Abstract
After Salmonella Enteritidis and S. Typhimurium, S. 4,[5],12:i:- is the most reported serovar in human clinical cases. During the past 20 years, many tools have been used for its typing and second-phase flagellar deletion characterization. Currently, whole genome sequencing (WGS) and different bioinformatic programs have shown the potential to be more accurate than earlier tools. To assess this potential, we analyzed by WGS and in silico typing a selection of 42 isolates of S. 4,[5],12:i:- and S. Typhimurium with different in vitro characteristics. Comparative analysis showed that SeqSero2 does not differentiate fljB-positive S. 4,[5],12:i:- strains from those of serovar Typhimurium. Our results proved that the strains selected for this work were non-clonal S. 4,[5],12:i:- strains circulating in Spain. Using WGS data, we identified 13 different deletion types of the second-phase flagellar genomic region. Most of the deletions were generated by IS26 insertions, showing orientation-dependent conserved deletion ends. In addition, we detected S. 4,[5],12:i:- strains of the American clonal line that would give rise to the Southern European clone in Spain. Our results suggest that new S. 4,[5],12:i:- strains are continuously emerging from different S. Typhimurium strains via different genetic events, at least in swine products.
Keywords: Salmonella, monophasic, deletion, IS26 insertion, typing, whole genome sequencing
1. Introduction
Salmonella enterica subs. enterica consist of more than 2600 serovars [1]. Nontyphoidal Salmonella serovars are common causative zoonotic agents of bacterial food-borne disease worldwide. After S. Enteritidis and S. Typhimurium, the monophasic variant S. 4,[5],12:i:- is the most frequently reported in clinical human infections, and is responsible for about 4.7% of total reported cases [2]. The monophasic variant S. 4,[5],12:i:- is antigenically similar to S. Typhimurium (which has the antigenic formula 4,5,12:i:1,2) but does not express the second-phase flagellar antigen, which is identified as 1,2 in the S. Typhimurium antigenic formula. The first described monophasic variant of S. Typhimurium emerged in Spain in 1997 [3] and became the fourth most common serovar in clinical isolates in 1998 [4]. Thereafter, the emergence of multiple clones of monophasic variant of S. Typhimurium has been reported worldwide [5]. According to data from seven European countries, reported to The European Surveillance System (TESSy), the clinical isolates of S. 4,[5],12:i:- increased from 360 in 2007 to 1416 in 2009 [5], consequently, this serovar was included in the European Salmonella control program. In 2018, 2553 cases were confirmed, associated mainly with the consumption of broiler (43.4%) and pig-derived products (39.6%) [2]. Specifically in Spain, S. 4,[5],12:i:- is still the third most common serovar with 126 cases reported in 2016 [6].
Given the high incidence of the S. 4,[5],12:i:- serovar, many research groups have carried out analyses of the deletions involving the second-phase flagellar genes, suggesting that multiple independent clones may be emerging in the United States and Europe [7]. Spanish isolates belong to the first described clone, which were: (i) U302 phage type, (ii) multi-resistant to ampicillin, chloramphenicol, sulphonamide, gentamicin, streptomycin, tetracycline and sulfamethoxazole-trimethoprim (ACSuGSTSxT profile), (iii) defective in 16 to 54 genes upstream of the iroB gene (including the lack of fljAB operon) and (iv) positive in the presence of the insertion sequence IS26 [4,8,9]. By 2002 and 2003, additional phage types began to be recognized, suggesting divergence from the original clone or introduction of new S. 4,[5],12:i:- lineages around the world, evolved through multiple independent events, most likely from S. Typhimurium ancestors [4,7,8,10,11,12,13].
A second clonal line was proposed by Soyer et al. in the United States (US), in 2009 [10]. These isolates lacked 77 genes from STM2692 to STM2772 and harbored a 7 kb fragment insertion composed of two partial Fels-2 genes (STM2704 and STM2706) and three genes homologous to STM1054, STM1053 and STM1997 (umuC), which encode two Gifsy-2 prophage genes and a component of DNA polymerase V (umuC) [10]. Later, in 2014, Mourão et al. proposed the Southern Europe clone to classify Portuguese origin strains that contained the same particular genomic deletion as the US strains described by Soyer et al. but showed resistance to chloramphenicol, streptomycin, sulfamethoxazole, tetracycline and trimethoprim (CSSuTTm) caused by the acquisition of IncR plasmids [14]. The authors suggested a common evolutionary origin for the US clone and Southern European clone, with the acquisition of IncR plasmids by the latter, possibly driven by antibiotic selective pressure and availability of IncR in the European metagenome [14].
Another clonal line is composed of a R-ASSuT monophasic strains, with the deletion of the fljB gene and which was detected in other European countries [7,12,15,16,17]. Moreover, in the latter clone, the presence of IS26 in the same position as the fljAB operon in many monophasic strains has been reported, supporting the hypothesis that the insertion sequence IS26 recognizes a hotspot in the second-phase flagellar genomic region [16,18]. Furthermore, it has been seen that this area has a lower average GC content in comparison with the Salmonella core genome (45 vs. 52.2%), supporting the idea that it could be an integration hotspot for foreign DNA [18]. Even so, little is known about the preference of IS26 towards that hotspot located in the second-phase flagellar genomic region. In addition to these clones that have the entire fljB deletion, strains possessing a fljB coding sequence (fljB-positives) have also been described in several European countries in recent years [19,20,21]. The emergence of these strains has been the result of other deletions, insertions or mutations in the fljAB operon [21].
To date, next generation sequencing (NGS) techniques have not been used for genetic typing of the S. 4,[5],12:i:- strains isolated in Spain. The genetic study of the second-phase flagellar region of S. 4,[5],12:i:- strains will allow us to determine if the characterized monophasic strains belong to some of the clones described above, or if the emerging variations suggest multiple clonal lineages. Therefore, the aims of this study were to analyze the Salmonella 4,[5],12:i:- serotype by whole genome sequencing (WGS) and bioinformatic tools, to characterize the deletion of the second-phase flagellar genomic region and to establish the genetic relationship between S. 4,[5],12:i:- and S. Typhimurium strains.
Here, we used WGS data and various bioinformatic tools that allowed us to recapitulate many of the results of standard microbiological typing assays. Additionally, our findings suggest that the genetic events leading to the emergence of the S. 4,[5],12:i:- monophasic serotype involved several lineages and not the expansion of a single clone. This research shows how new S. 4,[5],12:i:- monophasic strains are emerging from S. Typhimurium with no close phylogenetic relationship. Furthermore, this research is a clear example of the usefulness of NGS techniques to carry out a complete characterization of S. 4,[5],12:i:- and it shows that these techniques can be useful for the monitoring of S. 4,[5],12:i:- strains circulating both in Spain and worldwide.
2. Materials and Methods
2.1. Isolate Collection
A total of 42 Salmonella enterica isolates collected from 1999 to 2015, from different matrixes and Spanish locations, were selected for this study (Table A1). The selection was made based on the different origins and characteristics studied in vitro with the aim of reflecting the genetic variations among the monophasic strains circulating in recent years in Spain. Briefly, the isolates were: (i) 13 S. 4,[5],12:i:- from unrelated gastroenteric infection cases; (ii) 4 S. 4,[5],12:i:- from pork sausages; (iii) 15 S. 4,[5],12:i:- of asymptomatic pigs, of which 13 were from the intestinal content (IC) and 2 from mesenteric lymph nodes (MLNs); and (iv) 10 S. Typhimurium, of which 9 were from MLNs and 1 was from the IC of asymptomatic pigs. All the isolates were provided with the serotyping determined by the Kauffmann–White scheme [1], the antimicrobial susceptibility determined by the Kirby–Bauer disc diffusion test [22] and phage type defined according to Anderson et al. [23].
2.2. Whole Genome Sequencing and in Silico Genotyping
Genomic DNA from the 42 isolates was extracted and purified using the NucleoSpin Tissue DNA purification kit (Macherey-Nagel, Duren, Germany), according to the manufacturer’s instructions. Sequencing libraries were prepared using the NexteraXT library preparation kit and WGS was performed on the Illumina MiSeq platform, generating 250 bp paired-end reads. The sequences were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the project accession number PRJEB37694. Raw reads were assembled into contigs using the INNUca pipeline (https://github.com/theInnuendoProject/INNUca), which consists of several modules [24]. Firstly, INNUca calculates whether the sample raw data fulfil the expected coverage (minimum 15X). Then, INNUca uses FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to perform a read quality analysis and Trimmomatic [25] to trim the reads. After subjecting reads to quality analysis using FastQC again, INNUca proceeds to de novo draft genome assembly with SPAdes [26]. Subsequently, coverage filtering is performed using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) and Samtools (http://www.htslib.org/doc/samtools.html). Next, Pilon [27] improves the draft genome, removing very poorly represented sequences, correcting bases, fixing misassemblies and filling gaps. Finally, the INNUca workflow ends with species confirmation and MLST prediction of seven genes using mlst2 (https://github.com/tseemann/mlst).
Serovar and antibiotic resistance prediction was performed using SeqSero2 [28] and ResFinder 4.0 (95% ID threshold, 60% minimum length) [29], available as a web service at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org), and was then compared with those provided by classical microbiology.
Pathogenicity island, plasmid and prophage sequence prediction was performed using SPIFinder 1.0 (95% ID threshold) (https://cge.cbs.dtu.dk/services/SPIFinder), PlasmidFinder 2.1 (95% ID threshold) [30] and PHASTER (90% ID threshold) [31], respectively. The IS26 insertion sequence was detected by in silico PCR simulation [32].
The core genome MLST profile was analyzed using cgMLSTFinder 1.1 (https://cge.cbs.dtu.dk/services/cgMLSTFinder/) based on the Enterobase scheme [33] consisting of a core of 3002 genes.
2.3. Characterization of the fljAB Operon Deletions by WGS
The presence or absence and the deletion ends of the fljAB operon in S. 4,[5],12:i:- were determined on contigs by in silico PCR simulation [32]. Salmonella Typhimurium LT2 strain (GeneBank accession number AE006468.2) was used as a reference. Conventional PCRs were performed to corroborate the in silico characterization of the fljAB operon deletions. All the detected insertions were identified and annotated using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
3. Results
3.1. Characterization of S. 4,[5],12:i:- and S. Typhimurium by WGS and Bioinformatic Tools
WGS-based genotyping results obtained by INNUca MLST, SeqSero2, ResFinder, PlasmidFinder, PHASTER, IS26 in silico PCR and cgMLSTFinder are shown in Table 1. Molecular serotyping by SeqSero2 software confirmed phenotypical serotype in 39/42 (92.86%) strains in the blind study (Table 2). The remaining 3 isolates were serotyped as S. Typhimurium (4,[5],12:i:1,2) by SeqSero2, while they were serotyped as S. 4,[5],12:i:- by agglutination serology, suggesting a lack of flagellar antigen expression in vitro in the serotyping conditions.
Table 1.
Genotyping results from the open source bioinformatic tools on the 42 Salmonella isolates sequenced by whole genome sequencing (WGS).
| Isolate Code | SeqSero2 | INNUca MLST | cgMLSTFinder | ResFinder profile | SPIFinder | PlasmidFinder | PHASTER | In Silico IS26 PCR |
|---|---|---|---|---|---|---|---|---|
| 692 | 4,[5],12:i:- | 19 | 123420 | S | SPI-1, SPI-3, SPI-5, SPI-13, C63PI | IncFIB(S), IncFII(S) | G2, EGF2, F2 | − |
| 693 | 4,[5],12:i:1,2 | 34 | 84985 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, HP2, Ephi20 | + |
| 694 | 4,[5],12:i:1,2 | 34 | 78574 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1 | + |
| 695 | 4,[5],12:i:- | 34 | 141108 | ST | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | S3, G1 | + | |
| 696 | 4,[5],12:i:1,2 | 34 | 21377 | SSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1, IncFII | S3, G1, HP2, SfI | + |
| 697 | 4,[5],12:i:- | 19 | 85377 | CSSuTTm | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncR | G2, EGF2 | + |
| 698 | 4,[5],12:i:- | 34 | 31310 | AST | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | S3, G1, HP2, SfI | + | |
| 699 | 4,[5],12:i:- | 34 | 3719 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1, Col156 | S3, G1, HP2, SfI | + |
| 701 | 4,[5],12:i:- | 34 | 141108 | ST | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, C63PI | S3, G1 | + | |
| 702 | 4,[5],12:i:- | 19 | 85377 | CSSuTTm | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncR | G2, EGF2 | + |
| 703 | 4,[5],12:i:- | 34 | 132646 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | G2, HP2 | + | |
| 704 | 4,[5],12:i:- | 34 | 132646 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | G1, HP2 | + |
| 705 | 4,[5],12:i:- | 34 | 6912 | ST | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | G1 | + | |
| 711 | 4,[5],12:i:- | 19 | 156249 | CSSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncI1, IncA/C2 | G1, HP2 | + |
| 712 | 4,[5],12:i:- | 19 | 156249 | CSSuTTmNxCip | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | Col(BS512), IncA/C2 | G1, G2, HP2, EST104 | + |
| 713 | 4,[5],12:i:- | 19 | 156249 | ACSSuTTm | SPI-1, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | Col(BS512), IncA/C2 | G1, EST104 | + |
| 714 | 4,[5],12:i:- | 19 | 156249 | ACSSuTTm | SPI-1, SPI-2, SPI-3, SPI-5, SPI-14, C63PI | Col(BS512), IncA/C2 | G1, G2, HP2, EST104 | + |
| 743 | 4,[5],12:i:- | 34 | 132646 | ASSu | SPI-1, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, G2, HP2 | + |
| 744 | 4,[5],12:i:- | 34 | 43443 | ASSu | SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, G2, HP2 | + |
| 745 | 4,[5],12:i:- | 34 | 165159 | ASSuT | SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, G2, HP2, SfI | + |
| 746 | 4,[5],12:i:- | 34 | 132646 | ASSu | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14 | IncQ1 | G1 | + |
| 747 | 4,[5],12:i:- | 34 | 100540 | ASSuT | SPI-1, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1, Col440l | G1, Ep460 | + |
| 748 | 4,[5],12:i:- | 34 | 26728 | S | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | G1, EfiAA91, SfII | + | |
| 749 | 4,[5],12:i:- | 34 | 26728 | ASSu | SPI-1, SPI-2, SPI-3, SPI-5, SPI-9, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, EfiAA91, SfI | + |
| 750 | 4,[5],12:i:- | 34 | 26728 | ASSu | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | G1, EfiAA91, E186, SfI | + |
| 751 | 4,[5],12:i:- | 34 | 29699 | CS | SPI-1, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncI1 | G1, E186, EfiAA91, SfII | + |
| 752 | 4,[5],12:i:- | 34 | 89891 | CS | SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncI1 | S3, G1, G2, E186, EfiAA91, ESfV | + |
| 753 | 4,[5],12:i:- | 34 | 144738 | SSu | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, G2, EST104, Epro147 | + |
| 754 | 4,[5],12:i:- | 34 | 89891 | S | SPI-1, SPI-3, SPI-5, SPI-13, SPI-14 | IncI1 | S3, G1, E186, EfiAA91, Ep460 | + |
| 755 | 4,[5],12:i:- | 34 | 84985 | ASSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | S3, G1, G2, HP2 | + |
| 757 | 4,[5],12:i:- | 34 | 29699 | CS | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncI1 | G1, G2 | + |
| 758 | 4,[5],12:i:- | 34 | 144738 | SSu | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncQ1 | G1, EST104, Epro147 | + |
| 739 | 4,[5],12:i:1,2 | 19 | 35732 | S | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | ColpVC, IncFIB(S), IncFII(S), IncX1 | S3, G1, G2 | − |
| 756 | 4,[5],12:i:1,2 | 19 | 45281 | ACSSuT | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncFIB(S), IncFII(S) | EST104 | − |
| 759 | 4,[5],12:i:1,2 | 19 | 78568 | ACSSuT | SPI-1, SPI-5, SPI-13, SPI-14, C63PI | IncX1, IncFII(S) | G1, G2, EST104 | + |
| 760 | 4,[5],12:i:1,2 | 19 | 20179 | ACSSuTNxCip | SPI-1, SPI-2, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncFII(S) | G1, G2, EST104, HP2 | + |
| 761 | 4,[5],12:i:1,2 | 19 | 35732 | S | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | ColpVC | S3, G1, G2 | − |
| 767 | 4,[5],12:i:1,2 | 19 | 45281 | ACSSuT | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncFIB(S), IncFII(S) | G1, G2, EST104 | − |
| 773 | 4,[5],12:i:1,2 | 19 | 45281 | ACSSuT | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncFIB(S), IncFII(S) | S3, G1, G2, EST104 | − |
| 775 | 4,[5],12:i:1,2 | 19 | 45281 | ACSSuT | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | IncFIB(S), IncFII(S) | S3, G1, G2, EST104 | − |
| 778 | 4,[5],12:i:1,2 | 19 | 35732 | S | SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-13, SPI-14, C63PI | ColpVC, IncFIB(S), IncFII(S), IncX1 | G1, G2 | − |
| 779 | 4,[5],12:i:1,2 | 34 | 95263 | ASSuTCol | SPI-1, SPI-3, SPI-5, SPI-13, SPI-14, C63PI | IncQ1, IncHI2, IncHI2A | S3, G1 | + |
N/A: The predicted antigenic profile does not exist in the White–Kauffmann–LE Minor Scheme; A: Amoxyciline (beta-lactamic); C: Chloramphenicol (phenicol); S: Streptomycin (aminoglycoside); Su: Sulphonamide; T: Tetracycline; Tm: Trimethoprim; Nx: Nalidixic acid (quinolone); Cip: Ciprofloxacin (fluoroquinolone); Col: Colistin (polymyxine); EGF2: Edward_GF2; E186: Entero_186; EST104: Entero_ST104; EfiAA91: Entero_fiAA91; Ep460: Entero_mEp460; ESfV: Entero_SfV; Ephi20: Entero_phi20; Epro147: Escher_pro147; F2: Fels_2; G1: Gifsy1; G2: Gifsy2; HP2: Haemoph_HP2; S3: Sal3; SfI: Shigel_SfI; SfII: Shigel_SfII; +: amplified; -: not amplified.
Table 2.
Comparative results between SeqSero2 and classical serotyping, and between ResFinder genes and classical antimicrobial susceptibility tests.
| Isolate Code | SeqSero2 | Serotyping | ResFinder | Antibiogram Resistance Profile | |
|---|---|---|---|---|---|
| Antibiotic Resistance Genes and Mutations | Profile | ||||
| 692 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6′)-laa | S | Susceptible |
| 693 | 4,[5],12:i:1,2 | 4,[5],12:i:- | blaTEM-1B; aac(6’)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASISuT |
| 694 | 4,[5],12:i:1,2 | 4,[5],12:i:- | blaTEM-1B; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 695 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6´)-laa; tet(B) | ST | T |
| 696 | 4,[5],12:i:1,2 | 4,[5],12:i:- | aac(6´)-laa; aph(3″)-lb; Sul2; tet(B) | SSuT | SSuT |
| 697 | 4,[5],12:i:- | 4,[5],12:i:- | cmlA1; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; aadA1; aadA2; Sul3; dfrA12; tet(B) | CSSuTTm | T |
| 698 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6´)-laa; tet(B) | AST | AT |
| 699 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 701 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6´)-laa; tet(B) | ST | SIT |
| 702 | 4,[5],12:i:- | 4,[5],12:i:- | cmlA1; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; aadA1; aadA2; Sul3; dfrA12; tet(B) | CSSuTTm | SISuT |
| 703 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 704 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6´)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 705 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6´)-laa; tet(B) | ST | ASSuT |
| 711 | 4,[5],12:i:- | 4,[5],12:i:- | cmlA1; aac(6′)-Iaa; aadA8b; Sul1; Sul2; Sul3; tet(A) | CSSuT | T |
| 712 | 4,[5],12:i:- | 4,[5],12:i:- | cmlA1; aac(6′)-Iaa; Sul1; Sul2; Sul3; dfrA12; tet(A); gyrB p.E466D mutation | CSSuTTmNxCip | SuT |
| 713 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; cmlA1; aac(3)-IV; aac(6′)-Iaa; Sul1; Sul2; Sul3; dfrA12; tet(A) | ACSSuTTm | ASSuT |
| 714 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; cmlA1; aac(3)-IV; aac(6′)-Iaa; Sul1; Sul2; Sul3; dfrA12; tet(A) | ACSSuTTm | ASSuT |
| 743 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | ASSu | ASSu |
| 744 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | ASSu | ASSu |
| 745 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 746 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | ASSu | ASSu |
| 747 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 748 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6′)-laa | S | Susceptible |
| 749 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | ASSu | ASSu |
| 750 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | ASSu | A |
| 751 | 4,[5],12:i:- | 4,[5],12:i:- | floR; aac(3)-IV; aac(6′)-laa; aph(3″)-lb; aph(4)-la; aph(6)-ld | CS | ACS |
| 752 | 4,[5],12:i:- | 4,[5],12:i:- | floR; aac(3)-IV; aac(6′)-laa; aph(3″)-lb; aph(4)-la; aph(6)-ld | CS | ACSSu |
| 753 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | SSu | ACSISuT |
| 754 | 4,[5],12:i:- | 4,[5],12:i:- | aac(3)-IV; aac(6′)-laa; aph(3″)-lb; aph(4)-la; aph(6)-ld | S | AS |
| 755 | 4,[5],12:i:- | 4,[5],12:i:- | blaTEM-1B; aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2; tet(B) | ASSuT | ASSuT |
| 757 | 4,[5],12:i:- | 4,[5],12:i:- | floR; aac(3)-IV; aac(6′)-laa; aph(3″)-lb; aph(4)-la; aph(6)-ld | CS | CS |
| 758 | 4,[5],12:i:- | 4,[5],12:i:- | aac(6′)-laa; aph(3″)-lb; aph(6)-ld; Sul2 | SSu | SSu |
| 739 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | aac(6′)-laa | S | Susceptible |
| 756 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaCARB-2; floR; aac(6′)-laa; aadA2; Sul1; tet(G) | ACSSuT | ACSSuT |
| 759 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaOXA-1; catA1; aac(6′)-laa; aadA1; Sul1; tet(B) | ACSSuT | ACSSuT |
| 760 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaOXA-1; catA1; aac(6′)-laa; aadA1; Sul1; tet(B); gyrA p. D87N mutation | ACSSuTNxCip | ACSSuTNx |
| 761 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | aac(6′)-laa | S | Susceptible |
| 767 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaCARB-2; floR; aac(6′)-laa; aadA2; Sul1; tet(G) | ACSSuT | ACSSuT |
| 773 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaCARB-2; floR; aac(6′)-laa; aadA2; Sul1; tet(G) | ACSSuT | ACSSuT |
| 775 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaCARB-2; floR; aac(6′)-laa; aadA2; Sul1; tet(G) | ACSSuT | ACSSuT |
| 778 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | aac(6′)-laa | S | Susceptible |
| 779 | 4,[5],12:i:1,2 | 4,[5],12:i:1,2 | blaTEM-1B; blaCTX-M-9; aac(6′)-laa; aadA2; aph(3″)-lb; aph(6)-ld; Sul1; Sul2; dfrA16; tet(A); mcr-9 | ASSuTCol | ASSuTNxCfx |
N/A: The predicted antigenic profile does not exist in the White–Kauffmann–LE Minor Scheme; A: Amoxyciline (beta-lactamic); C: Chloramphenicol (phenicol); S: Streptomycin (aminoglycoside); Su: Sulphonamide; T: Tetracycline; Tm: Trimethoprim; Nx: Nalidixic acid (quinolone); Cip: Ciprofloxacin (fluoroquinolone); Col: Colistin (polymyxine); Cfx: Cefotaxime (third generation cephalosporin); SI: Intermediate susceptibility to streptomycin.
ResFinder allowed the detection of at least one antibiotic resistance gene in all S. 4,[5],12:i:- and S. Typhimurium strains (Table 2). The aminoglycoside family genes (i.e., aac, aad and/or aph) were the most common, with the cryptic gene aac(6′)-laa being found in all the isolates studied (Table 2). In addition, 14 different genotypic antimicrobial profiles were determined and 66.67% of the strains showed resistance genes to at least 3 antibiotics. The most frequent genotypic multi-resistance profile was ASSuT (19.05%). Interestingly, all the strains with this tetra-resistance profile were S. 4,[5],12:i:-, coinciding with the reported European monophasic clone. ResFinder correctly identified 92.12% (117/127) of the antibiotic resistances found phenotypically. Comparing with classical antibiograms, ResFinder detected the same antibiotic resistances or more in 85.71% (36/42) of the isolates. In the remaining 6 isolates, ResFinder detected fewer antibiotic resistances than in classical antibiograms.
SPIFinder found 8 different Salmonella pathogenicity islands (SPI-1, SPI-2, SPI-3, SPI-4, SPI-5, SPI-9, SPI-13 and SPI-14) and the Centisome 63 pathogenicity island (C63PI) (Table 1). SPI-5, which encodes the effector proteins for SPI-1 and SPI-2, was the only common pathogenicity island detected in all strains. SPI-1 and/or SPI-2 were detected together with SPI-5 in 95.24% of the strains. In two strains, neither SPI-1 nor SPI-2 were detected, even when SPI-5 was present. SPI-9, which encodes a type I secretory apparatus and a large repeats-in-toxin (RTX)-like protein, was detected in only one monophasic strain (code 749) from swine intestinal content. In addition, 73.81% of the strains showed at least 7 different pathogenicity islands.
PlasmidFinder detected plasmid replicons in most (85.71%) of the strains (Table 1). The replicon most frequently identified among monophasic strains was IncQ1 (46.88%). In contrast, among S. Typhimurium strains, the replicon most frequently identified was IncFII(S) (70.00%). All the genomes analyzed showed at least one prophage sequence by PHASTER analysis (Table 1). A total of 15 different prophage sequences were found, Gifsy1 (88.10%), Gifsy2 (50.00%) and Sal3 (47.62%) being the most frequent (Table 1). In silico PCR simulation detected the presence of the insertion sequence IS26 in 96.87% of the monophasic strains and in 30.00% of S. Typhimurium strains (Table 1).
Using a traditional Salmonella MLST scheme, formed by 7 housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA and thrA), the isolates were classified into 2 different sequence types (Table 1). On the one hand, most of the S. 4,[5],12:i:- (78.13%) were classified in ST-34 and the rest (21.87%) in ST-19. On the other hand, 90.00% of the S. Typhimurium strains were classified in ST-19 and the other 10.00% in ST-34. In contrast, by the cgMLST scheme consisting of 3002 genes, the isolates were classified into 23 different sequence types. None of the S. 4,[5],12:i:- strains shared the same cgMLST type with the S. Typhimurium strains, even though they were strains isolated from the same pig farm.
3.2. Characterization of the fljAB Operon Deletion Types by WGS
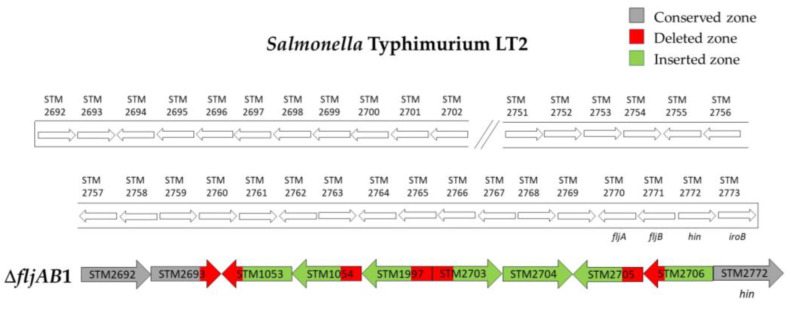
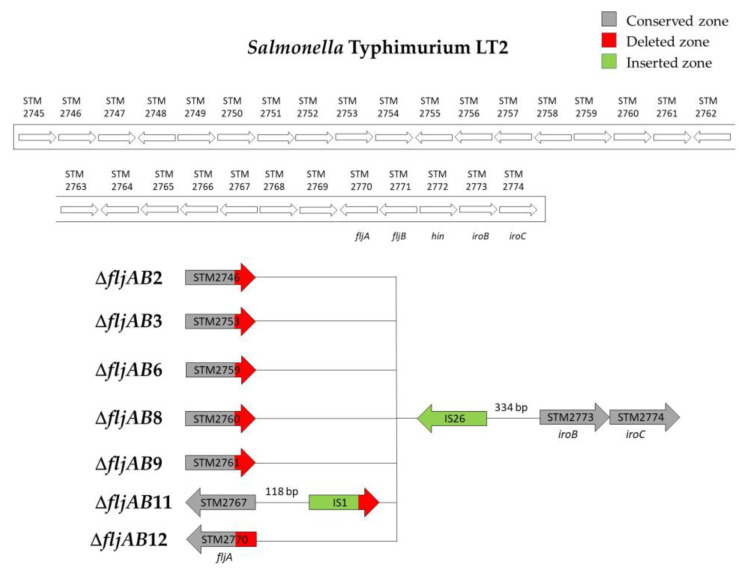
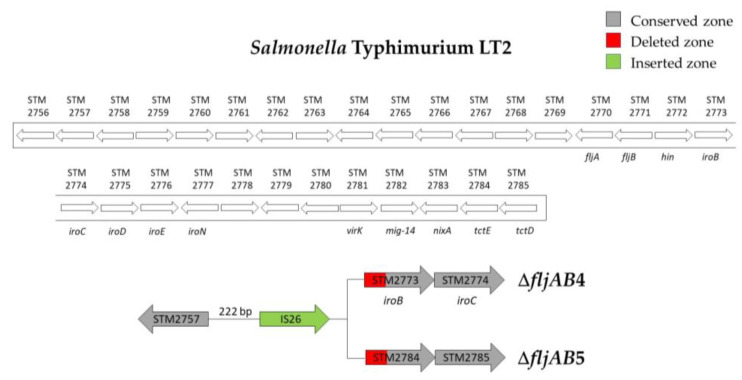
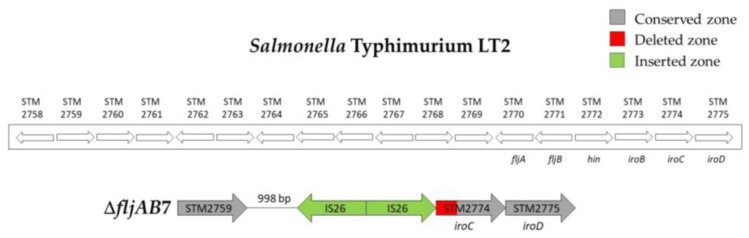
As mentioned above, the fljAB operon (fljA, fljB and hin genes) and the flanking genes from the S. Typhimurium LT2 genome were used as a reference to characterize the deletions. The area searched began at STM2693 and ended at STM2774. Thirteen different deletion types were characterized (Table 3 and Figure A1, Figure A2, Figure A3, Figure A4, Figure A5 and Figure A6). Twelve fljB-negative deletion types (ΔfljAB1-12) were sorted according to the deletion length (ΔfljAB1 being the longest and ΔfljAB12 the shortest). One fljB-positive deletion type was detected with different variations included (ΔfljAB13).
Table 3.
Description of the fljAB operon deletion types of the 32 S. 4,[5],12:i:-analyzed. In fljB-negative types (ΔfljAB1–12) the starting and ending points of the deletions have been specified.
| Deletion Type | Subtype | No. of Strains | Starting Point | Ending Point | Inserted Fragment † |
|---|---|---|---|---|---|
| ΔfljAB1 | 4 | 98 bp of STM2693 | 10 bp downstream of STM2771 (fljB) | 5654 bp (see Figure A1) | |
| ΔfljAB2 | ΔfljAB2-A | 1 | 1201 bp of STM2746 | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) |
| ΔfljAB2-B | 1 | 1263 bp of STM2746 | |||
| ΔfljAB3 | ΔfljAB3-A | 1 | 177 of STM2753 | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) |
| ΔfljAB3-B | 1 | 207 bp of STM2753 | |||
| ΔfljAB3-C | 1 | 353 bp of STM2753 | |||
| ΔfljAB4 | 3 | 222 bp downstream of STM2757 | 571 bp of STM2773 (iroB) | 820 bp (one IS26) | |
| ΔfljAB5 | 1 | 222 bp downstream of STM2757 | 848 bp of the STM2784 | 820 bp (one IS26) | |
| ΔfljAB6 | ΔfljAB6-A | 1 | 1079 bp of STM2759 | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) |
| ΔfljAB6-B | 2 | 142 bp downstream of the STM2759 | |||
| ΔfljAB7 | 1 | 998 bp downstream of STM2759 | 475 bp of STM2774 (iroC) | 1640 bp (two IS26) | |
| ΔfljAB8 | 7 | 88 bp of STM2760 | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) | |
| ΔfljAB9 | 1 | 175 bp of the STM2761 | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) | |
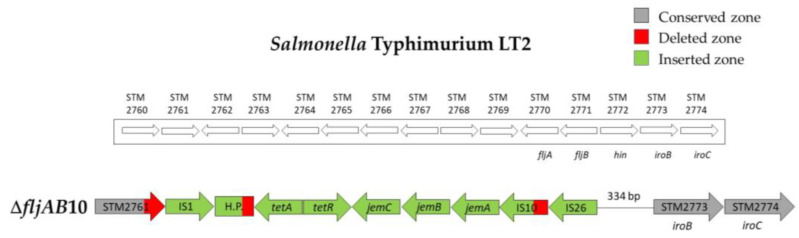
| ΔfljAB10 | 2 | 1125 bp of the STM2761 | 334 bp upstream from STM2773 (iroB) | 7648 bp (see Figure A5) | |
| ΔfljAB11 | 1 | 118 bp downstream of STM2767 | 334 bp upstream from STM2773 (iroB) | 1455 bp (see Figure A2) | |
| ΔfljAB12 | 1 | 155 bp of the fljA gene | 334 bp upstream from STM2773 (iroB) | 820 bp (one IS26) | |
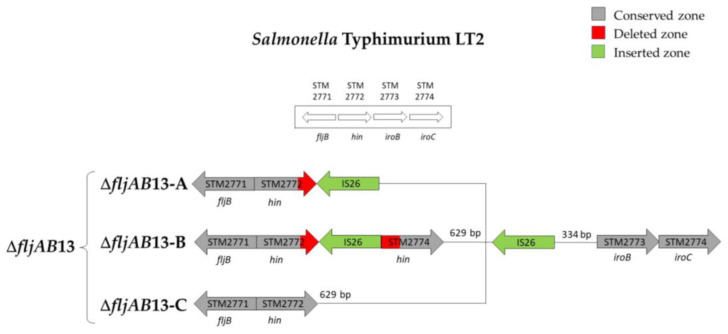
| ΔfljAB13 | ΔfljAB13-A | 1 | fljB-positive | 1640 bp (two IS26) | |
| ΔfljAB13-B | 1 | ||||
| ΔfljAB13-C | 1 | 820 bp (one IS26) | |||
The ΔfljAB1 type was the longest deletion (77 genes) and showed an insertion of 5654 bp, which contained different fragments encoding for Gyfsy-2 prophage proteins, UMUC protein and two fragments encoding for three Fels-2 prophage proteins (Figure A1). The deletion types ΔfljAB2-6, ΔfljAB8-9 and ΔfljAB12 varied in the length of their deletions, and they were all characterized by the insertion of one IS26 copy (Figure A2 and Figure A3). In the deletion ΔfljAB7, two IS26 copies were inserted (Figure A4). The deletion ΔfljAB10 had the largest insertion (7663 bp), which consisted of three insertion sequences (IS1, IS10 and IS26) and other additional genes, including tetracycline resistance genes (Figure A5). The main characteristic of ΔfljAB11 was the insertion of a truncated IS1 and a complete IS26 copy (Figure A2). Finally, in the deletion type ΔfljAB13, three variants of fljB-positive strains were included. These strains showed an insertion of one or two copies of IS26, generating a partial deletion, an interruption of the hin gene or were located after it (Figure A6). The deletion types belonging to ΔfljAB2-3, ΔfljAB6 and ΔfljAB8-13 had the IS26 copy, in the 3′-5′ direction, inserted in the same nucleotide of the intergenic region between the hin and iroB genes (the same ending point). However, the ΔfljAB4 and ΔfljAB5 deletions had the IS26 copy, in the 5′-3′ direction, inserted 222 bp downstream of the STM2757 gene (the same starting point).
4. Discussion
The emergence of the 4,[5],12:i:- monophasic variant of Salmonella Typhimurium demonstrates its evolutionary success. It has rapidly become one of the most prevalent serovars in humans in numerous countries worldwide [34]. The loss of the second-phase flagella has not prevented the emergence and worldwide spread of S. 4,[5],12:i:- monophasic variant strains. Flagella (H antigen) on the surface of S. Typhimurium had been characterized as a virulence factor that helps the bacteria move toward and adhere to host cells. However, Lockman and Curtiss [35] concluded that independent Tn10 insertions that were mapped to different flagellar genes did not affect the virulence of S. Typhimurium for mice and suggested that motility might be irrelevant as a virulence factor for an invasive, facultative intracellular pathogen.
Classification of Salmonella by serotyping is generally performed by accredited National Reference Centers, as an essential epidemiological tool. In case of S. 4,[5],12:i:-, this procedure is crucial, since the non-expression or non-detection of the first and second flagellar antigens leads to the erroneous typing of S. Typhimurium as its monophasic variant. To solve this problem, the bacteria should be sequentially subcultivated for a new serotyping and, in case of negative results, an additional multiplex PCR should be completed [5]. Since this method is highly time-consuming and entails an unnecessary manipulation of the pathogen, multiplex PCR is routinely used. This PCR amplifies the fljB-fljA intergenic region of the flagellin gene cluster [5] but is unable to differentiate the monophasic fljB-positive variant from S. Typhimurium [36]. In this study, we found that WGS can prevent the amplification of the long-inserted fragments in the fljAB operon. Since most of the monophasic variant studies seldomly search for the deletion of the second flagellar phase, a complete strain characterization often requires applying multiple techniques. However, as we have verified through this work, NGS technologies allow a complete characterization of Salmonella strains within a few days. As an alternative to WGS, in 2018, a liquid bead array was proposed for the identification and characterization of S. 4,[5],12:i:- variants to achieve results in a rapid and simple data analysis [36]. In this work, we demonstrate that WGS can also be a rapid method that enhances traditional profiling efforts for the characterization of the monophasic variant, including the prediction of clinically relevant phenotypic traits such as antibiotic resistance genes, plasmids or virulence genes. On the other hand, the specific matrix of bead arrays only allows us to discriminate between S. Typhimurium and S. 4,[5],12:i:-, whilst we observed that WGS allows a detailed characterization of the second flagellar phase genetic deletions involved in this serovar.
To achieve full molecular description of strains, the development of efficient, standardized and molecular-guided laboratory surveillance is necessary and a high priority [37]. As presented in this work, WGS and freely available web services and bioinformatic tools can be extremely useful for public health laboratories and epidemiological surveillance. For instance, the bioinformatic tools used in this work allowed the achievement of the complete typing of S. 4,[5],12:i:-. Regarding the serotype, SeqSero2 has been considered more reliable for the serological prediction of the monophasic variant of S. Typhimurium, compared to other tools such as MOST and SISTR [38]. Even so, our results indicate that SeqSero2 does not correctly predict the serotype of S. 4,[5],12:i:- fljB-positive strains. In our study, S. 4,[5],12:i:- fljB-negative strains were classified 100% correctly by SeqSero2, but all fljB-positive monophasic strains were erroneously classified as S. Typhimurium. These results were verified by complete characterization of the fljAB operon based on genome assemblies. Similar results have been reported in other studies when assessing the serotyping of S. 4,[5],12:i:- strains using this program [39,40]. This limitation of SeqSero2 may be crucial for public health laboratories since S. Typhimurium and S. 4,[5],12:i:- are the most frequent human clinical infections after S. Enteritidis [2]. For this reason, as suggested by the comparative study done by Banerji et al. [39], we find it necessary to include additional factors to the fljB gene that determine the integrity of the second-phase flagellar antigen to detect S. 4,[5],12:i:- fljB-positive strains.
The in silico typing done in this work showed that, although WGS analysis seems expensive and complex, bioinformatic tools have transformed it into a cost-effective tool. Furthermore, large amounts of time and materials would be required if all typing had to be done through classic microbiology. To ensure the success of in silico typing, it is essential to generate high-quality contigs, which in turn requires evaluating sequence quality and the existence of possible technical errors by establishing quality control measures. Assessing genome assembly quality is significant in this process because poor-quality assemblies hamper downstream analyses, resulting in incorrect interpretations [37]. As such, it is critical to identify, evaluate and minimize technical errors occurring during sample isolation, DNA preparation sequencing and genome assembly.
Regardless of the epidemiological information, the study of the second flagellar phase deletions could provide further insight into their origin, the genetic events yielding them and the characterization of the inserted fragment. Through WGS carried out in this work, 13 different deletion types and subtypes of the second-phase flagellar genomic region were found in 32 S. 4,[5],12:i:- strains. Genetic diversity observed in the deletion types and in the in silico typing achieved by bioinformatic tools (ResFinder, SPIFinder, PlasmidFinder, PHASTER, in silico PCR and cgMLSTFinder) prove that the selection of strains analyzed in this work is a representation of non-clonal monophasic strains circulating in Spain.
The deletion ΔfljAB1, where 77 genes were absent compared to the genome of S. Typhimurium LT2 strain, showed an insertion of 5654 bp. The sequence of this insertion is very similar to the insertion described by Soyer et al. in the US strains [10], although some differences could be detected, namely, a 5654 bp fragment inserted instead of a 7 kb fragment, and the presence of the complete STM2704 gene, which is partially deleted on US strains. Of the total number of strains analyzed, four strains of clinical origin had this deletion; nevertheless, they varied in phenotypic characteristics. Strain 692 had the aminoglycoside resistance gene aac (6′)-laa that is a cryptic gene in Salmonella and IncFIB(S) and IncFII(S) plasmids. In strain 705, the cryptic gene aac(6′)-laa and the tetracycline resistance gene tet(B) were detected but no plasmids. Given the information provided by WGS, we considered that these strains could belong to the same lineage as the American strains described by Soyer et al. in the US. However, the other two strains with this same deletion (697 and 702) had these characteristics: cmlA, aac(6′)–laa, aph(3″)-lb, aph(6)-ld, aadA1, aadA2, Sul3, tet(B) and dfrA12 resistance genes (CSSuTTm multi-resistance profile) and the presence of IncR plasmids. Interestingly, these two strains have the same characteristics as the Southern Europe clone described by Mourão et al. [14]. The results suggest that the strains 692 and 705 of the American clonal line are the ancestors of the Southern Europe clone in Spain. Moreover, the strains 697 and 702 with the American deletion, IncR plasmids and CSSuTTm multi-resistance profile are the representation of the Southern Europe clone in Spain, a result of the acquisition of IncR plasmids.
In 2016, Garcia et al. described an S. 4,[5],12:i:- clonal lineage widespread in Germany, Switzerland and Italy, carrying a ASSuT tetra-resistance induced by IncH1 plasmids, which replaced the second-phase flagellar genomic region [18]. We found eight strains S. 4,[5],12:i:- with an ASSuT tetra-resistance but none of these strains had the multidrug resistance plasmid described by Garcia et al. In six of these strains, only one or two copies of IS26 were detected in the second-phase flagellar genomic region (deletions ΔfljAB6, ΔfljAB7, ΔfljAB9, ΔfljAB12 and ΔfljAB13) and the remaining two S. 4,[5],12:i:- ASSuT strains were classified as ΔfljAB10 deletion type, showing a 7663 bp fragment between STM2761 and iroB genes. This fragment was composed of IS1, IS26 and a truncated IS10, tetA and tetR genes, which are implicated in tetracycline resistance, as well as of genes that codified a hypothetical protein or JemC, JemB and JemA products. Similar genetic composition has been described on an STM plasmid (pSRC27-H) and in the S. Typhimurium genome (T000240 strain). The putative roles of these insertion sequences would be the following: IS1 would drag the genes that appear on the T000240 strain; IS10, possibly located on the pSRC27-H plasmid, would be inserted by recognizing the tetA, tetR and jemC genes, and thus partially deleting the hypothetical protein of the T000240 strain; lastly, IS26 would interrupt IS10.
To date, two different models have been proposed to explain why the insertion sequence IS26 generates deletions in the second-phase flagellar region of S. 4,[5],12:i:- strains. On the one hand, it is suggested that most of the fljB-negative S. 4,[5],12:i:- strains observed globally could have emerged from a common ancestor containing an IS26 copy at that specific position [41]. On the other hand, it is theorized that several independent insertions may have occurred in different genetic events where an IS26 copy recognizes a hotspot [16,18]. In our study, S. 4,[5],12:i:- strains isolated in Spain containing at least one IS26 copy in the second-phase flagellar deletion have shown a genetic variability both in the region adjacent to the 3′-end of IS26 and in the genotyping based on WGS (presence of resistance genes, pathogenicity islands, plasmids, prophages and cgMLST). The great diversity found shows that S. 4,[5],12:i:- strains isolated in Spain and containing an IS26 copy do not belong to a single clone.
In this research, 85.71% of strains containing an IS26 in the second flagellar phase deletion had this insertion sequence in the 3′-5′ direction. In these deletions, the region adjacent to the 3′-end of IS26 (deletion starting point) varied between strains, while the region adjacent to the 5′-end (deletion ending point) was conserved. The IS26 insertion was found in the same position (i.e., 334 nucleotides upstream from the iroB gene) in fljB-negative strains isolated in the USA, South Korea and some European countries [18,36]. It is noteworthy that three strains of our work were fljB-positives (i.e., ΔfljAB13), even though they had IS26 inserted. The remaining 14.29% of strains had an IS26 in the 5′- 3′ direction and they all had the same deletion starting point. Interestingly, in a previous work with 60 strains of the S. 4,[5],12:i:- Spanish clone, 93.60% of the isolates shared the same starting point but different deletion ending points [9]. All these deletions had an IS26 inserted in the 5′-3′ direction in nucleotide no. 1444 of the gene STM2758 [9], very close to the starting point of the ΔfljAB4 and ΔfljAB5 deletions. WGS results show that the 5′-end of IS26 generates conserved deletion ends whilst creating significant variability in the genomic region adjacent to the 3′-end of the IS26 insertions. This finding suggests that the IS26 insertion sequence has a recognition end at 5′ that can be inserted in certain areas depending on its direction. Furthermore, we propose that S. 4,[5],12:i:- strains are evolving from different S. Typhimurium strains with no close phylogenetic relationship through different genetic events in which at least one IS26 was involved, promoting a pool of monophasic variants that share a similar deletion due to the IS26 5′-end recognition.
Several research groups further characterized S. 4,[5],12:i:- strains, reporting the existence of non-clonal S. 4,[5],12:i:- circulating in other European countries, such as Belgium, Italy, France and Poland [19,20,42]. However, other studies have observed S. 4,[5],12:i:- clonal lineages in the United Kingdom, Italy, Germany and Switzerland [7,16,18]. The detection of clonal and non-clonal strains may depend on the objectives of the research carried out in the above works. Although greatly expanded S. 4,[5],12:i:- clonal lines have been reported worldwide, it is possible that in the future, new S. 4,[5],12:i:- strains will be detected with different deletions of the second-phase flagellar genomic region. New S. 4,[5],12:i:- strains will be generated from S. Typhimurium strains in different genetic events, especially genomic rearrangements mediated by IS26. Pigs have been the main animal reservoir for S. 4,[5],12:i:- for years [34,43]. Therefore, it can be deduced that the genetic events causing the deletion of the second flagellar phase of the Salmonella strains analyzed in our study probably occurred within pigs. In fact, S. 4,[5],12:i:- was reported amongst the three most frequent serotypes in pigs in 2017, together with S. Typhimurium and S. Derby [34], in agreement with previous European guidelines [5]. Even so, in 2018, S. 4,[5],12:i:- strains causing human salmonellosis were associated mainly with pig (39.6%) and broiler (43.4%) sources [2]. This indicates that there has been a considerable expansion of S. 4,[5],12:i:- colonization of other animal niches. Based on these data, in our opinion, the emergence in the coming years of new S. 4,[5],12:i:- strains carrying deletions of the second flagellar phase should be expected from pig products and cannot be neglected from other animal sources.
5. Conclusions
In conclusion, our study demonstrates that the availability of sequencing technologies and the development of bioinformatic tools turn NGS into a realistic alternative to traditional methods for the characterization of S. 4,[5],12:i:- strains. In addition, these tools were essential to study the genetic bases of the monophasic phenotype and to identify S. 4,[5],12:i:- American clonal line strains in Spain that would give rise to the Southern Europe clone due to the acquisition of the IncR plasmid. Therefore, these tools were useful in determining the implication of the insertion sequence IS26 when generating new deletions and to establish the genetic link between S. 4,[5],12:i:- and S. Typhimurium strains. The results obtained in our study suggest that Salmonella monophasic variants are evolving from different S. Typhimurium strains through independent genetic events that may have taken place in swine. Within these genetic events, at least one IS26 was inserted whose 5′-end recognized a hotspot in the second-phase flagellar genomic region and generated conserved deletion ends. Finally, we consider that the genetic diversity observed in S. 4,[5],12:i:- strains analyzed in this study proves that non-clonal monophasic strains are circulating in Spain. Further studies are needed to analyze the recognition mechanism of the insertion sequence IS26 in the second-phase flagellar genomic region of S. 4,[5],12:i:-. This finding would help in the understanding of the mechanism by which new S. 4,[5],12:i:- strains are continuing to emerge.
Acknowledgments
We thank the National Centre for Microbiology, Majadahonda-Madrid, Spain, the National Centre for Animal Salmonellosis, Algete-Madrid, Spain, and the Public Health Laboratory, Zamudio-Vizcaya, Spain for their kind collaboration on Salmonella typing. Additionally, we thank SGIKER service (UPV/EHU), Spain for their technical sequencing procedure. We also acknowledge John Wild for his kind contribution to reviewing the English grammar.
Appendix A
Table A1.
Salmonella isolates selected to this work.
| Isolate Code | Year | Source | Origin | Serovar | Resistance Profile | Phage Type |
|---|---|---|---|---|---|---|
| 692 | 2008 | Human feces | NCM | 4,[5],12:i:- | Susceptible | 104b |
| 693 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASISuT | 193 |
| 694 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASSuT | 195 |
| 695 | 2008 | Human feces | NCM | 4,[5],12:i:- | T | 138 |
| 696 | 2008 | Human feces | NCM | 4,[5],12:i:- | SSuT | NRP |
| 697 | 2008 | Human feces | NCM | 4,[5],12:i:- | T | NRP |
| 698 | 2008 | Human feces | NCM | 4,[5],12:i:- | AT | 104b |
| 699 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASSuT | 193 |
| 701 | 2008 | Human feces | NCM | 4,[5],12:i:- | SIT | 138 |
| 702 | 2008 | Human feces | NCM | 4,[5],12:i:- | SISuT | NRP |
| 703 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASSuT | 7 |
| 704 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASSuT | NRP |
| 705 | 2008 | Human feces | NCM | 4,[5],12:i:- | ASSuT | NRP |
| 711 | 1999 | Chicken sausage | PHL | 4,[5],12:i:- | T | ND |
| 712 | 2000 | Chicken sausage | PHL | 4,[5],12:i:- | SuT | ND |
| 713 | 2000 | Chicken sausage | PHL | 4,[5],12:i:- | ASSuT | ND |
| 714 | 2000 | Chicken sausage | PHL | 4,[5],12:i:- | ASSuT | ND |
| 743 | 2012 | Swine IC | IdAB | 4,[5],12:i:- | ASSu | U311 |
| 744 | 2012 | Swine MLN | IdAB | 4,[5],12:i:- | ASSu | ND |
| 745 | 2012 | Swine MLN | IdAB | 4,[5],12:i:- | ASSuT | ND |
| 746 | 2012 | Swine IC | IdAB | 4,[5],12:i:- | ASSu | ND |
| 747 | 2012 | Swine IC | IdAB | 4,[5],12:i:- | ASSuT | ND |
| 748 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | Susceptible | ND |
| 749 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | ASSu | ND |
| 750 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | A | ND |
| 751 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | ACS | ND |
| 752 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | ACSSu | ND |
| 753 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | ACSISuT | ND |
| 754 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | AS | ND |
| 755 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | ASSuT | ND |
| 757 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | CS | ND |
| 758 | 2015 | Swine IC | IdAB | 4,[5],12:i:- | SSu | ND |
| 739 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | Susceptible | NRP |
| 756 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ACSSuT | ND |
| 759 | 2012 | Swine IC | IdAB | 4,[5],12:i:1,2 | ACSSuT | ND |
| 760 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ACSSuTNx | 104b |
| 761 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | Susceptible | NRP |
| 767 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ACSSuT | U302 |
| 773 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ACSSuT | 104b |
| 775 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ACSSuT | 104b |
| 778 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | Susceptible | ND |
| 779 | 2012 | Swine MLN | IdAB | 4,[5],12:i:1,2 | ASSuTNxCfx | ND |
NCM: Spanish National Centre for Microbiology (Majadahonda, Spain); PHL: Public Health Laboratory (Zamudio, Spain); IdAB: Institute of Agrobiotechnology (Navarra, Spain); IC: Intestinal content; MLN: Mesenteric lymph nodes; A: Amoxyciline (beta-lactamic); C: Chloramphenicol (phenicol); S: Streptomycin (aminoglycoside); Su: Sulphonamide; T: Tetracycline; Nx: Nalidixic acid (quinolone); Cfx: Cefotaxime (third generation cephalosporin); SI: Intermediate susceptibility to streptomycin; NRP: Non-recognizable pattern; ND: Not determined.
Figure A1.
Structure of the ΔfljAB1 deletion type. The ΔfljAB1 deletion (strains 692, 697, 702 and 705) started at the STM2693 gene, where only the last two nucleotides were lacking, and ended at the intergenic region between fljB and hin, specifically 10 nucleotides downstream of the fljB gene. Sequencing showed an insertion of 5654 bp (colored in green) which entirely matched with the Salmonella Typhimurium 08-1736 strain (GeneBank accession number CP006602), containing different fragments encoding for Gyfsy-2 prophage proteins (841 bp of STM1053 and STM1054 genes), UMUC protein (540 bp of STM1997 gene) and two fragments encoding for three Fels-2 prophage proteins (777 bp from STM2703 to STM2705, and 292 bp of STM2706 gene).
Figure A2.
Structures of the ΔfljAB2, ΔfljAB3, ΔfljAB6, ΔfljAB8, ΔfljAB9, ΔfljAB11 and ΔfljAB12 deletion types. These deletion types had different starting points, but the same ending point of the deletion at the intergenic zone between hin and iroB genes (334 nucleotides upstream of iroB) and at least one IS26 inserted (colored in green). The ΔfljAB2 deletion started at the STM2746 gene. In the variant ΔfljAB2-A (strain 753), the deletion started at nucleotide 1201 (the last 87 nucleotides of STM2746 were deleted), and the variant ΔfljAB2-B (strain 758) at nucleotide 1263 (the last 25 nucleotides of STM2746 were deleted). The ΔfljAB3 deletion started at the STM2753 gene. In the variant ΔfljAB3-A (strain 743), the deletion began at nucleotide 177 (the last 844 nucleotides of STM2753 were deleted), the variant ΔfljAB3-B (strain 746) began at nucleotide 207 (the last 814 nucleotides of STM2753 were deleted) and the variant ΔfljAB3-C (strain 744) began at nucleotide 353 (the last 668 nucleotides of STM2753 were deleted). The ΔfljAB6 deletion started at the STM2759 gene. In the variant ΔfljAB6-A (strain 695), the deletion started at nucleotide 1079 (the last 179 nucleotides of STM2759 were deleted) and in the variant ΔfljAB6-B (strains 698 and 699), the deletion started 142 bp downstream of the STM2759 gene. The ΔfljAB8 deletion (strains 748, 749 750, 751, 752, 754 and 757) started at nucleotide 88 of the STM2760 gene (the last 64 nucleotides were deleted). The ΔfljAB9 deletion (strain 755) started at nucleotide 175 of the STM2761 gene. The ΔfljAB11 deletion (strain 701) started at 118 bp downstream of the STM2767 gene. In the inserted fragment (colored in green), a truncated IS1 lacking 133 nucleotides and a completed IS26 were detected. Finally, the ΔfljAB12 deletion (strain number 747) started at nucleotide 155 of the fljA gene (the last 385 nucleotides were deleted).
Figure A3.
Structures of the ΔfljAB4 and ΔfljAB5 deletion types. These deletion types had the same starting point of the deletion at the intergenic zone between STM2757 and STM2758 (222 bp downstream from the STM2757) but different ending points. The ΔfljAB4 deletion (strains 712, 713, 714) ended at nucleotide 571 of the iroB gene. The ΔfljAB5 deletion (strain 711) ended at nucleotide 848 of the STM2784 gene. The insertion sequence IS26 (colored in green) was detected in the middle of the deletions.
Figure A4.
Structure of the ΔfljAB7 deletion type. The ΔfljAB7 (strain 745) deletion started 998 bp downstream of the STM2759 gene and ended at nucleotide 475 of the iroC gene. Two IS26 insertion sequences in opposite directions (colored in green) were detected in the middle of the deletion.
Figure A5.
Structure of the ΔfljAB10 deletion type. The ΔfljAB10 deletion (strains 703 and 704) started at nucleotide 1125 of the STM2761 gene and ended at the intergenic zone between hin and iroB genes (334 nucleotides upstream of iroB). The inserted fragment (colored in green) started with an IS1, 354 nucleotides from the protein COG1309 described in the S. Typhimurium T000240 strain, and after that the tetA, tetR, jemC, jemB and jemA genes appear and a truncated IS10 lacking 973 nucleotides by an IS26.
Figure A6.
Structures of the ΔfljAB13 deletion type. The ΔfljAB13 deletion had the entire fljB gene present. In all variants, at least one IS26 (colored in green) was detected between hin and iroB, 334 nucleotides upstream of the iroB gene. That IS26 had a flanking 8 bp duplication (ATCAATAC). In the variant ΔfljAB13-A (strain 696), the last 33 bp of hin were deleted by a second IS26. In the variant ΔfljAB13-B (strain 694), hin was interrupted by a second IS26 with a flanking 8 bp duplication (GTCGAGCG). In the variant ΔfljAB13-C (strain 693), all genes belonging to the operon fljAB (fljA, fljB and hin) were complete. The only inserted IS26 was the one that was shared with the other two variants.
Author Contributions
Conceptualization, M.J.G., I.M.-B., L.L., J.G. and J.B.; Formal analysis, A.A.-G., A.A.-L. and V.G.; Methodology, A.A.-G., A.A.-L. and V.G.; Writing—original draft, A.A.-G. and A.A.-L.; Writing—review & editing, A.A.-G., A.A.-L., V.G., M.J.G., I.M.-B., L.L., J.G. and J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by EFSA through the Innuendo project, grant agreement GP/EFSA/AFSCO/2015/01/CT2; and the Basque Government grant agreements PA16/01 and PA20/03. A.A-G. and A.A.-L.’s contracts were supported by UPV/EHU and the Basque Government, respectively.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grimont P., Weill F. Antigenic Formulae of the Salmonella Serovars. 9th ed. WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur; Paris, France: 2007. [Google Scholar]
- 2.EFSA. ECDC The European Union one health 2018 zoonoses report. EFSA J. 2019;17 doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Echeita M.-A., Aladueña A., Cruchaga S., Usera M.A. Fast-track communications emergence and spread of an atypical Salmonella enterica subsp. Enterica Soc. 1999;37:28220. doi: 10.1128/jcm.37.10.3425-3425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echeita M.A., Herrera S., Usera M.A. Atypical, fljB-negative Salmonella enterica subsp. enterica of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. Society. 2001;39:2981–2983. doi: 10.1128/JCM.39.8.2981-2983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.EFSA Panel on Biological Hazards (BIOHAZ) Scientific opinion on monitoring and assessment of the public health risk of “Salmonella Typhimurium-like” strains. EFSA J. 2010;8 doi: 10.2903/j.efsa.2010.1888. [DOI] [Google Scholar]
- 6.Centro Nacional de Epidemiología. CIBER Epidemiología y Salud Pública (CIBERESP) Instituto de Salud Carlos III . Resultados de la Vigilancia Epidemiológica de las Enfermedades Trasmisibles. Informe Anual 2016. Gobierno de España (Ministerio de Ciencias e Innovación y Universidades); Madrid, Spain: 2018. [Google Scholar]
- 7.Petrovska L., Mather A.E., Abuoun M., Branchu P., Harris S.R., Connor T., Hopkins K.L., Underwood A., Lettini A.A., Page A., et al. Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005–2010. Emerg. Infect. Dis. 2016;22:617–624. doi: 10.3201/eid2204.150531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garaizar J., Porwollik S., Echeita A., Rementeria A., Herrera S., Wong R.M.-Y., Frye J., Usera M.A., Mc Clelland M. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 2002;40:2074–2078. doi: 10.1128/JCM.40.6.2074-2078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laorden L., Herrera-León S., Martínez I., Sanchez A., Kromidas L., Bikandi J., Rementeria A., Echeita A., Garaizar J. Genetic evolution of the Spanish multidrug-resistant Salmonella enterica 4,5,12:i:- monophasic variant. J. Clin. Microbiol. 2010;48:4563–4566. doi: 10.1128/JCM.00337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soyer Y., Switt A.M., Davis M.A., Maurer J., McDonough P.L., Schoonmaker-Bopp D.J., Dumas N.B., Root T., Warnick L.D., Gröhn Y.T., et al. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 2009;47:3546–3556. doi: 10.1128/JCM.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Switt A.I.M., Soyer Y., Warnick L.D., Wiedmann M. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:- Foodborne Pathog. Dis. 2009;6:407–415. doi: 10.1089/fpd.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins K.L., Kirchner M., Guerra B., Granier S.A., Lucarelli C., Porrero M.C., Jakubczak A., Threlfall E.J., Mevius D.J. Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: A new pandemic strain? Euro Surveill. 2010;15 doi: 10.2807/ese.15.22.19580-en. [DOI] [PubMed] [Google Scholar]
- 13.Cito F., Baldinelli F., Calistri P., Di Giannatale E., Scavia G., Orsini M., Iannetti S., Sacchini L., Mangone I., Candeloro L., et al. Outbreak of unusual Salmonella enterica serovar Typhimurium monophasic variant 1,4,[5],12:i:-, Italy, June 2013 to September 2014. Euro Surveill. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.15.30194. [DOI] [PubMed] [Google Scholar]
- 14.Mourão J., Machado J., Novais C., Antunes P., Peixe L. Characterization of the emerging clinically-relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- (monophasic variant of S. Typhimurium) clones. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:2249–2257. doi: 10.1007/s10096-014-2180-1. [DOI] [PubMed] [Google Scholar]
- 15.Dionisi A.M., Graziani C., Lucarelli C., Filetici E., Villa L., Owczarek S., Caprioli A., Luzzi I. Molecular characterization of multidrug-resistant strains of Salmonella enterica serotype Typhimurium and monophasic variant (S. 4,[5],12:i:-) isolated from human infections in Italy. Foodborne Pathog. Dis. 2009;6:711–717. doi: 10.1089/fpd.2008.0240. [DOI] [PubMed] [Google Scholar]
- 16.Lucarelli C., Dionisi A.M., Filetici E., Owczarek S., Luzzi I., Villa L. Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J. Antimicrob. Chemother. 2012;67:111–114. doi: 10.1093/jac/dkr391. [DOI] [PubMed] [Google Scholar]
- 17.Garcia P., Hopkins K.L., Garcia V., Beutlich J., Mendoza M.C., Threlfall J., Mevius D., Helmuth R., Rodicio M.R., Guerra B. Diversity of plasmids encoding virulence and resistance functions in Salmonella enterica subsp. enterica serovar Typhimurium monophasic variant 4,[5],12:i:- strains circulating in Europe. PLoS ONE. 2014;9:e89635. doi: 10.1371/journal.pone.0089635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García P., Malorny B., Rodicio M.R., Stephan R., Hächler H., Guerra B., Lucarelli C. Horizontal acquisition of a multidrug-resistance module (R-type ASSuT) is responsible for the monophasic phenotype in a widespread clone of Salmonella serovar 4,[5],12:i:- Front. Microbiol. 2016;7:1–7. doi: 10.3389/fmicb.2016.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasyl D., Hoszowski A. Occurrence and characterization of monophasic Salmonella enterica serovar Typhimurium (1,4,[5],12:i:-) of non-human origin in Poland. Foodborne Pathog. Dis. 2012;9:1037–1043. doi: 10.1089/fpd.2012.1154. [DOI] [PubMed] [Google Scholar]
- 20.Barco L., Longo A., Lettini A.A., Cortini E., Saccardin C., Minorello C., Olsen J.E., Ricci A. Molecular characterization of “inconsistent” variants of Salmonella Typhimurium isolated in Italy. Foodborne Pathog. Dis. 2014;11:497–499. doi: 10.1089/fpd.2013.1714. [DOI] [PubMed] [Google Scholar]
- 21.Boland C., Bertrand S., Mattheus W., Dierick K., Wattiau P. Molecular typing of monophasic Salmonella 4,[5]:i:- strains isolated in Belgium (2008–2011) Vet. Microbiol. 2014;168:447–450. doi: 10.1016/j.vetmic.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 22.CLSI (Clinical and Laboratory Standards Institute) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. 4th ed. CLSI; Wayne, PA, USA: 2018. [Google Scholar]
- 23.Anderson E.S., Ward L.R., Saxe M.J., de Sa J.D. Bacteriophage-typing designations of Salmonella Typhimurium. J. Hyg. (Lond.) 1977;78:297–300. doi: 10.1017/S0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machado M., Halkilahti J., Jaakkonen A., Silva D., Mendes I., Nalbantoglu Y., Borges V., Ramirez M., Rossi M., Carriço J. GitHub-B-UMMI/INNUca: INNUENDO Quality Control of Reads, De Novo Assembly and Contigs Quality Assessment, and Possible Contamination Search. [(accessed on 10 November 2020)]; Available online: https://github.com/B-UMMI/INNUca.
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S., den Bakker H.C., Li S., Chen J., Dinsmore B.A., Lane C., Lauer A.C., Fields P.I., Deng X. SeqSero2: Rapid and improved Salmonella serotype determination using Whole-Genome Sequencing Data. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.01746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020 doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Millán R.M., Martínez-Ballesteros I., Rementeria A., Garaizar J., Bikandi J. Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Res. Notes. 2013;6:2–5. doi: 10.1186/1756-0500-6-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alikhan N.-F., Zhou Z., Sergeant M.J., Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.EFSA. ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16 doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lockman H.A., Curtiss R. Salmonella Typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect. Immun. 1990;58:137–143. doi: 10.1128/IAI.58.1.137-143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boland C., Van Hessche M., Mahillon J., Wattiau P. A liquid bead array for the identification and characterization of fljB-positive and fljB-negative monophasic variants of Salmonella Typhimurium. Food Microbiol. 2018;71:17–24. doi: 10.1016/j.fm.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Llarena A., Ribeiro-Gonçalves B.F., Nuno Silva D., Halkilahti J., Machado M.P., Da Silva M.S., Jaakkonen A., Isidro J., Hämäläinen C., Joenperä J., et al. INNUENDO: A cross-sectoral platform for the integration of genomics in the surveillance of food-borne pathogens. EFSA Support. Publ. 2018;15 doi: 10.2903/sp.efsa.2018.EN-1498. [DOI] [Google Scholar]
- 38.Uelze L., Borowiak M., Deneke C., Szabo I., Fischer J., Tausch S.H., Malorny B. Performance and accuracy of four oppen-source tools for in silico serotyping of Salmonella spp. based on Whole-Genome Short-Read Sequencing Data. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02265-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerji S., Simon S., Tille A., Fruth A., Flieger A. Genome-based Salmonella serotyping as the new gold standard. Sci. Rep. 2020;10:1–10. doi: 10.1038/s41598-020-61254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diep B., Barretto C., Portmann A.C., Fournier C., Karczmarek A., Voets G., Li S., Deng X., Klijn A. Salmonella serotyping; comparison of the traditional method to a Microarray-based method and an in silico platform using Whole Genome Sequencing Data. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boland C., Bertrand S., Mattheus W., Dierick K., Jasson V., Rosseel T., Van Borm S., Mahillon J., Wattiau P. Extensive genetic variability linked to IS26 insertions in the fljB promoter region of atypical monophasic variants of Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2015;81:3169–3175. doi: 10.1128/AEM.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bugarel M., Vignaud M.-L., Moury F., Fach P., Brisabois A. Molecular identification in monophasic and nonmotile variants of Salmonella enterica serovar Typhimurium. Microbiologyopen. 2012;1:481–489. doi: 10.1002/mbo3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Román B., Garrido V., Sanchez S., Martinez-Ballesteros I., Garaizar J., Mainar-Jaime R.C., Migura-Garcia L., Grilló M.J. Relationship between Salmonella infection, shedding and serology in fattening pigs in low-moderate prevalence areas. Zoonoses Public Health. 2018;65:481–489. doi: 10.1111/zph.12453. [DOI] [PubMed] [Google Scholar]