Abstract
The growing appreciation of human genetics and genomics in cardiovascular disease (CVD) accompanied by the technological breakthroughs in genome editing, particularly the CRISPR-Cas9 technologies, has presented an unprecedented opportunity to explore the application of genome editing tools in cardiovascular medicine. The ever-growing genome-editing toolbox includes an assortment of CRISPR-Cas systems with increasing efficiency, precision, flexibility, and targeting capacity. Over the past decade, the advent of large-scale genotyping technologies and genome-wide association studies (GWAS) has provided powerful tools to identify genotype-phenotype associations for diseases with complex traits. Notably, a growing number of loss-of-function mutations have been associated with favorable CVD risk-factor profiles that may confer protection. Combining the newly gained insights into human genetics with recent breakthrough technologies, such as the CRISPR technology, holds great promise in elucidating novel disease mechanisms and transforming genes into medicines. Nonetheless, translating genetic insights into novel therapeutic avenues remains challenging, and applications of “in body” genome editing for CVD treatment and engineering cardioprotection remain mostly theoretical. Here we highlight the recent advances of the CRISPR-based genome editing toolbox and discuss the potential and challenges of CRISPR-based technologies for translating GWAS findings into genomic medicines.
Introduction
Cardiovascular diseases (CVD) remain the leading cause of deaths in the United States and worldwide. CVDs comprise a broad range of disorders, including cardiomyopathies, valvular diseases, conduction disorders, and vascular diseases. While some of these disorders are Mendelian disorders caused by rare monogenic mutations, the most prevalent CVDs—including coronary artery disease (CAD), atrial fibrillation, heart failure, hypertension, and stroke—are complex traits driven by multiple genetic variants and reflecting the interplay of genetic and environmental factors (1, 2). Over the past decade, the advent of large-scale genotyping technologies combined with genome-wide association studies (GWAS) has enabled a deeper understanding of the human genome, providing insights into the potential implications of genetic variations in complex diseases such as CVDs. However, genetic association does not equate causation. Understanding the molecular basis of complex phenotypes associated with human disease requires disruptive technologies for functional validation of genotypes, which is vital for the eventual translation of genetic knowledge into medicines.
Technological breakthroughs, such as engineered and programmable nucleases, have revolutionized the field of genome editing. In particular, the advent and continued advancement of CRISPR-Cas9 technologies enable manipulations of the human genome with increasing precision and efficiency (3). CRISPR-Cas9 genome editing is a powerful tool to validate GWAS findings on disease-associated or -protective genetic variants. Functionally validated variants not only have the potential to reveal novel mechanistic players in CVD pathogenesis and risk reduction but also represent promising therapeutic targets. As the CRISPR-Cas9 toolbox and its packing and delivery system continue to be refined, in vivo genome editing has the potential to translate into clinical application.
In this review, we provide a summary of the technical advantages and limitations of the CRISPR-Cas9 technology. We highlight its application for functional validation of GWAS-identified pathogenic or protective genetic variants, as well as the opportunities and challenges of translating CRISPR-Cas9 genome editing into genomic medicines for CVDs. Finally, we discuss the ethical challenges for gene therapy and future research.
Versality and Limitations of CRISPR-Cas9 Genome Editing
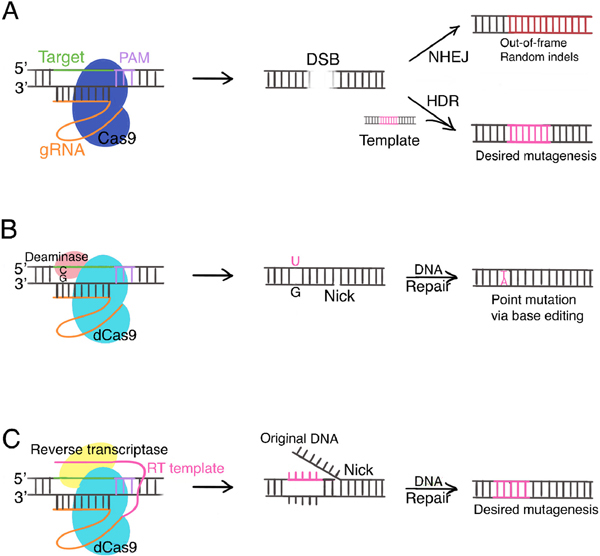
CRISPR stands for clustered regularly interspaced short palindromic repeat DNA sequences. They were discovered in bacteria to constitute an adaptive immune system together with the CRISPR-associated (Cas) proteins. While the details of the initial generation of the CRISPR-Cas9 system have been reviewed extensively elsewhere (4), here we provide an overview of the basics and highlight the strengths and limitations of the CRISPR-Cas9 toolbox along with recent developments. Specifically, we will compare and contrast three related technologies in the CRISPR-Cas9-based toolbox: conventional CRISPR-Cas9, base editing, and prime editing (Figure 1, Table 1).
Figure 1:
CRISPR-Cas9 based genome editing technologies. A) Conventional CRISPR-Cas9. Cas9 nuclease creates DSB at location determined by gDNA and PAM. NHEJ generates random indels, often leading to premature stop codons and gene knockout. HDR allows desired mutagenesis according to donor template. B) Base editors. dCas9 is fused to cytidine deaminase (cytidine base editors) or adenosine deaminase (adenine base editors). Base editors introduce point mutations by deamination without creating DSBs. Cytidine base editor is shown as an example. C) Prime editing. dCas9 is fused to an engineered reverse transcriptase. Prime editing gRNA includes an RT template, allowing direct rewriting of DNA. Abbreviations: PAM, protospacer adjacent motif; gRNA, guide RNA; DSB, double-strand break; NHEJ, non-homologous end joining; HDR, homology-directed repair; dCas9, catalytically impaired Cas9; RT, reverse transcriptase.
Table 1:
CRISPR-Cas9 toolbox comparison
| Pros | Cons | |
|---|---|---|
| Conventional CRISPR-Cas9 | NHEJ convenient for gene KO; HDR allows insertion, deletion, point mutation by incorporation of exogenous donor templates. | DSBs are associated with permanent undesired outcomes, including complex mixtures of products and deletion and translocations of large genomic regions. HDR has extremely low efficiency in terminally differentiated, quiescent cells, such as adult cardiomyocytes. Subjected to constraints of PAM sequences. |
| CRISPR-Cas9 base editors (cytidine base editor and adenine base editor) | Install point mutations with higher editing efficiency, better product purity, lower random indel rates, and fewer off-target events in both dividing and non-dividing cells. No DSB. | Limited to four types of single base pair edits (C→T, G→A, A→G, and T→C). Cannot install insertions, deletions, or other point mutations. Subjected to constraints of PAM sequences. |
| Prime editing | Highly versatile. “Search and replace” strategy directly writes new genetic information into a specified DNA site. Not constrained by PAM sequences | Roughly twice the size of conventional CRISPR-Cas9 system, particularly challenging for in vivo delivery. |
Abbreviations: NHEJ, non-homologous end joining; HDR, homology-directed repair; KO, knockout; DSB, double-strand break; PAM, protospacer adjacent motif.
First, the conventional CRISPR-Cas9 system takes advantage of the endonuclease activity of Cas9 proteins. The Cas9 nuclease is programed with a single strand guide RNA (gRNA) complementary to the target DNA sequence. In mammalian cells, it creates a double strand break (DSB) at the genomic location determined by target DNA sequence and a short motif called protospacer adjacent motif (PAM) juxtaposed to the target DNA sequence (5) (Figure 1a). Notably, the PAM sequence serves as an essential DNA binding signal for the Cas9 nuclease and varies depending on bacteria species. The DSB is repaired via either the efficient but error-prone non-homologous end joining (NHEJ) pathway or the less efficient but high-fidelity homology-directed repair (HDR) pathway (Figure 1a). The random insertions or deletions, or random indels, introduced in NHEJ often result in out-of-frame reading and premature stop codons, and therefore can lead to gene knockout. In contrast, HDR allows the incorporation of exogenous donor templates to guide DSB repair for precise mutagenesis. Depending on the donor template, HDR can either correct or introduce point mutations as well as insertions and deletions.
Second, CRISPR-Cas9 base editors utilizes an engineered, catalytically impaired Cas9 (dCas9) (6, 7). With dCas9 fused to cytidine or adenosine deaminase, base editors enable the direct, irreversible conversion of one target DNA base into another in a programmable manner, without requiring DSB, HDR, or donor templates (Figure 1b) (6, 7). Compared to HDR in conventional CRISPR-Cas9 genome editing, base editing can install point mutations with higher editing efficiency, precision, minimal error rates, and fewer off-target events in both dividing and non-dividing cells.
Third, the newest addition to the CRISPR-Cas9-based toolbox is “prime editing”. In prime editing, dCas9 is fused to an engineered reverse transcriptase (RT), programmed with a prime editing guide RNA that includes an RT template (Figure 1c) (8). Prime editing directly writes new genetic information into a specified DNA site and therefore can mediate insertion and deletion of various sizes in addition to all types of point mutations. In theory, the „search - and-replace‟ prime editing technology could correct up to 89% of known genetic variants associated with human diseases (8).
Compared to earlier engineered nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR-Cas9 is much simpler to construct, easier to program, and more flexible to use (4). Meanwhile, each of the aforementioned CRISPR-Cas9-based tools also has limitations, including off-target mutagenesis and restricted targetability; however, researchers have made efforts to address these issues (Table 1, 2).
Table 2:
Limitations of the CRISPR-Cas9 system and corresponding technical advancement
| Problem | Corresponding technical advancement | Reference |
|---|---|---|
| Double strand break | - CRISPR-Cas9 base editors using dCas9 - Prime editing using dCas9 |
Gaudelli et al 2017; Komor etal 2016 (6, 7) Anzalone et al 2019 (8) |
| Off-target mutagenesis | - Development of computational algorithms for optimized gRNA design - Use of high-fidelity Cas9 variants |
Doench et al 2016; Hsu et al 2013 (11, 12) Kleinstiver et al 2016 (13) |
| PAM restriction | - Discovery and application of different natural Cas9 species with different PAM sequences - Engineering of new variants of Cas9 nucleases and base editors with expanded target compatibility. - Development of near-PAMless Cas9 variants with little restriction |
Ran et al 2013; Friedland et al 2015; Hou et al 2013; Muller et al 2016 (14–17) Hu et al 2018; Thuronyi et al 2019; Miller et al 2020 (18–20) Walton et al 2020 (21) |
Abbreviations: gRNA, guide RNA; dCas9, catalytically impaired Cas9; PAM, protospacer adjacent motif.
The conventional CRISPR-Cas9 system involves the generation of a double strand break (DSB) by the Cas9 endonuclease. While homology-directed repair of DSBs can accomplish desired mutagenesis, the endogenous homologous recombination machinery is only present in dividing cells. Hence, HDR has extremely low efficiency in terminally differentiated, quiescent cells, such as adult cardiomyocytes. Furthermore, from a therapeutic perspective, DSBs are associated with permanent undesired outcomes, including complex deletion and translocations of large genomic regions (9). These issues are unique to the conventional CRISPR-Cas9 system, as the recently developed base editing and prime editing effectively bypass DSBs by using a catalytically impaired Cas9 (dCas9).
Off-target mutagenesis remains a major problem as the lack of precision raises safety concerns for clinical applications. Early investigations show that the off-target sites may harbor up to five mismatches at the gRNAs-DNA complimentary binding interface and many were mutagenized with frequencies comparable to (or higher than) those observed at the intended on-target site (10). Meanwhile, many computational algorithms for on- and off-target activity prediction have since been developed to support and optimize computational design of gRNAs to maximize on-target activity and minimize off-target effects (11, 12). High-fidelity Cas9 variants have also been developed to reduce non-specific DNA contacts and reduce off-target events (13).
Finally, another limitation of the CRISPR-Cas9 system concerns targeting capacity. Because PAM is required for Cas9 recognition and binding, a DNA sequence is targetable only if a PAM sequence is present adjacent to it. To address this constriction, researchers have discovered and explored the application of different natural Cas9 species with different PAMs (14–17) and engineered new variants of Cas9 nucleases and base editors with expanded target compatibility (18–20). Most recently, Walton et al utilized structure-guided engineering to develop near-PAMless Cas9 variants with little restriction (21).
In summary, since the discovery of the CRISPR-Cas9 system, ongoing efforts continue to tackle the limitations and refine the technology. A family of CRISPR-based tools with increasing efficiency, precision, and targeting capacity have been generated, each with its own strengths and weaknesses (Table 1). Notably, in silico predictive modeling of CRISPR-Cas9 editing efficiency does not necessarily translate to efficacious and specific genome editing in practice. Given the complexity of the mammalian genome and organ systems, it is essential to extensively validate the effectiveness and precision of genome editing approaches in vitro and preclinical models in vivo before any clinical application may take place.
Identification of Genetic Loci Associated with CVDs
Historically, case reports on familial aggregation of diseases have provided evidence of genetic etiology to many cardiovascular diseases. Pedigree mapping of index families and linkage analysis between affected and healthy family members are used to identify putative pathogenic genes. This method identified several genetic loci implicated in CVDs, including MYH7 in familial hypertrophic cardiomyopathy (22, 23), H-RAS and HERG in long QT syndrome (24, 25), and LDLR and PCSK9 in familial hypercholesterolemia (26–28). Linkage analysis is especially helpful in studying private mutations found only in few families and represents a reliable method for identifying disease-causing mutations for disorders with Mendelian hereditary pattern, otherwise known as monogenic disorders.
However, most common CVDs, such as coronary artery disease and myocardial infarction, are complex diseases that are driven by multiple variants of small to moderate effects (2). Because variants implicated in complex traits are found across the genome, they cannot be studied with pedigree mapping or linkage analysis. Over the past decade, the advent of large-scale genotyping technologies, genome-wide association studies (GWAS), and more recently, phenome-wide association studies (PheWAS), has provided much more powerful tools to identify genotype-phenotype associations. GWAS detects associations between a variety of genetic variants with a particular physiological or clinical phenotype; PheWAS explores the relationship between a wide range of phenotypes and a specific genetic variant (29). These approaches have made genetic studies possible for diseases with complex traits, allowing identification of genetic variants with modest yet significant effects on disease risk. GWAS for clinical CVD events has identified gene/locus features associated with hypertension, coronary artery disease, myocardial infarction, stroke, and aneurysms (30–33). Notably, disease-associated variants are often found in non-coding regions (34). For example, GWAS has identified multiple variants in ANRIL, or antisense noncoding RNA in the INK4 locus, associated with cardiometabolic disease traits, including myocardial infarction, coronary artery disease, and type 2 diabetes (34).
In addition to genetic predispositions to disease, these population-scale genetic analyses are also able to identify genetic variants that confer protection against disease risk (Table 3). Humans with rare, naturally occurring genetic variations, particularly those with putative loss-of-function (pLoF) alleles associated with lipoprotein transport and lipid metabolism, may be at lower risk of CVDs. In a recent study integrating GWAS and PheWAS with existing electronic medical records of US veteran participants, Klarin et al. identified protective effects of pLoF mutations in PCSK9 for abdominal aortic aneurysm, in ANGPTL4 for type 2 diabetes, and in PDE3B for coronary disease (35). Additional genetic variations associated with lower CVD risk identified by other studies include pLoF mutations in ANGPTL3, NPC1L1, and APOC3 (36–39).
Table 3:
pLoF variants associated with reduced risk of complex cardiovascular disorders
| Gene | Example pLoF variant(s) | Allele frequency | Functional effect1 | Disease protective effect1 | Reference; database |
|---|---|---|---|---|---|
| ANGPTL3 | 13 distinct pLoF variants including p.Ser17Ter, p.Asn121fs, p.Asn147fs, c.495+6T>C | 1 in 237 DiscovEHR study participants of European ancestry | 27% lower triglyceride levels (P=2.5×10−21); 9% lower LDL-C levels (P=2.8×10−5); 4% lower HDL-C levels (P=0.02) | 41% lower odds of CAD (OR 0.59; 95% CI 0.41 to 0.85; P=0.004) | Dewey et al 2017(36); DiscovEHR |
| ANGPTL4 | p.Cys80fs; p.Gln133Ter; p.Arg161Ter; c.547+1G>A; p.Gly313fs; p.Trp350Ter; p.Gln362fs; p.Tyr363Ter; p.Gln369Ter; p.Trp383Ter | 0.2% among 13,758 study participants | 35% reduced triglyceride levels (P=0.003) | 53% reduced risk of MI (OR 0.47; P=0.04) | Stitziel 2016 (73) |
| p.Glu40Lys | 4.0% among 176, 913 White veterans | triglyceride-lowering; HDL-C-raising | 16% reduced risk of CAD (OR 0.84; 95% CI 0.79 to 0.90; P=2.90 × 10−8); 12% reduced risk of type 2 diabetes (OR 0.88; 95% CI 0.83 to 0.93; P=2.50 × 10−6) | Klarin 2018 (35); MVP | |
| APOC3 | p.Arg19Ter; c.55+1G>A; c.179+1G>T; p.Ala43Thr | 1 in 150 study participants | 39% lower triglyceride levels (P<1 × 10−20) | 40% lower risk of CAD (OR 0.60; 95% CI 0.47 to 0.75; P=4×10−6) | Crosby 2014 (39) |
| GPR151 | p.Arg95Ter | 0.8% in European ancestry | Reduced BMI (−0.36 kg/m2, −0.07σ, P=4.9 × 10−8) | 14% lower odds of type 2 diabetes (OR 0.86; P=0.006); 9% lower odds of CAD (OR 0.91; P=0.01) | Emdin 2018 (74); UK Biobank |
| NPC1L1 | 15 distinct pLoF variants including p.Arg406Ter | 1 in 650 study participants | Lower LDL-C level (−12 mg dl−1, P=0.04) | 53% lower risk of CAD (OR 0.47; 95% CI 0.25 to 0.87; P=0.008) | Stitziel 2014 (37) |
| PCSK9 | p.Tyr142Ter, p.Cys679Ter | 0.8% and 1.8%, respectively, among 3,363 Black study participants | 28% reduced mean plasma LDL-C level (−38 mg dl−1, P<0.001) | 88% reduced risk of CAD (hazard ratio 0.11, 95% CI 0.02 to 0.81; P=0.03) | Cohen 2006 (75); ARIC |
| p. Arg46Leu | 3.1% among 176, 913 White veterans | LDL-C-lowering | 17% reduced risk of CAD (OR 0.83; 95% CI 0.78 to 0.88; P=1.38 × 10−13); 28% reduced risk of AAA (OR 0.72; 95% CI 0.62 to 0.83; P=2.05 × 10−6) | Klarin 2018 (35); MVP | |
| PDE3B | p.Arg783Ter | 1 in 625 | Higher blood HDL-C levels (4.72 mg dl−1, 0.41σ, P<2.8 × 10−16); lower blood triglyceride levels (−43.3 mg dl−1, −0.27σ, P=7.5 × 10−8) | 24% decreased risk of CAD2 (OR 0.76; 95% CI 0.65 to 0.90; P=0.0015) | Klarin 2018 (35); MVP, UK Biobank, MIGen, PMBB, and DiscovEHR |
Abbreviations and notes: OR, odds ratio; CI, confidence interval; LDL-C, low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; CAD, coronary artery disease; MI, myocardial infarction; AAA, abdominal aortic aneurysm; ARIC, Atherosclerosis Risk in Communities study; MVP, Million Veteran Program; MIGen, Myocardial Infarction Genetics Consortium; PMBB, Penn Medicine Biobank.
The functional effect and disease protective effect of each gene are calculated with all listed pLoF variants aggregated.
Because damaging mutations are individually rare, for association analysis with CAD, the study aggregated 47 rare damaging mutations in PDE3B.
Although genome-wide association studies have uncovered natural genetic variants that influence risks of common, complex diseases and traits, how these variants affect disease processes is far from clear. Determining the role and significance of genetic variants in CVDs has been a slow process as their functional validation is challenging. CRISPR-Cas9-based genome editing has opened a range of new opportunities to easily introduce and systematically characterize the effects of genetic variants identified in GWAS with cellular and animal models.
Functional Validation of Genetic Variants with CRISPR-Cas9
Several factors have made it difficult to bridge the gap between the statistical associations linking loci and traits and a functional understanding of the biology underlying disease risks (40). First, it is challenging to distinguish causal variants from co-inherited ones. Co-inherited variants may lack their own independent effects on disease risk despite their strong linkage disequilibrium with sentinel disease-associated variants. Second, disease-associated variants predominantly locate in non-coding regions of the genome, often enriched in regulatory elements such as enhancers. It is challenging to dissect the functional consequences of variants found in these regions because the regulatory elements often regulate more than one target gene and could be several hundred kilobases or even megabases from the genes they regulate (41). Last but not least, because complex diseases, by definition, are driven by multiple variants of small to moderate effects (2), the functional effect of any one particular variant could be subtle. Hence, functional validation requires experiments that can deliver high sensitivity and statistical power. CRISPR-Cas9-based genome editing allows the modification of a select genetic locus while preserving the background genome. This technique provides isogenic cellular and animal models that differ only at the candidate locus and eliminates confounding factors for studying the functional effect of a candidate variant. Here we provide several examples to demonstrate the use of CRISPR-Cas9-based models for functional validation of GWAS findings.
STXBP5, or syntaxin binding protein 5, is a regulator of platelet secretion discovered through GWAS (42). Human studies show that the non-synonymous coding single-nucleotide polymorphism SNP rs1039084 minor allele G is associated with decreased von Willebrand factor (VWF) levels, decreased venous thrombosis risk, and increased bleeding (43). Zhu et al introduced the rs1039084 minor allele of human STXBP5 into the orthologous mouse Stxbp5 locus by CRISPR-Cas9-mediated germline genome editing (44). Consistent with phenotypes observed in human carriers of the rs1039084 minor allele, genome-edited mice exhibited lower plasma VWF, prolonged bleeding, and decreased thrombosis. These results provided strong functional evidence for the regulatory role of a GWAS-discovered variant on human bleeding and thrombosis phenotype, suggesting that variation within STXBP5 has functional consequences in the genetic risk for venous thromboembolic disease.
CRISPR-Cas9 has also been used to functionally validate genetic variants in non-coding sequences of the genome. It is thought that noncoding variants reside within or near regulatory elements controlling expression of distal target genes through long-range DNA interactions (45). Hypothesizing that many GWAS loci associated with vascular diseases modulate endothelial functions, Lalonde et al mapped CAD-associated variants onto DNA long-range interactions between regulatory elements and expressed genes in immortalized human aortic endothelial cells (46). The authors identified AIDA as the putative target gene of an upstream regulatory region harboring a CAD-associated variant. Upon CRISPR-Cas9-mediated deletion of this regulatory region, AIDA expression roughly halved when stimulated by inflammatory cytokine tumor necrosis factor-α (TNFα). These results not only supported the notion that this regulatory element—and presumably the genetic variant(s) that it contains—can control AIDA expression, but also implicated TNFα-induced dysregulation of endothelial AIDA expression as a novel candidate mechanism for CAD pathogenesis.
Similarly, using CRISPR-Cas9, Zhang et al generated a mouse model for studying the functional effect of a regulatory region located on chromosome 4q25 upstream of the PITX2 gene, a non-coding region known to harbor GWAS variants associated with atrial fibrillation (AF) (45). Zhang‟s study identified a murine Pitx2 enhancer in 4q25 and demonstrated that its deletion with CRISPR-Cas9 resulted in reduced Pitx2 expression in the mouse left atrium and increased susceptibility to AF upon programmed intracardiac stimulation. These results, for the first time, directly linked the 4q25 noncoding region to PITX2 expression and AF, providing mechanistic insight into noncoding AF variants identified through GWAS.
These studies demonstrate the feasibility of combining CRISPR-Cas9 genome editing with in vitro and in vivo models to validate GWAS findings located in both protein coding and non-coding regions of the genome. Such validation is important for several reasons. First, it provides concrete evidence for novel genotype-phenotype causal relationships and a better understanding of the hereditary components of many CVDs previously known as idiopathic. It is worth mentioning that GWAS-derived genetic insights have been used to construct polygenic risk scores (PRS) that aggregates genetic influences of many common genetic variants to capture an individual‟s genetic predisposition to disease. Because the accuracy of PRS is inevitably influenced by the precision of variant association estimates from GWAS (47), functional validation of allele effects with CRISPR-Cas9 has the potential to improve PRS construction and its clinical utility. Second, functionally validated GWAS findings provide a reservoir of potential therapeutic targets for gene therapy.
Translating CRISPR-Cas9 Modalities into Genomic Medicines
In the realm of cardiovascular medicine, there have not been any human trials exploring the use of CRISPR-Cas9 genome editing for therapeutic purposes. The complex nature of CVD genetics complicates the selection of potential therapeutic targets. Even in a prototypic monogenic disorder such as familial hypertrophic cardiomyopathy (HCM), pathogenic mutations exist significant expressivity, variable penetrance, as well as allelic heterogeneity (48, 49). While the genotype-phenotype causal relationship between mutations in MYH7, the gene encoding beta-myosin heavy chain, and HCM is well established (23), there are almost 500 known variants in MYH7 (48), over 100 classified as pathogenic in ClinVar. Similar extreme allelic heterogeneity, or diversity of variants, is characterized in other causal genes as well, such as the HERG gene associated with long QT syndrome (50) and LDLR associated with familial hypercholesterolemia (51). Therefore, even if it is technically feasible to target and correct all pathogenic variants to wildtype allele with the CRISPR-Cas9 system, the disease course may not afford the time for the design and validation of individual agents. The cost of such personalized genome editing strategies would also be overwhelming.
In comparison to developing gene therapy for monogenic CVDs, the application of genome editing tools for the prevention of complex CVDs may be achievable in the clinic in a nearer future. As discussed above, GWAS has identified a number of naturally occurring pLoF mutations associated with reduced plasma lipid levels and lower risks of CVD (Table 3), including those found in PCSK9 and ANGPTL3 (35, 36, 52). As carriers of these genetic variants are healthy individuals with reduced CVD risk, these naturally occurring pLoF mutations represent putative genetic targets for long-term protection against complex CVDs. While using CRISPR-Cas9-mediated genome editing for CVD prevention remains a theoretical and explorative concept, pre-clinical models have shown promising results.
As a proof of concept of PCSK9 inhibition by permanent gene editing, Ding et al. used adenovirus to express CRISPR-Cas9 along with guide RNA targeting Pcsk9 in mouse liver, theorizing that non-homologous end-joining repair upon Cas9-mediated double-strand break (DSB) would lead to gene disruption and lipid-lowering. The authors found that the mutagenesis rate of Pcsk9 in the liver was as high as over 50% within days of virus administration and that this resulted in decreased plasma PCSK9 levels, increased hepatic low-density lipoprotein receptor (LDLR) levels, and decreased plasma cholesterol levels by 35 to 40% (53). These findings were corroborated by a separate study utilizing the aforementioned CRISPR-Cas9 base editing system. Chadwick et al delivered the cytidine base editor into the mouse liver to introduce an artificial stop codon, accomplishing 50% reduction in plasma PCSK9 protein levels and 30% reduction in plasma cholesterol levels (54). Using a similar in vivo base editing approach, the group successfully targeted the Angptl3 gene to reduce blood triglyceride and LDL cholesterol levels in mice (55). Collectively, these studies are encouraging examples of the utility of CRISPR-Cas9 toolkit for in vivo genome editing as a prophylactic measure against complex CVDs.
Challenges for “in body” CRISPR-Cas9 Genome Editing
Any potential use of CRISPR-Cas9 for “in body” human genome editing undoubtedly require close scrutiny to maximize efficacy and prevent adverse events. In particular, the difficulty of efficient and targeted delivery of CRISPR-Cas9 components to the desired organs and cells presents remarkable challenges to the clinical translation of CRISPR-based therapies.
Delivery vehicles can be classified into three general groups: physical delivery, viral vectors, and non-viral vectors. Physical delivery vehicles, most commonly microinjection and electroporation, are typically only suitable for in vitro and ex vivo genome editing (56). Non-viral vectors, though actively explored, have not yet shown delivery efficiency and/or organ specificity comparable to that of viral vectors. Therefore, for in vivo work, viral vectors remain the most common CRISPR/Cas9 delivery vectors (56).
Among viral vectors, adeno-associated virus (AAV) has been used extensively (57). Unlike lentivirus and adenovirus, AAVs are not known to cause any diseases in humans and provoke less immune response and associated toxicity (56). Nonetheless, AAV vectors have a relatively limited packaging capacity. As a result, delivery of all CRISPR-Cas9 components, including the gene that encodes Cas9 along with a donor template and gRNA, often requires concomitant delivery of multiple vectors in somatic cells - a formidable challenge. This hurdle is particularly prohibitive for the newly developed base editors and prime editors, as they are considerably larger than the conventional Cas9 system (58). These challenges are addressed by engineering trans-splicing vectors (59, 60), using Cas9 species of smaller size (61), and the development of novel delivery vehicles, such as non-viral lipid nanoparticles (62). Another technical challenge facing AAV delivery is a lack of tissue specificity. Although different serotypes have relatively different tissue tropism, it is often still too broad to achieve desired specificity. For example, AAV serotype 9 (AAV9) has a high cardiac tropism, yet it can also transduce skeletal muscle, liver, and the central nervous system (60, 63).
Finally, another important consideration when deploying CRISPR-Cas9 medicines is the human immune response. AAV-mediated delivery of the CRISPR-Cas9 components can persist indefinitely in cells (56). Such prolonged expression of the bacterial Cas9 protein could elicit immune responses as observed in mice (64, 65). Charlesworth et al have demonstrated that humans harbor pre-existing humoral and cell-mediated adaptive immune responses to Cas9 orthologs derived from bacterial species (66). There is also evidence showing the high prevalence of pre-existing neutralizing antibodies to AAV vectors in humans (67). These immune responses to the Cas9 protein and/or the delivery vehicle may jeopardize the safety and efficacy of the clinical use of CRISPR-Cas9 agents as they may result in significant organ toxicity.
Ethical Concerns
In June 2016, the US NIH approved the first human trial using CRISPR gene editing (68). In November 2018, He Jiankui announced that CRISPR-Cas9 had been used in the germline editing of twin girls at risk of HIV transmission born earlier that month in China (69). His action violated the near-universal guideline prohibiting germline editing and received widespread condemnation from the medical and scientific communities and beyond. On the one hand, the technical challenges discussed above remain prohibitive for many clinical applications of CRISPR-Cas9 genome editing. On the other hand, the rapid technological advancement accompanied by lagged regulatory policymaking poses serious ethical and safety concerns.
In a recent report from the National Academy of Sciences and National Academy of Medicine, genome editing is described as widely used for basic science research in laboratories; is in the early stages of development of clinical applications that involve somatic (i.e., nonreproductive) cells; and in the future might be usable for clinical applications involving reproductive cells, which would produce heritable changes (70). The report states that genome editing in the context of basic research and somatic gene therapy is valuable and adequately regulated. In contrast, heritable genome editing needs more research before it might be ready to be tried. Notably, it states that somatic therapy should be used only for treatment and prevention of disease and disability; it should not be tried for enhancement at this time.
Conclusion and Future Directions
CRISPR-Cas9, a technological breakthrough, has the potential to revolutionize the future of cardiovascular medicine. Despite its success, the technology is far from refined. The development of CRISPR-based therapies for CVD treatment and prevention faces technical challenges in agent design, delivery, and off-target effects. However, the continued technical advances, evident by the recent development of base editing and prime editing technologies, will likely address these concerns in the future. The recently discovered anti-CRISPRs (Acrs), small proteins that inhibit CRISPR-Cas enzymes, are anticipated to add new dimensions of control into the CRISPR toolbox and enable further fine-tuning of genome editing with their potent and novel mechanisms of inhibition (71). Furthermore, RNA editing, previously overshadowed by the CRISPR-Cas9-mediated DNA editing, is emerging as a potentially safer and more practical alternative to enable functional rescue at the messenger RNA. Bypassing the safety and ethical concerns presented by permanent genome editing, RNA editing may open up a new world for therapeutic editing (72). Although there is more work to be done, CRISPR technologies have progressed and will continue to improve. It seems almost certain that there will be novel medical applications and groundbreaking developments in the near future. CRISPR could one day be an effective way to treat cardiovascular diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Musunuru K, Kathiresan S. Genetics of Common, Complex Coronary Artery Disease. Cell. 2019;177(1):132–45. [DOI] [PubMed] [Google Scholar]
- 2.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169(7):1177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110(39):15644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, et al. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24(3):636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thuronyi BW, Koblan LW, Levy JM, Yeh WH, Zheng C, Newby GA, et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol. 2019;37(9):1070–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller SM, Wang T, Randolph PB, Arbab M, Shen MW, Huang TP, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321(20):1372–8. [DOI] [PubMed] [Google Scholar]
- 23.Solomon SD, Geisterfer-Lowrance AA, Vosberg HP, Hiller G, Jarcho JA, Morton CC, et al. A locus for familial hypertrophic cardiomyopathy is closely linked to the cardiac myosin heavy chain genes, CRI-L436, and CRI-L329 on chromosome 14 at q11-q12. Am J Hum Genet. 1990;47(3):389–94. [PMC free article] [PubMed] [Google Scholar]
- 24.Keating M Linkage analysis and long QT syndrome. Using genetics to study cardiovascular disease. Circulation. 1992;85(6):1973–86. [DOI] [PubMed] [Google Scholar]
- 25.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80(5):795–803. [DOI] [PubMed] [Google Scholar]
- 26.Ott J, Schrott HG, Goldstein JL, Hazzard WR, Allen FH, Falk CT, et al. Linkage studies in a large kindred with familial hypercholesterolemia. Am J Hum Genet. 1974;26(5):598–603. [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein JL, Brown MS. The LDL receptor locus and the genetics of familial hypercholesterolemia. Annu Rev Genet. 1979;13:259–89. [DOI] [PubMed] [Google Scholar]
- 28.Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–6. [DOI] [PubMed] [Google Scholar]
- 29.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12):1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49(3):403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattson DL, Liang M. Hypertension: From GWAS to functional genomics-based precision medicine. Nat Rev Nephrol. 2017;13(4):195–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arking DE, Chakravarti A. Understanding cardiovascular disease through the lens of genome-wide association studies. Trends Genet. 2009;25(9):387–94. [DOI] [PubMed] [Google Scholar]
- 33.Arnett DK. Genetics of CVD in 2015: Using genomic approaches to identify CVD-causing variants. Nat Rev Cardiol. 2016;13(2):72–4. [DOI] [PubMed] [Google Scholar]
- 34.Giral H, Landmesser U, Kratzer A. Into the Wild: GWAS Exploration of Non-coding RNAs. Front Cardiovasc Med. 2018;5:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50(11):1514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewey FE, Gusarova V, Dunbar RL, O’Dushlaine C, Schurmann C, Gottesman O, et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med. 2017;377(3):211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371(22):2072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371(1):32–41. [DOI] [PubMed] [Google Scholar]
- 39.Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO, Lange LA, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371(1):22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: From Association to Function. Am J Hum Genet. 2018;102(5):717–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–86. [DOI] [PubMed] [Google Scholar]
- 42.Smith NL, Chen MH, Dehghan A, Strachan DP, Basu S, Soranzo N, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Loon JE, Leebeek FW, Deckers JW, Dippel DW, Poldermans D, Strachan DP, et al. Effect of genetic variations in syntaxin-binding protein-5 and syntaxin-2 on von Willebrand factor concentration and cardiovascular risk. Circ Cardiovasc Genet. 2010;3(6):507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu QM, Ko KA, Ture S, Mastrangelo MA, Chen MH, Johnson AD, et al. Novel Thrombotic Function of a Human SNP in STXBP5 Revealed by CRISPR/Cas9 Gene Editing in Mice. Arterioscler Thromb Vasc Biol. 2017;37(2):264–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, Hill MC, Kadow ZA, Suh JH, Tucker NR, Hall AW, et al. Long-range Pitx2c Enhancer-Promoter Interactions Prevent Predisposition to Atrial Fibrillation. Proc Natl Acad Sci U S A. 2019;116(45):22692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalonde S, Codina-Fauteux VA, de Bellefon SM, Leblanc F, Beaudoin M, Simon MM, et al. Integrative analysis of vascular endothelial cell genomic features identifies AIDA as a coronary artery disease candidate gene. Genome Biol. 2019;20(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aragam KG, Natarajan P. Polygenic Scores to Assess Atherosclerotic Cardiovascular Disease Risk: Clinical Perspectives and Basic Implications. Circ Res. 2020;126(9):1159–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roma-Rodrigues C, Fernandes AR. Genetics of hypertrophic cardiomyopathy: advances and pitfalls in molecular diagnosis and therapy. Appl Clin Genet. 2014;7:195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marian AJ, van Rooij E, Roberts R. Genetics and Genomics of Single-Gene Cardiovascular Diseases: Common Hereditary Cardiomyopathies as Prototypes of Single-Gene Disorders. J Am Coll Cardiol. 2016;68(25):2831–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102(10):1178–85. [DOI] [PubMed] [Google Scholar]
- 51.Day IN, Whittall RA, O’Dell SD, Haddad L, Bolla MK, Gudnason V, et al. Spectrum of LDL receptor gene mutations in heterozygous familial hypercholesterolemia. Hum Mutat. 1997;10(2):116–27. [DOI] [PubMed] [Google Scholar]
- 52.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–5. [DOI] [PubMed] [Google Scholar]
- 53.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chadwick AC, Wang X, Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol. 2017;37(9):1741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chadwick AC, Evitt NH, Lv W, Musunuru K. Reduced Blood Lipid Levels With In Vivo CRISPR-Cas9 Base Editing of ANGPTL3. Circulation. 2018;137(9):975–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glass Z, Lee M, Li Y, Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018;36(2):173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Platt RJ. CRISPR tool enables precise genome editing. Nature. 2019;576(7785):48–9. [DOI] [PubMed] [Google Scholar]
- 59.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36(6):536–9. [DOI] [PubMed] [Google Scholar]
- 60.Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 2020;4(1):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chamberlain K, Riyad JM, Weber T. Cardiac gene therapy with adeno-associated virus-based vectors. Curr Opin Cardiol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25(3):427–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther. 2010;21(6):704–12. [DOI] [PubMed] [Google Scholar]
- 68.Reardon S First CRISPR clinical trial gets green light from US panel. Nat News. 2016. [Google Scholar]
- 69.The Lancet Haematology. CRISPR-Cas9 gene editing for patients with haemoglobinopathies. Lancet Haematol. 2019;6(9):e438. [DOI] [PubMed] [Google Scholar]
- 70.National Academies of Sciences Eg, and Medicine, Medicine NAo, Sciences NAo, Committee on Human Gene Editing: Scientific Md, and Ethical Considerations. Human Genome Editing: Science, Ethics, and Governance. 2017. [PubMed] [Google Scholar]
- 71.Watters KE, Shivram H, Fellmann C, Lew RJ, McMahon B, Doudna JA. Potent CRISPR-Cas9 inhibitors from. Proc Natl Acad Sci U S A. 2020;117(12):6531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reardon S Step aside CRISPR, RNA editing is taking off. Nature. 2020;578(7793):24–7. [DOI] [PubMed] [Google Scholar]
- 73.Stitziel NO, Stirrups KE, Masca NG, Erdmann J, Ferrario PG, König IR, et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374(12):1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Emdin CA, Khera AV, Chaffin M, Klarin D, Natarajan P, Aragam K, et al. Analysis of predicted loss-of-function variants in UK Biobank identifies variants protective for disease. Nat Commun. 2018;9(1):1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–72. [DOI] [PubMed] [Google Scholar]