Abstract
In this study, we examined five previously synthesized compounds and checked their binding affinity towards the SARS-CoV-2 main protease (Mpro) by molecular docking study, and compared the data with three FDA approved drugs, i.e., Remdesivir, Ivermectine and Hydroxychlorochine. In addition, we have investigated the docking study against the main protease of SARS-CoV-2 (Mpro) by using Autodock 4.2 software package. The results suggested that the investigated compounds have property to bind the active position of the protein as reported in approved drugs. Hence, further experimental studies are required. The formation of intermolecular interactions, negative values of scoring functions, free binding energy and the calculated binding constants confirmed that the studied compounds have significant affinity for the specified biotarget. These studied compounds were passed the drug-likeness criteria as suggested by calculating ADME data by SwissADME server. Moreover, the ADMET properties suggested that the investigated compounds to be orally active compounds in human. Furthermore, density functional computations (DFT) were executed by applying GAUSSIAN 09 suit program. In addition, Quantitative Structure-Activity Relationship (QSAR) was studied by applying HyperChem Professional 8.0.3 program.
Keywords: SARS-CoV-2, DFT, QSAR, In silico toxicity analysis, Docking studies
1. Introduction
The appearance of highly transmitted pathogen SARS-CoV-2 has given gives birth to the pandemic COVID-19, causing anxiety among the people, and has significantly influenced the global economy (Mohapatra et al., 2020a, Mohapatra et al., 2020b). The unwanted COVID-19 outbreak driven by this highly infectious virus SARS-CoV-2 has spread its tentacles over the entire world and has taken millions of people under its cover (Lipsitch et al., 2020, Mohapatra and Rahman, 2020). Everyday huge number of people loses their lives due to this deadly disease. The number of infected patients is spiraling exponentially despite adequate measures taken by the respective governments. Besides, there is a social stigma attached to the disease too. The household where any member contracts the disease gets ostracized by the neighbors. This outbreak has overturned the normal living of human being worldwide and turned on the most critical universal health disaster of this century due to cross country transmission. (Almendros, 2020, Tiwari et al., 2020). As a preventive measure, people are forced to use masks, gloves, sanitizers, tissue papers during their daily activities resulting a huge amounts of medical waste (Ramteke et al., 2019, Saadat et al., 2020). The medical waste management would soon emerge as a big challenge before the governments in the wake of the rapid transmission of the disease, and the large scale use of masks and other accessories thereof. As there is no efficient approved treatment or drugs, the computational strategy is a promising way and plays a significant role in pharmaceutical industries to bring new drugs (Baildya et al., 2020, Mohapatra et al., 2020, Milenkovic et al., 2020, Cardoso and Mendanha, 2021, Kodchakorn et al., 2020).
We examined the binding affinity of the five previously synthesized compounds (Sahu et al., 2020) towards the SARS-CoV-2 main protease (Mpro) by molecular docking study and compared the data with three FDA approved drugs, i.e. Remdesivir, Ivermectine and Hydroxychlorochine. The SARS-CoV-2 main protease (Mpro) is a basic CoV enzyme playing a key role in mediating viral replication and transcription. Using computer-aided drug design the crystal structure of the COVID-19 virus (Mpro) in the complex with the N3 ligand (PDB ID: 6LU7) was determined; candidates for clinical trials were found in the virtual screening of more than 10,000 compounds (Jin et al., 2020). Thus, docking research was aimed at this particular target. On the basis of results of molecular docking, we have calculated the scoring function indicating the enthalpy contribution to the value of free binding energy (Affinity DG) for the best conformational positions; values of free binding energy and binding constants (EDoc kcal/mol and Ki uM (micromolar)) for a fixed conformational position of the ligand allowed to measure the stability of complexes formed between ligands and the corresponding target. Furthermore, the studied compounds passed the drug-likeness criteria as suggested by calculating ADME data by SwissADME server.
2. DFT investigations
The studied compounds were first optimized and then analyzed using Gauss View 6.0.16 program (GaussView 6.0, (Gaussian Inc., Wallingford, CT, USA), 2019). The most reliable theoretical method, i.e., density functional computations (DFT) were performed by using GAUSSIAN 09 suit programs (Becke, 1993, Frisch, 2009). The calculations were executed with the help of B3LYP and exchange correlation functional with 6–31 G (d, p) basis set for carbon, nitrogen, oxygen, hydrogen atoms.
2.1. Computational investigation (DFT studies)
DFT calculations were measured on GAUSSIAN 09 platform to interpret the atomic arrangement of studied compound (Figs. S1–S5), and were optimized using B3LYP/ 6–31 + G (d, p) basis set to establish the geometry theoretically. The values for dipole moment (D) and single point energy of the compounds is tested by applying DFT/B3LYP 6–31 + G (d, p) basis sets (Table 1 ), revealing that compound 3 keeps grater single point energy as compared to other compounds (Mahapatra et al., 2013, Sarangi et al., 2020, El-ajaily et al., 2019). However, compound 5 assumed to have less energy means greater stability, whereas the compound 3 has higher dipole moment value as compared to other compounds. These values clearly confirm that all the compounds possess dipole–dipole interaction.
Table 1.
Properties of the studied compounds.
| Compounds | DFT/B3LYP 6.31G+ (d, P) |
|
|---|---|---|
| Single point energy (kcal/mol) | Dipole moment (D) | |
| C16H20O (Comp. 1) | −4.3735 × 105 | 0.139 |
| C17H20O2 (Comp. 2) | −5.0844 × 105 | 0.466 |
| C16H20O (Comp. 3) | −4.3727 × 105 | 0.950 |
| C16H22O3 (Comp. 4) | −5.3249 × 105 | −0.987 |
| C19H22O4S (Comp. 5) | −9.0126 × 105 | 0.393 |
2.2. Frontier molecular orbitals (FMO) studies
From the FMO studies it is clear to understand the reaction of the compound and to envisage the active site located in the conjugate system, whereas the EHOMO and ELUMO of the invetigated compounds clarified the global reactivity descriptors, viz., chemical hardness, chemical potential, and electrophilicity. However, the negative values of EHOMO and ELUMO confirmed that the investigated compounds are stable (Yousef et al., 2012, Yousef et al., 2013). The electron cloud of the compound 1 is confined to O17 for the active atomic sites susceptible to the attack of nucleophile. The difference in bond energy distinctly explains the chemical reactivity and chemical stability of the active molecules (Govindarajan et al., 2012). The low [EHOMO - ELUMO) value supports that compound 2 is highly reactive among all compounds. But in reverse the stability of compound-1 is more (Fig-S3).
The FMO studies explained the chemical reactivity and the selection of the active sites in the molecular system. The values for the band energy and energy difference (LUMO-HOMO) describe the charge transfer (CT) interaction. However, significant chemical reactivity parameters, such as electronegativity (χ), chemical potential (μ), global hardness (η), global softness (S) and global electrophilicity index (ω) (Pearson, 1989, Padmanabhan et al., 2007) are listed in Table 2 .
Table 2.
Quantum chemical parameters of the compounds.
| Compounds | HOMO (eV) | LUMO (eV) | ΔE (eV) | χ (Pauling) | η (eV) | σ | μ (eV) | S | ω (eV) |
|---|---|---|---|---|---|---|---|---|---|
| C16H20O (Comp. 1) | −0.2221 | −0.0256 | 0.1965 | −0.1238 | 0.0982 | 10.18 | 0.1238 | 5.091 | 0.196 |
| C17H20O2 (Comp. 2) | −0.2241 | −0.0411 | 0.183 | −0.1326 | 0.0915 | 10.92 | 0.1326 | 5.464 | 0.095 |
| C16H20O (Comp. 3) | −0.2276 | −0.0408 | 0.1868 | −0.1342 | 0.0934 | 10.70 | 0.1342 | 5.353 | 0.096 |
| C16H22O3 (Comp. 4) | −0.2353 | −0.0427 | 0.1926 | −0.139 | 0.0963 | 10.38 | 0.139 | 5.192 | 0.1 |
| C19H22O4S (Comp. 5) | −0.2492 | −0.0662 | 0.1832 | −0.1577 | 0.0915 | 10.92 | 0.1577 | 5.464 | 0.135 |
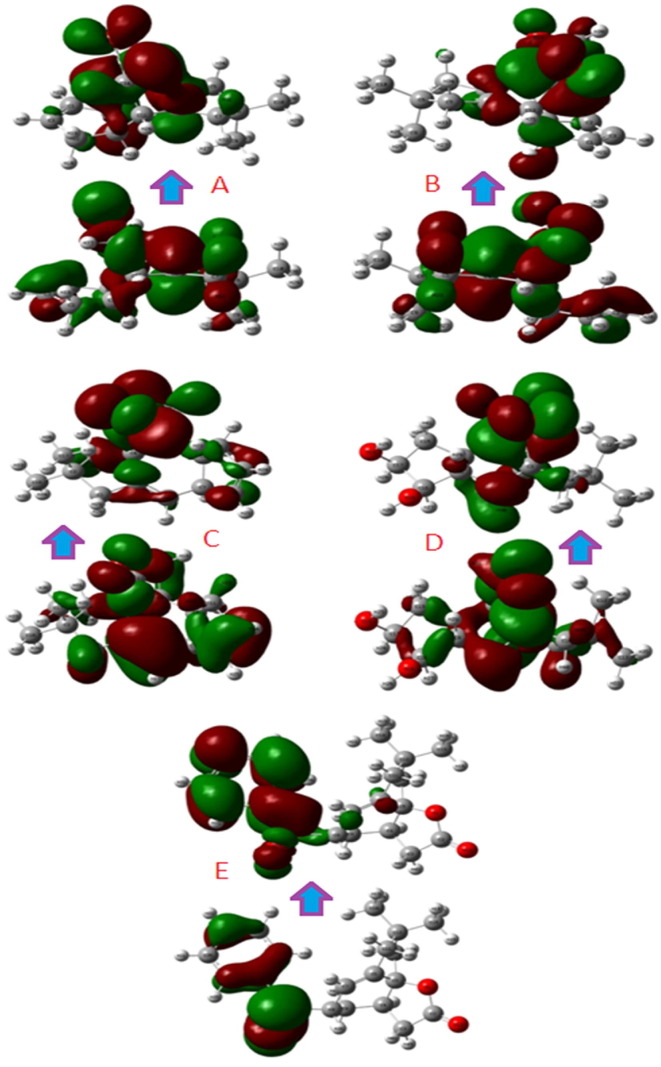
The EHOMO and ELUMO plots (Fig. 1 ) of the compounds established a precise orbital energy gap within the mapped molecular orbitals. The less chemical softness (S) for the compound 1 indicates its greater stability than the other compounds. However, some essential parameters, i.e., electrophilicity (), as a positive quantity, measures the tendency to accept electron from the surrounding. The whole study reveals that the low electrophilicity value for compound 1 makes it the most stable than the other investigated compounds (Table 2).
Fig. 1.
Comparison of HOMO-LUMO energy of the compounds (1–5).
2.3. Quantitative Structure-Activity Relationship (QSAR) studies
The QSAR technique anticipate the reactivity and properties of the investigated compounds. All the computations were obtained using HyperChem Professional 8.0.3 program. However, (MM+) force field with semi-empirical PM3 methods were applied to optimize the structures, whereas the energy minimization method was used with Fletcher-Reeves conjugate gradient algorithm. The partition coefficient (log P) value, which is more for compound 5, plays a key role in measuring the permeability of the investigated compound into the cell membrane (Padmanabhan et al., 2007). Some other physical parameters, i.e., volume, surface area, mass, refractivity, hydration energy, polarizability, free energy, total energy and RMS Gradient also suggest the action of the investigated compounds listed in Table 3 .
Table 3.
QSAR rating for optimized compounds.
| Function | Comp. 1 | Comp. 2 | Comp. 3 | Comp. 4 | Comp. 5 |
|---|---|---|---|---|---|
| Surface area (Approx) (Å2) | 328.09 | 334.43 | 327.49 | 352.68 | 467.38 |
| Surface area (Grid) (Å2) | 433.13 | 453.32 | 426.52 | 456.62 | 437.25 |
| Volume (Å3) | 721.14 | 767.44 | 719.88 | 768.74 | 938.40 |
| Hydration energy (Kcal/mole) | 0.64 | −0.75 | 0.56 | −7.42 | −4.78 |
| Log P | 5.09 | 4.23 | 5.28 | 3.26 | 8.68 |
| Refractivity (Å3) | 50.58 | 54.54 | 50.37 | 66.11 | 60.60 |
| Polarizability (Å3) | 26.74 | 28.44 | 26.74 | 27.62 | 33.64 |
| Mass (amu) | 228.33 | 256.34 | 228.33 | 262.35 | 346.44 |
| Total energy (kcal/mol) | −57607.3 | −67071.6 | −57595.7 | −71860.0.5 | −91108 |
| Dipole Moment (Debye) | 2.878 | 3.355 | 2.496 | 3.602 | 7.496 |
| Free energy (kcal/mol) | −57607.3 | −67071.6 | −57595.7 | −71860.0.5 | −91108 |
| RMS Gradient (kcal/Å mol) | 0.08536 | 0.09712 | 0.09696 | 0.08229 | 0.09258 |
2.4. Molecular docking study
The receptor-oriented flexible docking was carried out using Autodock 4.2 software package. The ligands were prepared by using MGL Tools 1.5.6 program and optimized with Avogadro program. The calculations were performed as reported previously (Mohapatra et al., 2020, Alam et al., 2019). The active macromolecule center of COVID-19 main protease (Mpro) (PDB ID:6LU7) obtained formhe Protein Data Bank was used as the biological target for the docking analysis. The receptor maps were designed with AutoGrid programs and MGL Tools. Water molecules, ions, and the ligand were detached from the PDB file ID: 6LU7. The following docking parameters were determined: the maximum RMS tolerance for the conformational cluster analysis – 2 Å; the free energy coefficient for torsional degrees of freedom – 0.2983; the cluster tolerance – 2 Å; the external grid energy – 1000; the maximum initial energy – 0; the maximum number of retries – 10000; the number of individuals in the population – 150; the maximum number of energy evaluations – 2500000; the maximum number of generations – 27,000; the number of top individuals to survive to the next generation – 1; the rate of gene mutation – 0.02; the rate of crossover – 0.8; the crossover mode – arithmetic; the α-parameter of Gauss distribution – 0; the β-parameter of Gauss distribution – 1. The visual analysis of complexes of substances in the active center of the COVID-19 main protease (Mpro) (PDB ID: 6LU7) was performed using the Discovery Studio Visualizer program.
3. Results of the docking study
Computer prediction of the antiviral activity was performed for molecules and drugs used in formularies for the treatment of COVID-19 (Jans and Wagstaff, 2020, Al-Tawfiq et al., 2020, Monforte et al., 2020). The scoring function (Affinity DG) for the best conformational positions; values of free binding energy and binding constants (EDoc kcal/mol and Ki uM (micromolar)) for a fixed conformational position of the ligand have been calculated on the basis of molecular docking analyses (Table 4 ).
Table 4.
The values for affinity DG, free binding energy, and binding coefficients of the test compounds in the complex with the COVID-19 virus (Mpro) protease (PDB ID: 6LU7).
| Compounds | 6LU7 |
||
|---|---|---|---|
| Affinity DG(kcal/mol) | EDoc (kcal/mol) | Ki (uMmicromolar) | |
| 1 | −6.4 | −5.19 | 155.75 µM |
| 2 | −6.2 | −4.56 | 453.00 µM |
| 3 | −6.5 | −6.22 | 27.69 µM |
| 4 | −6.4 | −5.39 | 112.79 µM |
| 5 | −7.3 | −6.25 | 26.20 µM |
| Hydroxychlorochine | −6.1 | −3.55 | 2.49 µM |
| Remdesivir | −6.5 | −2.53 | 14.00 µM |
| 22,23-dihydroavermectin B1a | −9.3 | −3.48 | 2.79 µM |
| 22,23-dihydroavermectin B1b | −8.3 | −6.21 | 28.28 µM |
The inhibitory activity of the tested molecules against COVID-19 virus (Mpro) protease may be due to the formation of stable complexes resulting from the energetically favorable geometric arrangement of ligands in the active site, and the formation of hydrogen bonds, donor–acceptor and intermolecular electrostatic interactions between them. The thermodynamic probability of such binding is established by the negative AffinityDG (kcal/mol) scoring function, calculated free binding energy EDoc (kcal/mol), and binding constants Ki (uM). As can be seen from the results, among the test compounds the leader is the molecule M16 (Affinity DG = −7.3 kcal/mol, EDoc = −6.25, Ki = 26.20 uM), but it is significantly inferior by the values of binding constants to the reference drugs Hydroxychlorochine (Ki = 2.49 µM) and 22,23-dihydroavermectin B1a (Ki = 2.79 µM) (Table 4).
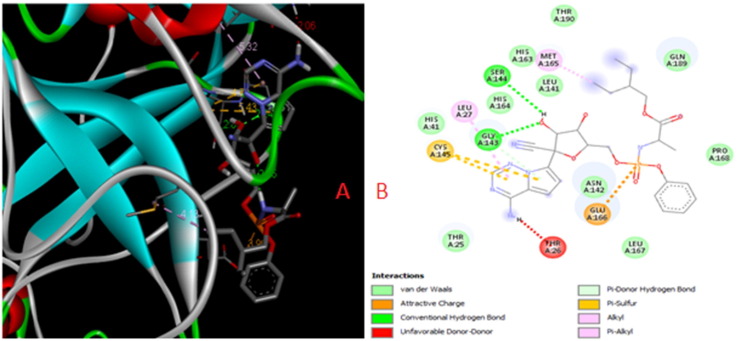
The next step in molecular docking is a thorough investigation of the geometrical arrangement of the studied molecules and the reference drugs in the active site of the viral protease. This will provide a complete understanding which molecular fragments are involved in binding to a biotarget, and allow us to give clear recommendations for the rational design of future candidates. Remdesivir with the viral protease (PDB ID: 6LU7) forms a complex due to hydrogen bonds between the oxygen atom of the hydroxyl group of the tetrahydrofuran fragment and the residues of amino acids Ser144 and Gly143. The attractive charge occurs between the phosphorus atom of the orthophosphoric acid residue and the Glu166 residue.
The formation of the complex is facilitated by the unfavorable donor–donor interaction, which occurs between the hydrogen proton of the substituted amino group of the pyrrolo (Mohapatra et al., 2020a, Mohapatra et al., 2020b, Mohapatra and Rahman, 2020) triazin cycle with the residue of amino acid Thr26. The π-H π-Sulphur interaction takes place between the pyrrolo (Mohapatra et al., 2020a, Mohapatra et al., 2020b, Mohapatra and Rahman, 2020) triazin cycle and the residues of Gly143 and Cys145, respectively. The complex of π-Alkand Alk interactions between triazine, ethyl fragments with the residues of leucine Leu27 and methionine Met165 is also stabilized (Fig. 2 ).
Fig. 2.
The superposition of the Remdesivir molecule and the intermolecular interactions in the complex with the COVID-19 virus (Mpro) protease (PDB ID: 6LU7).
Ivermectin is composed of two molecules – 22,23-dihydroavermectin B1a and 22,23-dihydroavermectin B1b. The complex of 22,23-dihydroavermectin B1a with the viral protease is formed by four hydrogen bonds. These bonds appear due to three oxygen atoms and a hydrogen proton of the hydroxyl group with the residues of amino acids His246, Glu240, Gln110. Carbon-hydrogen bonds occur between two ethoxyl substituents and the residues of glycine Gln107 and aspartic acid Asp245. The π-Alk and Alk interactions between methyl substituents of the molecule and the residues of His246 Ile249 and Phe294 contribute to the stabilization of the complex (Fig. S6a). The molecule of 22,23-dihydroavermectin B1b, unlike 22,23-dihydroavermectin B1a, has a different binding site. It forms a complex with a biotarget by hydrogen bond between the oxygen of the carbonyl group and the residue of leucine Leu287.
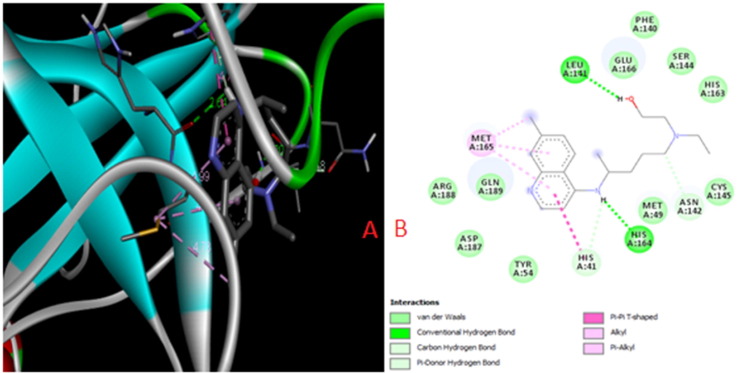
The carbon hydrogen bond between the ethoxyl group and the residue of Asp197 also contributes to the appearance of the complex. Intermolecular Alk interactions and the unfavorable acceptor–acceptor interaction with the residues of amino acids Met276, Leu286, Lys137, and Val171 contribute to the stabilization of the complex (Fig. S6b). Hydroxychlorochine with the viral protease forms a complex with the participation of hydrogen bonds between hydrogen protons of the hydroxyl, amino groups and the residues of leucine Leu141 and histidine His164. The hydrogen carbon and π-H bonds appear between the hydrogen proton of the amino and methyl groups with the residues of amino acids Asn142 and His41, respectively. Additionally, the complex of π-π, π-Alk and Alk interactions with the residues of amino acids Met165, His41 is stabilized (Fig. 3 ).
Fig. 3.
The superposition of the Hydroxychlorochinemolecule and the intermolecular interactions in the complex with the COVID-19 virus (Mpro)protease(PDB ID: 6LU7).
The values of interatomic distances in the active site of the COVID-19 virus (Mpro) protease (PDB ID: 6LU7) between fragments of molecules of the reference drugs and amino acid residues are shown in the diagrams (Figs. S2, S3, S6), categories and types of intermolecular interactions are given in Table S1.
Due to the oxygen atom in the active site of the COVID-19 virus (Mpro) protease, Compound 1 forms hydrogen bonds with the amino acid residues of threonine Thr292 and glycine Gln110. Complex is stabilized by π-Alk and Alk interactions with the residues of Val104, Phe294, Ile106 (Fig. S7). The formation of a complex with Compound 2 is provided by a hydrogen bond between the oxygen of the molecule and the residue of asparagusate Asn151. Further, the π-Alk interaction complex with the phenylalanine residue Phe294 is stabilized (Fig. S8).
The complex with the viral protease of Compound 3 is formed by π-Alk and Alk interactions between methyl substituents and the condensed cyclobuta[d]s-indacen-2-one system with the residues of amino acids of phenylalanine Phe 294 and valine Val104 (Fig. S9). Compound 4 forms a complex with the viral protease due to hydrogen bonds occurring between oxygen and hydrogen atoms of hydroxyl groups with the residues of Lys97, Asn95, and Gly15. The π-Alk and Alk interactions with the residues of amino acids Val73, Pro96, Ala70, and Trp31 contribute to the stabilization of the complex (Fig. S10).
The formation of a complex of Compound 5 with the COVID-19 virus (Mpro) protease is facilitated by hydrogen and carbon hydrogen bonds occurring between the oxygen atoms of the sulfo- and keto groups of the molecule studied with the residues of Asp151 and Asp153. The π-σ bond is formed between the phenyl fragment and the residue of isoleucine Ile106. The complex π-Alk is stabilized by interactions between methyl, cyclopentane, and phenyl fragments of the molecule with the residues of Phe294 and Val104, respectively (Fig. S11).
The values of interatomic distances in the active site of the COVID-19 virus (Mpro) protease (PDB ID: 6LU7) between fragments of the compounds (1–5) and amino acid residues shown in the diagrams (Figs. S7–S11), whereas types of intermolecular interactions are given in Table S2. On the basis of detailed analysis of the location of the tested molecules in the binding sites with the Mpro protease (PDB ID: 6LU7), formation of a number of intermolecular interactions, the negative scoring functions, free binding energy and the calculated binding constants, we can conclude that the studied molecules have affinity for the specified biotarget. As per the docking results, the test compounds have the same binding site with 22,23-dihydroavermectin B1a, which has the best values of scoring functions, free energy, and binding constants, indicating a high efficiency and direction of action of the studied molecules.
4. In silico toxicity analysis
The ADME data screening was carried out and identified the studied compounds with good drug-likeness (Amin et al., 2020, Daina et al., 2017, El-Saadi et al., 2015). Ccompounds (1–5) passed the drug-likeness criteria (Fig. S12) as suggested by the calculated ADME data by SwissADME server (http://www.swissadme.ch/ (as accessed on 30th Oct., 2020)). Radar plot of the compounds suggesting the drug-likeness are important to identify compounds with poor absorption and permeation, and address a portion of the range of obstacles, which a compound must show its druggability. The tested ccompounds (1–5) showed excellent drug-likeness, while the reference compounds displayed compromised data (Fig. S13). However, the flexibility of the molecules is poor. Additionally, these potential ccompounds (1–5) were considered for further Lipinski-Veber test (Lipinski et al., 2001, Veber et al., 2002) in Discover Studio 2016 (https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (as accessed on 30th Oct., 2020)). In order to accurately check the molecules, drug-like indices were utilized for comparing Lipinski’s RO5, Veber’s selective criteria for oral bioavailable drugs (Amin et al., 2018) (Table 5 ).
Table 5.
Results of the Lipinski-Veber test.
| Index | Name | HBALip | HBDLip | MW | ALogP | RB | MPSA | HBA | HBD |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Comp. 5 | 4 | 0 | 346.441 | 3.145 | 2 | 68.81 | 4 | 0 |
| 2 | Comp. 4 | 3 | 2 | 262.344 | 0.494 | 0 | 57.53 | 3 | 2 |
| 3 | Comp. 3 | 1 | 0 | 228.329 | 2.762 | 0 | 17.07 | 1 | 0 |
| 4 | Comp. 2 | 2 | 0 | 256.339 | 2.261 | 0 | 29.6 | 2 | 0 |
| 5 | Comp. 1 | 1 | 0 | 228.329 | 2.791 | 0 | 17.07 | 1 | 0 |
| 6 | Hydroxychlorochine | 4 | 2 | 335.872 | 3.457 | 9 | 48.39 | 4 | 2 |
Num_H_Acceptors_Lipinski, HBALip; Num_H_Donors_Lipinski, HBDLip; Molecular_Weight, MW; Num_RotatableBonds, RB; Molecular_Polar SurfaceArea, MPSA; Num_H_Acceptors, HBA; Num_H_Donors, HBD.
Each designed compounds met Lipinski’s RO5 criteria. Besides RO5, MW independent rules can accurately predict oral bioavailability (Veber et al., 2002). In addition, ADMET_BBB_Level, ADMET_EXT_CYP2D6#Prediction, ADMET_EXT_Hepatotoxic#Prediction, ADMET_EXT_PPB#Prediction, ADMET_AlogP98 and ADMET_PSA_2D properties were also checked for these molecules (https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (as accessed on 30th Oct., 2020)), and depicted in Table 6 for comparing with known compounds such as Remdesivir, Ivermectine_b1b, Ivermectine_b1a and Hydroxychlorochine.
Table 6.
Comparison of ADMET properties of the mentioned compounds.
| Index | Name | ADMET_BBB_Level | ADMET_EXT_CYP2D6#Prediction | ADMET_EXT_Hepatotoxic#Prediction | ADMET_EXT_PPB#Prediction | ADMET_AlogP98 | ADMET_PSA_2D |
|---|---|---|---|---|---|---|---|
| 1 | Remdesivir | 4 | FALSE | TRUE | FALSE | 1.588 | 205.713 |
| 2 | Comp. 5 | 2 | FALSE | FALSE | TRUE | 3.145 | 60.832 |
| 3 | Comp. 4 | 3 | FALSE | TRUE | FALSE | 0.494 | 58.931 |
| 4 | Comp. 3 | 1 | FALSE | TRUE | TRUE | 2.762 | 17.3 |
| 5 | Comp. 2 | 1 | FALSE | FALSE | TRUE | 2.261 | 26.23 |
| 6 | Comp. 1 | 1 | FALSE | FALSE | TRUE | 2.791 | 17.3 |
| 7 | Ivermectine_b1b | 4 | FALSE | TRUE | FALSE | 4.287 | 169.048 |
| 8 | Ivermectine_b1a | 4 | FALSE | TRUE | FALSE | 4.743 | 169.048 |
| 9 | Hydroxychlorochine | 1 | TRUE | TRUE | FALSE | 3.457 | 48.239 |
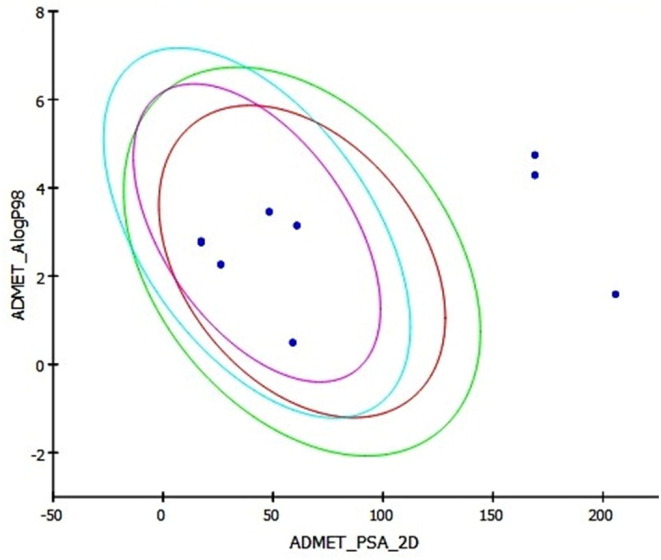
Moreover, the ADMET_AlogP98vsADMET_PSA_2D plot also suggested that Compounds 1–5 were within the applicability domain (Fig. 4 ), whereas Remdesivir, Ivermectine_b1b, Ivermectine_b1a were found to be outlier. Hence, the high compliance with in silico ADMET suggested that our compounds (1–5) were likely to be orally active compounds in human.
Fig. 4.
ADMET_AlogP98vsADMET_PSA_2D plot of these molecules.
5. Conclusion
In this work, we have investigated the molecular docking study of five previously synthesized compounds for their binding affinity towards the SARS-CoV-2 main protease (Mpro) and the data are compared with three FDA approved drugs (Remdesivir, Ivermectine and Hydroxychlorochine). As per DFT study, compound 1 is most stable because it possess less electrophilicity value corresponding to the others. According to QSAR study, the evaluated log P values for the compound 5 are greater than other compounds, and explain the biological activity of the reported synthesized compounds. These studied ccompounds also passed the drug-likeness criteria as suggested by calculating ADME data by SwissADME server. Moreover, the ADMET properties suggested that the investigated compounds are likely to orally active in human. As per Radar plot, the compounds showed excellent drug-likeness, while the reference compounds displayed compromised data. However, the flexibility of the molecules is poor. On the whole, we hope this work should help the researchers in this field to develop potential vaccines and therapeutics against the novel CoV. The formation of intermolecular interactions, negative scoring functions, free binding energy and the calculated values of binding constants confirmed that the studied compounds have significant affinity for the specified biotarget. As per the docking results, the test compounds have the same binding site with 22,23-dihydroavermectin B1a, which has the best values of scoring functions, free energy, and binding constants. This indicates a high efficiency and direction of action of the studied molecules. We hope this work should help the researchers to develop potential drugs and therapeutics for combating the novel CoV-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the financial support through Researchers Supporting Project number (RSP-2020/147), King Saud University, Riyadh, Saudi Arabia. Authors are also thankful to Mr. Sk. Abdul Amin of Jadavpur University, India for his cooperation in computational studies.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2020.101315.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alam M., Kim Y., Park S. Synthesis, characterization (IR, 1H, 13C & 31P NMR), fungicidal, herbicidal and molecular docking evaluation of steroid phosphorus compounds. Open Chem. 2019;17:621–628. [Google Scholar]
- Almendros A. Can companion animals become infected with Covid-19? Vet Rec. 2020;186(12):388–389. doi: 10.1136/vr.m1194. [DOI] [PubMed] [Google Scholar]
- Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med. Infect. Dis. 2020;34:101615. doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S.A., Adhikari N., Jha T. Diverse classes of HDAC8 inhibitors: in search of molecular fingerprints that regulate activity. Future Med Chem. 2018;10(13):1589–1602. doi: 10.4155/fmc-2018-0005. PMID: 29953251. [DOI] [PubMed] [Google Scholar]
- Amin SA, Ghosh K, Gayen S, Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J Biomol Struct Dyn. 2020, 1-10. doi: 10.1080/07391102.2020.1780946. PMID: 32568618; PMCID: PMC7332872. [DOI] [PMC free article] [PubMed]
- Baildya N., Ghosh N.N., Chattopadhyay A.P. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: An insight from MD-simulation studies. J. Mol. Struct. 2020;1219 doi: 10.1016/j.molstruc.2020.128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke A.D. J. Chem. Phys. 1993;98(7):5648–5652. [Google Scholar]
- Cardoso W.B., Mendanha S.A. Molecular dynamics simulation of docking structures of SARS-CoV-2 main protease and HIV protease inhibitors. J. Mol. Struct. 2021;1225:129143. doi: 10.1016/j.molstruc.2020.129143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-ajaily M.M., Sarangi A.K., Mohapatra R.K., Hassan S.S., Eldaghare R.N., Mohapatra P.K., Raval M.K., Das D., Mahal A., Cipurkovic A. Chem. Select. 2019;4:9999–10005. [Google Scholar]
- El-Saadi M.W., Williams-Hart T., Salvatore B.A., Mahdavian E. Use of in-silico assays to characterize the ADMET profile and identify potential therapeutic targets of fusarochromanone, a novel anti-cancer agent. Silico Pharmacol. 2015 Dec;3(1):6. doi: 10.1186/s40203-015-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.J., et al. Gaussian Inc; Wallingford CT: 2009. GAUSIAN 09. [Google Scholar]
- GaussView 6.0, (Gaussian Inc., Wallingford, CT, USA) 2019.
- Govindarajan M., Periandy S., Carthigayen K. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2012;97:411. doi: 10.1016/j.saa.2012.06.028. [DOI] [PubMed] [Google Scholar]
- http://www.swissadme.ch/ (as accessed on 30th Oct., 2020).
- https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (as accessed on 30th Oct., 2020).
- Jans D.A., Wagstaff K.M. The broad spectrum host-directed agent ivermectin as an antiviral for SARS-CoV-2? Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Kodchakorn K., Poovorawan Y., Suwannakarn K., Kongtawelert P. Molecular modelling investigation for drugs and nutraceuticals against protease of SARS-CoV-2. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/s0169-409x(00)00129-0. PMID: 11259830. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Swerdlow D.L., Finelli L. Defining the Epidemiology of Covid-19 – Studies Needed. New England J. Med. 2020;382(13):1194–1196. doi: 10.1056/NEJMp2002125. [DOI] [PubMed] [Google Scholar]
- Mahapatra B.B., Mishra R.R., Sarangi A.K. J. Saudi Chem. Soc. 2013;20(6):635–643. [Google Scholar]
- Milenkovic D.A., Dimic D.S., Avdovic E.H., Markovic Z.S. Several coumarin derivatives and their Pd(II)complexes as potential inhibitors of the mainprotease of SARS-CoV-2, an in silico approach. RSC Adv. 2020;10:35099. doi: 10.1039/d0ra07062a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra R.K., Elajaily M.M., Alassbaly F.S., Sarangi A.K., Das D., Maihub A.A., BenGweirif S.F., Mahal A., Suleiman M., Perekhoda L., Azam M., AlNoor T.H. DFT, anticancer, antioxidant and molecular docking investigations of some ternary Ni(II) complexes with 2[(E)[4(dimethylamino) phenyl]methyleneamino]phenol. Chem. Pap. 2020 doi: 10.1007/s11696-020-01342-8. [DOI] [Google Scholar]
- Mohapatra R.K., Das P.K., Kandi V. Challenges in Controlling COVID-19 in Migrants in Odisha, India. Diabetes Metab. Syndr.: Clin. Res. Rev. 2020;14(6):1593–1594. doi: 10.1016/j.dsx.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan K. Mohapatra V.P. Saikishore Mohammad Azam Susanta K. Biswal Synthesis and physico-chemical studies of a series of mixed ligand transition metal complexes and their molecular docking investigations against Coronavirus main protease, Open 18 1 2020 1495 1506
- Mohapatra R.K., Rahman M. Is it possible to control the outbreak of COVID-19 in Dharavi, Asia’s largest slum situated in Mumbai? Anti-Infective Agents. 2020;18 doi: 10.2174/2211352518999200831142851. [DOI] [Google Scholar]
- Mohapatra R.K., Pintilie L., Kandi V., Sarangi A.K., Das D., Sahu R., Perekhoda L. The recent challenges of highly contagious COVID-19; causing respiratory infections: Symptoms, diagnosis, transmission, possible vaccines, animal models and immunotherapy. Chem. Biol. Drug Des. 2020;96(5):1187–1208. doi: 10.1111/cbdd.v96.510.1111/cbdd.13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte A.A., Tavelli A., Bai F., Marchetti G., Cozzi-Lepri A. Effectiveness of hydroxychloroquine in COVID-19 disease: A done and dusted deal? Int. J. Infect. Diseases. 2020;99:75–76. doi: 10.1016/j.ijid.2020.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan J., Parthasarathi R., Subramanian V., Chattaraj P. J. Phys. Chem. A. 2007;111:1358. doi: 10.1021/jp0649549. [DOI] [PubMed] [Google Scholar]
- Pearson R.G. J. Org. Chem. 1989;54:1423. [Google Scholar]
- S. Ramteke B.L. Sahu Novel coronavirus disease (COVID-19) pandemic: considerations for the biomedical waste sector in India Case Studies in Chemical and Environmental Engineering 2020 2019 10.1016/j.cscee.2020.100029 [DOI] [PMC free article] [PubMed]
- Saadat S., Rawtani D., Hussain C.M. Environmental perspective of COVID-19. Sci. Total Environ. 2020;728:138870. doi: 10.1016/j.scitotenv.2020.138870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R. Sahu R.K. Mohapatra S.I. Al-Resayes D. Das P.K. Parhi L. Pintilie M. Azam An Efficient Synthesis Towards the Core of Crinipellin and Alliacol-B Along With Their Docking Studies Preprints 2020120206 2020 doi: 10.20944/preprints202012.0206.v1
- A.K. Sarangi B.B. Mahapatra R.K. Mohapatra S.K. Sethy D. Das L. Pintilie M. K.-E-Zahan, M. Azam, H. Meher, Applied Organometallic Chemistry 34 e5693 2020 10.1002/aoc.5693
- Tiwari R., Dhama K., Sharun K., Iqbal Yatoo M., Malik Y.S., Singh R., Michalak I., Sah R., Bonilla-Aldana D.K., Rodriguez-Morales A.J. COVID-19: Animals, veterinary and zoonotic links. Veterinary Quarter. 2020;40(1):169–182. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D.F. Veber S.R. Johnson H.Y. Cheng B.R. Smith K.W. Ward K.D. Kopple Molecular properties that influence the oral bioavailability of drug candidates J Med Chem. 45 12 2002 2615 23 10.1021/jm020017n PMID: 12036371 [DOI] [PubMed]
- Yousef T.A., Abu El-Reash G.M., El Morshedy R.M. Polyhedron. 2012;45:71. [Google Scholar]
- Yousef T.A., Abu El-Reash G.M., El Morshedy R.M. J. Mol. Struct. 2013;1045:145. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.