHMES, SH-SY5Y and differentiating neural stem cells (Tong et al. 2017). SH-SY5Y cells were most sensitive to killing by CAP, CR2 and TER, and HUVEC cells were most sensitive to killing by HCP. Cytotoxicity was then compared with biomarker gene expression assayed by qRT-PCR.
Fig. 2.
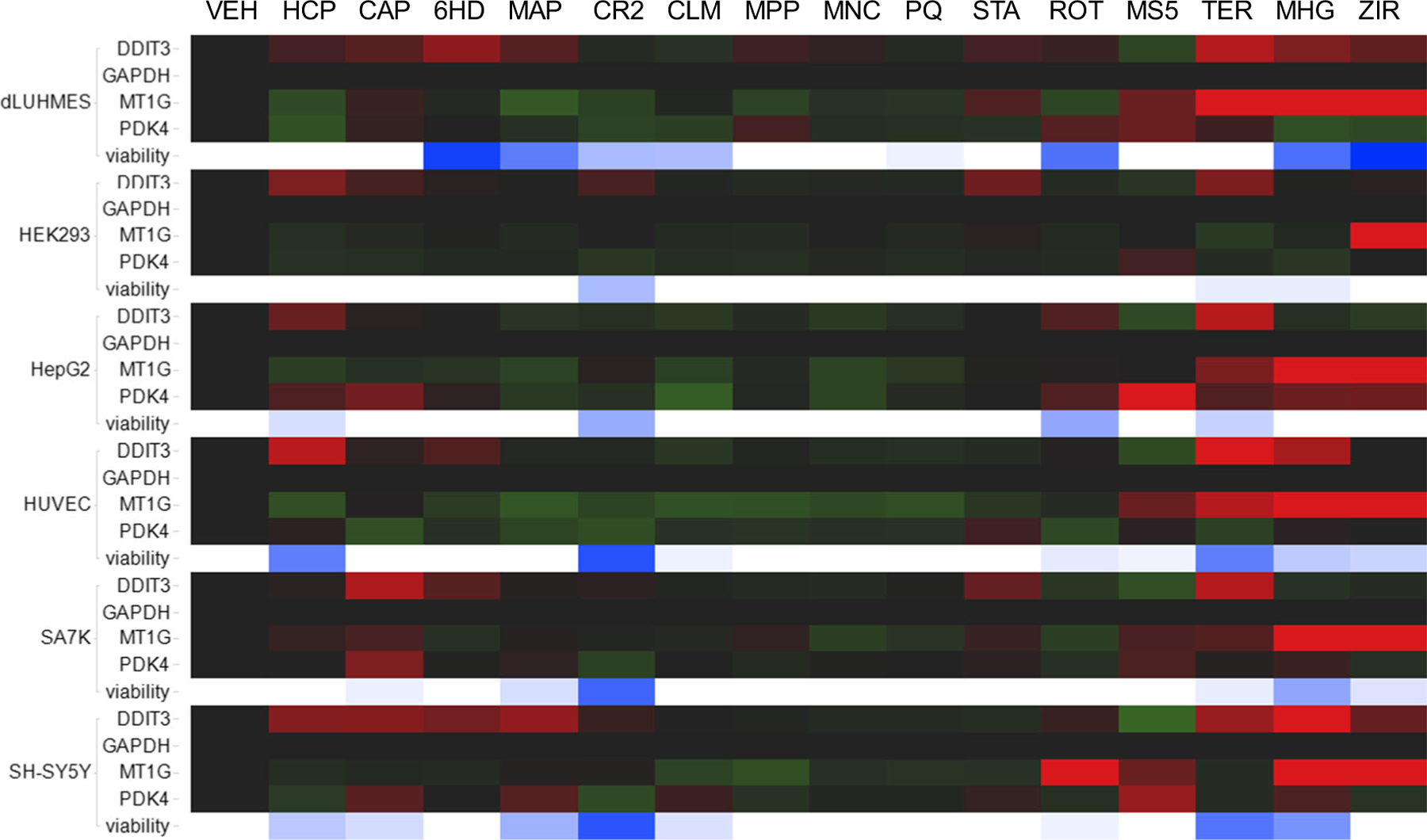
Transcriptional responses of three biomarker genes to 15 toxicants in six cell lines. The six cell lines indicated at left were treated with vehicle or one of 15 toxicants. Transcriptional responses by DDIT3, MT1G, PDK4, and GAPDH were quantified by qRT-PCR at 6 h, and cell viability was assessed at 24 h as intracellular [ATP]. Below the four genes for each cell line is a row of cells that indicate cell viability. White indicates no detectable loss of viability whereas blue color saturation indicates the % of cell death 24 h post-treatment. Toxicant treatments were HCP, Hexachlorophene 10 μM; CAP, captan 10 μM; 6HD, 6-hydroxydopamine 3 μM; MAP, 4-(methylamino)phenol hemisulfate salt 5 μM; CR2, sodium dichromate 5 μM; CLM, chlorambucil 10 μM; MPP, 1-methyl-4-phenylpyridinium 10 μM; MNC, manganese chloride 10 μM; PQ, paraquat 10 μM; STA, staurosporine 2 μM; ROT, rotenone 3 μM; MS5, MS275 10 μM; TER, terfenadine 10 μM; MHG, methylmercury chloride 2 μM; ZIR, ziram, 6 μM
qRT-PCR was used to survey expression of MT1G, DDIT3, PDK4, and GAPDH in these six cell lines. DDIT3 expression was increased in most or in all six cell lines by treatment with HCP, CAP, 6HD, TER, MHG, and ziram (Fig. 2). Clearly, the DDIT3 gene was responsive to particular toxicants in all six cell lines. Curiously, MS275 downregulated DDIT3 in all six cell lines, perhaps by inhibiting histone deacetylases (HDACs) that regulate transcription downstream of transcription factors. This is consistent with reports that MS275 and other nonselective inhibitors of histone deacetylases may exacerbate cytotoxicity by inhibiting DNA repair responses (Zhou et al. 2020).
PDK4 gene transcripts were variously increased or decreased by toxicants in dLUHMES, HepG2, SA7K, and SH-SY5Y cell lines. PDK4 encodes pyruvate dehydrogenase kinase 4, an enzyme that promotes glycolysis and inhibits aerobic pyruvate metabolism. PDK4 responses were most dynamic in HepG2 cells, consistent with the central role of hepatocytes in regulating the bodies’ glucose anabolism and catabolism. It was intriguing to see that PDK4 was decreased as well as increased by various toxicant-cell combinations; however, PDK4 transcripts were less dynamic than DDIT3 or MT1G. Since PDK4 responses to toxicants were hypodynamic, and highly variable by toxicant, PDK4 was not pursued further as a biomarker of toxicity.
MT1G was increased dynamically in each of the six cell lines by various toxicants (Fig. 2). MT1G responded to ziram in all six cell lines, to MHG in all cell lines except HEK293, to MS275 in dLUHMES, SH-SY5Y, HUVEC, and SA7K, and to TER in dLUHMES, HepG2, and SA7K. In HEK293 cells the MT1G gene responded only to ziram; in fact, HEK293 cells showed the fewest transcript responses for all three biomarker genes, suggesting that this cell line may lack several cell stress response pathways. The MT1G response to ROT was observed only in SH-SY5Y cells. Scrutiny revealed that the MT1G transcripts were not detected by qRT-PCR in vehicle-treated dLUHMES cells, and the fold-change responses of MT1G to toxicants by dLUHMES were 10–100-fold lower as measured by qRT-PCR relative to RNA-seq. This is unsurprising in light of the extremely low transcript numbers seen in vehicle-treated dLUHMES cells by deep RNA sequencing (Fig. 1b) and the reduced dynamic range observed for MT1G by qRT-PCR (Fig. 1c); however, this implies that MT1G transcript responses less than 10–100-fold were likely to be missed using qRT-PCR. Samples from the RNA-seq experiment shown in Fig. 1 were assayed for MT1G by qRT-PCR. Again, MT1G transcripts were below the threshold of detection (36 cycles) by qRT-PCR in vehicle-treated dLUHMES cells, and the fold-change responses of MT1G to toxicants were much lower by qRT-PCR relative to RNA-seq. Although it is possible that more efficient cDNA synthesis reactions would have improved the sensitivity of the qRT-PCR method, we conclude that deep RNA sequencing was much more sensitive than qRT-PCR in detecting rare transcripts and their responses.
The most important result from Fig. 2 was that for many toxicants, biomarker genes responded at 6 h at concentrations that did not cause measurable cytotoxicity at 24 h. For example, DDIT3 increased without cytotoxicity in HCP-treated HEK293 cells; in CAP-treated dLUHMES, HEK293, and SA7K cells; in 6HD-treated HUVEC and SA7K cells; in MPP-treated dLUHMES cells; in STA-treated dLUHMES, HEK293, and SA7K cells; in TER-treated dLUHMES and SA7K cells; and in ziram-treated SH-SY5Y cells. Similarly, qRT-PCR revealed MT1G increases without cytotoxicity in CAP-treated SA7K cells; in ROT-treated SH-SY5Y cells; in MS275-treated dLUHMES, HUVEC, SA7K, and SH-SY5Y cells; in TER-treated dLUHMES cells; in MHG-treated HepG2 cells; and in ziram-treated HEK293, HepG2, and SH-SY5Y cells. Thus, measuring dynamic biomarker responses may provide increased sensitivity relative to measuring cytotoxicity. In other cases either the toxicant concentration used provoked neither biomarker changes nor cytotoxicity, or both events, such that the relative sensitivities of biomarkers and cytotoxicity were unresolved.
Cytotoxicity and MT1G Induction in Neural Cell Lines by Metal Chelates
The dynamic biomarker and cytotoxic responses provoked in dLUHMES cells by ziram treatment spurred us to investigate the basis of these responses. Ziram is a pesticide composed of dithiocarbamate chelator DDC chelating Zn2+ ions in a 2:1 stoichiometry. The DDC chelator is also used without metals in the pesticide thiram, raising the question whether DDC is itself toxic, or the zinc ions carried by DDC render ziram toxic. We treated LUHMES and dLUHMES cells with twofold increasing concentrations of DDC and were somewhat surprised to see that DDC alone was cytotoxic to all four cell types (Fig. 3a). It was also apparent that differentiated dLUHMES neurons were more sensitive to DDC than undifferentiated LUHMES cells, consistent with many other neurotoxicants (Tong et al. 2017).
Fig. 3.
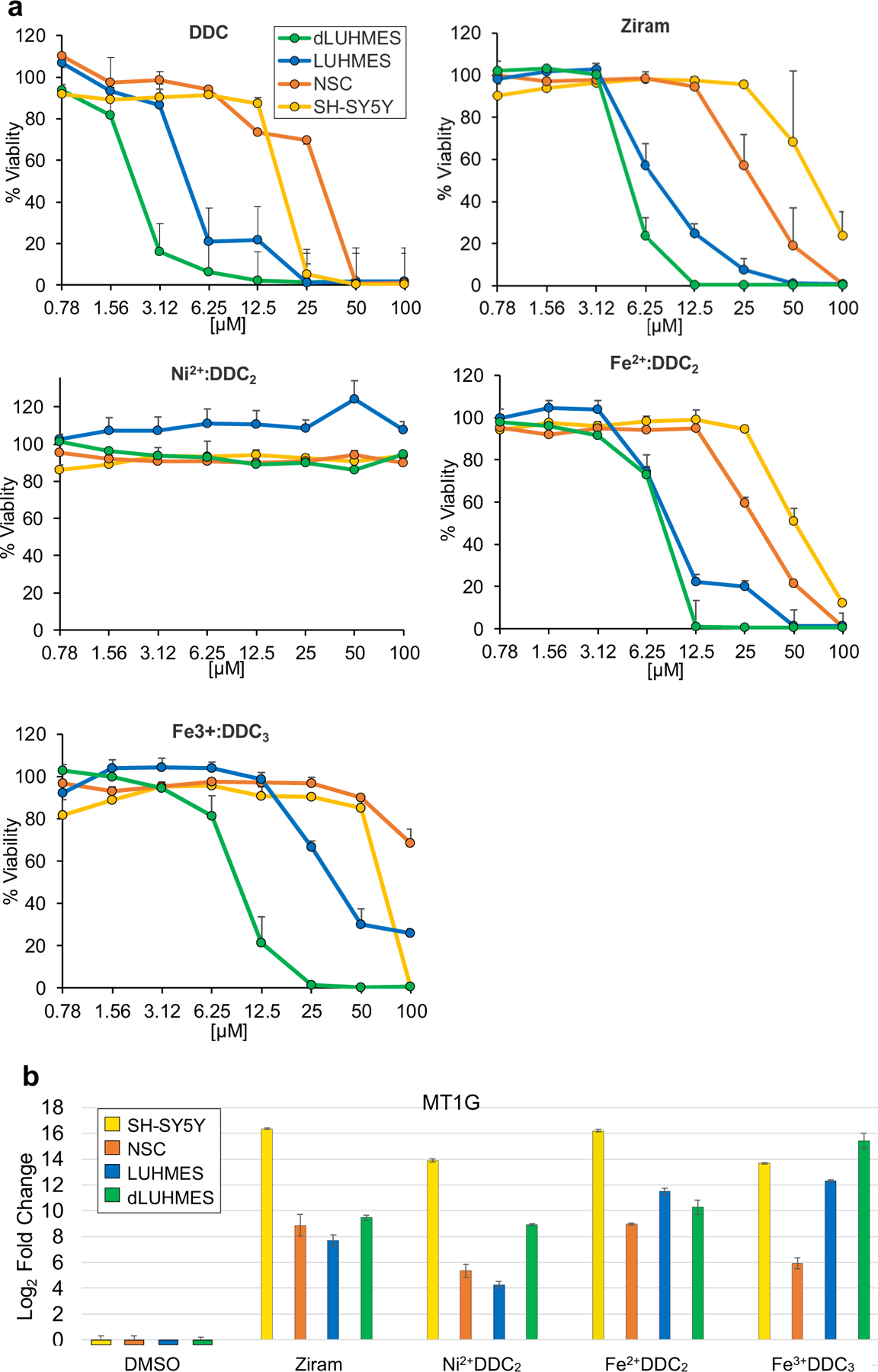
Metal chelates cytotoxicity to neural cell lines and MT1G induction. a. Each cell line was treated for 24 h and % viability was measured as intracellular [ATP] relative to the vehicle-treated control. b. Cells were treated with 5 μM each chemical for 6 h, and MT1G transcripts were quantified by qRT-PCR in the indicated cell types and expressed as Log2 fold change relative to the vehicle-treated control
DDC is formulated not only with zinc as ziram, but also as Fe3+DDC3 in the pesticide Ferbam, or in industrial chemical complexes as Ni2 +DDC2 (sankel), or Fe2 +DDC2 (AC1LAOJX). Figure 3b shows an evaluation of the cytotoxicities of DDC-metal chelates Ni2+DDC2, ziram, Fe2+DDC2 or Fe3+DDC3 to neural cell lines SH-SY5Y, human neural stem cells (hNSC), LUHMES and dLUHMES cells. Comparison of DDC to ziram revealed that DDC alone is slightly more toxic than ziram (Fig. 1). Again, dLUHMES were more sensitive than LUHMES cells to ziram. Fe2+DDC2 and Fe3+DDC3 were similarly more cytotoxic to dLUHMES than LUHMES cells, and hNSC and SH-SY5Y were markedly less sensitive than LUHMES cells. dLUHMES showed identical sensitivity to Fe2+DDC2 and Fe3+DDC3, whereas LUHMES cells were far less sensitive to Fe3+DDC3 than dLUHMES cells (Fig. 3b). Since Fe2+ is thought to cause cytotoxicity by producing hydroxyl radicals, we hypothesized that dLUHMES cells are deficient in limiting redox cycling of Fe3+ to Fe2+, whereas LUHMES cells prior to differentiation have some way to minimize re-oxidation or sequester Fe3+.
Collectively, Fig. 3a suggested that the DDC chelator drives cytotoxicity, rather than the zinc or iron, since DDC was more toxic than any of the metal complexes. We conclude that DDC interferes with normal metabolism of metal(s), likely iron, zinc, or copper that are needed in small quantities as cofactors for various enzymes. Interestingly, the nickel chelate of DDC, Ni2+DDC2, was not cytotoxic to any of the four cell lines at concentrations up to 100 μM, although Ni2+DDC2 was a potent inducer of MT1G (Fig. 3b). This result reveals that regulation of MT1G is separable from cytotoxicity. It is important to note that nickel has been found to be neurotoxic in vivo, so the lack of toxicity of Ni2+DDC2 in vitro should not be taken as evidence that nickel exposure is benign (Sulinskiene et al. 2019). It was also notable that 5 μM Fe2+DDC2 and Fe3+DDC3 caused dynamic increases in MT1G transcription at 6 h (Fig. 3b) despite minimal toxicity at 24 h to dLUHMES or LUHMES (Fig. 3a), and no apparent toxicity to SH-SY5Y or NSC. Thus, MT1G mRNA increase is a very sensitive indicator of disrupted metal metabolism, but this is not synonymous with cytotoxicity. We conclude that dLUHMES cells are exquisitely sensitive to disruption of metal metabolism by the chelator DDC or various DDC-metal chelates. Why would neurons be so dependent on metal metabolism? One theory is that neurons in vivo are depend on lactate from astrocytes for energy(Mason 2017), so are particularly dependent on the mitochondrial electron transport chain in oxidative metabolism. The electron transport complexes require iron-sulfur centers, iron-heme centers, as well as copper as a cofactor for cytochrome oxidase.
NRF2- and MTF1-Inhibitors and MT1G Induction
The selective cytotoxicity of DDC and DDC-metal chelates toward dLUHMES neurons led us to further investigate transcriptional increase of MT1G as a possible biomarker of disrupted metal metabolism, and two transcription factors that might mediate MT1G regulation. Studies in non-neural cells indicate that the human MT1- and MT2A genes as well as mouse Mt1 and Mt2 genes are dynamically regulated by the metal-responsive transcription factor MTF1 and by oxidative stress via NRF2 (Fujie et al. 2019; Sharma and Ebadi 2014; Wu et al. 2015). Figure 4 shows the MT1G mRNA responses of LUHMES and dLUHMES cells to treatment with MHG, copper(II), DDC, or DDC complexes of nickel(II) and zinc(II). Each treatment with metal, chelator, or chelator-metal complex was also performed following addition of inhibitors of MTF1 (APTO-235, (Tsai et al. 2018) or NRF2 (ML385, (Singh et al. 2016). HgCl2, CuCl2, and especially DDC and ziram were potent inducers of MT1G mRNA in both LUHMES and dLUHMES cells. Treatments with APTO-235 or ML385 however, had no effect on MT1G induction, except that APTO-235 seemed to augment MT1G induction by CuCl2 in both cell types (Fig. 4). Although ML385 and APTO-235 showed maximal activities at 5 μM (Singh et al. 2016; Tsai et al. 2018), we tried further increasing concentrations of ML385 and APTO-235 to 10 μM, but saw no effect on MT1G induction (not shown).
Fig. 4.
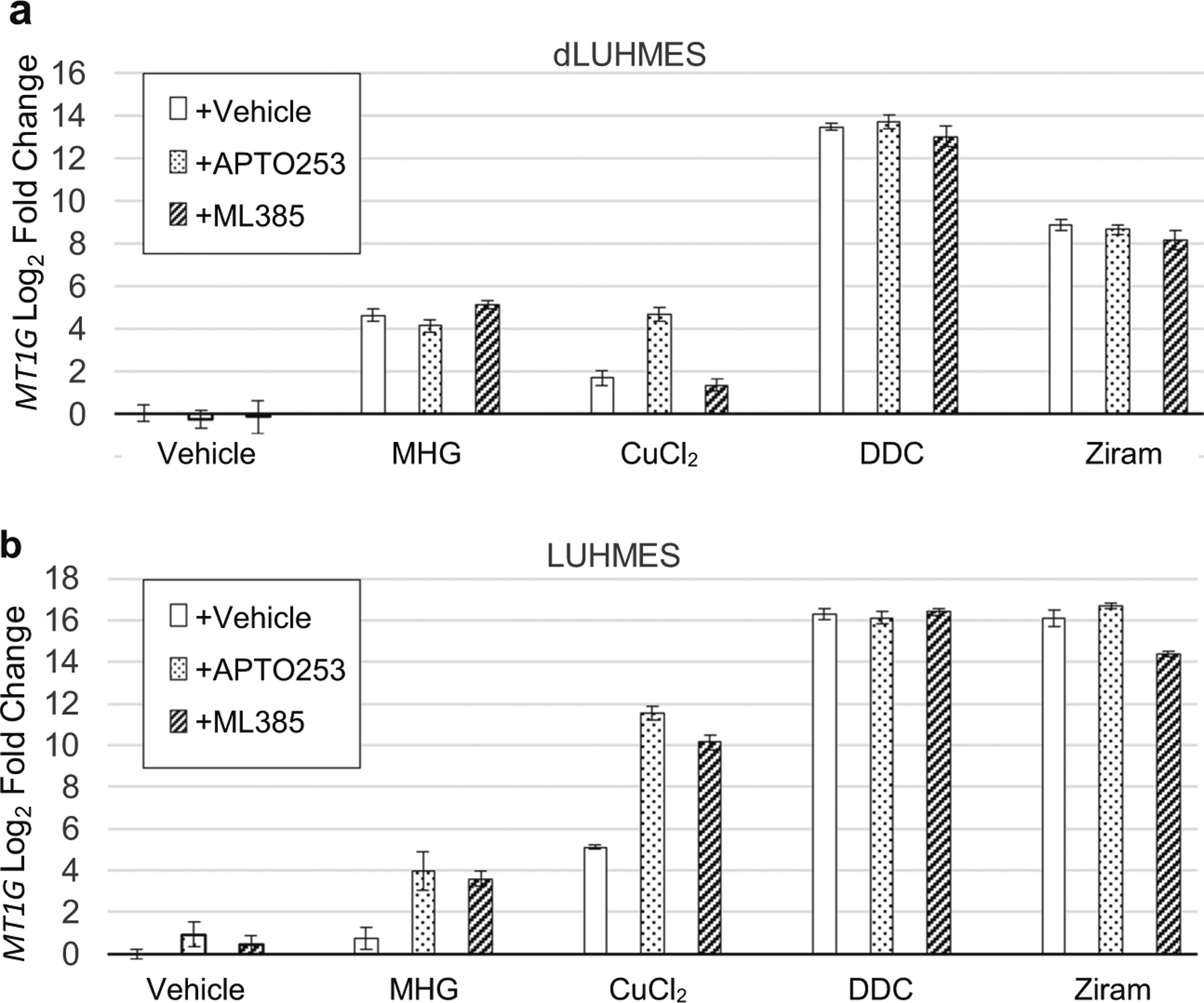
MT1G mRNA expression responses to chelated and unchelated metals. a dLUHMES cells were treated for 6 h with toxicants MHG, CuCl2, DDC, or ziram; and with vehicle, white bars; APTO-253 5 μM, speckled bars; or ML385 5 μM, dark hatched bars. MT1G expression was measured by qRT-PCR. Treatments were MHG 2 μM; CuCl2 5 μM; DDC 5 μM; NiDDC2 5 μM; ziram 5 μM; b undifferentiated LUHMES cells subjected to the same treatments
The failure of NRF2 or MTF1 inhibitors to mitigate MT1G increases spurred examination of the transcription factors expressed in LUHMES and dLUHMES cells using RNA sequencing (Fig. 1 data and unpublished data). These RNA-Seq results showed that LUHMES and dLUHMES cells do express mRNAs for MTF1 and NRF2. We hypothesized that LUHMES and dLUHMES might express additional transcription factors that repressed metallothionein expression. Further examination of RNA-seq data revealed that LUHMES and dLUHMES cells also expressed genes for MTF2 and NRF1; transcription factors that bear protein domains that resemble their paralogs MTF1 and NRF2. Specifically, MTF2 encodes protein sequences that include six zinc fingers like the MTF1 protein, but MTF2 is poorly characterized. NRF1 protein domain structure resembles the structure of NRF2; however, in several cell types, NRF2 is the predominant responder to oxidative stress via interaction with the oxidation sensor protein KEAP1 (Fujie et al. 2019; Wu et al. 2015). We speculate that MTF2 and NRF1 might also participate in MT1G regulation in dLUHMES neurons, however further work is necessary to resolve the unexpected lack of inhibition of APTO-235 or ML385 on MT1G induction by metals or DDC.
Conclusions
Genome-wide gene expression in dLUHMES neurons was surveyed for potential neurotoxicity biomarkers using RNA-Seq (Fig. 1), revealing that transcripts of MT1G and other MT genes were increased by all seven toxicant treatments. Additional gene transcripts increased by multiple toxicants included SLC30A1 and SLC30A2 zinc secretion genes, SLC7A11 encoding the xCT cystine/glutamate antiporter important for glutathione synthesis, DDIT3 (DNA damage inducible transcript 3), and secreted growth factors FIBIN, APLN, and CXCL12. Figs. 2 and 3 illustrate that MT1G and DDIT3 respond to a variety of toxicants, at sub-cytotoxic concentrations, like the proverbial “canary in a coal mine.” Metal chelator DDC or several metal-DDC chelates were notably cytotoxic and provoked dynamic transcript increases in MT1G by a mechanism that also leads to cell death. These results indicate that neurons, including dLUHMES neurons in vitro, are characteristically sensitive to depletion of glutathione or to disruption of cellular metal distribution. The former observation is consistent with the low concentration of glutathione observed in cultured primary neurons (Raps et al. 1989) and CNS neurons (Sun et al. 2006). The sensitivity of dLUHMES neurons to DDC and metal chelates of DDC is consistent with prior work indicating roles for iron in catalyzing formation of hydroxyl radical and the protective roles of metallothioneins to mouse models of PD (Ebadi et al. 2005). The sensitivity of dLUHMES neurons to disrupted glutathione and metal pathways calls for additional research to identify environmental toxicants that may cause neurodegenerative diseases such as PD or AD, or neurodevelopmental diseases such as schizophrenia or autism.
Supplementary Material
Acknowledgments
We thank the DNA Sequencing and Genomics Core at NHLBI, NIH for supporting RNA library preparation and sequencing. We also thank the Compound Management group at NCATS for expert support in screening compounds.
Funding This research was supported by the Intramural Research Program of the National Institutes of Health, including the National Heart Lung and Blood Institute, the intramural research program of the National Center for Advancing Translational Sciences, and the National Toxicology Program of the National Institute of Environmental Health Sciences.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s12640-020-00272-3) contains supplementary material, which is available to authorized users.
Data Availability RNA-Seq data are deposited in the Gene Expression Omnibus. Accession # GSE150005.
Conflict of Interest The authors declare that they have no conflict of interest.
Ethics Approval Approved by NCATS manuscript clearance.
Code Availability No novel code was used.
References
- Chu PH et al. (2020) Stem cell-derived endothelial cell model that responds to tobacco smoke like primary endothelial cells. Chem Res Toxicol 33:751–763. 10.1021/acs.chemrestox.9b00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RS, West AK (2004) A role for extracellular metallothioneins in CNS injury and repair. Neuroscience 123:595–599. 10.1016/j.neuroscience.2003.10.019 [DOI] [PubMed] [Google Scholar]
- Donato MT, Tolosa L, Gomez-Lechon MJ (2015) Culture and Functional characterization of human hepatoma HepG2 cells. Methods Mol Biol 1250:77–93. 10.1007/978-1-4939-2074-7_5 [DOI] [PubMed] [Google Scholar]
- Dong G, Chen H, Qi M, Dou Y, Wang Q (2015) Balance between metallothionein and metal response element binding transcription factor 1 is mediated by zinc ions (review). Mol Med Rep 11:1582–1586. 10.3892/mmr.2014.2969 [DOI] [PubMed] [Google Scholar]
- Ebadi M, Brown-Borg H, El Refaey H, Singh BB, Garrett S, Shavali S, Sharma SK (2005) Metallothionein-mediated neuroprotection in genetically engineered mouse models of Parkinson’s disease. Brain Res Mol Brain Res 134:67–75. 10.1016/j.molbrainres.2004.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie T, Takenaka F, Yoshida E, Yasuike S, Fujiwara Y, Shinkai Y, Kumagai Y, Yamamoto C, Kaji T (2019) Possible mechanisms underlying transcriptional induction of metallothionein isoforms by tris(pentafluorophenyl)stibane, tris(pentafluorophenyl)arsane, and tris(pentafluorophenyl)phosphane in cultured bovine aortic endothelial cells. J Toxicol Sci 44:327–333. 10.2131/jts.44.327 [DOI] [PubMed] [Google Scholar]
- Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, Quake SR (2016) Cellular taxonomy of the mouse striatum as revealed by single-cell RNA-Seq. Cell Rep 16:1126–1137. 10.1016/j.celrep.2016.06.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Herz KT (2015) Trace elements as paradigms of developmental neurotoxicants: lead, methylmercury and arsenic. J Trace Elem Med Biol 31:130–134. 10.1016/j.jtemb.2014.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardyman JE et al. (2016) Zinc sensing by metal-responsive transcription factor 1 (MTF1) controls metallothionein and ZnT1 expression to buffer the sensitivity of the transcriptome response to zinc. Metallomics 8:337–343. 10.1039/c5mt00305a [DOI] [PubMed] [Google Scholar]
- Hozumi I, Inuzuka T, Tsuji S (1998) Brain injury and growth inhibitory factor (GIF)–a minireview. Neurochem Res 23:319–328. 10.1023/a:1022401315721 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Sumino A, Kajioka D, Shibagaki F, Yamamuro A, Yoshioka Y, Maeda S (2017) Apelin protects against NMDA-induced retinal neuronal death via an APJ receptor by activating Akt and ERK1/2, and suppressing TNF-alpha expression in mice. J Pharmacol Sci 133:34–41. 10.1016/j.jphs.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Janssens R, Struyf S, Proost P (2018) Pathological roles of the homeostatic chemokine CXCL12. Cytokine Growth Factor Rev 44:51–68. 10.1016/j.cytogfr.2018.10.004 [DOI] [PubMed] [Google Scholar]
- Jauhiainen A et al. (2012) Distinct cytoplasmic and nuclear functions of the stress induced protein DDIT3/CHOP/GADD153. PLoS One 7: e33208 10.1371/journal.pone.0033208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao J, Huang R, Steiner T, Bourner M, Mitchell M, Thompson DC, Zhao B, Xia M (2017) Development and application of human renal proximal tubule epithelial cells for assessment of compound toxicity. Curr Chem Genom Transl Med 11:19–30. 10.2174/2213988501711010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotharius J, Falsig J, van Beek J, Payne S, Dringen R, Brundin P, Leist M (2005) Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J Neurosci 25: 6329–6342. 10.1523/JNEUROSCI.1746-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S (2017) Lactate shuttles in neuroenergetics-homeostasis, allostasis and beyond. Front Neurosci 11:43 10.3389/fnins.2017.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki I et al. (2011) Astrocyte-derived metallothionein protects dopaminergic neurons from dopamine quinone toxicity. Glia 59:435–451. 10.1002/glia.21112 [DOI] [PubMed] [Google Scholar]
- Odelstad L et al. (1981) Neuron-specific enolase in relation to differentiation in human neuroblastoma. Brain Res 224:69–82. 10.1016/0006-8993(81)91117-3 [DOI] [PubMed] [Google Scholar]
- Oskarsson A (1987) Comparative effects of ten dithiocarbamate and thiuram compounds on tissue distribution and excretion of lead in rats. Environ Res 44:82–93. 10.1016/s0013-9351(87)80088-9 [DOI] [PubMed] [Google Scholar]
- Ottestad-Hansen S et al. (2018) The cystine-glutamate exchanger (xCT, Slc7a11) is expressed in significant concentrations in a subpopulation of astrocytes in the mouse brain. Glia 66:951–970. 10.1002/glia.23294 [DOI] [PubMed] [Google Scholar]
- Pape M, Doxakis E, Reiff T, Duong CV, Davies A, Geissen M, Rohrer H (2008) A function for the calponin family member NP25 in neurite outgrowth. Dev Biol 321:434–443. 10.1016/j.ydbio.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Raps SP, Lai JC, Hertz L, Cooper AJ (1989) Glutathione is present in high concentrations in cultured astrocytes but not in cultured neurons. Brain Res 493:398–401 [DOI] [PubMed] [Google Scholar]
- Rashidi-Nezhad A, Talebi S, Saebnouri H, Akrami SM, Reymond A (2014) The effect of homozygous deletion of the BBOX1 and Fibin genes on carnitine level and acyl carnitine profile. BMC Med Genet 15:75 10.1186/1471-2350-15-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildknecht S et al. (2009) Requirement of a dopaminergic neuronal phenotype for toxicity of low concentrations of 1-methyl-4-phenylpyridinium to human cells. Toxicol Appl Pharmacol 241: 23–35. 10.1016/j.taap.2009.07.027 [DOI] [PubMed] [Google Scholar]
- Schweigel-Rontgen M (2014) The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr Top Membr 73:321–355. 10.1016/B978-0-12-800223-0.00009-8 [DOI] [PubMed] [Google Scholar]
- Sharma S, Ebadi M (2014) Significance of metallothioneins in aging brain. Neurochem Int 65:40–48. 10.1016/j.neuint.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL (2002) Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 16:869–871. 10.1096/fj.01-0995fje [DOI] [PubMed] [Google Scholar]
- Singh A et al. (2016) Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem Biol 11:3214–3225. 10.1021/acschembio.6b00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulinskiene J, Bernotiene R, Baranauskiene D, Naginiene R, Staneviciene I, Kasauskas A, Ivanov L (2019) Effect of zinc on the oxidative stress biomarkers in the brain of nickel-treated mice. Oxidative Med Cell Longev 2019:8549727 10.1155/2019/8549727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Shih AY, Johannssen HC, Erb H, Li P, Murphy TH (2006) Two-photon imaging of glutathione levels in intact brain indicates enhanced redox buffering in developing neurons and cells at the cerebrospinal fluid and blood-brain interface. J Biol Chem 281:17420–17431. 10.1074/jbc.M601567200 [DOI] [PubMed] [Google Scholar]
- Tanner CM, Goldman SM, Ross GW, Grate SJ (2014) The disease intersection of susceptibility and exposure: chemical exposures and neurodegenerative disease risk. Alzheimers Dement 10:S213–S225. 10.1016/j.jalz.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Tong ZB, Hogberg H, Kuo D, Sakamuru S, Xia M, Smirnova L, Hartung T, Gerhold D (2017) Characterization of three human cell line models for high-throughput neuronal cytotoxicity screening. J Appl Toxicol 37:167–180. 10.1002/jat.3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Huang R, Wang Y, Klumpp-Thomas CA, Braisted JC, Itkin Z, Shinn P, Xia M, Simeonov A, Gerhold DL (2018) The toxmatrix: chemo-genomic profiling identifies interactions that reveal mechanisms of toxicity. Chem Res Toxicol 31:127–136. 10.1021/acs.chemrestox.7b00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY et al. (2018) APTO-253 is a new addition to the repertoire of drugs that can exploit DNA BRCA1/2 deficiency. Mol Cancer Ther 17:1167–1176. 10.1158/1535-7163.MCT-17-0834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishta A, Hetman M (2014) Inhibitors of histone deacetylases enhance neurotoxicity of DNA damage. NeuroMolecular Med 16: 727–741. 10.1007/s12017-014-8322-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Costello S, Cockburn M, Zhang X, Bronstein J, Ritz B (2011) Parkinson’s disease risk from ambient exposure to pesticides. Eur J Epidemiol 26:547–555. 10.1007/s10654-011-9574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H et al. (2015) Metallothionein plays a prominent role in the prevention of diabetic nephropathy by sulforaphane via up-regulation of Nrf2. Free Radic Biol Med 89:431–442. 10.1016/j.freeradbiomed.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang X, Zhou X, Cheng B, Li G, Bai B (2017) Temporal expression of apelin/apelin receptor in ischemic stroke and its therapeutic potential. Front Mol Neurosci 10:1 10.3389/fnmol.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y et al. (2020) Discovery of Peptide boronate derivatives as histone deacetylase (HDAC) and proteasome dual inhibitors for overcoming bortezomib resistance of multiple myeloma. J Med Chem. 10.1021/acs.jmedchem.9b02161 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


