Abstract
Simple Summary
Uterine leiomyomas are benign smooth muscle tumors affecting millions of women globally. On a molecular level, leiomyomas can be classified into three main subtypes, each characterized by mutations affecting either MED12, HMGA2, or FH. Leiomyomas are still widely regarded as a single entity, although early observations suggest that different subtypes behave differently, in terms of both clinical outcomes and therapeutic requirements. The majority of classification studies on leiomyomas have been performed using fresh frozen tissue. Archival formalin-fixed paraffin-embedded (FFPE) tissue represents an invaluable source of biological material that can be studied retrospectively. Methods capable of generating high-quality data from FFPE material are in high demand. Here, we show that 3′RNA sequencing can accurately classify leiomyomas that have been stored as FFPE tissue in hospital archives for years. A targeted 3′RNA sequencing panel could provide researchers and clinicians with a cost-effective and scalable diagnostic tool for classifying smooth muscle tumors.
Abstract
Uterine leiomyomas are benign smooth muscle tumors occurring in 70% of women of reproductive age. The majority of leiomyomas harbor one of three well-established genetic changes: a hotspot mutation in MED12, overexpression of HMGA2, or biallelic loss of FH. The majority of studies have classified leiomyomas by complex and costly methods, such as whole-genome sequencing, or by combining multiple traditional methods, such as immunohistochemistry and Sanger sequencing. The type of specimens and the amount of resources available often determine the choice. A more universal, cost-effective, and scalable method for classifying leiomyomas is needed. The aim of this study was to evaluate whether RNA sequencing can accurately classify formalin-fixed paraffin-embedded (FFPE) leiomyomas. We performed 3′RNA sequencing with 44 leiomyoma and 5 myometrium FFPE samples, revealing that the samples clustered according to the mutation status of MED12, HMGA2, and FH. Furthermore, we confirmed each subtype in a publicly available fresh frozen dataset. These results indicate that a targeted 3′RNA sequencing panel could serve as a cost-effective and robust tool for stratifying both fresh frozen and FFPE leiomyomas. This study also highlights 3′RNA sequencing as a promising method for studying the abundance of unexploited tissue material that is routinely stored in hospital archives.
Keywords: 3′RNA sequencing, QuantSeq, fibroids, FFPE, MED12, HMGA2, FH
1. Introduction
Uterine leiomyomas, or fibroids, are benign tumors originating from the smooth muscle cells of the myometrium. Leiomyomas are one of the most common human neoplasms, affecting 70% of women of reproductive age [1]. Although considered benign, leiomyomas frequently cause a variety of symptoms including pressure upon adjacent organs, abnormal uterine bleeding, pelvic pain, and impaired fertility [2]. Leiomyomas are the leading indication for hysterectomy worldwide and pose a significant socio-economic impact [3]. Approximately 10% of leiomyomas display nonconventional histopathology or abnormal growth patterns, some of which have been associated with specific molecular features [4].
Recent studies have revealed the existence of various molecular leiomyoma subtypes [5]. Indeed, 80−90% of leiomyomas harbor one of three genetic changes: a hotspot mutation in mediator complex subunit 12 (MED12), a chromosomal aberration resulting in significant overexpression of high mobility group AT-hook 2 (HMGA2), or biallelic loss of fumarate hydratase (FH). Leiomyomas with FH deficiency constitutes a rare but clinically relevant subtype, as pathogenic germline variants in FH predispose to hereditary leiomyomatosis and renal cell cancer (HLRCC). This syndrome is characterized by an early onset of multiple symptomatic uterine leiomyomas as well as renal cell cancer and cutaneous leiomyomas [6]. The majority of FH-deficient uterine leiomyomas are, however, sporadic. Other less frequent aberrations have been reported in leiomyomas, but most of these have been detected as subclonal alterations that co-occur with the other established driver aberrations [5]. For example, high mobility group AT-hook 1 (HMGA1) and PLAG1 zinc finger (PLAG1) rearrangements have been detected in both tumors with and without a MED12 mutation [7,8,9]. Early observations suggest that different molecular subtypes behave differently, in terms of both clinical outcomes and therapeutic requirements [6,10,11]. For example, leiomyomas of the MED12 subtype are typically more numerous, smaller in size, and subserosal [11]; leiomyomas of the HMGA2 subtype are larger, isolated, and have a high vasculature density making them potentially sensitive to angiogenesis inhibitors [6,10]. Leiomyomas of the FH subtype have been associated with variant histopathology, including some histologic features associated with malignancy [6].
While the strongest evidence of each molecular subtype may be derived from whole-genome sequencing, it is not always the most feasible and cost-effective method for classifying leiomyomas. Most studies have therefore utilized cytogenetics, immunohistochemistry, qPCR, and Sanger sequencing [12,13,14]. Indeed, Sanger sequencing is the most common method for detecting hotspot mutations in exons 1 and 2 of MED12, whereas cytogenetics, immunohistochemistry, and qPCR have all been utilized for detecting HMGA2 aberrations. Biallelic loss of FH can be accurately detected by immunohistochemistry with an antibody against S-(2-succinyl)cysteine, which is a chemical modification generated by high levels of fumarate [15].
We and others have previously shown that MED12, HMGA2, and FH aberrations result in distinct global gene expression patterns, suggesting that leiomyomas can be classified by gene expression profiling [7,14]. However, the majority of molecular studies on leiomyomas have been performed using fresh frozen tissue. While fresh frozen tissue might be ideal for most molecular analyses, archival formalin-fixed paraffin-embedded (FFPE) tissue represents an invaluable source of biological material that is routinely stored in hospital archives [16]. FFPE tissue has many advantages, including long-term storage at room temperature, abundance and availability, and connection to valuable clinical data such as histopathology and patient outcomes. However, FFPE-derived RNA is degraded and chemically modified, limiting its use in molecular studies [17]. Methods capable of generating high-quality transcriptome data from archival FFPE tissues are in high demand [16].
RNA sequencing has become an essential tool in cancer research, drug development, and cancer diagnostics [18]. One bias of standard whole-transcript RNA sequencing is that longer transcripts are represented by more reads than shorter transcripts [19]. Many recent RNA sequencing technologies, such as TagSeq and QuantSeq, have been developed to minimize this bias [20,21]. These techniques capture and sequence short fragments at the 3′ end of poly-adenylated RNA, generating only one read per transcript. 3′RNA sequencing has low RNA input and quality requirements, making it suitable for use with FFPE samples. Unlike gene expression microarrays, these newer technologies have the ability to utilize low amounts of RNA to generate transcriptome data that are comparable between FFPE and fresh frozen samples [22]. In this study, we have evaluated the potential of 3′RNA sequencing in analyzing archival leiomyoma material. In addition, we have assessed whether a set of 80 previously reported gene expression biomarkers could function as a targeted RNA sequencing panel to classify both fresh frozen and FFPE leiomyomas.
2. Results
2.1. Principal Component Analysis Confirms Different Molecular Subtypes
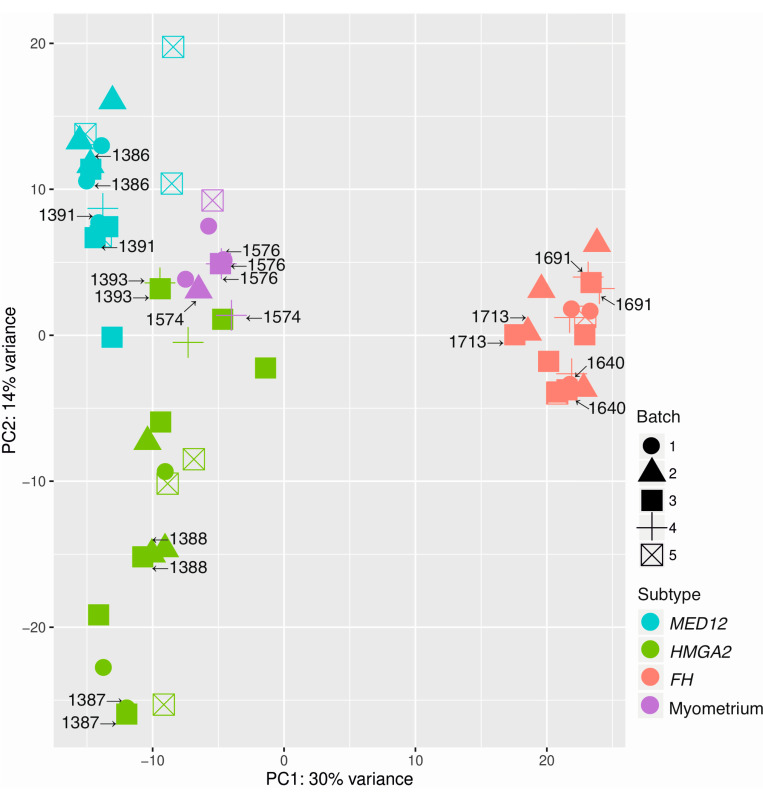
Principal component analysis using the 3′RNA sequencing data revealed that the 49 (44 leiomyomas + 5 myometrium) samples and 11 technical replicates grouped according to the predetermined mutation status of MED12, HMGA2, and FH (Figure 1). The age of the tissue material ranged from 4 to 31 years. The samples were sequenced in five batches, and the dataset included eleven technical replicates. We found a weak negative relationship (r = −0.27) between the age of the block and total read counts (Table S1). Taken together, these observations show high reproducibility and no obvious batch effect (Figure 1).
Figure 1.
Principal component analysis shows a high degree of reproducibility. Principal component analysis using all genes revealed high reproducibility and no batch effect among the 49 samples and 11 technical replicates. The technical replicates clustered next to each other and the samples grouped according to the mutation status of MED12, HMGA2, and FH.
2.2. Differential Expression Analysis Confirms Distinct Gene Expression Patterns
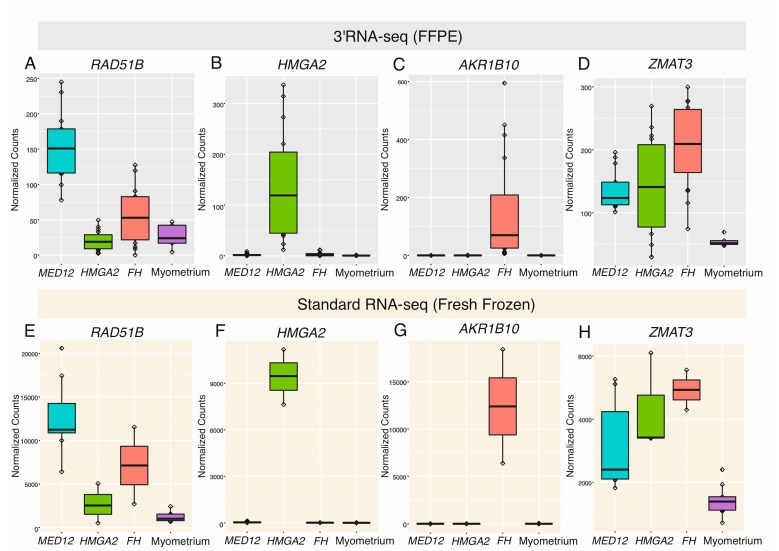
We compared the 44 leiomyomas against the 5 myometrium samples, revealing 328 differentially expressed genes (q < 0.05; |FC| > 2, Table S2). We also compared each leiomyoma subtype against the myometrium samples, revealing 671 differentially expressed genes in leiomyomas of the MED12 subtype, 905 in leiomyomas of the HMGA2 subtype, and 1537 in leiomyomas of the FH subtype (q < 0.05; |FC| > 2, Table S2). See Figure S1 for a Venn diagram summarizing the number of shared and uniquely expressed genes among the different subtypes. We confirmed the expression pattern of several previously highlighted biomarkers (Figure 2A–D), including upregulation of RAD51 paralog B (RAD51B) in leiomyomas with a MED12 mutation (q-value = 6.20 × 10−13, FC = 4.76), upregulation of HMGA2 in leiomyomas with positive staining for HMGA2 (q-value = 4.84 × 10−19, FC = 28.44), upregulation of aldo-keto reductase family 1 member B10 (AKR1B10) in leiomyomas with positive staining for 2SC (q = 1.54 × 10−07, FC = 11.47), and upregulation of zinc finger matrin-type 3 (ZMAT3) in all leiomyoma subtypes (q-value = 2.90 × 10−04, FC = 2.69). The same expression pattern was also seen in an independent and publicly available fresh frozen dataset of 13 leiomyoma and 9 matching myometrium samples that were sequenced using standard RNA sequencing (Figure 2E–H). However, high expression of AKR1B10 was observed in two fresh frozen samples that were originally classified as HMGA1 overexpressing leiomyomas (Figure 2G).
Figure 2.
Highlighted genes previously shown to be dysregulated in leiomyomas. (A) Using 3′RNA sequencing of formalin-fixed paraffin-embedded (FFPE) tissue, we confirmed significant upregulation of RAD51B in leiomyomas with a MED12 mutation. (B) We also confirmed significant upregulation of HMGA2 in leiomyomas displaying positive IHC staining for HMGA2. (C) High expression levels of AKR1B10 were only seen in leiomyomas displaying positive IHC staining for 2SC. (D) Upregulation of ZMAT3 was seen in the majority of leiomyomas, regardless of subtype. (E–H) Similar observations were seen in an independent dataset of fresh frozen tissue that was sequenced using standard RNA sequencing.
2.3. Supervised Hierarchical Clustering Accurately Classifies Leiomyomas
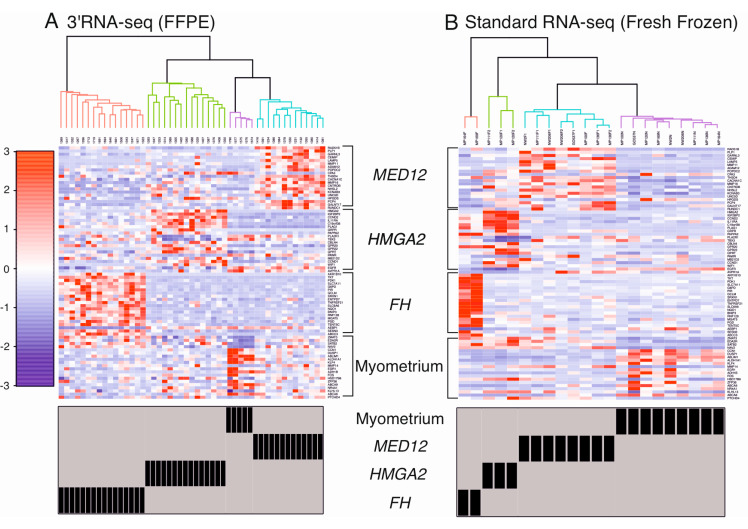
Supervised hierarchical clustering analysis using a set of 80 genes previously shown to be dysregulated in leiomyomas revealed that the samples clearly clustered into four distinct groups, representing the three most common leiomyoma subtypes and the normal myometrium controls (Figure 3A). The same clustering analysis was performed with the fresh frozen dataset. Again, the samples clustered clearly into four similar groups, and the two HMGA1 overexpressing leiomyomas displayed clear expression patterns of leiomyomas of the FH subtype (Figure 3B).
Figure 3.
Supervised hierarchical clustering analysis using a set of 80 selected genes. (A) Hierarchical clustering analysis revealed that the 49 FFPE samples clustered into four distinct groups, representing the three most common leiomyoma subtypes and myometrium controls. (B) The same clustering analysis was performed with an independent dataset of 22 fresh frozen samples that were sequenced using standard RNA sequencing [14], revealing four similar groups. Two of the fresh frozen samples, originally classified as HMGA1 overexpressing leiomyomas (MP164F, MP169F), displayed expression patterns of leiomyomas of the FH subtype.
2.4. Exome Sequencing Reveals Biallelic Loss of FH in Two HMGA1 Overexpressing Leiomyomas
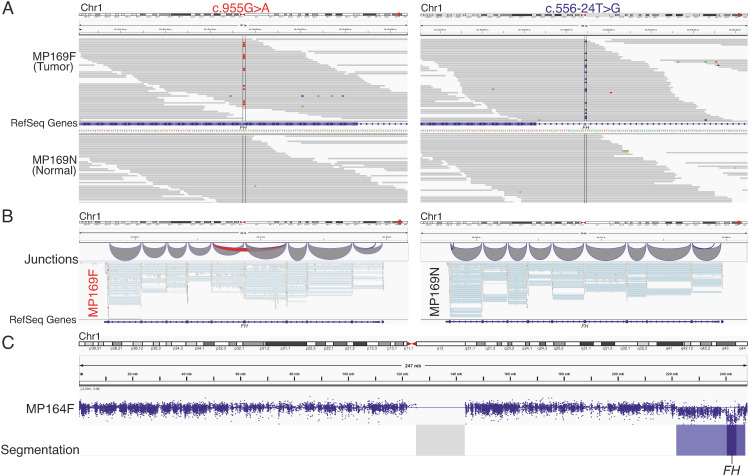
Two samples, originally classified as HMGA1 overexpressing leiomyomas [14], displayed clear expression patterns of leiomyomas of the FH subtype. Exome sequencing revealed a somatic missense variant FH c.955G > A, p.(Asp319Asn) and a somatic intronic variant FH c.556-24T > G in one of the samples (MP169F, Figure 4A). The missense variant was predicted deleterious with a Combined Annotation-Dependent Depletion (CADD) score of 28 [23], and the intronic variant was predicted to result in loss of a natural splice acceptor site according to spliceAI with a delta score of 0.56 [24]. Indeed, the intronic variant resulted in skipping of exon 5 according to the RNA sequencing data (Figure 4B). Both of the mutations were also present in the RNA sequencing data of the tumor and absent in the normal myometrium. Copy number analysis of the exome sequencing data revealed a homozygous deletion of FH in the other sample (MP164F, Figure 4C).
Figure 4.
Biallelic loss of FH in two samples originally classified as HMGA1 overexpressing tumors. (A) One HMGA1 overexpressing tumor (MP169F) harbored a somatic missense variant c.955G > A, p.(Asp319Asn) in FH and a somatic substitution c.556-24T > G in intron 4 of FH. (B) The intronic variant resulted in RNA transcripts lacking exon 5. (C) Copy number analysis revealed a homozygous deletion encompassing FH in the other sample (MP164F).
2.5. HMGA1 and PLAG1 Are Upregulated in Leiomyomas of the FH Subtype
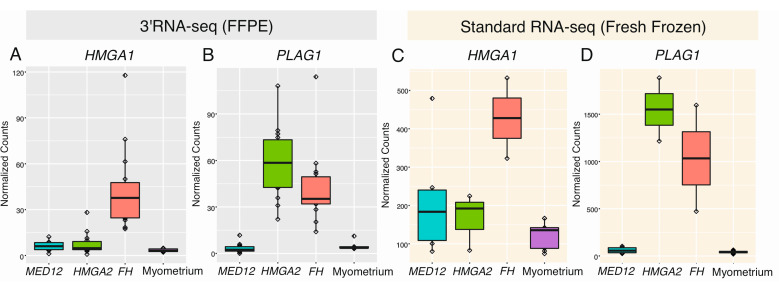
As we detected biallelic loss of FH in two leiomyomas previously reported to overexpress HMGA1, we explored the expression pattern of HMGA1 in our dataset as well. We also explored the expression pattern of PLAG1 in both datasets, since both HMGA1 and HMGA2 have been proposed to regulate the expression of this gene [7,25]. HMGA1 was significantly upregulated in only leiomyomas of the FH subtype (q-value = 5.64 × 10−14, FC = 9.00), whereas PLAG1 was significantly upregulated in both leiomyomas of the HMGA2 (q-value = 4.84 × 10−19, FC = 9.13) and FH subtypes (q-value = 7.81 × 10−14, FC = 6.45) (Figure 5).
Figure 5.
Expression of HMGA1 and PLAG1 in different leiomyoma subtypes and myometrium samples. (A,C) In both the FFPE and fresh frozen datasets, significant upregulation of HMGA1 was only seen in the leiomyomas of the FH subtype, (B,D) whereas significant upregulation of PLAG1 was seen in both leiomyomas of the HMGA2 and FH subtypes, but not in leiomyomas of the MED12 subtype nor the myometrium samples.
3. Discussion
Uterine leiomyomas are benign smooth muscle tumors that are still widely regarded as a single entity despite well-known heterogeneity in histology and emerging evidence of potential differences in clinical characteristics and response to therapies. Recent studies have also revealed that the majority of leiomyomas can be classified into three main molecular subtypes, reflecting mutations in MED12, HMGA2, and FH. Furthermore, we and others have shown that each subtype is characterized by a distinct global gene expression pattern [7,14], suggesting that leiomyomas could be classified by gene expression profiling. However, RNA-based studies on leiomyomas have largely been limited to fresh frozen tissue specimens. Archival FFPE tissue represents an invaluable source of biological material that could be used for retrospective studies. Using 3′RNA sequencing, we sequenced 49 FFPE samples in multiple batches and included 11 technical replicates, revealing high reproducibility and no noticeable batch effect. We also confirmed several previously reported subtype-specific patterns in two independent datasets and demonstrated that 3′RNA sequencing can accurately classify FFPE leiomyomas. This method of sequencing is relatively cost-effective and can be performed with low quantities of tissue material. Indeed, our samples were sequenced with only 80 ng of RNA and included FFPE material that was as old as 31 years.
We have previously highlighted a set of dysregulated genes in leiomyomas that could assist in their classification [7]. Here, we confirm that FFPE leiomyomas can similarly be classified into the three most common subtypes using the 80 most significant of these genes. In addition, we confirmed the accuracy of these 80 genes in a second independent dataset comprising fresh frozen samples that were sequenced using standard RNA sequencing. Furthermore, this led to the identification of biallelic loss of FH in two tumors that were originally classified as HMGA1 overexpressing tumors by George et al. [14]. This motivated us to explore the expression pattern of HMGA1 in our FFPE dataset, revealing significant upregulation of HMGA1 in leiomyomas of the FH subtype, but not in leiomyomas of the MED12 and HMGA2 subtypes. HMGA2 has been shown to directly regulate the expression of the oncogene PLAG1, suggesting that the homologue HMGA1 may have a similar role in tumorigenesis [7]. As expected, we detected significant upregulation of PLAG1 in leiomyomas of the HMGA2 and FH subtypes, but not in leiomyomas of the MED12 subtype. It is tempting to speculate that leiomyomas of the FH subtype may in part promote tumorigenesis through dysregulation of HMGA1.
There are also some limitations of the study. While all of the samples in both datasets clustered accurately according to the mutation status, the sample size is still relatively small. In addition, although the great majority of leiomyomas can be explained by defects in MED12, HMGA2, or FH, there is a small subset of leiomyomas where the driver alteration is unknown. We do not expect the inclusion of such samples to have a strong effect on the clustering, but this needs to be confirmed in further studies with larger sample sizes. Possible challenges related to clinical translation include the requirement of bioinformatics knowledge and the time required for data production and analysis.
Taken together, these observations demonstrate that our set of gene expression biomarkers are highly effective in classifying leiomyomas, regardless of whether the samples are fresh frozen or FFPE. These results also highlight 3′RNA sequencing as a promising method for studying the abundance of tissue material that is routinely stored in hospital archives.
4. Materials and Methods
4.1. Study Material and Sample Selection
The research has been approved by The Ethics Review Board of Hospital District of Helsinki and Uusimaa, Helsinki, Finland (ethical codes: 88/13/03/03/2015 and 24/13/03/03/2015). All samples were collected with the authorization from the National Supervisory Authority for Welfare and Health (Valvira) or with a written informed consent from the patients. The study material consisted of 49 archival FFPE tissue samples and corresponding hematoxylin-eosin stained slides that were obtained from the Department of Pathology, Helsinki University Hospital, Helsinki, Finland. Histopathological re-examination of the tumor and normal tissue slides was performed by a pathologist (RB or AP). The age of the tissue material ranged from 4 to 31 years. The mutation statuses of MED12, HMGA2, and FH were determined in five previously published studies [13,26,27,28,29]. See Table S3 for an overview of the mutation status of each sample. In brief, the mutation status of MED12 was determined by Sanger sequencing, and the overexpression of HMGA2 and the loss of FH were determined by immunohistochemistry using antibodies against HMGA2 and 2SC, respectively. The 49 FFPE samples comprised 13 leiomyomas with a MED12 mutation, 15 with significant overexpression of HMGA2, 16 with FH deficiency, and 5 myometrium samples that served as normal tissue controls. The samples entered 3′RNA sequencing in 5 batches and included 11 technical replicates from 10 samples. In addition, we analyzed publicly available sequencing data of 13 fresh frozen leiomyomas and 9 corresponding myometrium samples. The tumors in this dataset were originally classified by George et al. [14] as 8 leiomyomas with a MED12 mutation, 3 with significant overexpression of HMGA2, and 2 with significant overexpression of HMGA1.
4.2. RNA Extraction and Sequencing
Total RNA was extracted from 2 to 4 macrodissected 10 um tissue sections. The RNA yields ranged from 2175−41,940 ng with an average yield of 11,070 ng. RNA was extracted and purified using the RNeasy® FFPE Kit (QIAGEN, Hilden, Germany) and the deparaffinization solution (QIAGEN) according to the manufacturer’s protocol. The concentration and purity of the extracted RNA were analyzed using the LabChip GX Touch HT RNA Assay Reagent Kit (PerkinElmer, Waltham, MA, USA) and the Qubit RNA BR kit (Thermo Fisher Scientific, Waltham, MA, USA). Genomic DNA contamination was measured using the Qubit DNA BR kit (Thermo Fisher Scientific). DNase treatment was performed to eliminate genomic DNA fragments.
Dual-indexed mRNA libraries were prepared from 80 ng of total RNA with QuantSeq 3′mRNA-Seq Library Prep Kit FWD (Lexogen Gmbh, Vienna, Austria) according to the manufacturer’s instructions. During second strand synthesis, 6 bp unique molecular identifiers (UMI) were introduced with the UMI Second Strand Synthesis Module (Lexogen Gmbh) for detection and removal of PCR duplicates. The quality of the libraries was measured with LabChip GX Touch HT DNA High Sensitivity Reagent Kit (PerkinElmer). The libraries were multiplexed and sequenced using the NovaSeq 6000 System (Illumina, San Diego, CA, USA) at the Institute for Molecular Medicine Finland (FIMM) with a read length of 2 × 101 base pairs and a minimum target coverage of 15 M reads for each library. Total number of aligned reads obtained for each sample are listed in (Table S1).
4.3. RNA Sequencing Data Analysis
FASTQ preprocessing was performed with default parameters using the QuantSeq 3′mRNA-Seq Integrated Data Analysis Pipeline version 2.3.1 FWD UMI (Lexogen Gmbh) implemented on the Bluebee® Genomics platform. In brief, the reads were trimmed using BBDuk, aligned against the Genome Reference Consortium human build 38 (GRCh38) reference genome using Spliced Transcripts Alignment to a Reference (STAR) [30], and counted using HTSeq [31]. Principal component analysis (PCA) and differential expression analysis were performed using DESeq2 [32] implemented on the Chipster platform [33]. As no batch effect was observed in the PCA analysis, the read counts of the technical replicates were merged for the subsequent analyses. Additional raw RNA sequencing data (FASTQ) of 13 leiomyoma and 9 corresponding myometrium samples from the discovery dataset of George et al. [14] were retrieved from the Sequence Read Archive (SRA) database (BioProject PRJNA498292). The samples were aligned against the GRCh38 reference genome using STAR [34], and the reads were counted using HTSeq [31].
Raw read counts were normalized using DESeq2. Supervised hierarchical clustering analysis using Spearman’s rank correlation was performed with 80 genes previously shown to be differentially expressed in leiomyomas (Table S4) [6]. These 80 genes represent the 20 most uniquely expressed genes in leiomyomas of the MED12, HMGA2, and FH subtypes as well as the 20 most significantly differentially expressed genes in normal myometrium compared to all leiomyomas (q < 0.05; |FC| > 2).
4.4. Whole-Exome Sequencing
Raw whole-exome sequencing (WES) data (FASTQ) of two HMGA1 overexpressing leiomyomas and eight myometrium samples from the discovery dataset of George et al. [14] were retrieved from the Sequence Read Archive (SRA) database (BioProject PRJNA494823). Adapter and read trimming were performed using Trimmomatic [35]. Data preprocessing was performed according to Genome Analysis Toolkit 4 best practices [36]. In brief, the samples were aligned against the GRCh38 reference genome using Burrows-Wheeler Aligner (BWA-MEM) [37], duplicate reads were removed using Mark Duplicates, and base quality score recalibration was performed using BaseRecalibrator. Somatic variant calling was performed against the corresponding normal using Mutect2 [38]. Somatic variants affecting the exonic and intronic regions of FH were evaluated further using BasePlayer [39]. Somatic copy number alterations were called using CNVkit against a pooled reference generated with the eight myometrium samples [40].
5. Conclusions
This proof-of-principle study demonstrates that 3′RNA sequencing is highly suitable for analyzing and classifying archival smooth muscle tissue material. Future work utilizing this method will not only provide insights into underlying defects of the remaining uncharacterized leiomyomas, but also allow for detection of biomarkers that can differentiate leiomyomas from their malignant counterparts: leiomyosarcomas. 3′RNA sequencing may provide researchers with an accurate tool to retrospectively study the transcriptome of rare tumor subtypes and cancers that are difficult to obtain. To conclude, our observations indicate that a targeted 3′RNA sequencing panel could provide researchers and clinicians with a cost-effective and scalable diagnostic tool for stratifying smooth muscle tumors. Millions of women suffer from leiomyomas, and the ability to accurately stratify each lesion should pave the way towards personalized treatments.
Acknowledgments
We express gratitude to the patients for their contributions. We thank Lotta Katainen and Anne Mäkelä for technical assistance. We acknowledge CSC, IT Center for Science, Finland, for computational resources and the Sequencing Unit of the Institute for Molecular Medicine Finland (FIMM), Finland for their services.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/12/3839/s1, Figure S1: Venn diagram demonstrating shared and uniquely expressed genes in leiomyomas of the different subtypes. Table S1: Age of the samples and number of reads generated by RNA sequencing. Table S2: Differentially expressed genes in each leiomyoma subtype. Table S3: Mutation status of uterine leiomyoma samples used in 3′ RNA sequencing. Table S4: A set of 80 significantly dysregulated genes in uterine leiomyomas.
Author Contributions
Conceptualization, M.M., P.V., T.H., T.A., and S.K.; methodology, M.M. and S.K.; formal analysis, M.M.; investigation, M.M., P.V., S.K., and T.A.; resources, A.Ä., A.P., R.B., P.P., and P.H.; writing—original draft preparation, M.M.; writing—review and editing, M.M., P.V., T.H., T.A., S.K., A.Ä., P.P., A.P., R.B., and P.H.; visualization, M.M. and S.K.; supervision, P.V. and M.M.; funding acquisition, P.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Academy of Finland, grant number 307773, Sigrid Jusélius Foundation, Cancer Foundation Finland, and the iCAN Digital Precision Cancer Medicine Flagship.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stewart E.A., Cookson C.L., Gandolfo R.A., Schulze-Rath R. Epidemiology of uterine fibroids: A systematic review. BJOG Int. J. Obs. Gynaecol. 2017;124:1501–1512. doi: 10.1111/1471-0528.14640. [DOI] [PubMed] [Google Scholar]
- 2.Sabry M., Al-Hendy A. Medical treatment of uterine leiomyoma. Reprod. Sci. 2012;19:339–353. doi: 10.1177/1933719111432867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart E.A. Uterine fibroids. N. Engl. J. Med. 2015;372:1646–1655. doi: 10.1056/NEJMcp1411029. [DOI] [PubMed] [Google Scholar]
- 4.Kurman R.J., Carcangiu M.L., Herrington S., Young R.H. WHO Classification of Tumours of Female Reproductive Organs. 4th ed. IARC Publications; Lyon, France: 2014. [Google Scholar]
- 5.Mehine M., Mäkinen N., Heinonen H., Aaltonen L.A., Vahteristo P. Genomics of uterine leiomyomas: Insights from high-throughput sequencing. Fertil. Steril. 2014;102:621–629. doi: 10.1016/j.fertnstert.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 6.Lehtonen H.J. Hereditary leiomyomatosis and renal cell cancer: Update on clinical and molecular characteristics. Fam. Cancer. 2011;10:397–411. doi: 10.1007/s10689-011-9428-z. [DOI] [PubMed] [Google Scholar]
- 7.Mehine M., Kaasinen E., Heinonen H., Mäkinen N., Kämpjärvi K., Sarvilinna N., Aavikko M., Vähärautio A., Pasanen A., Butzow R., et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc. Natl. Acad. Sci. USA. 2016;113:1315–1320. doi: 10.1073/pnas.1518752113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panagopoulos I., Gorunova L., Brunetti M., Agostini A., Andersen H.K., Lobmaier I., Bjerkehagen B., Heim S. Genetic heterogeneity in leiomyomas of deep soft tissue. Oncotarget. 2017;8:48769–48781. doi: 10.18632/oncotarget.17953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markowski D.N., Bartnitzke S., Loning T., Drieschner N., Helmke B.M., Bullerdiek J. MED12 mutations in uterine fibroids–Their relationship to cytogenetic subgroups. Int. J. Cancer. 2012;131:1528–1536. doi: 10.1002/ijc.27424. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Qiang W., Griffin B.B., Gao T., Chakravarti D., Bulun S., Kim J.J., Wei J. HMGA2-mediated tumorigenesis through angiogenesis in leiomyoma. Fertil. Steril. 2020;114:1085–1096. doi: 10.1016/j.fertnstert.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinonen H., Pasanen A., Heikinheimo O., Tanskanen T., Palin K., Tolvanen J., Vahteristo P., Sjöberg J., Pitkänen E., Butzow R., et al. Multiple clinical characteristics separate MED12-mutation-positive and-negative uterine leiomyomas. Sci. Rep. 2017;7:1015. doi: 10.1038/s41598-017-01199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg A.A. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: Leiomyoma. Cancer Genet. Cytogenet. 2005;158:1–26. doi: 10.1016/j.cancergencyto.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 13.Mäkinen N., Kämpjärvi K., Frizzell N., Butzow R., Vahteristo P. Characterization of MED12, HMGA2, and FH alterations reveals molecular variability in uterine smooth muscle tumors. Mol. Cancer. 2017;16:101. doi: 10.1186/s12943-017-0672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George J.W., Fan H., Johnson B., Carpenter T.J., Foy K.K., Chatterjee A., Patterson A.L., Koeman J., Adams M., Madaj Z.B., et al. Integrated epigenome, exome, and transcriptome analyses reveal molecular subtypes and homeotic transformation in uterine fibroids. Cell Rep. 2019;29:4069–4085.e6. doi: 10.1016/j.celrep.2019.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bardella C., El-Bahrawy M., Frizzell N., Adam J., Ternette N., Hatipoglu E., Howarth K., O’Flaherty L., Roberts I., Turner G., et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 2011;225:4–11. doi: 10.1002/path.2932. [DOI] [PubMed] [Google Scholar]
- 16.Kokkat T.J., Patel M.S., McGarvey D., LiVolsi V.A., Baloch Z.W. Archived formalin-fixed paraffin-embedded (FFPE) blocks: A valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv. Biobank. 2013;11:101–106. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Ahlfen S., Missel A., Bendrat K., Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS ONE. 2007;2:e1261. doi: 10.1371/journal.pone.0001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buzdin A., Sorokin M., Garazha A., Glusker A., Aleshin A., Poddubskaya E., Sekacheva M., Kim E., Gaifullin N., Giese A., et al. RNA sequencing for research and diagnostics in clinical oncology. Semin. Cancer Biol. 2020;60:311–323. doi: 10.1016/j.semcancer.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Stark R., Grzelak M., Hadfield J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 20.Moll P., Ante M., Seitz A., Reda T. QuantSeq 3’ mRNA sequencing for RNA quantification. Nat. Methods. 2014;11:i–iii. doi: 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- 21.Lohman B.K., Weber J.N., Bolnick D.I. Evaluation of TagSeq, a reliable low-cost alternative for RNAseq. Mol. Ecol. Resour. 2016;16:1315–1321. doi: 10.1111/1755-0998.12529. [DOI] [PubMed] [Google Scholar]
- 22.Turnbull A.K., Selli C., Martinez-Perez C., Fernando A., Renshaw L., Keys J., Figueroa J.D., He X., Tanioka M., Munro A.F., et al. Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: Comparison of gene expression profiling approaches. BMC Bioinform. 2020;21:30. doi: 10.1186/s12859-020-3365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Klemke M., Muller M.H., Wosniok W., Markowski D.N., Nimzyk R., Helmke B.M., Bullerdiek J. Correlated expression of HMGA2 and PLAG1 in thyroid tumors, uterine leiomyomas and experimental models. PLoS ONE. 2014;9:e88126. doi: 10.1371/journal.pone.0088126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mäkinen N., Vahteristo P., Kämpjärvi K., Arola J., Butzow R., Aaltonen L.A. MED12 exon 2 mutations in histopathological uterine leiomyoma variants. Eur. J. Hum. Genet. 2013;21:1300–1303. doi: 10.1038/ejhg.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kämpjärvi K., Mäkinen N., Mehine M., Välipakka S., Uimari O., Pitkänen E., Heinonen H., Heikkinen T., Tolvanen J., Ahtikoski A., et al. MED12 mutations and FH inactivation are mutually exclusive in uterine leiomyomas. Br. J. Cancer. 2016;114:1405–1411. doi: 10.1038/bjc.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahvenainen T.V., Mäkinen N.M., von Nandelstadh P., Vahteristo M.E.A., Pasanen A.M., Bützow R.C., Vahteristo P.M. Loss of ATRX/DAXX expression and alternative lengthening of telomeres in uterine leiomyomas. Cancer. 2018;124:4650–4656. doi: 10.1002/cncr.31754. [DOI] [PubMed] [Google Scholar]
- 29.Äyräväinen A., Pasanen A., Ahvenainen T., Heikkinen T., Pakarinen P., Härkki P., Vahteristo P. Systematic molecular and clinical analysis of uterine leiomyomas from fertile-aged women undergoing myomectomy. Hum. Reprod. 2020;35:2237–2244. doi: 10.1093/humrep/deaa187. [DOI] [PubMed] [Google Scholar]
- 30.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S., Pyl P.T., Huber W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallio M.A., Tuimala J.T., Hupponen T., Klemelä P., Gentile M., Scheinin I., Koski M., Käki J., Korpelainen E.I. Chipster: User-friendly analysis software for microarray and other high-throughput data. BMC Genom. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katainen R., Donner I., Cajuso T., Kaasinen E., Palin K., Mäkinen V., Aaltonen L.A., Pitkänen E. Discovery of potential causative mutations in human coding and noncoding genome with the interactive software BasePlayer. Nat. Protoc. 2018;13:2580–2600. doi: 10.1038/s41596-018-0052-3. [DOI] [PubMed] [Google Scholar]
- 40.Talevich E., Shain A.H., Botton T., Bastian B.C. CNVkit: Genome-Wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.