Abstract
Simple Summary
Mutation of the APC gene is a common early event in colorectal cancer, however lower rates have been reported in younger cohorts of colorectal cancer patients. In sporadic cancer, mutations are typically clustered around a mutation cluster region, a narrowly defined hotspot within the APC gene. In this study we used a sequencing strategy aimed at identifying mutations more widely throughout the APC gene in patients aged 50 years or under. We found high rates of APC mutation in our young cohort that were similar to rates seen in older patients but the mutations we found were spread throughout the gene in a pattern more similar to that seen in inherited rather than sporadic mutations. Our study has implications both for the sequencing of the APC gene in early-onset colorectal cancer and for the etiology of this disease.
Abstract
While overall colorectal cancer (CRC) cases have been declining worldwide there has been an increase in the incidence of the disease among patients under 50 years of age. Mutation of the APC gene is a common early event in CRC but is reported at lower rates in early-onset colorectal cancer (EOCRC) than in older patients. Here we investigate the APC mutation status of a cohort of EOCRC patients in New Zealand using a novel sequencing approach targeting regions of the gene encompassing the vast majority of known APC mutations. Using this strategy we find a higher rate (72%) of APC mutation than previously reported in EOCRC with mutations being spread throughout the gene rather than clustered in hotspots as seen with sporadic mutations in older patients. The rate of mutations falling within hotspots was similar to those previously seen in EOCRC and as such our study has implications for sequencing strategies for EOCRC patients. Overall there were low rates of both loss of heterozygosity and microsatellite instability whereas a relatively high rate (40%) of APC promoter methylation was found, possibly reflecting increasing exposure of young people to pro-oncogenic lifestyle factors.
Keywords: early-onset, colorectal cancer, APC, sequencing, DNA methylation, diet
1. Introduction
Over recent decades there has been an overall drop in the worldwide incidence rates of colorectal cancer (CRC) worldwide, primarily due to increased rates of colonoscopy screening which leads to the early detection and removal of precancerous polyps [1,2]. However, over the same period there has been a worldwide increase in incidence of early-onset colorectal cancer (EOCRC) [3,4]. While hereditary syndromes such as Lynch Syndrome [5] and familial adenomatous polyposis (FAP) [6] account for a small proportion of cases [7], and an even smaller proportion arise due to long-standing inflammatory bowel disease [8], the majority of these cancers are considered to be sporadic [9]. Whereas the majority of EOCRC patients are not found to have an inherited disorder, they are more likely to have a family history of disease compared to older patients [10]. Against this complex background, the underlying causative factors that initiate the genetic mutations required to drive the adenoma-carcinoma sequence in these individuals are not well understood.
Mutation of the tumour-suppressor adenomatous polyposis coli (APC) gene is the most frequent early genetic event in the progression of colorectal carcinogenesis. APC mutations are found in the earliest microscopic adenomas [11], and at the same rate in early adenomas as in sporadic tumours [12]. APC is a multi-functional protein in epithelial cells, having roles in Wnt signalling [13], cell structure [14], epithelial cell polarity [15], and apoptosis [16]. Consequently, the loss of a fully functioning APC protein can result in cells that display a number of pro-carcinogenic characteristics.
Both alleles of APC require inactivation for carcinogenesis to occur [11]. This may involve mutation of both alleles, or the second allele may be silenced by loss of heterozygosity (LOH) [17]. However, the worldwide increase in early onset CRC hints more at the involvement of environmental and/or lifestyle factors [1], and this is reinforced by studies on people who migrate from low- to high-risk areas of the world [18]. Accordingly, epigenetic modification of the promoter region of this gene can also result in altered APC expression in the absence of mutated DNA sequence, as recently reported in a mouse model of obesity [19].
Despite mutations of the APC gene being well documented in colorectal cancers as a whole, the incidence of mutations is reported at notably lower rates in EOCRC than in older patients [20,21], adding to speculation that sporadic EOCRC may be a subtype of CRC with distinct clinical and molecular features [10,22,23] that reflect alternative pathways for the development of CRC. For example, 15 to 30% of CRC cases develop from serrated polyps [24], a subset of CRCs that rarely present with APC mutations but commonly have mutations in BRAF and evidence of microsatellite instability [25]. However, another possibility may also exist. The APC gene is located on chromosome 5q21-q22 and consists of 15 exons, with the final exon making up more than 75% of the coding sequence of the gene. It is this final exon where the bulk of mutations reportedly occur, specifically within a mutation cluster region (MCR) between codons 1286 and 1513 [26]. Consequently, it is this hotspot where many studies investigating APC mutation have focused their efforts [27,28,29,30], raising the possibility those APC mutations that fall outside this region are not detected.
The aim of this study was to explore the molecular genotype of a cohort of tumour tissues collected from EOCRC patients in Canterbury, New Zealand. The first objective was to identify whether the rate of APC mutation in this cohort was similar to that reported elsewhere [20,21]. For this study however a sequencing approach was developed to maximise coverage of the APC gene, enabling identification of both the presence and location of mutations throughout the APC gene as a whole. Further investigation was undertaken to determine whether loss of heterozygosity [17] and/or methylation of the APC gene promoter region [31] may have contributed to silencing of the second APC gene allele in these early-onset colorectal cancers. The occurrence, type, and location of APC mutations are reported, as is evidence of microsatellite instability, loss of heterozygosity, and APC promoter methylation, and these findings are compared to clinical features of the tumours.
2. Results
The cohort of 25 early-onset CRC cases consisted of 14 (56%) women and 11 (44%) men aged 50 years or under at diagnosis (median 44 years, range 28–50 years) (Table 1). The tumours were predominantly distal (17/25, 68%). All but two tumours were classed as adenocarcinomas (92%), with a single adenosquamous carcinoma (4%) and a single signet-ring cell carcinoma (4%) completing the cohort (Table 1).
Table 1.
Clinical features of the early-onset CRC cohort.
| Number | % | |
|---|---|---|
| Total | 25 | |
| Gender | ||
| Male | 11 | 44 |
| Female | 14 | 56 |
| Age at diagnosis (years) | ||
| <30 | 1 | 4 |
| 31–40 | 6 | 24 |
| 41–50 | 18 | 72 |
| Tumour site | ||
| Caecum | 1 | 4 |
| Ascending colon | 6 | 24 |
| Hepatic flexure | 1 | 4 |
| Splenic flexure | 1 | 4 |
| Descending colon | 3 | 12 |
| Sigmoid colon | 12 | 48 |
| Rectosigmoid | 1 | 4 |
| Pathology | ||
| Adenocarcinoma | 23 | 92 |
| Adenosquamous carcinoma | 1 | 4 |
| Signet-ring cell carcinoma | 1 | 4 |
Twenty four mutations meeting the inclusion criteria for absolute (>10) and relative (>20%) read number were found in 18/25 (72%) tumour samples and confirmed by Sanger sequencing. Twelve tumours had a single APC mutation while six tumours had two mutations. The mutations consisted of 14 nonsense, six frameshift, three missense, and one splice region mutation (Table 2). Sequencing of matched normal tissues revealed that while the majority of mutations were sporadic, two APC mutations were determined to be germline as they were also present in matched normal tissue.
Table 2.
Molecular characteristics of EOCRC patients.
| Patient Number | Microsatellite Instability | Mutation | Mutational Outcome | Mutational Assessment 1 | APC Methylation | LOH | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| DNA | rsID | Protein | |||||||
| 10006 | MSI | c.2626C>T | rs121913333 | p.R876X | Nonsense | Damaging | U | - | [32] |
| 10765 | MSS | c.835-8A>G | rs1064793022 | Splice variant | Truncation 2 | Damaging | U | - | [33] |
| 12294 | MSS | c.1660C>T | rs137854573 | p.R554X | Nonsense | Damaging | M | - | [34] |
| 12961 | MSS | c.2853T>G | rs575406600 | p.Y951X | Nonsense | Damaging | M | - | [34] |
| 13046 | MSS | c.4488delT | n/a | p.P1497fsX | Frameshift | Damaging | U | - | This Study |
| c.2805C>A | rs137854575 | p.Y935X | Nonsense | Damaging | [35] | ||||
| 13501 | MSS | c.2636delA | n/a | p.Q879fsX | Frameshift | Damaging | U | - | This Study |
| 13564 | MSS | c.4033G>T | rs1211642532 | P.E1345X | Nonsense | Damaging | U | LOH | [36] |
| 14459 | MSS | c.4463delT | rs1114167577 | p.L1488fsX | Frameshift | Damaging | M | - | [37] |
| c.3826delT | n/a | p.S1276fsX | Frameshift | Damaging | This Study | ||||
| 15052 | MSS | - | - | - | - | - | M | - | - |
| 15296 | MSS | - | - | - | - | - | U | - | - |
| 15310 | MSS | c.4216C>T | rs587782518 | p.Q1406X | Nonsense | Damaging | U | LOH | [38] |
| c.3949G>C | rs1801166 | p.E1317Q | Missense | Benign | [39] | ||||
| 15471 | MSS | - | - | - | - | - | M | - | - |
| 16872 | MSS | - | - | - | - | - | U | - | - |
| 16993 | MSS | c.4717G>T | rs878853217 | p.E1573X | Nonsense | Damaging | U | - | [40] |
| c.2821G>T | n/a | p.E941X | Nonsense | Damaging | [37] | ||||
| 17068 | MSS | - | - | - | - | - | M | - | - |
| 17871 | MSS | c.4485delT | n/a | p.S1459fsX | Frameshift | Damaging | U | - | This Study |
| c.2626C>T | rs121913333 | p.R876X | Nonsense | Damaging | [32] | ||||
| 18090 | MSS | c.3925G>T | n/a | p.E1309X | Nonsense | Damaging | M | LOH | [27] |
| 19199 | MSS | c.646C>T | rs62619935 | p.R216X | Nonsense | Damaging | U | - | [36] |
| 19513 | MSS | c.3740C>T | n/a | p.A1247V | Missense | Benign | M | - | This Study |
| c.3340C>T | rs121913331 | p.R1114X | Nonsense | Damaging | [40] | ||||
| 19906 | MSS | - | - | - | - | - | U | - | - |
| 20039 | MSS | c.4230C>A | n/a | p.C1410X | Nonsense | Damaging | U | - | This Study |
| 20085 | MSI | c.694C>T | rs397515734 | p.R232X | Nonsense | Damaging | M | - | [35] |
| 20187 | MSS | c.3374T>C | rs377278397 | p.V1125A | Missense | Benign | U | - | [41] |
| 21025 | MSI | c.2563_2564dupGA | n/a | p.R856fsX | Frameshift | Damaging | M | - | This Study |
| 21082 | MSS | - | - | - | - | - | U | - | - |
1 Missense mutations were considered damaging if 3/5 or more software tools predicted the mutation to be possibly or probably damaging. Nonsense or frameshift mutations were all considered damaging without predictive tools. 2 The splice variant c.835-8A>G results in a frameshift that ultimately leads to premature truncation of APC. Mutations in bold denote germline status. MSS, microsatellite stable; MSI, microsatellite instability; M, methylated APC promoter; U, Unmethylated APC promoter; LOH, Loss of heterozygosity at the APC locus; n/a, these mutations have no rsID.
One of the germline mutations (a c.3374T>C variant) results in a substitution at codon 1125 of a valine to an alanine (p.V1125A) and is regarded as a conservative change as they are closely related amino acids. Coupled with the occurrence of the variant in the general population [41], as well as its position in a poorly conserved region of the gene, the c.3374T>C variant is considered benign.
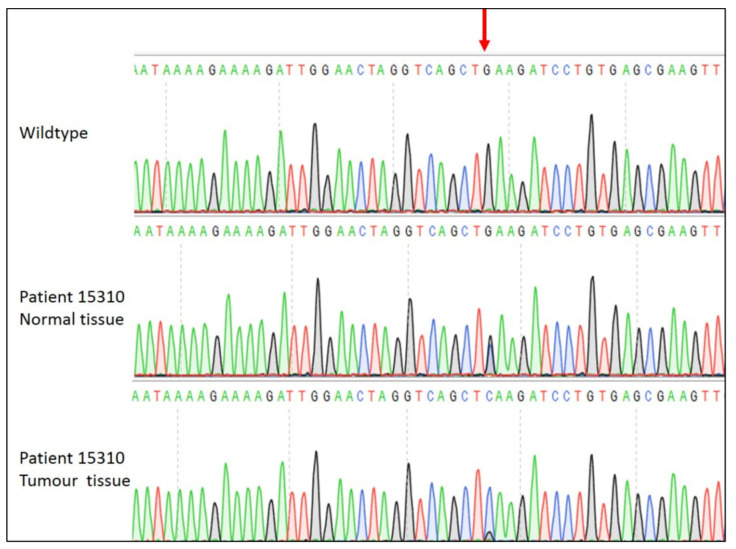
However, the second germline mutation, c.3949G>C, which results in a glutamic acid to glutamine change at codon 1317 of APC (p.E1317Q), was accompanied by loss of the wild type allele in the tumour tissue (Figure 1). This mutation was found in a 36 year old female (Patient 15310) with a stage 2 adenocarcinoma with no lymph node involvement or metastasis. Subsequent colonoscopies have shown the patient to be disease-free up to eight years post-surgery with no evidence of polyps despite the germline APC mutation. There was no reported family history of bowel cancer.
Figure 1.
Sanger sequencing of APC c.3949G>C mutation. The guanine base indicated by the red arrow in the wild type APC sequence is mutated to a cytosine in one allele in the normal tissue of the patient to give a heterozygous sequence. In the tumour tissue the wild type guanine allele is barely visible.
3. Distribution of APC Mutations
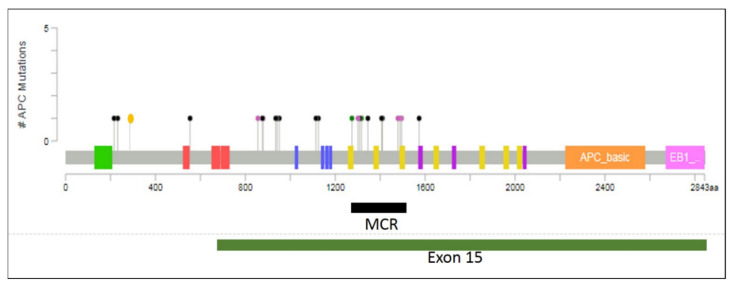
The mutations found in the early-onset cohort were distributed widely across the first 1600 codons of APC. While only 8/25 (32%) of patients had mutations located within the mutation cluster region [26], a total of 14/25 (56%) of patients had one or two mutations in the large final exon of APC and that figure rose to 18/25 (72%) throughout the whole gene (Figure 2).
Figure 2.
APC mutations found in EOCRC samples mapped to their position within the protein. Each lollipop represents the position of a mutation with the colours of the lollipop representing different types of mutation: black, nonsense mutations; green, missense mutation; pink, frameshift mutation; yellow, splice variant. While a high proportion of mutations occur within the mutation cluster region (MCR, black bar), mutations are found throughout the first 1600 codons of APC including some upstream of exon 15 (green bar). Protein domains: green, oligomerisation; red, armadillo repeats; blue, 15 amino acid repeats; yellow, 20 amino acid repeats; purple, SAMP motif. The plot was generated using Mutation Mapper (www.cbioportal.org) and manually curated.
4. Microsatellite Instability
Of the 24 samples that were stained for mismatch repair proteins, 21 stained for all four mismatch repair proteins and were therefore considered microsatellite stable. All three of the remaining samples displayed microsatellite instability, with one negative for MSH6 staining and two negative for PMS2 staining. The three mismatch repair deficient tumours did not cluster at any location in the colon, as they were found in the ascending colon, splenic flexure and the sigmoid colon, respectively. Neither was there any association of microsatellite instability with age or gender.
One of the 21 samples that stained positively for all four mismatch repair genes was reported to have a BRAF mutation. This sample was the only adenosquamous carcinoma in the cohort. This combination of a BRAF mutation in a microsatellite stable adenosquamous colon cancer has been reported before [42].
5. Loss of Heterozygosity is Associated with APC Mutation Near Codon 1300
Loss of heterozygosity at the APC locus was found in 3/25 (12%) samples in the cohort. Strikingly, the three samples with LOH all carried an APC mutation clustered around codon 1300, a phenomenon previously reported in sporadic CRC [43]. As with microsatellite stability, no association was seen between LOH and any of age, gender or tumour location.
6. Methylation of the APC Promoter
Methylation analysis of the CpG island at the APC 1A promoter, responsible for the predominant APC isoform, was detected in 10/25 (40%) of tumours, a higher rate than previously reported in CRC [31], although there has been at least one recent report of higher APC promoter methylation in CRC [44]. There was no association of APC promoter methylation with age (mean age 43.5 years vs 44 years for the whole cohort), gender (60% vs. 56% female) or tumour location (70% vs. 68% distal).
7. Discussion
In this study we found a higher than anticipated rate of APC gene mutation in a cohort of EOCRC tumours, with sporadic mutations occurring throughout the gene rather than clustering in one region as expected. The APC gene is long, encompassing 8529 bases of coding sequence over multiple exons covering over 100 kilobases of chromosome 5, making targeted sequencing strategies cumbersome and expensive. Therefore, previous studies have focused on mutation hotspots in the gene, particularly the mutation cluster region between amino acids 1286 and 1513 [26] or more widely within the final, large exon which encompasses the majority of APC mutations [45]. Although targeted sequencing is seen as cost-effective, it has the potential to miss mutations that fall outside these regions. Accordingly, we developed a sequencing strategy to maximize the mutations identified while maintaining a manageable amount of sequencing.
Regions of APC with very low rates of mutation in sporadic CRCs were identified using the Catalogue of Somatic Mutation In Cancer (COSMIC) database, and excluded from our sequencing strategy. In this study, exons 5, 6, 8, 9, 11, 12 and 13, as well as exon 15 from amino acids 788 to 1593 were targeted. These exons encompass the vast majority of non-synonymous mutations of the APC gene in the COSMIC database, and each was sequenced. This approach identified 24 mutations in 18/25 (72%) tumour samples across these regions of the APC gene, a notably higher rate than some previous studies looking at APC mutation in early-onset CRCs [20,21] that may, in part, reflect the extent of APC gene coverage by the methodology used here. Importantly, if we had only sequenced the mutation cluster region, this number would have dropped to eight out of 25 tumours (32%). Likewise, mutations in the final exon of the gene were identified in only 14/25 (56%) of the samples. These findings convincingly highlight that protocols applied to study APC mutations are of critical importance, and that studies limited to “hotspot” regions of the gene may significantly under-report the mutation rate.
Despite known familial colorectal cancer syndromes being excluded, two patients were found to have germline APC mutations. While the p.V1125A variant was deemed to be benign, the p.E1317Q variant has been associated with colorectal cancer previously [39]. Notably, p.E1317Q is a missense mutation, resulting in a glutamic acid to glutamine change rather than the more common truncated APC proteins that result from APC mutations in FAP [46]. While the matched normal tissue showed both mutant and wild type alleles, the tumour tissue showed only mutant allele present (Figure 1) and this sample was one of three to display LOH at the APC locus. Accordingly, the lack of family history might be due to the single mutant allele alone not being sufficient for carcinogenesis, suggesting a second genetic hit is required for the pathogenicity of the p.E1317Q mutation to occur, an example of the damaging potential caused by loss of heterozygosity. It was noted that this patient did not present with polyps and would therefore be unlikely to be considered as a possible case of FAP and therefore not tested for APC mutation. As such, although we cannot rule out the possibility that the mutation has arisen de novo in this patient, it is likely that this patient has an atypical FAP phenotype and our finding may have implications for FAP testing.
We found that 8/24 (33%) mutations occurred within the MCR as defined by Miyoshi and colleagues [26]. Removing the two germline mutations made little difference to the proportion of sporadic mutations in the MCR (7/22, 32%). This is lower than might have been expected based on previous studies of CRC [45] where the majority of somatic mutations occur within the MCR.
Mutations in APC predominantly result in truncated proteins that retain some function essential for the survival of the cells [47]. The location of these truncating APC mutations affects the properties of the resultant protein and the tumour phenotype [48]. While familial APC mutations are scattered across the 5′ half of the gene, with notable hotspots at codons 1061 and 1309, somatic mutations tend to occur within the MCR [45]. This region contains a number of 20-amino acid repeats that act as β-catenin binding sites. The location of a mutation within the MCR determines how many of these repeats are included in the truncated protein.
Kohler and colleagues showed that mutations in the MCR resulted in the disruption of the third 20-amino acid repeat region while maintaining the first 20aa repeat. They also showed by co-immunoprecipitation that the second 20 amino acid repeat region, unlike the other two, had no β-catenin binding capacity therefore all the mutations within the MCR produced truncated proteins with similar β-catenin binding efficiency [49]. Our finding of an increased proportion of APC mutations occurring outside the MCR in EOCRC suggests that the β-catenin binding capacity of truncated APC is of less importance in EOCRC than in older patients, and that alternative functions of APC may be at play in these tumours.
Three decades ago biological differences were identified in tumours located in either the proximal (right-sided) or distal (left-sided) colon, leading to the proposal of distinct categories of colorectal cancer based on tumour location [50]. In particular, proximal tumours had characteristics similar to Lynch Syndrome, while distal tumours displayed characteristics more familiar to FAP. Moreover, EOCRC patients present more often with distal tumours than older CRC patients. One study had 32% of patients aged 35–39 diagnosed with having tumours in their rectum, with the percentage in subsequent age groups decreasing, down to 1% in those aged over 85 years. Conversely, only 9.3% of the 35–39 years age group had tumours in the caecum, rising to 23.2% in the over 85 year olds [51]. Further to Bufill’s categorisation of CRC by tumour location, it has been suggested that EOCRC should be similarly categorised by location [52].
This leads to an intriguing hypothesis. APC mutations in the MCR that result in 2–3 intact 20-amino acid repeats are more likely to be found in proximal tumours, while mutations leaving only 1 or no 20-amino acid repeats tend to be present in distal CRC [48]. Inherited mutations in FAP occur more widely across the 5′ half of the APC gene than somatic mutations, of which a large proportion are found in the MCR [45]. Our finding that APC mutations in EOCRC more closely resembled the pattern of APC mutations seen in FAP fits with the observation that EOCRC tumours are predominantly distal. In support of this hypothesis is the observation that the majority of EOCRC tumours do not show microsatellite instability and those that do are usually due to Lynch Syndrome [53]. In population-based studies MSI is found in between 7% and 17% of CRC cases under the age of 50 years [54]. Our finding of MSI in three out of 24 (12.5%) tumours for which MMR staining was available fits with these previous findings.
The CpG island methylator phenotype (CIMP), where multiple tumour suppressor genes are silenced by promoter methylation, is reportedly found less in EOCRC patients than in older-onset CRC patients, with one study reporting the CIMP to be absent in all 47 EOCRCs, but present in 15/49 (31%) of CRCs in over 60 year olds [10]. However, we found promoter methylation of the APC gene in 40% of EOCRC tumours, a higher rate than reported in overall CRC [31], suggesting that specific methylation at the APC gene promoter may play an important role in EOCRC, despite lower methylation more generally. A previous study of patients presenting with CRC or colorectal polyps found APC methylation present in 8/50 (16%) samples [55]. When this cohort was stratified by age however the rate increased to 6/11 samples (55%) in those patients under 60 years old, reinforcing the idea that APC promoter methylation may play a significant role in EOCRC.
While diet is a major environmental factor proposed to influence DNA methylation in the colon [56], a finding of promoter methylation of the APC gene does not inform a direct role for diet in the genesis of early onset CRC. Moreover, whether this effect is the result of early maternal nutrition that can markedly affect the epigenetic patterning in the fetus is also beyond the scope of this study. However, our finding of tumour-specific APC promoter methylation in 40% early-onset CRC, with or without retention of a homozygous APC gene, highlights growing awareness that environmental factors need to be taken into consideration as we try to find ways to reduce the number of young people developing this disease.
8. Methods
8.1. Cohort
Samples from 25 patients with early-onset colorectal cancer, diagnosed at or under the age of 50 years, were obtained from the Cancer Society Tissue Bank, Christchurch, New Zealand. Patients with known hereditary syndromes, HNPCC or FAP were excluded. The samples consisted of frozen tumour tissue as well as matched normal tissue taken at the time of surgery. The study was granted ethics approval by the University of Otago Human Ethics Committee (H18/143).
8.2. DNA Extraction
DNA was extracted from up to 25 mg of frozen tissue using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Prior to extraction the tissue was homogenized in a Precellys Evolution homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) with 180 µL lysis buffer using 2.8 mm zirconium oxide beads in reinforced 2 mL microtubes. DNA was then extracted according to the manufacturer’s instructions.
8.3. Sequencing Library Preparation
The APC gene sequence of the tumour samples and matched normal tissue was determined by running pooled, indexed PCR products on an Illumina MiSeq instrument following the previously described protocol [57]. Briefly, dual-indexed amplicon sequencing libraries for APC were generated using a two-step PCR approach. In the first PCR step, regions of the APC gene, accounting for the vast majority of known cancer-associated mutations, were amplified. APC-specific primers were designed with an additional 18 bp of known non-specific sequence that was used as a priming site for the second PCR reaction (Table S1). PCR products from the same study participant and sample were pooled and purified using HighPrep beads (MagBio, Gaithersburg, MD, USA). In the second PCR step, pooled PCR products were amplified using a unique pair of indexed primers designed to add sequences necessary for multiplex sequencing on an Illumina MiSeq (Illumina, San Diego, CA, USA) (Tables S2 and S3).
To prepare the amplicon library for sequencing, sample-specific libraries were pooled, purified using HighPrep beads, and quantified with the Qubit dsDNA HS Assay Kit (Thermo Fisher (Waltham, MA, USA). Sequencing libraries were run on a DNA7500 Bioanalyzer chip (Illumina, San Diego, CA, USA) to determine the average library size. Libraries were sequenced on an Illumina MiSeq using V2-500 cycle reagent kits (Illumina, San Diego, CA, USA).
8.4. Sequence Analysis and Annotation
Raw paired end reads were cleaned with Trimmomatic v.0.35 [58]. Cleaned reads were aligned to the human reference genome (GRCh37/hg19) using the Burrows-Wheeler Aligner v.0.7.10 [59]. Amplicons were sequenced to a minimum depth of 40 reads. Single nucleotide variants and insertion/deletion variants were called using ‘The Genome Analysis Toolkit’ (GATK) v.3.6 [60]. The effects of variants were predicted using SnpEff v.4.2 [61]. All putative mutations were visually inspected using the Integrative Genomics Viewer [62]. The functional consequences of missense variants were predicted in silico using five different models that included SIFT [63], Provean [64], Mutation Assessor [65], and PolyPhen2 (using both HumDiv and HumVar datasets) [66]. Mutations predicted to be deleterious by at least 3/5 predictive models were considered of interest. Mutations with low absolute (<10) or relative (<20%) allele counts were excluded. Mutations discovered by this method were confirmed by Sanger sequencing.
8.5. LOH at APC Locus
The APC sequences obtained above were used to detect loss of heterozygosity (LOH) by comparing levels of each allele at four single nucleotide polymorphisms (SNPs) within the APC gene. SNPs showing heterozygosity, by presence of both alleles, in normal tissue were considered to have LOH if the read count of one allele was reduced in relation to the alternate allele in tumour tissue. A low number of alleles present for the missing allele likely reflected non-tumour cells, such as lymphocytes, within the tumour tissue. The four SNPs analysed were rs2229992, rs351771, rs1801166 and rs41115. If all the informative SNPs within the APC gene in a single patient showed loss of an allele in the tumour tissue then the tumour was considered to display LOH at the APC locus.
Sanger Sequencing
Mutations detected using the method above were confirmed by Sanger sequencing. Briefly, purified PCR products were prepared for sequencing using BIG Dye chemistry (Applied Biosystems, Foster City, CA, USA) and sequenced on an ABI3700xl Genetic Analyser (Applied Biosystems, Foster City, CA, USA).
8.6. Microsatellite Instability
Slides of sectioned tumour tissue were available from 24 of the 25 patients, and these were stained for mismatch repair proteins to assess microsatellite instability [67]. Briefly, following incubation with antigen retrieval solution, consecutive slides were immunostained with primary antibodies to the mismatch repair proteins MSH2, MSH6, MLH1, and PMS2. Slides were then incubated with DAB to visualise primary antibody staining, followed by counterstaining with haematoxylin and finally bluing agent. All slides were scored by a clinical pathologist.
8.7. Bisulphite Treatment and Methylation Analysis
DNA was modified by sodium bisulphite using EZ DNA Methylation-Gold kit (Zymoresearch, Irvine, CA, USA) according to the manufacturer’s protocol. Methylation-specific PCR (MSP) was used to amplify methylated and unmethylated APC promoter 1A sequences using previously published conditions and primers [31].
9. Conclusions
Our study of a cohort of colorectal tumours from patients diagnosed under the age of 50 years identified APC mutations at a rate similar to that found in later-onset CRC, suggesting that similar mechanisms underlie tumour development in both age groups and that the earlier onset seen in the current cohort may be due to environmental factors. However, we found that the distribution of APC mutations more closely resembled that of familial APC mutations than sporadic later-onset CRC, despite the mutations being almost exclusively sporadic. This has implications for APC sequencing techniques as methodologies focusing on the mutation cluster region rather than the whole gene may miss a substantial number of APC mutations in sporadic EOCRC.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/12/3829/s1, Table S1: APC amplicon specific primers for next generation sequencing, Table S2: Forward adapter primers for creating next generation sequencing libraries, Table S3: Reverse adapter primers for creating next generation sequencing libraries.
Author Contributions
Conceptualisation, A.A. and J.I.K.; Investigation, A.A., C.H. and R.C.D.; Resources, H.R.M.; Funding acquisition, J.I.K. and F.A.F.; Writing- original draft, A.A.; Writing—review and editing, J.I.K. and F.A.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a University of Otago Research Grant (ORG 0119-0320). AA was supported by grants from the Christchurch Cancer Foundation and the Colorectal Surgical Society of Australia and New Zealand.
Conflicts of Interest
The authors report no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Pan J., Xin L., Ma Y.F., Hu L.H., Li Z.S. Colonoscopy Reduces Colorectal Cancer Incidence and Mortality in Patients with Non-Malignant Findings: A Meta-Analysis. Am. J. Gastroenterol. 2016;111:355–365. doi: 10.1038/ajg.2015.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey C.E., Hu C.Y., You Y.N., Bednarski B.K., Rodriguez-Bigas M.A., Skibber J.M., Cantor S.B., Chang G.J. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–010. JAMA Surg. 2015;150:17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hessami Arani S., Kerachian M.A. Rising rates of colorectal cancer among younger Iranians: Is diet to blame? Curr. Oncol. 2017;24:e131–e137. doi: 10.3747/co.24.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch H.T., Lanspa S., Smyrk T., Boman B., Watson P., Lynch J. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I & II). Genetics, pathology, natural history, and cancer control, Part I. Cancer Genet Cytogenet. 1991;53:143–160. doi: 10.1016/0165-4608(91)90093-a. [DOI] [PubMed] [Google Scholar]
- 6.Groden J., Thliveris A., Samowitz W., Carlson M., Gelbert L., Albertsen H., Joslyn G., Stevens J., Spirio L., Robertson M., et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 7.Fearon E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 8.Lasry A., Zinger A., Ben-Neriah Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016;17:230–240. doi: 10.1038/ni.3384. [DOI] [PubMed] [Google Scholar]
- 9.Connell L.C., Mota J.M., Braghiroli M.I., Hoff P.M. The Rising Incidence of Younger Patients with Colorectal Cancer: Questions About Screening, Biology, and Treatment. Curr. Treat. Options Oncol. 2017;18:23. doi: 10.1007/s11864-017-0463-3. [DOI] [PubMed] [Google Scholar]
- 10.Kirzin S., Marisa L., Guimbaud R., De Reynies A., Legrain M., Laurent-Puig P., Cordelier P., Pradère B., Bonnet D., Meggetto F., et al. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: A morphological, molecular and genetics study. PLoS ONE. 2014;9:e103159. doi: 10.1371/journal.pone.0103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinzler K.W., Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 12.Polakis P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Boman B.M., Fields J.Z. An APC: WNT Counter-Current-Like Mechanism Regulates Cell Division Along the Human Colonic Crypt Axis: A Mechanism That Explains How APC Mutations Induce Proliferative Abnormalities That Drive Colon Cancer Development. Front. Oncol. 2013;3:244. doi: 10.3389/fonc.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munemitsu S., Souza B., Muller O., Albert I., Rubinfeld B., Polakis P. The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 15.Bellis J., Duluc I., Romagnolo B., Perret C., Faux M.C., Dujardin D., Formstone C., Lightowler S., Ramsay R.G., Freund J.N., et al. The tumor suppressor Apc controls planar cell polarities central to gut homeostasis. J. Cell Biol. 2012;198:331–341. doi: 10.1083/jcb.201204086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cristofaro M., Contursi A., D’Amore S., Martelli N., Spaziante A.F., Moschetta A., Villani G. Adenomatous polyposis coli (APC)-induced apoptosis of HT29 colorectal cancer cells depends on mitochondrial oxidative metabolism. Biochim. Biophys. Acta. 2015;1852:1719–1728. doi: 10.1016/j.bbadis.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Lamlum H., Ilyas M., Rowan A., Clark S., Johnson V., Bell J., Frayling I., Efstathiou J., Pack K., Payne S., et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: A new facet to Knudson’s ‘two-hit’ hypothesis. Nat. Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 18.Flood D.M., Weiss N.S., Cook L.S., Emerson J.C., Schwartz S.M., Potter J.D. Colorectal cancer incidence in Asian migrants to the United States and their descendants. Cancer Causes Control. 2000;11:403–411. doi: 10.1023/A:1008955722425. [DOI] [PubMed] [Google Scholar]
- 19.Li R., Grimm S.A., Chrysovergis K., Kosak J., Wang X., Du Y., Burkholder A., Janardhan K., Mav D., Shah R., et al. Obesity, rather than diet, drives epigenomic alterations in colonic epithelium resembling cancer progression. Cell Metab. 2014;19:702–711. doi: 10.1016/j.cmet.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kothari N., Teer J.K., Abbott A.M., Srikumar T., Zhang Y., Yoder S.J., Brohl A.S., Kim R.D., Reed D.R., Shibata D. Increased incidence of FBXW7 and POLE proofreading domain mutations in young adult colorectal cancers. Cancer. 2016;122:2828–2835. doi: 10.1002/cncr.30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willauer A.N., Liu Y., Pereira A.A., Lam M., Morris J.S., Raghav K.P., Morris V.K., Menter D., Broaddus R., Meric-Bernstam F., et al. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 2019;125:2002–2010. doi: 10.1002/cncr.31994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perea J., Rueda D., Canal A., Rodríguez Y., Álvaro E., Osorio I., Alegre C., Rivera B., Martínez J., Benítez J., et al. Age at onset should be a major criterion for subclassification of colorectal cancer. J. Mol. Diagn. 2014;16:116–126. doi: 10.1016/j.jmoldx.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Raman R., Kotapalli V., Adduri R., Gowrishankar S., Bashyam L., Chaudhary A., Vamsy M., Patnaik S., Srinivasulu M., Sastry R., et al. Evidence for possible non-canonical pathway(s) driven early-onset colorectal cancer in India. Mol. Carcinog. 2014;53(Suppl. 1):E181–E186. doi: 10.1002/mc.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettington M., Walker N., Clouston A., Brown I., Leggett B., Whitehall V. The serrated pathway to colorectal carcinoma: Current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 25.De Palma F.D.E., D’Argenio V., Pol J., Kroemer G., Maiuri M.C., Salvatore F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers. 2019;11 doi: 10.3390/cancers11071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori Y., Nagse H., Ando H., Horii A., Ichii S., Nakatsuru S., Aoki T., Miki Y., Mori T., Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: Mutation cluster region in the APC gene. Hum. Mol. Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 27.Lüchtenborg M., Weijenberg M.P., Roemen G.M., de Bruïne A.P., van den Brandt P.A., Lentjes M.H., Brink M., van Engeland M., Goldbohm R.A., de Goeij A.F. APC mutations in sporadic colorectal carcinomas from The Netherlands Cohort Study. Carcinogenesis. 2004;25:1219–1226. doi: 10.1093/carcin/bgh117. [DOI] [PubMed] [Google Scholar]
- 28.Poncin J., Mulkens J., Arends J.W., de Goeij A. Optimizing the APC gene mutation analysis in archival colorectal tumor tissue. Diagn. Mol. Pathol. 1999;8:11–19. doi: 10.1097/00019606-199903000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Xu X.-M., Qian J.-C., Cai Z., Tang T., Wang P., Zhang K.-H., Deng Z.-L., Cai J.-P. DNA alterations of microsatellite DNA, p53, APC and K-ras in Chinese colorectal cancer patients. Eur. J. Clin. Investig. 2012;42:751–759. doi: 10.1111/j.1365-2362.2011.02641.x. [DOI] [PubMed] [Google Scholar]
- 30.Abdelmaksoud-Damak R., Miladi-Abdennadher I., Triki M., Khabir A., Charfi S., Ayadi L., Frikha M., Sellami-Boudawara T., Mokdad-Gargouri R. Expression and mutation pattern of beta-catenin and adenomatous polyposis coli in colorectal cancer patients. Arch. Med. Res. 2015;46:54–62. doi: 10.1016/j.arcmed.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Esteller M., Sparks A., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 32.Powell S.M., Zilz N., Beazer-Barclay Y., Bryan T.M., Hamilton S.R., Thibodeau S.N., Vogelstein B., Kinzler K.W. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 33.Fostira F., Yannoukakos D. A distinct mutation on the alternative splice site of APC exon 9 results in attenuated familial adenomatous polyposis phenotype. Fam. Cancer. 2010;9:395–400. doi: 10.1007/s10689-009-9317-x. [DOI] [PubMed] [Google Scholar]
- 34.Friedl W., Aretz S. Familial adenomatous polyposis: Experience from a study of 1164 unrelated german polyposis patients. Hered. Cancer Clin. Pract. 2005;3:95–114. doi: 10.1186/1897-4287-3-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simbolo M., Mafficini A., Agostini M., Pedrazzani C., Bedin C., Urso E.D., Nitti N., Turri G., Scardoni M., Fassan M., et al. Next-generation sequencing for genetic testing of familial colorectal cancer syndromes. Hered. Cancer Clin. Pract. 2015;13:18. doi: 10.1186/s13053-015-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gómez-Fernández N., Castellví-Bel S., Fernández-Rozadilla C., Balaguer F., Muñoz J., Madrigal I., Milà M., Graña B., Vega A., Castells A., et al. Molecular analysis of the APC and MUTYH genes in Galician and Catalonian FAP families: A different spectrum of mutations? BMC Med. Genet. 2009;10:57. doi: 10.1186/1471-2350-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyaki M., Konishi M., Kikuchi-Yanoshita R., Enomoto M., Igari T., Tanaka K., Muraoka M., Takahashi H., Amada Y., Fukayama M. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 38.Lagarde A., Rouleau E., Ferrari A., Noguchi T., Qiu J., Briaux A., Bourdon V., Remy V., Gaildrat P., Adélaïde J., et al. Germline APC mutation spectrum derived from 863 genomic variations identified through a 15-year medical genetics service to French patients with FAP. J. Med. Genet. 2010;47:721–722. doi: 10.1136/jmg.2010.078964. [DOI] [PubMed] [Google Scholar]
- 39.White S., Bubb V.J., Wyllie A.H. Germline APC mutation (Gln1317) in a cancer-prone family that does not result in familial adenomatous polyposis. Genes Chromosomes Cancer. 1996;15:122–128. doi: 10.1002/(SICI)1098-2264(199602)15:2<122::AID-GCC7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 40.Aceto G., Curia M.C., Veschi S., De Lellis L., Mammarella S., Catalano T., Stuppia L., Palka G., Valanzano R., Tonelli F., et al. Mutations of APC and MYH in unrelated Italian patients with adenomatous polyposis coli. Hum. Mutat. 2005;26:394. doi: 10.1002/humu.9370. [DOI] [PubMed] [Google Scholar]
- 41.Li F.-F., Liu Z., Yan P., Shao X., Deng X., Sam C., Chen Y.-G., Xu Y.-P., Wang X., Wang G.-Y., et al. Identification of a novel mutation associated with familial adenomatous polyposis and colorectal cancer. Int. J. Mol. Med. 2015;36:1049–1056. doi: 10.3892/ijmm.2015.2303. [DOI] [PubMed] [Google Scholar]
- 42.Ishida H., Yamaguchi T., Chiba K., Iijima T., Horiguchi S.I. A case report of ascending colon adenosquamous carcinoma with BRAF V600E mutation. Int. Cancer Conf. J. 2017;6:93–97. doi: 10.1007/s13691-017-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowan A.J., Lamlum H., Ilyas M., Wheeler J., Straub J., Papadopoulou A., Bicknell D., Bodmer W.F., Tomlinson I.P.M. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc. Natl. Acad. Sci. USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B.Q., Liu P.P., Zhang C.H. Correlation between the methylation of APC gene promoter and colon cancer. Oncol. Lett. 2017;14:2315–2319. doi: 10.3892/ol.2017.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fearnhead N.S., Britton M.P., Bodmer W.F. The ABC of APC. Hum. Mol. Genet. 2001;10:721–733. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 46.Leoz M.L., Carballal S., Moreira L., Ocana T., Balaguer F. The genetic basis of familial adenomatous polyposis and its implications for clinical practice and risk management. Appl. Clin. Genet. 2015;8:95–107. doi: 10.2147/TACG.S51484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandra S.H., Wacker I., Appelt U.K., Behrens J., Schneikert J. A common role for various human truncated adenomatous polyposis coli isoforms in the control of beta-catenin activity and cell proliferation. PLoS ONE. 2012;7:e34479. doi: 10.1371/journal.pone.0034479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christie M., Jorissen R.N., Mouradov D., Sakthianandeswaren A., Li S., Day F., Tsui C., Lipton L., Desai J., Jones I.T., et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene. 2013;32:4675–4682. doi: 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohler E.M., Derungs A., Daum G., Behrens J., Schneikert J. Functional definition of the mutation cluster region of adenomatous polyposis coli in colorectal tumours. Hum. Mol. Genet. 2008;17:1978–1987. doi: 10.1093/hmg/ddn095. [DOI] [PubMed] [Google Scholar]
- 50.Bufill J.A. Colorectal cancer: Evidence for distinct genetic categories based on proximal or distal tumor location. Ann. Intern. Med. 1990;113:779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 51.Davis D.M., Marcet J.E., Frattini J.C., Prather A.D., Mateka J.J., Nfonsam V.N. Is it time to lower the recommended screening age for colorectal cancer? J. Am. Coll. Surg. 2011;213:352–361. doi: 10.1016/j.jamcollsurg.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 52.Perea J., Cano J.M., Rueda D., García J.L., Inglada L., Osorio I., Arriba M., Pérez J., Gaspar M., Fernández-Miguel T., et al. Classifying early-onset colorectal cancer according to tumor location: New potential subcategories to explore. Am. J. Cancer Res. 2015;5:2308–2313. [PMC free article] [PubMed] [Google Scholar]
- 53.Ballester V., Rashtak S., Boardman L. Clinical and molecular features of young-onset colorectal cancer. World J. Gastroenterol. 2016;22:1736–1744. doi: 10.3748/wjg.v22.i5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salovaara R., Loukola A., Kristo P., Kääriäinen H., Ahtola H., Eskelinen M., Härkönen N., Julkunen R., Kangas E., Ojala S., et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J. Clin. Oncol. 2000;18:2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 55.Leung W.K., To K.-F., Man E.P.S., Chan M.W.Y., Hui A.J., Ng S.S.M., Lau J.Y.W., Sung J.J.Y. Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am. J. Gastroenterol. 2007;102:1070–1076. doi: 10.1111/j.1572-0241.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 56.Nystrom M., Mutanen M. Diet and epigenetics in colon cancer. World J. Gastroenterol. 2009;15:257–263. doi: 10.3748/wjg.15.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hakkaart C., Ellison-Loschmann L., Day R., Sporle A., Koea J., Harawira P., Cheng S., Gray M., Whaanga T., Pearce N., et al. Germline CDH1 mutations are a significant contributor to the high frequency of early-onset diffuse gastric cancer cases in New Zealand Maori. Fam. Cancer. 2019;18:83–90. doi: 10.1007/s10689-018-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S.B., Daly M.J., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118, iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 64.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.South C.D., Yearsley M., Martin E., Arnold M., Frankel W., Hampel H. Immunohistochemistry staining for the mismatch repair proteins in the clinical care of patients with colorectal cancer. Genet. Med. 2009;11:812–817. doi: 10.1097/GIM.0b013e3181b99b75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.