Abstract
Two metagenome-assembled genomes (MAGs), obtained from laboratory-scale enhanced biological phosphorus removal bioreactors, were analyzed. The values of 16S rRNA gene sequence identity, average nucleotide identity, and average amino acid identity indicated that these genomes, designated as RT and SSD2, represented two novel species within the genus Thiothrix, ‘Candidatus Thiothrix moscowensis’ and ‘Candidatus Thiothrix singaporensis’. A complete set of genes for the tricarboxylic acid cycle and electron transport chain indicates a respiratory type of metabolism. A notable feature of RT and SSD2, as well as other Thiothrix species, is the presence of a flavin adenine dinucleotide (FAD)-dependent malate:quinone oxidoreductase instead of nicotinamide adenine dinucleotide (NAD)-dependent malate dehydrogenase. Both MAGs contained genes for CO2 assimilation through the Calvin–Benson–Bassam cycle; sulfide oxidation (sqr, fccAB), sulfur oxidation (rDsr complex), direct (soeABC) and indirect (aprBA, sat) sulfite oxidation, and the branched Sox pathway (SoxAXBYZ) of thiosulfate oxidation to sulfur and sulfate. All these features indicate a chemoorganoheterotrophic, chemolithoautotrophic, and chemolithoheterotrophic lifestyle. Both MAGs comprise genes for nitrate reductase and NO-reductase, while SSD2 also contains genes for nitrite reductase. The presence of polyphosphate kinase and exopolyphosphatase suggests that RT and SSD2 could accumulate and degrade polyhosphates during the oxic-anoxic growth cycle in the bioreactors, such as typical phosphate-accumulating microorganisms.
Keywords: metagenome-assembled genome, biological phosphorus removal, colorless sulfur bacteria, Thiothrix
1. Introduction
Filamentous colorless sulfur bacteria (FCSB) have been a classic object of microbiology since the time of S.N. Winogradsky, who used them in the research that culminated in the discovery of chemosynthesis. Nevertheless, the genomics and metabolic potential of the FCSB from the genus Thiothrix remain poorly understood [1,2,3]. The bacteria retained in the genus after the last taxonomic revision [4] include five species: T. nivea, T. fructosivorans, T. unzii, T. caldifontis and T. lacustris. Despite the fact that the genomes of T. lacustris (GCF_000621325.1), T. caldifontis (GCF_900107695.1), and T. nivea (GCF_000260135.1) are available from databases, comprehensive analysis of the main metabolic pathways of sulfur, carbon, and nitrogen has not been reported. In addition, 25 metagenome-assembled genomes (MAGs) obtained from metagenomes of natural hydrogen sulfide biotopes are currently available. Two of these genomes were attributed to certain species, T. nivea DOLJORAL78_51_81 [5,6] and T. lacustris A8 [7], and 23 others were designated as Thiothrix sp. or uncultured Thiothrix sp. [8,9,10].
Due to the feature of energy metabolism, where hydrogen sulfide is the main donor of electrons during chemosynthesis, sulfur springs were considered to be a traditional habitat for Thiothrix, where filamentous bacteria often form massive fouling. There are reports of intense growth of Thiothrix in wastewater rich in reduced sulfur compounds [11,12,13,14]. An active growth of Thiothrix in wastewater treatment plants leads to the formation of fouling that can disrupt the operation of the entire system, degrading the quality of the wastewater and increasing the treatment time [15,16]. On the other hand, the ability of Thiothrix from activated sludge to accumulate polyphosphates and participate in the removal of phosphorus from wastewater was found along with other polyphosphate accumulating organisms (PAOs) [17,18,19], such as ‘Candidatus Accumulibacter phosphatis’, Tetrasphaera [20,21], and Halomonas [22].
The present study aims to analyse of genomic and metabolic potential of two recently obtained Thiothrix genomes—Thiothrix sp. RT and Thiothrix sp. SSD2, recovered from metagenomes of laboratory-scale enhanced biological phosphorus removal bioreactors (EBPR). Based on the phylogenetic analysis of MAGs, we propose two new species, ‘Candidatus Thiothrix moscovensis’ sp.nov. and ‘Candidatus Thiothrix singaporensis’ sp.nov.
2. Materials and Methods
2.1. PAO-Enriched Laboratory Culture for Thiothrix sp. RT
Enrichment and cultivation of the PAO microbial community in a lab-scale bioreactor operated at Research Center of Biotechnology (Moscow, Russia) was reported earlier [23]. Briefly, a 15 L sequencing batch laboratory bioreactor, inoculated with an activated sludge from Luberetskya wastewater treatment plant (Moscow, Russia), was supplied with the following medium: 670 mg/L CH3COONa·3H2O, 50 mg/L NH4Cl, 54.3 mg/L KH2PO4, 56.7 mg/L Na2HPO4, and 8.6 mg/L yeast extract. The bioreactor was operated at room temperature in a cyclic mode. A 6 h cycle of cultivation included the stages of filling the bioreactor with fresh medium (30 min), anaerobic growth (110 min), aerobic growth (180 min), precipitation of activated sludge (30 min), and discharge of the spent medium (10 min). Under these conditions, the PAO bioreactor stably removed 75–80% of phosphorus from the medium [23].
2.2. Metagenome Sequencing and Assembly of Thiothrix sp. RT
The total DNA was extracted from 1 g of sludge sample using a DNeasy PowerSoil DNA isolation kit (Qiagen, Hilden, Germany). A total of about 1.5 μg of DNA was obtained. A metagenomic DNA sample was sequenced using the Illumina (Illumina, San Diego, CA, USA) platform. The TruSeq DNA library was prepared using NEBNext® Ultra™ II DNA Library Prep Kit (New England Biolabs, Hitchin, UK) and sequenced on the Illumina HiSeq2500 instrument (250 nt single-end reads). Adapter sequences and low-quality reads were removed using Cutadapt [24] and Sickle (https://github.com/najoshi/sickle accessed on 10 November 2020), respectively. A total of about 50 million high-quality reads were obtained. The reads were de novo assembled into contigs using MetaSPAdes v. 3.7.1 [25]. The contigs were binned into MAGs using MetaBAT v. 2.12.1 [26]. Completeness of the MAGs and their possible contamination were estimated using the CheckM v. 1.0.13 tool [27]. The taxonomic assignment of the obtained MAGs was performed using the Genome Taxonomy Database Toolkit (GTDB-Tk) v. 1.1.1 [28] and Genome Taxonomy Database (GTDB) [29].
Gene search and annotation were performed using the RAST server 2.0 [30] followed by manual correction by the searches of predicted protein sequences against the National Center for Biotechnology Information (NCBI) databases.
2.3. Genome of Thiothrix sp. SSD2
MAG of Thiothrix sp. SSD2 was obtained from sludge from a full-scale wastewater treatment plant (PUB, Singapore) and assembled in a single contig with a total length of 4,541,730 [31]. The information about bioreactor operating conditions, samples preparation, DNA isolation, metagenome sequencing and assembly has been published [31]. Genome annotation revealed 16S rRNA gene, 43 tRNA genes and 4097 potential protein-coding genes [31]. For the present study, the Thiothrix sp. SSD2 genome was obtained from NCBI GenBank (CP059265).
2.4. Phylogenetic Analysis of MAGs
Average nucleotide identity (ANI) was calculated using an online resource (https://www.ezbiocloud.net/tools/ani) based on the OrthoANIu algorithm, using USEARCH [32]. Average amino acid identity (AAI) between the genomes was determined using the aai.rb script from the enveomics collection [33]. Digital DNA-DNA hybridization (dDDH) calculation was performed using the GGDC online platform (https://ggdc.dsmz.de/ggdc.php# accessed on 10 November 2020).
2.5. Nucleotide Sequence Accession Number
The annotated genome sequence of Thiothrix sp. RT bacterium was submitted to the NCBI GenBank database and is accessible under the number JAEHNX000000000.
3. Results
3.1. Assembly of the Genome of New Member of the Genus Thiothrix from EBPR Bioreactor
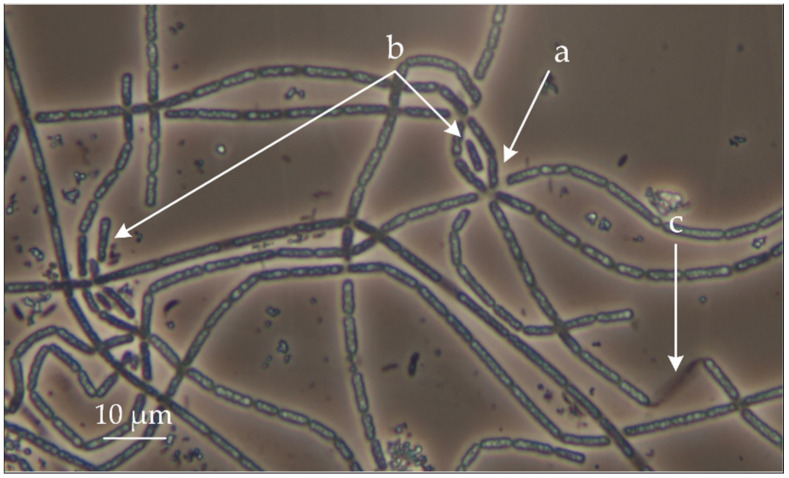
To obtain MAGs of microbial community members, we sequenced the metagenome of activated sludge from of the PAO bioreactor operated at Research Center of Biotechnology (Moscow, Russia) using Illumina technique. Analysis of the taxonomic affiliation of the obtained MAGs showed that one of them, designated Thiothrix sp. RT, belongs to the genus Thiothrix. Microscopic analysis of microorganisms from the PAO bioreactor also indicates the presence of cells morphologically similar to cultivated members of the genus Thiothrix: cells form rosettes, gonidia, and sheath (Figure 1).
Figure 1.
Thiothrix sp. as part of a phosphate-accumulating microbial community. Arrows show cells forming rosettes (a); detached cells that serve for reproduction (gonidia) (b); a sheath (c). Cell morphology was observed by using an Olympus CX 41 microscope equipped with a phase-contrast device.
The assembled Thiothrix sp. RT MAG consisted of 78 contigs (N50 size of 65,012 bp) with a total length of 3,693,042 bp; the CheckM-estimated completeness of this MAG as 99.8% with 0.92% possible contamination. Genome annotation revealed 16S rRNA gene, 38 tRNA genes, and 3645 potential protein-coding genes. The Thiothrix sp. RT 16S rRNA gene shares 97.9% sequence identity with Thiothrix nivea DSM 5205 and more than 99.5% identity with several environmental 16S rRNA gene sequences detected in activated sludge [34].
3.2. Phylogenetic Analysis of MAGs
Besides MAG of Thiothrix sp. RT, only one genome of Thiothrix sp. from EBPR bioreactors, Thiothrix sp. SSD2, is available [31]. Here we compared these genomes with each other and with genomes of known species of the genus Thiothrix, which have all been isolated from natural sulfidic environments.
The identity level of 16S rRNA genes of Thiothrix sp. RT and Thiothrix sp. SSD2 with genes of other members of the genus Thiothrix varies from 93.0 to 97.9%, while the 16S rRNAs of all described representatives of the genus are 93.3-98.9% identical to each other (Supplementary Materials Table S1). These values suggest assignment of Thiothrix sp. RT and Thiothrix sp. SSD2 to novel species within the same genus [35].
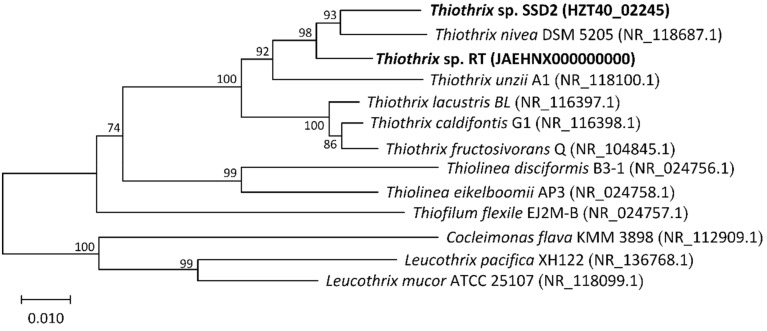
All species remaining in the genus Thiothrix after the last taxonomic revision [4] form two phylogenetic clusters on the tree, reconstructed on the basis of the nucleotide sequences of the 16S rRNA gene (Figure 2).
Figure 2.
Phylogenetic tree of 16S rRNA gene sequences reconstructed by the neighbor-joining method showing the position of novel strains of Thiothrix sp. RT and Thiothrix sp. SSD2. The sequences of bacteria from genera Leucothrix and Cocleimonas are included as an outgroup. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2000 replicates) are shown next to the branches. Bar, 0.01 substitutions per nucleotide position.
The first cluster includes Thiothrix sp. RT and Thiothrix sp. SSD2 together with Thiothrix nivea DSM 5205 and Thiothrix unzii A1. The second cluster includes T. lacustris BLT, T. caldifontis G1T, and T. fructosivorans QT.
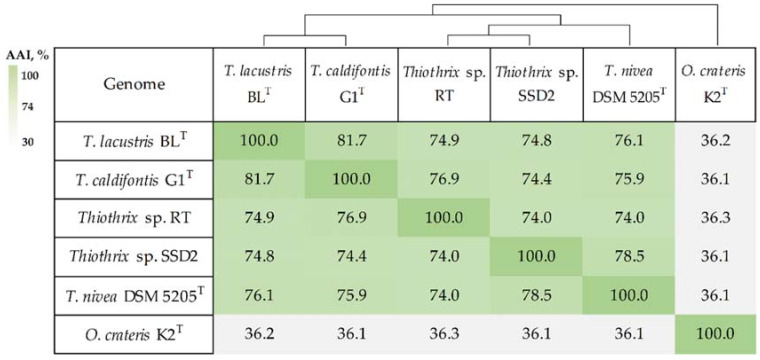
The AAI values between Thiothrix sp. RT and other Thiothrix genomes were from 74.0 to 76.9%, and 74.0–78.5% AAI was detected between Thiothrix sp. SSD2 and other species (Figure 3). These values are consistent with the assignment of Thiothrix sp. RT and Thiothrix sp. SSD2 to new species of the genus Thiothrix according to the thresholds proposed for genome-based taxonomic delineation [36]. Both of the two new genomes could also be assigned to novel species according to their ANI (<80.2%) and DDH (<30.2%) values (Table S2). The obtained values of AAI, ANI and dDDH are below the threshold for species delineation [37].
Figure 3.
Heatmap of pairwise amino acid identity (AAI) values in the genus Thiothrix using Oceanispirochaeta crateris K2T as an outgroup.
Based on results of phylogenomic analysis, we propose classifying strains RT and SSD2 as ‘Candidatus Thiothrix moscovensis’ sp.nov. and ‘Candidatus Thiothrix singaporensis’ sp.nov., respectively. ‘Ca. Thiothrix moscovensis’ sp.nov. (mos.co.ven’sis N.L. fem. adj. moscovensis pertaining to the city of Moscow, Russia, where the EBPR bioreactor used to obtain this genome operated). ‘Ca. Thiothrix singaporensis’ sp.nov. (sin.ga.po.ren´sis N.L. fem. adj. singaporensis pertaining to the city of Singapore, Singapore, where the EBPR bioreactor used to obtain this genome operated).
3.3. Comparative Analysis of Metabolic Pathways
3.3.1. Dissimilatory Metabolism of Sulfur Compounds
Members of the genus Thiothrix are known representatives of sulfur-oxidizing prokaryotes. Dissimilatory sulfur metabolism of the genus Thiothrix is represented by various sulfur-oxidizing complexes involved in the oxidation of hydrogen sulfide, thiosulfate, elemental sulfur, and sulfite.
Genes that encode enzymes involved in the oxidation of sulfide to elemental sulfur, sulfide:quinone oxidoreductase (SQR) and flavocytochrome c-sulfide dehydrogenase (FCSD) were identified in Thiothrix sp. RT and Thiothrix sp. SSD2.
The sqrA, sqrF, and fccAB genes were found in the genomes of Thiothrix sp. RT and Thiothrix sp. SSD2. The sqrD gene, phylogenetically close to sqrF, was additionally revealed in Thiothrix sp. SSD2 (Table 1, Table S3).
Table 1.
Genes for nitrogen, sulfur, phosphorus metabolism and key enzymes of the Calvin–Benson–Bassham cycle.
| Genes of Metabolic Pathways |
T. nivea DSM 5205T |
T. lacustris BLT |
T. caldifontis G1T |
Thiothrix sp. RT |
Thiothrix sp. SSD2 |
|---|---|---|---|---|---|
| Molecular nitrogen fixation | nifDKH | - | nifDKH | nifDKH | nifDKH |
| Dissimilatory nitrate reduction | - | narGHI | narGHI, nirS, cnorBC, |
narGHI,
cnorBC, |
narGHI, nirS, cnorBC |
| Dissimilatory reduction of NO3− to NO2−, nitrate reductase from the Nap family | napAB | - | - | - | - |
| Assimilatory reduction of NO2− to NH4+ | nirBD | nirBD, | nirBD, | nirBD, | nirBD |
| Dissimilatory sulfur metabolism |
soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB |
soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB |
soeABC, soxAXBYZ, aprAB, sat, dsr sqr, fccAB |
soeABC, soxAXBZY, aprAB, sat, dsr, sqr, fccAB |
soeABC, soxAXBYZ, aprAB, sat, dsr, sqr, fccAB |
| Calvin–Benson–Bassam cycle | IAc, IAq, II, prk(2) |
IAq, II, prk(2) |
IAc, IAq, II, prk(2) |
IAc, IAq, II, prk(2) |
IAc, IAq, II, prk(2) |
| Phosphorus metabolism | phoURB, ptsSACB, ppk, epp | phoURB, ptsSACB, ppk, epp |
phoURB, ptsSACB, ppk, epp |
phoURB, ptsSACB, ppk, epp |
phoURB, ptsSACB, ppk, epp |
| FAD-dependent malate: quinone oxidoreductase (EC 1.1.5.4) | mqo | mqo | mqo | mqo | mqo |
| Malate dehydrogenase (oxaloacetate–decarboxylating) (NADP) (EC 1.1.1.40) | maeB | maeB | maeB | maeB | maeB |
For all members of the genus, including Thiothrix sp. RT and Thiothrix sp. SSD2, the presence of genes of the SOX multienzyme system that oxidizes thiosulfate was revealed (Table 1, Table S3). A feature of the genus is the absence of genes for the dimeric protein SoxCD. The oxidation of thiosulfate is carried out by the branched Sox pathway (the SoxAXBYZ enzymatic complex), which oxidizes thiosulfate to sulfate and elemental sulfur, accumulated inside the cells.
Elemental sulfur can be further oxidized to sulfite using the reverse dissimilatory sulfite reductase rDsrAB. In Thiothrix sp. RT and Thiothrix sp. SSD2 genes of this pathway form one cluster consisting of 14 genes (dsrABCEFHLMKJOPRS) and 12 genes (dsrABCEFHLMKJOP), respectively (Table 1, Table S3). An accessory gene dsrR was not found in the genome of Thiothrix sp. SSD2, while Thiothrix sp. RT contains all genes of the dsr cluster.
Genomes of Thiothrix sp. RT and Thiothrix sp. SSD2 contains genes for membrane-bound cytoplasmic sulfite:quinone oxidoreductase SoeABC, which directly oxidizes sulfite to sulfate. Genes for indirect oxidation of sulfite to sulfate (the aprAB genes encoding APS reductase (AprAB) and the sat gene encoding ATP sulfurylase (Sat)) were also found. Oxidation of sulfite to sulfate occurs via the APS intermediate.
Thus, it can be assumed that Thiothrix sp. RT and Thiothrix sp. SSD2, like the well-known pure cultures of the genus Thiothrix, have pathways for the oxidation of reduced sulfur compounds, which provide the oxidation of hydrogen sulfide, elemental sulfur, sulfite, and thiosulfate.
3.3.2. Autotrophic CO2 Assimilation
All genes encoding enzymes enabling autotrophic assimilation of CO2 through the Calvin–Benson–Bassham cycle (except for the sedoheptulose-1,7-bisphosphatase) were found in all members of the genus Thiothrix, as well as in Thiothrix sp. RT and Thiothrix sp. SSD2 (Table 1, Table S3). It should be noted that the sedoheptulose-1,7-bisphosphatase activity in bacteria is often catalyzed by fructose-1,6-bisphosphatase with dual sugar specificity [38,39]. A key Calvin–Benson–Bassham cycle enzyme, RuBisCo, is represented by three types (IAc, IAq, II) in Thiothrix sp. RT and Thiothrix sp. SSD2. Another key enzyme of the Calvin–Benson–Bassham cycle, phosphoribulokinase, is encoded by two copies of the prk gene, which are phylogenetically distant from each other, which is typical for Thiothrix species.
3.3.3. Assimilation of N2
Genomes of Thiothrix sp. RT and Thiothrix sp. SSD2 contain almost a complete set of nif genes required for N2 fixation in nifASVB1XX2YENQVWMHDKZT and nifASVB1XX2B2ENQV WMHDKZTO operons. The composition of the genes of accessory nitrogenase subunits differed slightly. Genes for NifB2 which, along with NifB1, regulates the nitrogenase complex [40] and NifO which involved in synthesis of the iron-molybdenum cofactor of nitrogenase [41] were not found in the genome of Thiothrix sp. RT. The nifY gene, which promotes the maturation of the enzyme through its binding to the MoFe cofactor, was not found in the genome of Thiothrix sp. SSD2.
3.3.4. Dissimilatory Nitrate Reduction
Genes for the NarGHI membrane complex involved in the dissimilatory reduction of nitrate to nitrite were found in genomes of Thiothrix sp. RT and Thiothrix sp. SSD2. Genes nirBD that encode the NirBD enzyme involved in the removal of the toxic NO2- by assimilatory reduction to NH4+ were found in the genomes of all members of the genus Thiothrix, including Thiothrix sp. RT and Thiothrix sp. SSD2. This chain is realized in almost all representatives of the genus Thiothrix, with the exception of T. nivea, in which instead of narGHI, the periplasmic nitrate reductase encoded by napAB was found (Table 1, Table S3).
The presence of the nirS gene for dissimilatory reduction of NO2− to N2O was shown for Thiothrix sp. SSD2, unlike Thiothrix sp. RT. The genomes of both bacteria contain the cnorBC genes for respiratory nitric oxide reductase that reduce NO to N2O. The nosZ gene is missing in both genomes, like in other representatives of the genus Thiothrix. The presence of genes for dissimilatory nitrate reduction suggests the ability of Thiothrix sp. RT and Thiothrix sp. SSD2 to perform anaerobic respiration, where nitrate and nitrous oxide can function as terminal electron acceptors. Thiothrix sp. SSD2 was also predicted to reduce nitrite. We demonstrated previously the ability to perform anaerobic respiration in the presence of NO for T. lacustris AS, T. caldifontis, and T. unzii [42].
3.3.5. Phosphorus Accumulation
The accumulation of polyphosphate (poly-P) granules is known for members of the genus Thiothrix [17,43]. Both Thiothrix sp. RT and Thiothrix sp. SSD2 contained genes for polyphosphate kinase PPK (ppk), the main enzyme that catalyzes the transfer of ATP (ppk1) or GTP (ppk2) to the active site of the protein during the synthesis of the poly-P chain [44]. The ppk1 gene encoding PPK1 was found in the genomes of Thiothrix sp. RT and Thiothrix sp. SSD2. Since this reaction is reversible, in the case of PPK1, it could be used to make ATP from poly-P [44]. The gene for the exopolyphosphatase EPP (epp), which catalyzes the hydrolysis of inorganic phosphorus from the polyphosphate chain, was also found in the genomes of Thiothrix sp. RT and Thiothrix sp. SSD2. Genes for PPK and EPP are also present in other members of the genus Thiothrix.
Thus, both enzymes, PPK1 and EPP, can regulate the accumulation and degradation of polyphosphates in the cell. Genes phoURB, pstSACB which encodes systems for the regulation and transfer of inorganic phosphorus into the cell were also found in the genomes of Thiothrix sp. SSD2 and Thiothrix sp. RT. Along with participation in biosynthetic processes, polyphosphates are probably used for temporal ATP storage, which is realized through the activity of polyphosphate kinase PPK1 and exopolyphosphatase EPP in members of the genus Thiothrix.
3.3.6. Central Metabolic Pathways
Genome analysis of Thiothrix sp. RT and Thiothrix sp. SSD2 revealed the presence of a complete set of genes for the Embden–Meyerhof (EM) pathway, the tricarboxylic acid (TCA) cycle, and the glyoxylate cycle (Table S3).
TCA cycle has a remarkable feature in the known species of the genus Thiothrix, as well as in Thiothrix sp. SSD2 and Thiothrix sp. RT, —the absence of the mdh gene of the classic cytoplasmic nicotinamide adenine dinucleotide (NAD)-dependent malate dehydrogenase, which is found in the vast majority of prokaryotes. Instead, the mqo gene encoding membrane-bound FAD-dependent malate:quinone oxidoreductase was found in the genomes of representatives of the genus Thiothrix, including Thiothrix sp. RT and Thiothrix sp. SSD2.
Genomes of Thiothrix sp. RT and Thiothrix sp. SSD2 include all the genes of the EM pathway. The gene pckA for ATP-dependent phosphoenolpyruvate (PEP)-carboxykinase, which decarboxylates oxaloacetate to form pyruvate, was also revealed in the genomes of Thiothrix sp. SSD2 and Thiothrix sp. RT. The genomes contain genes for pyruvate carboxylase (pyc) and decarboxylating NADP-dependent malate dehydrogenase (maeB). The two latter enzymes likely shunt the reaction of malate conversion to oxaloacetate in the TCA cycle and glyoxylate cycle in order to ensure the production of sufficient oxaloacetate for gluconeogenesis and amino acid biosynthesis.
The release to constructive metabolism from the TCA cycle occurs at the level of oxaloacetate and 2-oxoglutarate. Oxaloacetate is involved in aspartate synthesis with the participation of aspartate aminotransferase (aspB), while the conversion of 2-oxoglutarate is carried out by glutamate synthase (glt) to glutamate.
3.3.7. Respiratory Electron Transport Chain
Genomes of Thiothrix sp. RT and Thiothrix sp. SSD2 encode all the major components of the electron transport chain required to generate energy through oxidative phosphorylation. Complex I, NADH:quinone oxidoreductase, is encoded by the genes forming the nuoABCDEFGHIJKLM cluster and separate nuoN gene in Thiothrix sp. SSD2. The complex is encoded by two gene clusters, nuoACDEFGHIJLM and nuoBKN, in Thiothrix sp. RT.
The succinate dehydrogenase II complex includes a cytochrome b556 subunit (sdhC), an iron-sulfur protein (sdhB), a flavoprotein subunit (sdhA), a hydrophobic membrane anchor protein (sdhD). The genes form a single sdhABCD cluster in the genome of Thiothrix sp. RT, and are located in two separate loci, sdhBCD and sdhA, in Thiothrix sp. SSD2.
Complex III, ubiquinol-cytochrome c reductase, consists of an iron-sulfur subunit (encoded by the rpi1 gene), a cytochrome b subunit (encoded by the cytB gene), and a cytochrome c 1 subunit (encoded by the cyc1 gene).
Complex IV is represented by five types of oxidases: six cbb3 type subunits (encoded by ccoNOPGQS), six cox subunits (encoded by coxABCD11,15), and four cytochrome d subunits of ubiquinol oxidase (encoded by cydABCDX). Conversion of ADP to ATP occurs via F-type ATP synthetase, encoded at the atpABCDEGH cluster, with a separate gene for the F-type ATP synthetase B subunit, atpF. The phosphates required for ATP synthetase to function are supplied by inorganic pyrophosphatase, encoded by the ppa gene, and polyphosphate kinase, encoded by ppk.
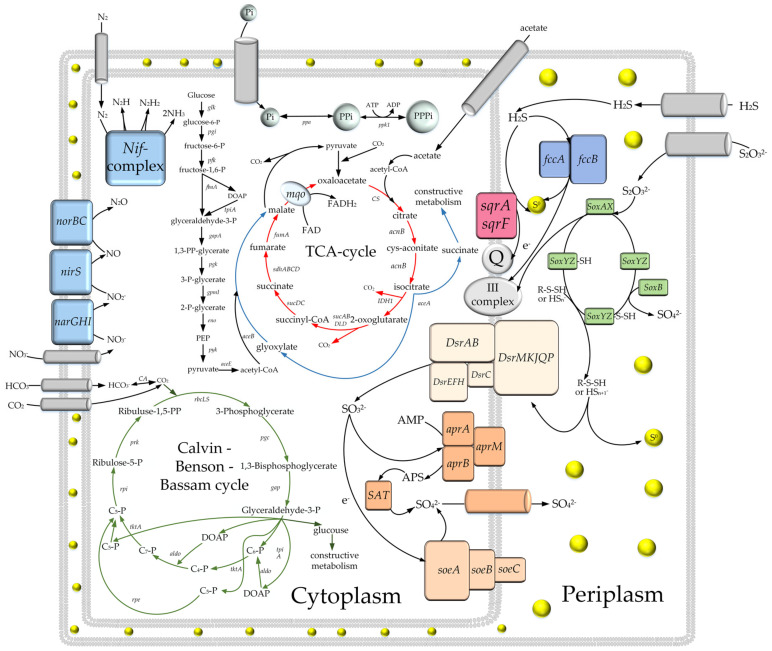
Overall, genome analysis of Thiothrix sp. SSD2 and Thiothrix sp. RT did not reveal any important differences in the composition of genes for metabolic pathways from other representatives of the genus Thiothrix. The scheme of potential metabolic pathways of representatives of the genus Thiothrix, based on our genome-based reconstructions, is shown in Figure 4.
Figure 4.
A hypothetical metabolic scheme for Thiothrix sp. RT and Thiothrix sp. SSD2 reconstructed from the received MAGs. The nirS gene encoding nitrite reductase was not found in the Thiothrix sp. RT genome.
4. Discussion
Phylogenetic analysis of Thiothrix sp. RT and Thiothrix sp. SSD2 and other members of the genus Thiothrix supported classification of the two new strains as new species ‘Candidatus Thiothrix moscovensis’ sp.nov. and ‘Candidatus Thiothrix singaporensis’ sp.nov., respectively.
The results of the analysis of Thiothrix sp. RT and Thiothrix sp. SSD2 genomes show that the studied organisms carry out the respiratory type of metabolism, and that they are facultative aerobes and facultative heterotrophs. Complete oxidation of organic substrates can occur through the TCA pathway with the formation of a transmembrane proton gradient, which is further used by ATP synthase for the synthesis of ATP. Thiothrix sp. RT and Thiothrix sp. SSD2 are potentially capable of assimilation of molecular nitrogen, anaerobic respiration on nitrates and incomplete denitrification.
Autotrophic growth in the presence of reduced sulfur compounds (hydrogen sulfide, elemental sulfur, sulfite, and thiosulfate) with the participation of enzymatic systems of dissimilatory sulfur metabolism is possible for representatives of the genus Thiothrix, including Thiothrix sp. RT and Thiothrix sp. SSD2, which ensure the lithotrophic growth of these organisms. A feature of thiosulfate oxidation is the presence of a branched Sox pathway (the SoxAXBYZ enzymatic complex) and the absence of soxCD genes, which leads to the oxidation of thiosulfate to sulfates and elemental sulfur.
A notable feature of Thiothrix sp. RT and Thiothrix sp. SSD2, as well as other representatives of the genus Thiothrix, is the absence of the classical cytoplasmic NAD-dependent malate dehydrogenase, which is found in the vast majority of prokaryotes. Instead, the mqo gene encoding a FAD-dependent malate:quinone oxidoreductase was found in the genomes of two MAGs, as well as in the genomes of other known representatives of the genus Thiothrix. The presence of only MQO, as an enzyme capable of directly oxidizing malate to oxaloacetate in TCA and in the glyoxylate cycle, is quite rare in prokaryotes [45]. In terms of energy potential (Gibbs free energy, Δ G ° ‘), this enzyme is much more efficient than MDH and, at the same time, its activity increases more than four times in the presence of acetate [45,46].
The two EBPR systems analyzed used a batch reactor with alternating aerobic and anaerobic conditions with acetate as a carbon and energy source. The fact that we managed to assemble MAGs representing new species of the genus Thiothrix may also be due to the fact that the samples were taken from EBPR systems. During organoheterotrophic growth, the presence of MQO in these organisms gave an advantage when growing on acetate relative to organisms with classical MDH. This capacity probably facilitated enrichment of Thiothrix species in the reactors, as well as their ability to accumulate and degrade polyphosphates as typical phosphate-accumulating microorganisms. It should be noted that the growth of a number of members of the genus Thiothrix (T. lacustris, T. caldifontis) is preferable under chemolithoheterotrophic conditions (cell biomass increases by 2–3 times). With batch cultivation under organoheterotrophic conditions other than the EBPR system, growth is ineffective and is accompanied by a slight increase in biomass, by 20–30% (our unpublished data).
Thiothrix usually grows in nature in organic-poor hydrogen sulfide biotopes or in organic-rich wastewater, but in the presence of sulfide. In this case, bacteria use a lithoautotrophic or lithoheterotrophic type of metabolism.
Periodic changes in the growth conditions realized in the EBPR system probably provided an advantage for the enrichment of new Thiothrix species that were not previously identified in natural biotopes and wastewater. There is now sufficient evidence that Thiothrix can participate in the removal of inorganic phosphorus [17,18,19]. Analysis of MAGs obtained from EBPR system metagenomes will probably be useful not only for identifying new Thiothrix species, but also for creating optimal conditions for the growth of Thiothrix as phosphate accumulating organisms in the EBPR system. In particular, the addition of nitrate could probably stimulate the growth of Thiothrix in the EBPR bioreactors, since they are able to survive anaerobic conditions by switching to anaerobic respiration on nitrates. The addition of acetate to the culture medium promotes the induction of MQO activity involved in the conversion of malate to oxaloacetate. This process is vital because oxaloacetate is an important precursor of gluconeogenesis and amino acid synthesis of the aspartate branch.
Acknowledgments
We thank Rohan B.H. Williams, Irina Bessarab, and Krithika Arumugam from Singapore Centre for Environmental Life Sciences Engineering for bringing our attention to Thiothrix sp. SSD2 genome and helpful discussions.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/12/2030/s1, Table S1: Identity level of 16S rRNA genes in representatives of the genus Thiothrix, %; Table S2: Molecular characteristics of representatives of the genus Thiothrix; Table S3: Genes encoding enzymes of metabolic pathways in Thiothrix sp. SSD2 and Thiothrix sp. RT.
Author Contributions
Conceptualization, N.V.R. and M.Y.G.; investigation, A.V.M., E.V.G., D.D.S., T.S.R., A.V.B., and N.D.M.; resources, Y.Y.B. and N.V.P.; data curation, A.V.B. and N.V.R.; writing—original draft preparation, N.V.R., D.D.S., T.S.R., M.V.G., and M.Y.G.; writing—review and editing, T.S.R., M.V.G., N.V.R., and M.Y.G.; supervision, N.V.R. and M.Y.G.; funding acquisition, M.Y.G. and A.V.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly funded by the Russian Science Foundation (project 20-14-00137, comparative genome analysis of Thiothrix) and the Russian Foundation for Basic Research (project 18-29-25016, metagenome sequencing and assembly of Thiothrix sp. RT genome).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Larkin J.M., Shinabarger D.L. Characterization of Thiothrix nivea. Int. J. Syst. Bacteriol. 1983;33:841–846. doi: 10.1099/00207713-33-4-841. [DOI] [Google Scholar]
- 2.Odintsova E.V., Wood A.P., Kelly D.P. Chemolithoautotrophic growth of Thiothrix ramose. Arch. Microbiol. 1993;160:152–157. doi: 10.1007/BF00288718. [DOI] [Google Scholar]
- 3.Unz R.F., Head I.M. Bergey’s Manual of Systematic Bacteriology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. Thiothrix; pp. 1–16. [DOI] [Google Scholar]
- 4.Boden R., Scott K.M. Evaluation of the genus Thiothrix Winogradsky 1888 (Approved Lists 1980) emend. Aruga et al. 2002: Reclassification of Thiothrix disciformis to Thiolinea disciformis gen. nov., comb. nov., and of Thiothrix flexilis to Thiofilum flexile gen. nov., comb nov., with emended description of Thiothrix. Int. J. Syst. Evol. Microbiol. 2018;68:2226–2239. doi: 10.1099/ijsem.0.002816. [DOI] [PubMed] [Google Scholar]
- 5.Dudek N.K., Sun C.L., Burstein D., Kantor R.S., Aliaga Goltsman D.S., Bik E.M., Thomas B.C., Banfield J.F., Relman D.A. Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome. Curr. Biol. 2017;27:3752–3762.e6. doi: 10.1016/j.cub.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Bik E.M., Costello E.K., Switzer A.D., Callahan B.J., Holmes S.P., Wells R.S., Carlin K.P., Jensen E.D., Venn-Watson S., Relman D.A. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 2016;7:10516. doi: 10.1038/ncomms10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharrar A.M., Flood B.E., Bailey J.V., Jones D.S., Biddanda B.A., Ruberg S.A., Marcus D.N., Dick G.J. Novel large sulfur bacteria in the metagenomes of groundwater-fed chemosynthetic microbial mats in the Lake Huron basin. Front. Microbiol. 2017;8:791. doi: 10.3389/fmicb.2017.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 9.Pachiadaki M.G., Brown J.M., Brown J., Bezuidt O., Berube P.M., Biller S.J., Poulton N.J., Burkart M.D., La Clair J.J., Chisholm S.W., et al. Charting the complexity of the marine microbiome through single-cell genomics. Cell. 2019;179:1623–1635. doi: 10.1016/j.cell.2019.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., Liu Y., Xu W., Pan J., Luo Z.-H., Li M. Genome- and community-level interaction insights into carbon utilization and element cycling functions of Hydrothermarchaeota in hydrothermal sediment. mSystems. 2020;5:e00795-19. doi: 10.1128/mSystems.00795-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sears K., Alleman J.E., Barnard J.L., Oleszkiewicz J.A. Impacts of reduced sulfur components on active and resting ammonia oxidizers. J. Ind. Microbiol. Biotechnol. 2004;31:369–378. doi: 10.1007/s10295-004-0157-2. [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre O., Moletta R. Treatment of organic pollution in industrial saline wastewater: A literature review. Water Res. 2006;40:3671–3682. doi: 10.1016/j.watres.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Lu H., Wu D., Jiang F., Ekama G.A., van Loosdrecht M.C.M., Chen G.-H. The demonstration of a novel sulfur cycle-based wastewater treatment process: Sulfate reduction, autotrophic denitrification, and nitrification integrated (SANI®) biological nitrogen removal process. Biotechnol. Bioeng. 2012;109:2778–2789. doi: 10.1002/bit.24540. [DOI] [PubMed] [Google Scholar]
- 14.Pikaar I., Sharma K.R., Hu S., Gernjak W., Keller J., Yuan Z. Water engineering. Reducing sewer corrosion through integrated urban water management. Science. 2014;345:812–814. doi: 10.1126/science.1251418. [DOI] [PubMed] [Google Scholar]
- 15.Wanner J., Kucman K., Ottová V., Grau P. Effect of anaerobic conditions on activated sludge filamentous bulking in laboratory systems. Water Res. 1987;21:1541–1546. doi: 10.1016/0043-1354(87)90139-4. [DOI] [Google Scholar]
- 16.Gonzalez-Gil G., Holliger C. Dynamics of microbial community structure of and enhanced biological phosphorus removal by aerobic granules cultivated on propionate or acetate. Appl. Environ. Microbiol. 2011;77:8041–8051. doi: 10.1128/AEM.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Graaff D.R., van Loosdrecht M.C., Pronk M. Biological phosphorus removal in seawater-adapted aerobic granular sludge. Water Res. 2020;172:115531. doi: 10.1016/j.watres.2020.115531. [DOI] [PubMed] [Google Scholar]
- 18.Meng Q., Zeng W., Wang B., Fan Z., Peng Y. New insights in the competition of polyphosphate-accumulating organisms and glycogen-accumulating organisms under glycogen accumulating metabolism with trace Poly-P using flow cytometry. Chem. Eng. J. 2020;385:123915. doi: 10.1016/j.cej.2019.123915. [DOI] [Google Scholar]
- 19.Rubio-Rincón F.J., Welles L., Lopez-Vazquez C.M., Nierychlo M., Abbas B., Geleijnse M., Nielsen P.H., van Loosdrecht M.C.M., Brdjanovic D. Long-term effects of sulphide on the enhanced biological removal of phosphorus: The symbiotic role of Thiothrix caldifontis. Water Res. 2017;116:53–64. doi: 10.1016/j.watres.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima T., Uda N., Okamoto M., Onuki M., Satoh H., Mino T. Abundance of Candidatus ‘Accumulibacter phosphatis’ in enhanced biological phosphorus removal activated sludge acclimatized with different carbon sources. Microbes Environ. 2007;22:346–354. doi: 10.1264/jsme2.22.346. [DOI] [Google Scholar]
- 21.Nguyen H.T., Le V.Q., Hansen A.A., Nielsen J.L., Nielsen P.H. High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol. Ecol. 2011;76:256–267. doi: 10.1111/j.1574-6941.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen H.T.T., Nielsen J.L., Nielsen P.H. ‘Candidatus Halomonas Phosphatis’, a novel Polyphosphate-Accumulating Organism in Full-Scale Enhanced Biological Phosphorus Removal Plants. Environ. Microbiol. 2012;14:2826–2837. doi: 10.1111/j.1462-2920.2012.02826.x. [DOI] [PubMed] [Google Scholar]
- 23.Kotlyarov R.Y., Beletsky A.V., Ravin N.V., Mardanov A.V., Kallistova A.Y., Dorofeev A.G., Nikolaev Y.A., Pimenov N.V. A Novel Phosphate-Accumulating Bacterium Identified in a Bioreactor for Phosphate Removal from Wastewater. Microbiology. 2019;88:751–755. doi: 10.1134/S0026261719060055. [DOI] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 25.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaumeil P.A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 30.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: Amodular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arumugam K., Bessarab I., Haryono M.A.S., Liu X., Zuniga-Montanez R.E., Roy S., Qiu G., Drautz-Moses D.I., Law Y.Y., Wuertz S., et al. Analysis procedures for assessing recovery of high quality, complete, closed genomes from Nanopore long read metagenome sequencing. bioRxiv. 2020 doi: 10.1101/2020.03.12.974238. [DOI] [Google Scholar]
- 32.Yoon S.H., Ha S.M., Lim J., Kwon S., Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-R L.M., Konstantinidis K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1. doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 34.Jiang Y., Chen Y., Zheng X. Efficient polyhydroxyalkanoates production from a waste-activated sludge alkaline fermentation liquid by activated sludge submitted to the aerobic feeding and discharge process. Environ. Sci. Technol. 2009;43:7734–7741. doi: 10.1021/es9014458. [DOI] [PubMed] [Google Scholar]
- 35.Yarza P., Yilmaz P., Pruesse E., Glöckner F.O., Ludwig W., Schleifer K.-H., Whitman W.B., Euzéby J., Yarza P., Rosselló-Móra R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014;12:635. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 36.Konstantinidis K.T., Rosselló-Móra R., Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 38.Tamoi M., Ishikawa T., Takeda T., Shigeoka S. Enzymic and molecular characterization of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC 7942: Resistance of the enzyme to hydrogen peroxide. Biochem. J. 1996;316:685–690. doi: 10.1042/bj3160685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flechner A., Gross W., Martin W.F., Schnarrenberger C. Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 1999;447:200–202. doi: 10.1016/S0014-5793(99)00285-9. [DOI] [PubMed] [Google Scholar]
- 40.Vernon S.A., Pratte B.S., Thiel T. Role of the nifB1 and nifB2 promoters in cell-type-specific expression of two Mo nitrogenases in the cyanobacterium Anabaena variabilis ATCC 29413. J. Bacteriol. 2017;199:e00674-16. doi: 10.1128/JB.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosch R., Rodrıćguez-Quiñones F., Imperial J. Identification of gene products from the Azotobacter vinelandii nifBfdxNnifOQ operon. FEMS Microbiol. Lett. 1997;157:19–25. doi: 10.1111/j.1574-6968.1997.tb12747.x. [DOI] [PubMed] [Google Scholar]
- 42.Trubitsyn I.V., Belousova E.V., Tutukina M.N., Merkel A.Y., Dubinina G.A., Grabovich M.Y. Expansion of ability of denitrification within the filamentous colorless sulfur bacteria of the genus Thiothrix. FEMS Microbiol. Lett. 2014;358:72–80. doi: 10.1111/1574-6968.12548. [DOI] [PubMed] [Google Scholar]
- 43.Rubio-Rincón F.J., Lopez-Vazquez C.M., Welles L., van Loosdrecht M.C.M., Brdjanovic D. Sulphide effects on the physiology of Candidatus Accumulibacter phosphatis type I. Appl. Microbiol. Biotechnol. 2017;101:1661–1672. doi: 10.1007/s00253-016-7946-1. [DOI] [PubMed] [Google Scholar]
- 44.Rao N.N., Gómez-García M.R., Kornberg A. Inorganic polyphosphate: Essential for growth and survival. Annu. Rev. Biochem. 2009;78:605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 45.Molenaar D., van der Rest M.E., Drysch A., Yücel R. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 2000;182:6884–6891. doi: 10.1128/JB.182.24.6884-6891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kretzschmar U., Rückert A., Jeoung J.H., Görisch H. Malate: Quinone oxidoreductase is essential for growth on ethanol or acetate in Pseudomonas aeruginosa The GenBank accession number for the Pseudomonas aeruginosa ATCC 17933 mqo sequence reported in this work is AY129296. Microbiology. 2002;148:3839–3847. doi: 10.1099/00221287-148-12-3839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.