Abstract
A series of hybrid indolo[3,2-c]isoquinoline (δ-carboline) analogs incorporating two pyrimidine and piperizine ring frameworks were synthesized. Intending biological activities and SAR we propose replacements of fluorine, methyl and methoxy of synthetic compounds for noteworthy antimicrobial, antioxidant, anticancer and anti-tuberculosis activities. Among these compounds 3a, 4a and 5e were progressively strong against E. coli and K. pneumonia. Whereas, compounds 4a, 5a and 6a with addition of various functional groups (OCH3, CH3) were excellent against S. aureus and B. subtilis. Compound 5c exhibited strong RSA and dynamic ferrous ion (Fe2+) metal chelating impact with IC50 of 7.88 ± 0.93 and 4.06 ± 0.31 µg/mL, respectively. Compound 5e was considerably cytotoxic against all cancer cells displaying activity better than the standard drug. Compounds 6b and 6e inhibited M. tuberculosis (MIC 1.0 mg/L) considerably. Molecular docking studies indicate that compounds 4d, 5a, 5b, 6b and 6f exhibited good interactions with 6LZE (COVID-19) and 6XFN (SARS-CoV-2) at active sites. The structure of the synthesized compounds were elementally analyzed using IR, 1H, 13C NMR and mass spectral information.
Keywords: Indolo[3,2-c]isoquinoline; Pyrimidine; Piperazine; COVID-19; Anti-TB
1. Introduction
Coronavirus disease 2019 (COVID-19), a fatal respiratory illness was reported in Dec 2019 from China. Initially the causal organism was named as 2019-nCoV. The earliest article in January 2020, related to 2019-nCoV revealed that 2019-nCoV belongs to the beta-coronavirus group, sharing ancestry with bat coronavirus HKU9-1, similar to SARS coronaviruses and that despite sequence diversity its spike protein interacts strongly with the human ACE2 receptor [1]. In February 2020, WHO renamed the virus as SARS-CoV-2 (severe acute respiratory syndrome corona virus 2). Several pre existing antiretroviral drugs showed potentiality against COVID-19 viz, oseltamivir [2], lopinavir [3], ritonavir [3], remdesivir [4], favipiravir [5], ribavirin [5], chloroquine and hydroxychloroquine.[6] alone or in combination. Out of these, ritonavir, remdesivir, chloroquine and hydroxychloroquine has shown efficacy at cellular level [7]. Nucleotide sequences of SARS-CoV-2 reveal it as a member of betacoronaviruses such as the SARS and MERS HCoVs [8], [9], [10]. HCoVs are positive-sense, long (30,000 bp), single-stranded RNA viruses SARS-CoV-2 genomic organization consists of a 50-untranslated region (UTR), a replicase complex (orf1ab) encoding non-structural proteins (nsps), a spike protein (S) gene, envelope protein (E) gene, amembrane protein (M) gene, a nucleocapsid protein (N) gene, 30-UTR, and several unidentified non-structural open reading frames [11].
Cancer and Tuberculosis (TB) are two significant reasons for death, because still no widespread strong medications taking care of them have been synthesized. Concurring, World Health Organization (WHO) to factual information that eight million people attribute new infection with T.B. annually [12,13]. Tuberculosis is a chronic bacterial contamination transmitted thru the air. This sickness is brought about by the bacteria Mycobacterium tuberculosis and primarily influences lungs (pulmonary TB). Mycobacterial cell membrane could initiate nitric oxide generation and reactive oxygen species (ROS) each involved in carcinogenesis [14]. National Cancer Institute review and reported that patients with pulmonary tuberculosis had more desirable danger of lung cancer and assessed a double rise in chance of lung cancer in males with tuberculosis [12,15]. Furthermore, Cancer is lethal illness with strengthening cause for mortality and measurements with anticipated annual 20 million cases globally by 2020 every year [16]. It is conceivable that endogenous free radical reaction started by ionization radiation, can also end result in tumors arrangement. One of the most basic relationships among reactive oxygen species and cancer is the expanded demise rate from leukemia and malignant neoplasia of the breast, ovaries and rectum because of more prominent effect of lipid peroxidation [17]. The pathological classification of the disease has ample evidence of changes in free radicals and ROS with inflammation, malignant tumors, myocardial infarction, rheumatism and neurodegenerative problems [18], [19], [20], [21], [22], [23]. ROS can also promote lipid peroxidation, integrated restriction of mitochondrial respiratory complexes and inactivation of GAP-DH(glyceraldehydre-3-phosphate dehydrogenase). Inhibition of Na+/K+-ATPase activity, sodium channel membrane inactivation and other oxidative modification in proteins. Totally these impacts are probably to play an imperative part in stock pathophysiology, inflammation and ischemia/reperfusion (I/R) injury [24].
The literature is flooded with reports variety of biological activities of α-, β- γ- and δ-carboline (pyrido-indole or indolo[3,2-c]isoquinoline) analogues [25], [26], [27], [28], [29]. Among them, δ-carbolines are the least studied class of compounds as compared to their α-, β- and γ- carboline analogues. Effective drugs have not been found yet among these class of heterocycles. Nevertheless, a rather broad spectrum of biological activity has been reported for several compounds of δ-carbolines such as antibacterial [30] , antiplasmodial [30], anti-malarial [31] and potent cancer therapeuticals (SYUIQ-5) [32]. In addition, tetracyclic isoquinolinone units fused with indole are established in the central structure of various effective bioactive compounds displaying antitumor [33], cytotoxic [34] , anticancer [35], and DNA cleavage [36]. An insight into literature survey showed that many hybridized α-, δ-carboline (indolo[2,3-c]isoquinoline, indolo[3,2-c]isoquinoline) analogs demonstrate diverse biological activities like bactericidal, fungicidal [37], anticancer [38], antineoplastic [39], antitumor [40], antihistaminic [41], oxytocic [42, 43], analgesic and anti-inflammatory activities [44]. In recent years, applications of indolo[3,2-c]isoquinoline include development of antimicrobial, antioxidant, anticancer and anti-TB [45], [46], [47] properties from our laboratory. Based on this speculation with a broader outlook in medicine citations, this area was considered. These results encouraged to continue the research work in the synthesis of biologically significant heterocyclic compounds, herein we have focused mainly synthesis of indolo[3,2-c]isoquinoline (δ-carboline) base hybrid analogs. All these compounds were screened for their in vitro antimicrobial, antioxidant and anticancer activities.
2. Materials and methods
2.1. Chemistry
Synthesis of marked compounds were accomplished as delineated in Scheme 1. The starting material ethyl 8-fluoro-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate 1a were synthesized as per the procedure [42]. The compound 1a was subjected to N-methylation with methyl iodide in dimethylformamide (DMF) using potassium carbonate (K2CO3) as base and as phase transfer catalyst tetra-n-butyl ammonium bromide (n-TBAB) was used to acquire 2a which has indicated a strong ingestion at 2954 cm−1 relating to N–CH3. Assimilation at 1715 and 1657 cm−1 relates to carbonyl stretching. 1H NMR spectrum of 2a has shown multiplets between δ 6.9 and δ 8.2 (m, 7H, Ar-H) relating to fifteen aromatic protons within the molecule. Peaks at δ 3.7 and δ 4.0 ppm are allotted for the methyl protons respectively. 13C NMR spectrum of 2a has shown a downfield signals at δ 165.2 and δ 154.5 peaks for two carbonyls by means of indolo[3,2-c]isoquinoline ring and δ 39.6 and δ 14.3 incorporated for methyl carbons. The mass spectrum of compound 2a has shown molecular ion peak at m/z 339 (M + H). The above spectral information confirms the arrangement of compound 2a. Spectral information of compounds 2b, 2c and 2d additionally upheld their structures. Further, the ester group hydrolysis of compound 2a yielded 3a as key intermediate. A strong retention at 2985 cm−1 corresponds to indolo[3,2-c]isoquinoline N–CH3. Assimilation at 1676 and 1638 cm−1 relates to C O stretching respectively. The 1H NMR spectrum of 3a which has displayed a singlet at δ 10.6 was assigned to hydroxyl group and multiplets between δ 7.1 and δ 8.3 ppm corresponds to seven aromatic protons present in the molecule. Methyl protons assigned peaks at δ 3.9 ppm. 13C NMR spectrum of 3a manifested signals at δ 164.7 and δ 160.5 integrated for two symmetric carbonyls by the indolo[3,2-c]isoquinolinone ring and δ 36.6 signals for methyl carbon. The mass spectrum of compound 3a has shown molecular ion peak at m/z 311 [M + H]+. An attempt to chloroethyl ester, 4a obtained from 3a by cyclization with dichloroethane and anhydrous K2CO3 in acetone, the reaction was refluxed for 6 h. Within the IR spectrum of 4a absorption at 2962 cm−1 relates to (N–CH3) an absorption at 1715 and 1657 cm−1 which corresponds to carbonyl stretch. The 1H NMR spectrum of 4a has multiplets in the range δ 7.0 and 8.3 ppm corresponds to seven aromatic protons present in the molecule. Peaks at δ 4.9 and 4.6 were attributed to the protons of oxy-ethane and chloro-ethane, respectively. 13C NMR spectrum for 4a showed signals in a falling field at δ 164.7 and less deshielded peak at δ 160.8 integrated for the C O, indolo [3,2-c]isoquinolinone ring and δ 39.5 integrated for N-methyl carbon. The mass spectrum of compound 4a showed a peak of the molecular ion at m/z 420 [M+ + 2]. The spectral data presented confirm the formation of compound 4a. Spectral data of the compounds 4c-4d confirm their structures.
Scheme 1.
Synthesis of indolo[3,2-c]isoquinoline base hybrid analogs 1-4(a-d) and 5,6 (a-h).
The compound 4a with substituted pyrimidine-2-thiol dry K2CO3 were introduced into DMF with stirring for 8–10 h to give 5a showing a strong absorption at 3154 cm−1 corresponding to OH and absorption at 2984 cm−1 corresponds to N–CH3. Absorptions at 1715, 1657 and 1540 cm−1 correspond to C O and C N stretching, respectively. Spectrum 1H NMR of 5a showed a singlet at δ 10.8 due to (s, 1H, OH) and multiplied peaks between δ 7.1–8.4 ppm (m, 8H, Ar-H) integrating for nineteen aromatic protons. A singlets at δ 3.9 and at δ 4.2 were for N–CH3 and S-CH2 for the protons. 13C NMR spectrum of 5a has showed a downfield signals at δ 161.5, 161.0 for C O carbon for indolo[3,2-c]isoquinoline ring and a less shielded peak at δ 159.8 and δ 154.8 integrated for the C N of the pyrimidine ring. The field signals at δ 65.4, 39.9, 36.4 and δ 14.4 correspond to the two carbons CH2, N–CH3 and CH3 carbons of pyrimidine and indolo[3,2-c]isoquinoline rings respectively. The mass spectrum of compound 5a showed a peak of molecular ion at m/z 479 [M+H]+ which corresponds to its molecular weight.
Finally, the compound 6a was synthesized by condensation of the carboxylic acid key intermediate 3a with appropriate amines (aryl-substituted-piperazines) in presence of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimidehydrochloride and hydroxybenzotriazole after stirring and reflux for 6h to obtained 6a. The IR spectrum of 6a has shown a strong absorption at 2959 cm−1 corresponding to N–CH3 and absorption at 1715, 1657 cm−1 corresponds to carbonyl stretching. 1H NMR spectrum of 6a showed singles for four carbon atoms (piperazine ring) at δ 5.3–4.2(m, 8H, CH2), singlet at δ 3.9 which was assigned for N–CH3 proton. The multiplets at δ 7.1–8.3 ppm range, corresponded to eleven aromatic protons in the molecule. 13C NMR spectrum of 6a has displayed a downfield signals at δ 164.7 and δ 160.4 for carbonyl carbon. Upfield signals at range δ 40.5–39.8 ppm and δ 37.6 were integrated for piperazine ring and N–CH3 carbons. The mass spectrum of compound 6a has shown molecular ion peak at m/z 473 [M + H]+. The above spectral data supports the formation of compound 6a. Spectral data of compounds 6b-6h also confirm their structures. Reactions were scrutinized by thin layer chromatography using silica gel-G coated aluminium plates (Merck). All synthesized compounds were characterized by spectrophotometric (IR, NMR 1H and 13C), mass spectra, elemental analysis was calculated (Anal. Calcd) and 1H, 13C NMR are given in supplementary information.
3. Results and discussion
3.1. Antimicrobial activity
The development and unfolding of antimicrobial resistance has become foremost serious concern across the globally with reference to public health. Antimicrobial resistance is flexibility to inactivate, exclude or block the inhibitory or lethal mechanism of the antimicrobial agents by microbes [48]. Electron-rich heteroatoms such as nitrogen, sulfur, halogen heterocycles have a essential role in diverse medicinal chemistry [49]. The synthesized compounds were investigated in vitro for antimicrobial activity against four strains for bacterial included Escherichia coli, Klebsiella pneumoniae (Gram-negative), Staphylococcus aureus and Bacillus subtilis (Gram-positive) and Aspergillus niger, Aspergillus oryzae, Candida albicans and Pencillium chrysogenum for fungal, by broth dilution method [50], [51], [52]. The minimal inhibitory concentrations (MIC) that hindered the development of the tried microorganisms for bactericidal and fungicidal activities had been observed. Ciprofloxan and fluconazole were chosen as reference drugs for antibacterial and antifungal activities, respectively and results are summarized in Table 1 .
Table 1.
Anti-bacterial and anti-fungal activities of compounds 2-4(a-d) and 5,6(a-h).
| MIC µg/mL |
||||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria |
Fungi |
|||||||
| Comps | Ec | Kp | Sa | Bs | An | Ao | Ca | Pc |
| 2a | 3.12 | 3.12 | 75 | 25 | 12.5 | 25 | 25 | 75 |
| 2b | 12.5 | 75 | 50 | 25 | 100 | 100 | 100 | 100 |
| 2c | 25 | 50 | 25 | 100 | 100 | 100 | 100 | 100 |
| 2d | 100 | 100 | 50 | 50 | 75 | 12.5 | 25 | 25 |
| 3a | 1.5 | 1.5 | 25 | 12.5 | 3.12 | 25 | 25 | 100 |
| 3b | 50 | 75 | 25 | 6.25 | 12.5 | 12.5 | 25 | 75 |
| 3c | 25 | 100 | 6.25 | 100 | 25 | 75 | ND | ND |
| 3d | 50 | 25 | 12.5 | 12.5 | 12.5 | 50 | 25 | ND |
| 4a | 1.5 | 1.5 | 6.25 | 12.5 | 1.5 | 1.5 | 1.5 | 1.5 |
| 4b | 6.25 | 25 | 50 | 50 | 3.12 | 3.12 | 50 | 12.5 |
| 4c | 75 | 100 | 12.5 | 3.12 | 12.5 | 12.5 | ND | 75 |
| 4d | 3.12 | 6.25 | 3.12 | 12.5 | 25 | 75 | 25 | 12.5 |
| 5a | 6.25 | 12.5 | 25 | 100 | 1.5 | 1.5 | 1.5 | 1.5 |
| 5b | 25 | 3.12 | 1.5 | 1.5 | 3.12 | 6.25 | 50 | 25 |
| 5c | 50 | 25 | 12.5 | 25 | 12.5 | 12.5 | 25 | 75 |
| 5d | 12.5 | 12.5 | 12.5 | 6.25 | 25 | 75 | 100 | 6.25 |
| 5e | 100 | 75 | 75 | 75 | 25 | 25 | 1.5 | 1.5 |
| 5f | 1.5 | 1.5 | 25 | 6.25 | 6.25 | 12.5 | 3.12 | 3.12 |
| 5g | 75 | 100 | 6.25 | 3.12 | 12.5 | 25 | 25 | 100 |
| 5h | 75 | 100 | 12.5 | 100 | 12.5 | 25 | 25 | 75 |
| 6a | 3.12 | 12.5 | 6.25 | 100 | 1.5 | 1.5 | 1.5 | 1.5 |
| 6b | 12.5 | 6.25 | 100 | 25 | 3.12 | 6.25 | 12.5 | 3.12 |
| 6c | 25 | 25 | 3.12 | ND | 50 | 75 | 25 | 12.5 |
| 6d | 25 | 25 | 25 | 50 | 1.5 | 1.5 | 75 | 100 |
| 6e | 3.12 | 12.5 | 50 | 75 | 12.5 | 25 | 12.5 | 50 |
| 6f | 25 | 3.12 | 1.5 | 1.5 | 12.5 | 6.25 | 50 | 75 |
| 6g | 25 | 50 | 1.5 | 1.5 | 50 | 75 | 25 | 3.12 |
| 6h | ND | 50 | 25 | 25 | 25 | 100 | 75 | 100 |
| Ciprofloxan | 1.5 | 1.5 | 3.12 | 1.5 | - | - | - | - |
| Fluconazole | - | - | - | - | 1.5 | 3.12 | 1.5 | 3.12 |
| DMSO | - | - | - | - | - | - | - | - |
Ec-Eshericha coli, Kp-Klebsiella pneumonia, Sa-Staphylococcus aureus, Bs-Bacillus subtilis, An-Aspergillus niger, Ao-Aspergillus oryzae, Ca-Candidia albicans, Pc-Pencillium chysogenum. ND-Not determined, MIC- Low concentration of drug, which completely inhibits the growth of bacteria and fungi. Ciprofloxan and fluconazole were used as reference drugs for anti-bacterial and anti-fungal activities, MIC values given in brackets. (−) did not show antimicrobial activity.
Based on the initial microbiological concept, it was found that the antibacterial activity of all compounds against pathogenic bacteria was moderately good, except for compounds with fluoro, hydrogen substitutes into in the indolo[3,2-c]isoquinoline. For pyrimidine rings of compounds 3a, 4a and 5e good activity was observed against E. coli and K. pneumonia. Establishment of OCH3, CH3 at various para positions in combinations of indolo[3,2-c]isoquinoline, pyrimidine and piperizine systems like compounds like 5b, 6f and 6g exhibited a higher antibacterial activity against S. aureus and B. subtilis. For antifungal activity, Compounds with fluorine and methyl substitutes (-F-indolo[3,2-c]isoquinoline, -CH3-pyrimidine and -F-piperazine rings), i.e 4a, 5a and 6a were excellent against A. niger, A. oryzae, C. albicans and P. chrysogenum irrespective of concentration taken. If only the fluoro substitute 5e was included in the indolo[3,2-c]isoquinoline ring, the activity improved and possessed very good activity against C. albicans and P. chrysogenum. A Similar activity was observed in compound 6d against A. niger and A. Oryzae. Other compounds presented moderate to least activity against the tested fungi.
With regard to structural activity relationship, the presentation of molecule atom rather than the methyl, methoxy groups at the para position of the indolo[3,2-c]isoquinoline, pyrimidine and piperizine rings was exclusively dependable for expanding in Gram-negative bacteria and antifungal activities. Similarly, replacing of fluorine group by methyl, methoxy groups affected the activity in gram positive bacteria. It has been found that the introduction of electron-withdrawing group significantly enhances the antibacterial (Gram-negative) and antifungal activities of the titled compounds. Presentation of electron donating methyl and methoxy groups showed a great potential against Gram-positive bacteria. SAR considers that presence of electron withdrawing and electron donating on indolo[3,2-c]isoquinoline, pyrimidine and piperizine ring systems intensified the antimicrobial activities.
3.2. Antioxidant activities
Increased free radicals in the body have been involved in harming human life [53,54]. Antioxidant agent properties are significant in checking the injurious impacts on nourishments and biological systems by free radicals [55]. All the synthesized compounds were evaluated for their in vitro antioxidant activity by DPPH and Metal-Chelating Activity (Fe2+) methods. The values of the effective concentration at which half of the radicals were scavenged (IC50) were tried for antioxidant activities. A lower IC50 value suggests greater antioxidant activity and IC50 values of μg/mL usually indicated best antioxidant properties.
3.3. 1, 1-Diphenyl-2-picryl hydrazyl (DPPH) radical scavenging activity (RSA)
Antioxidants scavenge free radicals by donating DPPH electrons [56]. DPPH, a static free radical, has an odd electron and therefore has a tough retention at 517 nm. As electron gets combined off to saturation, the consumption diminishes stoichiometrically in terms of the number of absorbing electrons or hydrogen particles. This reaction is significantly embraced to examine the ability of molecules as free radical scavengers. We evaluated the scavenging powers of synthetic compounds on DPPH radical. The effects were in contrast with the standards 2-ter-butyl-4-methoxy phenol (butylated hydroxyl anisole, BHA). All the examined compounds showed a decrease in the activity of the absorption of free radicals, when, in contrast to BHT (IC50, 5.77 ± 0.76 μg/mL). Consequences of free radical scavenging activity in percentage and IC50 are shown summarized in Table 2 . The consolidation of various heteroatoms into indolo[3,2-c]isoquinoline framework showed expansive range of results. In the tested analogs, it was found that compound 5c has a strong to RSA (73.25 ± 1.02, 76.68 ± 1.25, 88.86 ± 1.57 and 82.78 ± 0.32%) at 25, 50, 75, 100 μg/mL concentration respectively with IC50 values 5.78 ± 0.35 μg/mL. Compounds 5d (IC50, 7.10 ± 0.99 μg/mL), 5g (IC50, 7.88 ± 0.93 μg/mL) and 6h (IC50, 6.96 ± 0.79 μg/mL) exhibited good RSA. The minimum RSA activity was shown for compound 4a with IC50 which is 9.97 ± 0.24 μg/mL. The accelerated undertaking (5c, 5g and 6h) may be due to the presence of methyl and hydrogen substitutes in the para position of the indolo[3,2-c]isoquinoline and pyrimidine rings. Conversely, fluorine substituting indolo[3,2-c]isoquinoline systems 3a, 4a, 5a and 6a exhibited minimal activity as compared to standard.
Table 2.
DPPH activity of synthesized compounds.
| Comps | Concentrations (µg/mL) |
||||
|---|---|---|---|---|---|
| 25 | 50 | 75 | 100 | IC50 | |
| 3a | 32.65 ± 0.26 | 45.78 ± 1.18 | 57.75 ± .0.21 | 60.02 ± 0.78 | 9.05+0.86 |
| 3b | 30.62 ± 0.53 | 50.12 ± 1.17 | 53.52 ± 0.51 | 58.35 ± 0.79 | 9.32 ± 0.92 |
| 3c | 35.12 ± 1.47 | 54.56 ± 1.18 | 63.45 ± 1.18 | 64.23 ± .1.09 | 8.57 ± 0.79 |
| 3d | 43.25 ± 0.71 | 52.63 ± 1.06 | 62.13 ± 1.02 | 61.18 ± 1.62 | 8.45 ± 0.65 |
| 4a | 40.89 ± 1.12 | 65.02 ± 1.24 | 68.86 ± 1.17 | 70.05 ± 1.98 | 9.97 ± 0.24 |
| 4b | 35.32 ± 1.43 | 36.55 ± 0.84 | 50.45 ± 1.01 | 52.85 ± 1.03 | 8.50 ± 0.54 |
| 4c | 30.36 ± 1.01 | 52.42 ± 1.27 | 55.82 ± 1.18 | 64.46 ± 1.25 | 9.23 ± 0.49 |
| 4d | 35.21 ± 0.41 | 42.18 ± 1.02 | 54.05 ± 1.65 | 59.28 ± 1.86 | 8.34 ± 0.69 |
| 5a | 38.67 ± 0.51 | 50.21 ± 0.25 | 51.65 ± 1.25 | 62.13 ± 1.33 | 8.01 ± 0.88 |
| 5b | 40.12 ± 0.74 | 48.56 ± 0.61 | 60.18 ± 0.69 | 66.54 ± 1.47 | 9.44 ± 0.82 |
| 5c | 73.25 ± 1.02 | 76.68 ± 1.25 | 88.42 ± 1.57 | 82.78 ± 0.32 | 5.78 ± 0.35 |
| 5d | 67.12 ± 1.22 | 79.02 ± 1.07 | 80.92 ± 0.25 | 81.59 ± 1.02 | 7.10 ± 0.99 |
| 5e | 36.42 ± 0.91 | 49.48 ± 0.24 | 50.92 ± 0.94 | 62.25 ± 1.21 | 8.13 ± 0.24 |
| 5f | 34.47 ± 1.01 | 50.32 ± .0.68 | 59.15 ± 0.87 | 58.55 ± 1.08 | 9.41 ± 0.27 |
| 5g | 52.89 ± 1.04 | 62.14 ± 1.15 | 74.91 ± 0.61 | 80.72 ± 0.97 | 7.88 ± 0.93 |
| 5h | 57.47 ± 1.54 | 62.52 ± 1.54 | 69.24 ± 1.05 | 78.64 ± 1.38 | 8.61 ± 0.19 |
| 6a | 39.28 ± 0.57 | 56.02 ± 0.84 | 69.86 ± 1.29 | 72.05 ± 1.43 | 8.71 ± 0.37 |
| 6b | 46.68 ± 0.24 | 50.02 ± 1.37 | 57.71 ± 1.71 | 58.59 ± 1.87 | 8.93 ± 0.22 |
| 6c | 35.12 ± 1.03 | 43.56 ± 1.44 | 59.18 ± 1.05 | 62.34 ± 1.97 | 8.03 ± 0.78 |
| 6d | 38.28 ± 1.24 | 48.19 ± 1.02 | 56.82 ± 0.50 | 62.12 ± 1.26 | 8.96 ± 0.29 |
| 6e | 35.35 ± 1.07 | 47.25 ± 1.25 | 56.82 ± 1.04 | 65.25 ± 1.03 | 8.13 ± 0.68 |
| 6f | 41.42 ± .0.14 | 55.48 ± 0.85 | 53.92 ± 1.38 | 61.01 ± 1.05 | 9.19 ± 0.57 |
| 6g | 70.68 ± 0.72 | 79.78 ± 1.00 | 80.86 ± 0.25 | 82.88 ± 0.80 | 6.96 ± 0.79 |
| 6h | 42.12 ± 0.54 | 58.96 ± 0.97 | 66.19 ± 0.85 | 68.94 ± 0.83 | 9.46 ± 0.94 |
| BHA | 70.47 ± 0.28 | 85.52 ± 0.68 | 90.51 ± 0.04 | 92.64 ± 0.84 | 5.77 ± 0.76 |
3.4. Ferrous (Fe2+) ion metal chelating activity
Ferrous ion is one of the types of metal ions. Deficiency of ferrous ions protects the oxygen-related reaction and from lipid generation. Ferrous ion chelators evacuate ferrous ion (Fe2+) that will take part in generation of hydroxyl radical. The chelating impact of ferrous ion on synthesized compounds was resolved and results were contrasted with BHA as standard as mentioned in the method of Dinis et al. [57].
In this strategy, ferrozine can quantitatively form the ferrous ion complex. The chelating agents disrupt the complex system leading to a decrease in redness of the complex. Therefore, the evaluation of the colour reduction indicates chelation activity of the metals of coexisting chelators. Low retention points out high metal chelation activity. In the study, synthetic compounds hindered the development of ferrous and ferrozine complex. It is captivating to note that the compound 5c exhibited excellence activity at 25, 50, 75 and 100 µg/mL, concentrations with IC50 values of 4.06 ± 0.31 µg/mL and showed a greater metal chelating effect than BHT (IC50, 4.79 ± 0.91 µg/mL). Interestingly, compound 5g, was exhibiting promising metal chelating activity at IC50 values 5.67 ± 0.27. Some of the remaining compounds were moderately active and some less active. Screening indicated that all the synthesized indolo[3,2-c]isoquinoline analogues were active metal chelaters, with fluctuating degrees of potency by ‘CH3’ and ‘H’ replacements on indolo[3,2-c]isoquinoline and pyrimidine rings improve the action. The results obviously imply, the consolidation of methyl and hydrogen substituted at para position in the indolo[3,2-c]isoquinoline and pyrimidine rings may assume a significant role to act as a superior metal chelating activity antioxidant agent. The results are displayed in Table 3 . Structure activity relationship (SAR) has shown noteworthy RSA and metal chelating activity of the analogs with replacement or substitution on the aromatic rings of indoloisoquinoline, pyrimidine and piperizine with the CH3 group. Compounds 5c, 5b, 5g and 6h with electron-donating group methyl group at the para-position were better antioxidants than electron retreating fluorine moiety due to enhanced stability of the radical and metal chelating activities.
Table 3.
Ferrous (Fe2+) ion chelating activity of synthesized compounds.
| Comps | Concentrations (µg/mL) |
||||
|---|---|---|---|---|---|
| 25 | 50 | 75 | 100 | IC50 | |
| 3a | 45.21 ± 0.20 | 52.47 ± 0.56 | 60.12 ± 0.65 | 61.42 ± 1.02 | 8.45 ± 0.80 |
| 3b | 40.55 ± 1.14 | 49.82 ± 1.45 | 66.74 ± 0.57 | 70.59 ± 1.05 | 9.13 ± 0.70 |
| 3c | 46.12 ± 0.58 | 55.35 ± 0.85 | 66.49 ± 0.67 | 69.62 ± 1.91 | 8.64 ± 0.22 |
| 3d | 49.98 ± 1.24 | 58.99 ± 1.06 | 64.25 ± 1.45 | 71.58 ± 0.40 | 8.09 ± 0.17 |
| 4a | 52.73 ± 0.51 | 61.78 ± 1.21 | 70.54 ± 1.36 | 72.79 ± 0.75 | 7.90 ± 0.11 |
| 4b | 45.98 ± 0.84 | 55.85 ± 0.87 | 63.85 ± 1.25 | 68.58 ± 1.12 | 8.54 ± 0.65 |
| 4c | 65.02 ± 0.12 | 68.57 ± 1.12 | 79.45 ± 0.76 | 80.35 ± 0.91 | 9.35 ± 0.61 |
| 4d | 40.63 ± 1.01 | 51.36 ± 0.58 | 60.58 ± 1.07 | 65.84 ± 1.01 | 9.00 ± 0.07 |
| 5a | 46.55 ± 0.25 | 50.72 ± 10.2 | 59.85 ± 0.91 | 60.54 ± 0.67 | 8.55 ± 0.80 |
| 5b | 45.21 ± 1.24 | 52.47 ± 1.17 | 60.12 ± 1.32 | 61.42 ± 1.18 | 8.45 ± 0.80 |
| 5c | 60.71 ± 1.25 | 64.28 ± 0.84 | 70.98 ± 1.18 | 76.84 ± 1.49 | 8.92 ± 0.47 |
| 5d | 81.41 ± 0.17 | 89.14 ± 1.27 | 91.32 ± 1.01 | 92.42 ± 1.34 | 4.06 ± 0.31 |
| 5e | 36.48 ± 1.02 | 57.35 ± 1.20 | 60.99 ± 1.24 | 68.62 ± 1.71 | 8.49 ± 0.79 |
| 5f | 45.98 ± 1.31 | 55.89 ± 1.27 | 62.25 ± 1.37 | 68.58 ± 1.32 | 8.56 ± 0.08 |
| 5g | 78.52 ± 0.24 | 79.57 ± 1.65 | 84.75 ± 1.02 | 90.42 ± 0.64 | 5.67 ± 0.27 |
| 5h | 47.55 ± 0.21 | 58.12 ± 1.11 | 61.85 ± 1.22 | 68.98 ± 1.58 | 8.22 ± 0.43 |
| 6a | 45.98 ± 1.03 | 52.19 ± 1.23 | 63.25 ± 1.15 | 67.58 ± 0.69 | 8.68 ± 0.12 |
| 6b | 45.21 ± 1.2 | 52.47 ± 1.11 | 60.12 ± 1.57 | 61.42 ± 1.25 | 8.45 ± 0.80 |
| 6c | 60.02 ± 1.14 | 55.57 ± 0.87 | 63.45 ± 1.36 | 60.35 ± 0.92 | 8.55 ± 0.43 |
| 6d | 46.12 ± 1.03 | 55.35 ± 1.94 | 66.49 ± 1.79 | 69.62 ± 1.74 | 8.64 ± 0.22 |
| 6e | 40.55 ± 1.10 | 62.82 ± 0.37 | 69.74 ± 1.92 | 70.59 ± 1.09 | 7.39 ± 0.57 |
| 6f | 52.73 ± 0.90 | 61.78 ± 1.07 | 70.54 ± 1.47 | 72.79 ± 1.57 | 8.09 ± 0.17 |
| 6g | 45.98 ± 1.25 | 55.85 ± 1.54 | 63.85 ± 0.73 | 68.58 ± 1.36 | 8.54 ± 0.65 |
| 6h | 49.98 ± 0.54 | 58.99 ± 1.36 | 64.25 ± 0.94 | 71.58 ± 0.94 | 7.90 ± 0.11 |
| BHA | 80.01 ± 0.02 | 86.52 ± 0.12 | 91.24 ± 0.84 | 92.01 ± 0.61 | 4.79 ± 0.91 |
3.5. Cytotoxic activity
The cytotoxic activities of newly synthesized compounds 3-4(a-d) and 5,6(a-h) were assessed by an in vitro assay carried out against four human tumor cell lines, e.g. MCF-7 (breast), A549 (lung), HeLa (cervical) and Panc-1 (pancreas) using 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) [58] with doxorubicin as positive reference. The results are presented in Table 4 . Among these analogue compounds 5e MCF-7(IC50, 0.90 ± 0.17 µM), A-549 (IC50, 0.81 ± 0.80 µM), HeLa (IC50, 0.8 ± 0.54 µM) and Panc-1(IC50, 1.24 ± 0.90 µM) and compound 6d MCF-7 (IC50 0.79 ± 0.50 µM), A-549(IC50, 1.8 ± 1.29 µM), HeLa (1.5 ± 0.35 µM) Panc-1(IC50 1.12 ± 0.58 µM) both were effective cytotoxic against all cancer cell lines than the drug. Compound 6a against all the four cell lines MCF-7(IC50, 1.3 ± 0.92 µM), A-549 (IC50, 1.6 ± 0.60 µM), HeLa (IC50, 1.6 ± 0.14 µM) and Panc-1(IC50, 1.7 ± 0.91 µM). 3a against Panc-1 (IC50, 1.9 ± 1.20 µM), 4a against MCF-7(IC501.8 ± 0.51 µM), A-549(IC50, 1.9 ± 0.81 µM). 6b against A-549 (IC50, 1.01 ± 0.29 µM). Panc-1(IC50, 1.0 ± 0.72 µM) and 6e against MCF-7 (IC50, 1.91 ± 0.20), HeLa (IC50, 1.8 ± 0.90 µM) have shown good cytotoxicity. Of the rest, the compounds have mild to moderate processes and no performance against all the four cell lines. Structure activity relationship results reveal that indolo[3,2-c]isoquinoline, pyrimidine and piperazine rings with electron-withdrawing substitution exhibit significantly stronger cytotoxicity against all the four cell lines. Alternative compounds such as ‘para-fluorine’ and ‘para- hydrogen’ replacement has demonstrated few inclination to improve the activity than compared with para-CH3 and para-OCH3 substituted indolo[3,2-c]isoquinoline analogues.
Table 4.
In vitro cytotoxicity assay data activity of compounds 3-4(a-d) and 5,6(a-h).
| IC50 (µM)a |
|||||||
|---|---|---|---|---|---|---|---|
| Comps | R | R1 | R2 | MCF-7 | A-549 | HeLa | Panc-1 |
| 3a | F | - | - | 2.1 ± 0.41 | 6.7 ± 1.05 | 6.4 ± 1.06 | 1.9 ± 1.20 |
| 3b | MeO | - | - | 4.4 ± 2.14 | 6.1 ± 1.15 | 6.9 ± 1.71 | 5.9 ± 2.40 |
| 3c | Me | - | - | 5.3 ± 1.02 | 6.6 ± 1.16 | 4.1 ± 1.03 | 4.6 ± 1.20 |
| 3d | H | - | - | 9.1 ± 1.07 | 7.3 ± 2.01 | 6.2 ± 1.47 | 7.7 ± 1.50 |
| 4a | F | - | - | 1.8 ± 0.51 | 1.9 ± 0.81 | 5.9 ± 1.7 | 4.9 ± 2.40 |
| 4b | MeO | - | - | 9.1 ± 2.01 | 7.3 ± 2.21 | 6.2 ± 2.47 | 7.7 ± 0.05 |
| 4c | Me | - | - | 8.4 ± 1.04 | 8.4 ± 2.11 | 9.9 ± 2.07 | 7.2 ± 1.50 |
| 4d | H | - | - | 7.6 ± 2.17 | 7.9 ± 1.06 | 6.5 ± 2.04 | 7.4 ± 1.25 |
| 5a | F | Me | - | 2.9 ± 1.17 | 10.9 ± 1.19 | 1.5 ± 0.51 | 9.4 ± 2.21 |
| 5b | MeO | Me | - | 3.9 ± 1.10 | 2.3 ± 1.01 | 2.7 ± 1.04 | 2.5 ± 2.05 |
| 5c | Me | Me | - | 6.3 ± 2.04 | 8.1 ± 1.25 | 7.2v1.07 | 6.1 ± 2.04 |
| 5d | H | Me | - | 5.9 ± 3.00 | 6.1 ± 2.24 | 10.5 ± 1.05 | 9.7 ± 2.11 |
| 5e | F | H | - | 0.90 ± 0.17 | 0.81 ± 0.80 | 0.8 ± 0.54 | 1.24 ± 0.90 |
| 5f | MeO | H | - | 3.5 ± 0.73 | 5.0 ± 1.90 | 3.5 ± 1.14 | 5.5 ± 2.50 |
| 5g | Me | H | - | 6.52 ± 0.45 | 4.9 ± 0.52 | 10.2 ± 1.14 | 8.91 ± 0.20 |
| 5h | H | H | - | 4.4 ± 1.90 | 3.8 ± 0.57 | 4.7 ± 1.19 | 5.7 ± 1.60 |
| 6a | F | - | 4-FPh | 1.3 ± 0.92 | 1.6 ± 0.60 | 1.6 ± 0.14 | 1.7 ± 0.91 |
| 6b | MeO | - | 4-FPh | 4.1 ± 2.70 | 1.01 ± 0.29 | 7.6 ± 0.80 | 1.0 ± 0.72 |
| 6c | Me | - | 4-FPh | 4.7 ± 1.14 | 4.70 ± 0.66 | 6.45 ± 2.73 | 5.4 ± 1.12 |
| 6d | H | - | 4-FPh | 0.79 ± 0.50 | 1.8 ± 1.29 | 1.5 ± 0.35 | 1.12 ± 0.58 |
| 6e | F | - | 4- MeOPh | 1.91 ± 0.20 | 15.7 ± 0.50 | 1.8 ± 0.90 | 10.5 ± 0.22 |
| 6f | MeO | - | 4-MeOPh | 7.2 ± 1.35 | 11.4 ± 1.20 | 10.5 ± 1.81 | 9.7 ± 2.11 |
| 6g | Me | - | 4-MeOPh | 8.6 ± 1.32 | 9.9 ± 1.24 | 10.1 ± 1.42 | 9.4 ± 1.09 |
| 6h | H | - | 4-MeOPh | 3.6 ± 1.54 | 7.9 ± 2.19 | 8.1 ± 1.05 | 6.4 ± 1.05 |
| Doxorubicin - | 0.92 ± 0.50 | 1.02 ± 0.80 | 1.02 ± 0.72 | 1.41 ± 0.58 | |||
IC50 values are indicated as mean ± SD of three independent tests.
3.6. Anti-TB activity by MHB method
The in vitro anti-tuberculosis activity (anti-TB) was decided by the broth microtiter dilution method [59] against H37Rv strain M. tuberculosis (ATCC 27294). Further, minimum inhibition concentrations (MICs) with rifampicin as the standard drug was determined. All tested compounds illustrated great to excellent anti-tuberculosis activity. Compounds 8-Methoxy-6-{[4-(4-fluorophenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6b) and 8-Fluoro-6-{[4-(4-methoxyphenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6e) showed intense activity indicating MIC value of 1.0 mg/L against M. tuberculosis. Compounds 6b and 6e having fluorine and methoxy groups substitutions R and R2 at alternate para positions in the indolo[3,2-c]isoquinoline and piperazine rings indicated inhibitory impacts similar to the positive drugs, however less dynamic than rifampicin. The remaining synthesized compounds appeared to show moderate activity (MIC range 4–128 mg/L).
From the consequences of anti-tuberculosis examination, it is clear that introduction of substituents ‘CH3’ and ‘H’ on indolo[3,2-c]isoquinoline and piperazine rings as in 6c, 6d, 6g and 6h were less dynamic. Structure activity relationships make the impacts of the replacement designs on the indolo[3,2-c]isoquinoline and piperazine with `F' and `OCH3'substitutes scaffolds clear. The lipophilicity of compounds decide the effectiveness of anti-tuberculosis activity. The result are presented in Table 5 .
Table 5.
Anti-tuberculosis activity of synthesized compounds against M. tuberculosis by MHB method.
| MICa |
||
|---|---|---|
| Comps | Mg/L | µM |
| 3a | >128 | 353.9 |
| 3b | 64 | 66.2 |
| 3c | >128 | 344.8 |
| 3d | >128 | 336.5 |
| 4a | >128 | 359.8 |
| 4b | 64 | 63.5 |
| 4c | >128 | 322.1 |
| 4d | 128 | 298.5 |
| 5a | 4 | 11.2 |
| 5b | 4 | 13.7 |
| 5c | 8 | 20.8 |
| 5d | 64 | 54.8 |
| 5e | 4 | 12.8 |
| 5f | 4 | 13.2 |
| 5g | 8 | 21.2 |
| 5h | 16 | 53.7 |
| 6a | 2 | 5.9 |
| 6b | 1 | 2.6 |
| 6c | 4 | 12.6 |
| 6d | 8 | 20.6 |
| 6e | 1 | 2.9 |
| 6f | 4 | 14.5 |
| 6g | 8 | 21.6 |
| 6h | 16 | 58.4 |
| Rifampicin | 0.125 | - |
Minimum inhibition concentration (MIC) against H37Rv strain M. tuberculosis (ATCC 27294).
3.7. Drug likeness profile
For preliminary confirmation of the physio-chemical properties, ADMET and drug likeness for molecules is crucial in their underlying distinctive proof as a synthetic lead and establishes a benchmark, against which incorporated compounds were measured amid lead advancement. Stages of Absorption, Distribution, Metabolism, Excretion of the ligand molecules were screened using SwissADME software [60]. This investigation aims mainly to approximate the pharmacokinetics profile of the examined compounds intrigued. The drug likeness profiles of newly synthesized compounds were predicted and ADMET properties are illustrated in Table 6 .
Table 6.
Molecular properties of compounds 3-4(a-d) and 5-6(a-h).
| Comp. | MW | cLogp | FC | TPSA | MR | NHA | NHD | NRB | NV5R |
|---|---|---|---|---|---|---|---|---|---|
| 3a | 310.28 | 2.84 | 0.06 | 64.23 | 85.85 | 4 | 1 | 1 | 0 |
| 3b | 322.31 | 1.88 | 0.11 | 73.46 | 92.39 | 4 | 1 | 2 | 0 |
| 3c | 306.32 | 2.69 | 0.11 | 64.23 | 90.86 | 3 | 1 | 1 | 0 |
| 3d | 292.29 | 2.72 | 0.06 | 64.23 | 85.89 | 3 | 1 | 1 | 0 |
| 4a | 372.78 | 3.53 | 0.16 | 53.23 | 99.78 | 4 | 0 | 4 | 0 |
| 4b | 384.81 | 2.56 | 0.20 | 62.46 | 106.31 | 4 | 0 | 5 | 0 |
| 4c | 368.81 | 3.38 | 0.20 | 53.23 | 104.78 | 3 | 0 | 4 | 0 |
| 4d | 354.79 | 3.42 | 0.16 | 53.23 | 99.82 | 3 | 0 | 4 | 0 |
| 5a | 466.48 | 3.67 | 0.22 | 123.48 | 135.28 | 7 | 1 | 5 | 1 |
| 5b | 478.52 | 2.33 | 0.25 | 132.71 | 141.81 | 7 | 1 | 6 | 2 |
| 5c | 462.52 | 3.51 | 0.25 | 123.48 | 140.29 | 6 | 1 | 5 | 1 |
| 5d | 448.49 | 3.30 | 0.22 | 123.48 | 135.32 | 6 | 1 | 5 | 1 |
| 5e | 452.46 | 3.05 | 0.18 | 123.48 | 130.47 | 7 | 1 | 5 | 1 |
| 5f | 464.49 | 2.12 | 0.22 | 132.71 | 137.01 | 7 | 1 | 6 | 2 |
| 5g | 448.49 | 2.89 | 0.22 | 123.48 | 135.48 | 6 | 1 | 5 | 1 |
| 5h | 434.47 | 2.95 | 0.18 | 123.48 | 130.51 | 6 | 1 | 5 | 1 |
| 6a | 472.49 | 4.62 | 0.19 | 50.48 | 140.02 | 4 | 0 | 3 | 2 |
| 6b | 484.52 | 3.65 | 0.21 | 59.71 | 146.55 | 4 | 0 | 4 | 2 |
| 6c | 468.52 | 4.45 | 0.21 | 50.48 | 145.03 | 3 | 0 | 3 | 2 |
| 6d | 454.50 | 4.25 | 0.19 | 50.48 | 140.06 | 3 | 0 | 0 | 2 |
| 6e | 484.52 | 4.32 | 0.21 | 59.71 | 146.55 | 4 | 0 | 4 | 3 |
| 6f | 496.56 | 3.63 | 0.24 | 68.94 | 153.09 | 4 | 0 | 5 | 2 |
| 6g | 480.56 | 4.15 | 0.24 | 59.71 | 151.56 | 3 | 0 | 4 | 1 |
| 6h | 466.53 | 3.95 | 0.21 | 59.71 | 146.60 | 3 | 0 | 4 | 1 |
MW: molecular weight (g/mol); cLogp; Calculated octanol/water partition Coefficient; FC: Fraction Csp3 TPSA: Topological Polar Surface Area; MR: Molar Refractivity; NHA: Number of Hydrogen Acceptor; NHD: Number of Hydrogen Donor; NRB: Number of Rotatable Bonds; NV: Number of Violation of 5 Rules.
3.8. Molecular docking results
Molecular docking tool aids in recognizing novel drug-like compounds that display high binding affinity with specific targets and rational ADMET characters. Docking is the most widespread program for specification of protein–ligand interactions. Docking results bought for every ligand with the receptors had been scrutinized aside from docking energy and binding modes, interaction of every ligand with the functional residues of PDBs.
The receptor: 6LZE (Resolution: 1.50 Å) and 6XFN (Resolution: 1.70 Å) the crystal structure of COVID-19 and SARS-CoV-2 (COVID-19) main proteases. Among the synthesized compounds 5a and 6b interacted commendably with PDB: 6LZE. Whereas, compounds 4d, 5b and 6f showed the highest conventional hydrogen bonding interactions with 6XFN.
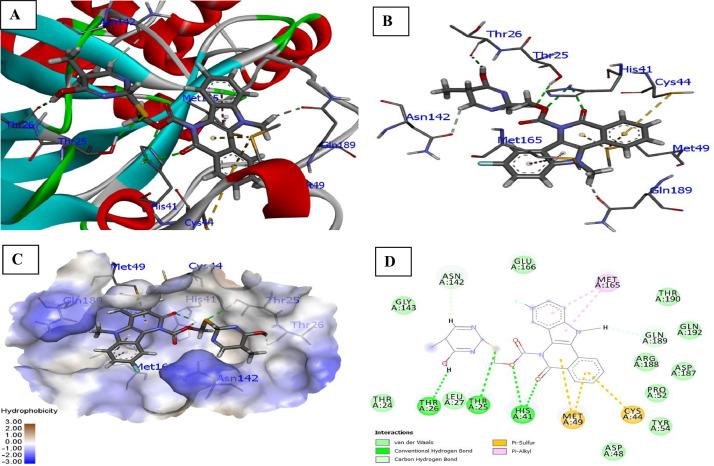
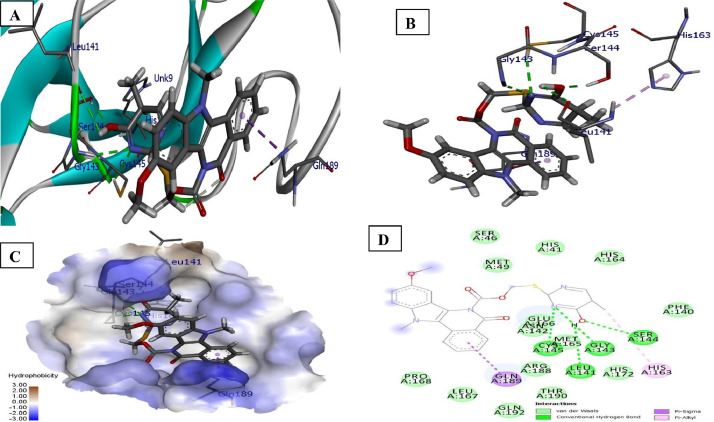
Detailed interactions between ligands and 6LZE: The interaction of 6LZE and ligand 5a with various amino acids present in the binding pockets of receptor is expressed in Fig. 1 . It was seen that compound 5a of indolo[3,2-c]isoquinoline carboxyl and acetate (oxygen atom) with NH-imidazole of His41 protease appeared to display two conventional hydrogen interaction at bond distance 2.79 and 2.97 Å. Furthermore, binding affinity of the sulphur atom of 5a with OH group of Thr25 (2.99 Å) and OH group of pyrimidine ring of ligand with Thr26 (2.41 Å) carbonyl, where conventional hydrogen bonding was noticed. The N–CH3 and pyrimidine were involved in carbon- hydrogen bonding with receptors Asn142 (2.41 Å) and Gln189 (2.59 Å). In addition to hydrophobic interactions, such as Pi-Alkyl (Met165) and Pi-Sulphur (Cys44, Met49) were found with ligand 5a.
Fig. 1.
Docked poses of compound 5a (stick figure) with COVID-19 main protease (PDB ID: 6LZE) (A) Protein backbone-ligand interactions; (B) Docked pose of ligand 5a (colour code for ligand: Black-C: White-H; Red-O; Blue-N) conventional hydrogen bonding interactions shown as dashed lines (Green) with 6LZE; (C) Lingard 5a with 6LZE (hydrophobicity surface) at the active binding site; (D) 2D view of interaction type of ligand 5a with surrounding amino acids of 6LZE. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
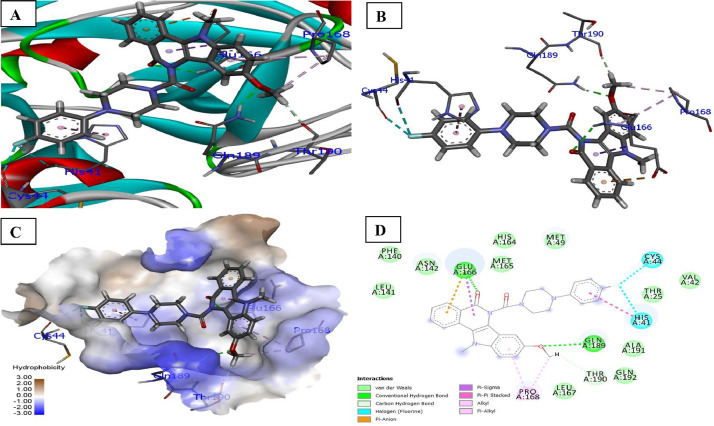
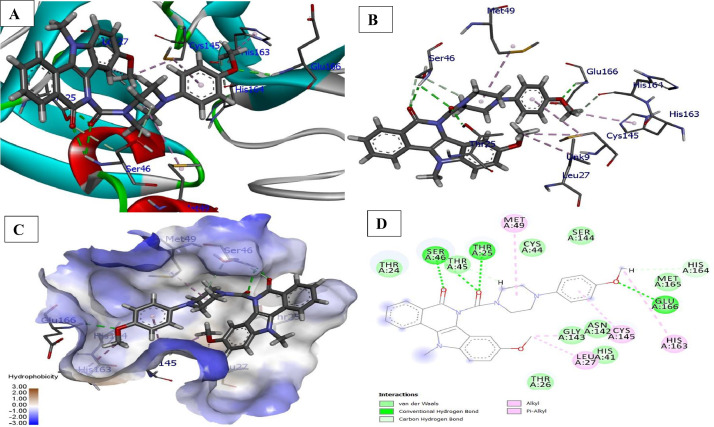
The docking of ligand 6e was performed with (COVID-19 main protease) in complex with a binding interaction of ligand 6e with 6LZE main protease of COVID-19 which revealed that indolo[3,2-c]isoquinoline carbonyl (C O) with NH of Glu166 (2.53 Å) receptor form conventional hydrogen bond interaction. Additionally, OCH3 (R2) of pyrimidine ring showed conventional hydrogen bond interaction with amino group (NH2) of Gln186 at bond distance 2.17 Å. The receptors Cys44, His44 of carbonyl groups halogen bound with florine atom at distance 3.03 and 3.08 Å. While carbon-hydrogen interaction (non-classic) with Thr190, the hydrophobic interactions of the ligand in the binding pocket such as Pi-Alkyl (His41, Pro168), Alkyl (Pro168) and Pi-Sigma (Glu166) were identified. The docked structure presented in Fig. 2 .
Fig. 2.
Docked poses of compound 6b (stick figure) with COVID-19 main protease (PDB ID: 6LZE) (A) Protein backbone-ligand interactions; (B) Docked pose of ligand 6b (colour code for ligand: Black-C: White-H; Red-O; Blue-N) conventional hydrogen bonding interactions shown as dashed lines (Green) with 6LZE; (C) Lingard 6b with 6LZE (hydrophobicity surface) at the active binding site; (D) 2D view of interaction type of ligand 6b with surrounding amino acids of 6LZE. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
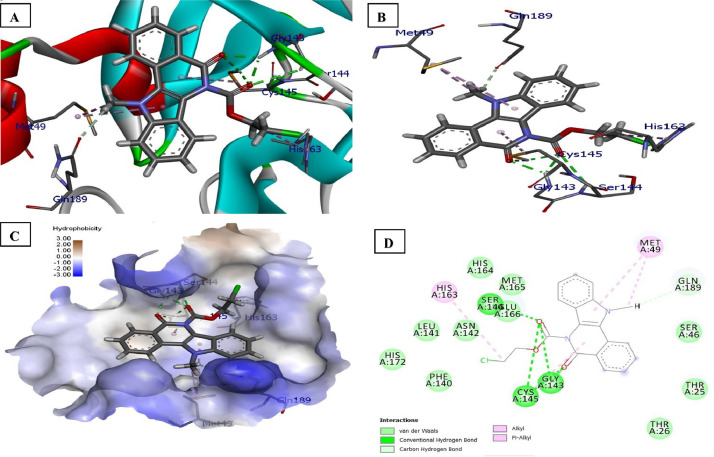
Detailed interactions between ligands and 6XFN: The carboxyl groups (C O) of indolo[3,2-c]isoquinoline ring and ester of the compound 4d forms five conventional hydrogen bond with the side chain residue of three amino acids namely Cys145, Gly143 and Ser144. Two bonds are formed by SH group of Cys145 (bond distance = 2.38 and 2,56 Å), anther NH group of Gly143(2.01 Å and 2.05 Å), one bond form by NH group of Ser144 (2.96 Å) with ester group. The carbon-hydrogen bond (non-classic) of Gln189 (carboxyl group) interacted with N–CH3 of ligand 4d. Other hydrophobic bond interaction of 4d includes pi-alkyl (Cys145, His163, Met49) and alkyl (Met49) at binding sites of the receptor (6XFN). The most fitting binding modes of 4d in the active site of 6XFN (COVID-19) are expressed in Fig. 3 . Further, six hydrogen bonding were identified between the protein and ligand 5b, four of which shows conventional hydrogen bonding with Cys145, Gly143, Ser144 of the receptor with an interatomic distance of 2.63, 2.63, 1.96 and 2.62 Å, respectively from 5b. In addition, His163 formed pi-alkyl bonds and Gln186 pi-sulfur bond with 5b. The docked structure presented in Fig. 4 . Correspondingly the carboxyl groups (C O) of indolo[3,2-c]isoquinoline ring and piperazine ring of compound 6f was found to be involved in two conventional hydrogen bonds by amide group of Ser46 at bond distance 2.31 and 2.81 Å. Similar, conventional hydrogen bonds were also formed between ligand and receptor i.e., piperazine C O with OH of Thr26(2.06 Å) and NH group of Glu166 (2.50 Å) with oxygen atom of OCH3 (R2). Further, indolo[3,2-c]isoquinoline carbonyl and piperazine ring forms carbon-hydrogen (C–H) interaction (non-classic) with Ser46, whereas OCH3 (R2) of ligand 6f involve in C–H interaction with carbonyl group of His164. Other hydrophobic interactions such as Pi-Alkyl (Cys145, His163), Alkyl (Met49, Leu27, Cys145) and Pi-Sigma (Ser46(2HB Thr26, Glu166) with 6f were identified. The results are illustrated in Fig. 5 . All synthesized compounds were found to inhibit the target proteins by completely occupying the active sites as shown in Tables 7 and 8 (please see supplementary information). Thus, it clearly suggests that compounds 4d, 5a, 5b, 6b and 6f display a positive range of drug likeness.
Fig. 3.
Docked poses of compound 4d (stick figure) with SARS-CoV-2 (COVID-19) main protease (PDB ID: 6XFN) (A) Protein backbone-ligand interactions; (B) Docked pose of ligand 4d (colour code for ligand: Black-C: White-H; Red-O; Blue-N) conventional hydrogen bonding interactions shown as dashed lines (Green) with 6XFN; (C) Lingard 4d with 6XFN (hydrophobicity surface) at the active binding site; (D) 2D view of interaction type of ligand 4d with surrounding amino acids of 6XFN. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Docked poses of compound 5b (stick figure) with SARS-CoV-2 (COVID-19) main protease (PDB ID: 6XFN) (A) Protein backbone-ligand interactions; (B) Docked pose of ligand 5b (colour code for ligand: Black-C: White-H; Red-O; Blue-N) conventional hydrogen bonding interactions shown as dashed lines (Green) with 6XFN; (C) Lingard 6b with 6XFN (hydrophobicity surface) at the active binding site; (D) 2D view of interaction type of ligand 5b with surrounding amino acids of 6XFN. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Docked poses of compound 6f (stick figure) with SARS-CoV-2 (COVID-19) main protease (PDB ID: 6XFN) (A) Protein backbone-ligand interactions; (B) Docked pose of ligand 6f (colour code for ligand: Black-C: White-H; Red-O; Blue-N) conventional hydrogen bonding interactions shown as dashed lines (Green) with 6XFN; (C) Lingard 6f with 6XFN (hydrophobicity surface) at the active binding site; (D) 2D view of interaction type of ligand 6f with surrounding amino acids of 6XFN. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Conclusion
In summary, the procedure of molecular hybridization has been effectively connected to the design of indolo[3,2-c]isoquinoline (δ-carboline) analogs bearing pyrimidine and piperazine moieties as a novel series. Synthesized compounds intended for biological activities. The results indicated that compounds 3a, 4a and 5e were strong against gram-negative bacteria, while compounds 5b, 6f and 6g antibacterial activity against gram-positive bacteria. Compounds 4a, 5a and 6a exhibit excellent antifungal activity. Moreover, the radical scavenging and ferrous ion (Fe2+) metal chelating impact of compound 5c demonstrated the board range in antioxidant activity. The expansion in anticancer action of compounds 5e and 6d can be credited to the electron-withdrawing groups such as `F' and `H' atoms. They cause active suppression of proliferation on human MCF-7, A549, HeLa and Panc-1 cell lines. Compounds 6b and 6e have good anti-tuberculosis action against H37Rv strain M. tuberculosis. Further studies are needed to realize the action mechanism of the compounds tested. Molecular docking was performed to predict the conceivable binding mode of compounds 4d, 5a, 5b, 6b and 6f with the PDBs: 6LZE (COVID-19) and 6XFN: SARS-CoV-2 (COVID-19) main proteases at active sites. Also, further investigations are required into biological validation (in-vitro and in-vivo) for more effects of the examined compounds. Some of the synthesized compounds would possibly represent preliminary leads for the design of more new potent multi-target therapeutic agents.
5. Experimental protocol
All reagents were commercially purchased and used without further purification. Melting points were determined using open capillaries and were not corrected. The purity of the compounds was verified by TLC using aluminum plates coated with silica gel G (Merck), hexane:ethyl acetate (3:1), chloroform:ethanol (5:1), benzene:methol (2:1) mixtures as solvent ratios and spots were visualized by exposing the dry plates in iodine vapours. The IR (KBr) spectra were recorded using a Perkin-Elmer Spectrum on FT-IR spectrometer. The 1H NMR (DMSO) spectra were recorded using Marcy Plus (Varian 400 MHz) and the chemical shifts were expressed in ppm (δ scale) and 13C NMR (125 MHz, DMSO) spectra recorded on Bruker NMR. Mass spectra were recorded using a ILS-CHU-C-41- VBV4 MS mass spectrometer. Elemental analysis was carried out using Flash EA 1112 series.
5.1. General procedure for the synthesis of ethyl 8-substituted-5-oxo-5H-indolo[3,2-c] isoquinoline-6(11H)-carboxylates (1a-1d) was by following literature procedure [42].
5.1.1. General procedure for the synthesis of ethyl 8-substituted-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (2a-2d).
To a combination of 1a-1d (0.1 mol), potassium carbonate (0.2 mol) in dimethylformamide (DMF) (40 mL), Tetra-n-butylammonium bromide (n-TBAB) as catalyst was added at 0 °C in mixture. Then, the mixture was stirred at room temperature for 30 min following which methyl iodide (0.3 mol) was added dropwise and continuously stirring for 12 h. The reaction progress was checked by using TLC. Then the reaction mixture was poured into ice-cold water and extracted with ethyl acetate. The organic layer was dried over anhydrous MgSO4 and evaporated under vacuum. The crude residue was purified by column chromatography (petroleum ether: ethyl acetate (4:1)) to obtain the pure compounds 2a-2d.
5.1.2. Ethyl 8-fluoro-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (2a)
Yellow shinning crystals, yield: 88%, mp 258–59 °C; Rf, 0.67; FTIR (KBr cm−1): 2954 (N–CH3), 1715, 1657 (C O); 1H NMR (DMSO-d6, δ, ppm): 6.9–8.2 (m, 7H, Ar-H), 4.7 (q, 2H, CH2), 4.0(s, 3H, N–CH3), 3.7 (t, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 165.2 (C O), 157.8 (C–F), 154.5(C19, C O), 134.1,131.1, 130.4, 129.5, 128.4, 128.3, 127.9, 126.4,120.2, 119.3, 119.2, 114.3, 106.3, 67.5 (CH2), 39.6(N–CH3), 14.3(CH3); MS: m/z 339 [M+H]+; Anal. Calcd. for C19H15N2O3F: C, 67.45; H, 4.47; N, 8.28. Found: C, 67.42; H, 4.44; N, 8.23%.
5.1.3. Ethyl 8-methoxy-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (2b)
Orange crystals, yield: 82%, m.p. 274–275 °C; Rf, 0.78; FTIR (KBr cm−1): 3014 (N–CH3), 1718, 1653 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–81 (m, 7H, Ar-H), 5.9 (s, 3H, OCH3), 4.3 (q, 2H, CH2), 3.9(t, 3H, N–CH3), 3.7 (s, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 165.7(C O), 155.1(C O), 135.2, 133.1, 132.3, 131.0, 129.9, 129.1, 127.8, 127.4, 127.1, 126.5, 125.2, 120.2, 114.3, 111.1, 57.1(CH2), 55.8 (OCH3), 36.1(N–CH3), 15.3(CH3); Anal. Calcd. for C20H18N2O4: C, 68.56; H, 5.18; N, 8.00;. Found: C, 68.52; H, 5.15; N, 8.05%.
5.1.4. Ethyl 8,11-dimethyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (2c)
Green shiny crystals, yield: 78%, m.p. 285–286 °C; Rf, 0.65; FTIR (KBr cm−1): 2947 (N–CH3), 1660, 1645 (C O); 1H NMR (DMSO-d6, δ, ppm): 6.7–7.9 (m, 7H, Ar-H), 4.2 (q, 2H, CH2), 3.9(s, 3H, N–CH3), 3.7 (t, 3H, CH3), 2.5 (s, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 166.5(C O), 156.5(C O), 136.2, 134.5, 133.2, 132.3, 130.0, 129.0, 127.2, 126.8, 126.5,124.5, 123.3, 121.2, 117.3, 109.2, 57.1(CH2), 38.2(N–CH3), 25.5(CH3), 19.1(CH3); Anal. Calcd. for C20H18N2O3: C, 71.84; H, 5.43; N, 8.38; Found: C, 71.82; H, 5.45; N, 8.35 %.
5.1.5. Ethyl 11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (2d)
Pale yellow solid, yield: 82%, m.p. 262–263 °C; Rf, 0.58; FTIR (KBr cm−1): 3051 (N–CH3), 1673, 1652 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 8H, Ar-H), 5.5 (q, 2H, CH2), 3.7(s, 3H, N–CH3), 2.9 (t, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 168.1(C O), 155.2(C O), 134.9, 134.3, 133.4, 132.6, 130.7, 129.7, 126.7, 125.8, 125.5, 124.3, 123.2, 120.2, 119.6, 110.4, 56.3(CH2), 36.7(N–CH3),16.2(CH3); Anal. Calcd. for C19H16N2O3: C, 71.24; H, 5.03; N, 8.74; Found: C, 71.21; H, 5.00; N, 8.71%.
5.2. General procedure for the synthesis of 8-Substituted-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acids (3a-3d)
To a solution of compounds 2a-d(0.01 mol) in ethanol (30 mL), 4N NaOH (30 mL) was added and heated under reflux for 3 h. The reaction mixture was cooled and acidified with 2N HCl. The completion of reaction was monitored by TLC using silica gel coated plates using solvents (ethyl acetate: petroleum ether 1:1) as eluent and observed in UV light. The solid residue was filtered off and washed with cold water, dried and purified from ethanol to get 3a-3d.
5.2.1. 8-Fluoro-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acid (3a)
White solid, yield: 87%, m.p. 271–272 °C; Rf, 0.58; FTIR (KBr cm−1): 2974 (N–CH3), 1676, 1638 (C O); 1H NMR (DMSO-d6, δ, ppm):10.6(1H, -OH), 7.1–8.3 (m, 7H, Ar-H), 3.9(s, 3H, N–CH3); 13C NMR (DMSO-d6, δ, ppm); 164.7(C O), 160.5(C O), 157.5 (C–F), 136.7, 135.9, 135.6, 134.3, 131.0, 130.6, 126.1, 125.0, 123.6, 123.4, 123.2, 119.4, 118.9, 36.6(N–CH3); MS: m/z 311 [M+H]+: Anal. Calcd. for C17H11N2O3F: C, 65.81; H, 3.57; N, 9.03; Found: C, 65.79; H, 3.55; N, 9.01 %.
5.2.2. 8-Methoxy-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acid (3b)
Greenish solid, yield: 81%, mp 262–263 °C; Rf, 0.73; FTIR (KBr cm−1): 2985 (N–CH3), 1675, 1635 (C O); 1H NMR (DMSO-d6, δ, ppm):10.2(1H, -OH), 6.9–8.0 (m, 7H, Ar-H), 3.9(s, 3H, N–CH3), 2.5 (s, 3H, OCH3); 13C NMR (DMSO-d6, δ, ppm); 163.1(C O), 152.2(C O), 134.7, 134.3, 132.7, 132.5, 132.2, 129.7, 128.1, 126.8, 125.5, 123.9, 122.7, 122.4, 120.1, 115.6, 55.4(OCH3), 38.2(N–CH3); Anal. Calcd. for C18H14N2O4: C, 67.07; H, 4.38; N, 8.69; Found: C, 67.05; H, 4.35; N, 8.70 %.
5.2.3. 8,11-Dimethyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acid (3c)
Yellow solid, yield: 75%, mp 291–292 °C; Rf, 0.59; FTIR (KBr cm−1): 2954 (indol,N–CH3), 1715, 1660 (C O); 1H NMR (DMSO-d6, δ, ppm):10.7(1H, -OH), 7.1–8.1 (m, 7H, Ar-H), 3.7(s, 3H, N–CH3), 2.9 (s, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 162.5(C O), 160.7(C O), 133.8, 133.5, 132.6, 132.4, 131.5, 130.5, 126.7, 126.8, 125.5,123.9, 122.7, 122.4, 120.1, 115.6, 39.4(CH3), 36.1(N–CH3); Anal. Calcd. for C18H14N2O3: C, 70.58; H, 4.61; N, 9.15; Found: C, 70.59; H, 4.59; N, 9.16%.
5.2.4. 11-Methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acid (3d)
Off-white crystal, yield: 75%, m.p. 274–275 °C; Rf, 0.61; FTIR (KBr cm−1): 3053 (N–CH3), 1710, 1651 (C O); 1H NMR (DMSO-d6, δ, ppm):10.2(1H, -OH), 7.1–8.1 (m, 8H, Ar-H), 3.7(s, 3H, N–CH3); 13C NMR (DMSO-d6, δ, ppm); 165.0(C O), 159.3(C O), 134.5, 133.7, 133.2, 132.7, 132.6, 131.2, 125.7, 124.8, 124.1,123.5, 123.2, 121.5, 120.4, 117.6, 32.7(N–CH3); Anal. Calcd. for C17H12N2O3: C, 69.86; H, 4.14; N, 9.58; Found: C, 69.85; H, 4.13; N, 9.57%.
5.3. General procedure for the synthesis of 2-Chloroethyl 8-substituted-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylates (4a-4d)
To a solution of 8-substituted-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylic acids (3a-3d) (1 mol), dichloroethane (2 mol) and anhydrous K2CO3 (2 mol) in acetone (40 mL) was added and resulting solution was refluxed for 6 h. The resulting residue was washed with petroleum ether and distilled water progressively and separated under reduced pressure. Then the residue was dried and purified by silica gel chromatography (petroleum ether: ethyl acetate (2:1)) as eluent to afford compounds 4a-4d.
5.3.1. 2-Chloroethyl 8-fluoro-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (4a)
Yellow crystals, yield: 78%, m.p. 301–302 °C; Rf, 0.77; FTIR (KBr cm−1): 2962 (N–CH3), 1715, 1657 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.3 (m, 7H, Ar-H), 4.9 (q, 2H, CH2), 4.6 (q, 2H, CH2Cl), 3.9(s, 3H, N–CH3); 13C NMR (DMSO-d6, δ, ppm); 164.7(C O), 160.8(C O), 157.5 (C–F), 137.1, 134.3, 132.7, 131.1, 130.9, 130.8, 130.5, 129.1,128.3, 127.6, 120.2, 117.3, 113.9, 72.6(CH2), 39.5(N–CH3), 29.4(CH2Cl); MS: m/z 420 [M++2]; Anal. Calcd. for C19H14N2O3FCl: C, 61.22; H, 3.79; N, 7.51;Found: C, 61.21; H, 3.74; N, 7.49%.
5.3.2. 2-Chloroethyl 8-methoxy-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (4b)
Orange crystals, yield: 72%, m.p. 298–299 °C; Rf, 0.73; FTIR (KBr cm−1): 2951 (N–CH3), 1712, 1655 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 7H, Ar-H), 4.5 (q, 2H, CH2), 4.1 (q, 2H, CH2Cl); 3.9 (s, 3H, OCH3), 3.7(s, 3H, N–CH3), 13C NMR (DMSO-d6, δ, ppm); 168.1(C O), 160.0(C O), 134.3, 134.0, 132.2, 132.0, 128.9, 128.1, 126.9, 126.4, 125.3,124.3, 124.2, 121.1, 116.2, 110.2, 65.3(CH2), 55.3 (OCH3), 36.1(N–CH3), 25.7(CH2Cl); Anal. Calcd. for C20H17N2O4Cl: C, 62.42; H, 4.45; N, 7.28; Found: C, 62.38; H, 4.40; N, 7.26 %.
5.3.3. 2-Chloroethyl 8,11-dimethyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (4c)
Green solid, yield: 71%, m.p. 295–296 °C; Rf, 0.69; FTIR (KBr cm−1): 2983 (N–CH3), 1670, 1652 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 7H, Ar-H), 4.8 (q, 2H, CH2), 4.2 (q, 2H, CH2Cl), 3.9(s, 3H, N–CH3), 2.9 (s, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 167.1(C O), 159.9(C O), 134.3, 134.2, 132.5, 132.2, 131.2, 129.4, 126.7, 125.6, 124.7,124.2, 123.3, 120.2, 119.3, 110.1, 63.1(CH2), 38.2(N–CH3), 29.2(CH2Cl), 25.6(CH3); Anal. Calcd. for C20H17N2O3Cl: C, 65.13; H, 4.65; N, 7.60; Found: C, 65.10; H, 4.63; N, 7.56 %.
5.3.4. 2-Chloroethyl 11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (4d)
Yellow crysstal, yield: 72%, m.p. 287–288 °C; Rf, 0.65; FTIR (KBr cm−1): 3001 (N–CH3), 1670, 1661 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 8H, Ar-H), 5.6 (q, 2H, CH2), 4.0 (q, 2H, CH2Cl); 3.9(s, 3H, N–CH3), 13C NMR (DMSO-d6, δ, ppm); 162.5(C O), 159.6(C O), 134.5, 134.2, 133.2, 132.5, 129.8, 129.5, 125.9, 125.5, 124.5,124.2, 123.2, 121.3, 118.7, 109.2, 60.2(CH2), 36.7(N–CH3),28.2(CH2Cl); Anal. Calcd. for C19H15N2O3Cl: C, 64.32; H, 4.26; N, 7.90; Found: C, 64.28; H, 4.20; N, 7.87 %.
5.4. General procedure for the synthesis of 2-(5-cyano-4-hydroxy-6-methoxypyrimidin-2-ylthio)ethyl 8-Substitued-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylates (5a-5h)
Compounds 4a-d (3 mol), substituted pyrimidine-2-thiol (3 mol) and dry K2CO3 (3 mol) were introduced to dry DMF (20 mL). The reaction mixture was stirred at room temperature for 8–10 h. When the reaction was completed (by means of TLC), the reaction mixture was poured into ice water (100 mL) and acidified with acetic acid. The obtained residue was filtered, washed with water, dried and purified by silica gel (petroleum ether: ethyl acetate (3:1)) to afford the compounds 5a–5h.
5.4.1. 2-(4-Hydroxy-5-methylpyrimidin-2-ylthio)ethyl 8-fluoro-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5a)
Yellow crystals, yield: 68%, m.p. 311–312 °C; Rf, 0.77; FTIR (KBr cm−1): 3154 (O–H), 2984 (N–CH3), 1715, 1657 (C O),1612, 1540(C N), 1153(C-O); 1H NMR (DMSO-d6, δ, ppm): 10.8(s,1H,OH),7.1–8.4 (m, 8H, Ar-H), 4.7 (t, 2H, CH2), 4.2(t, 2H, S-CH2), 3.9(s, 3H, N–CH3), 1.5(s,3H,CH3); 13C NMR (DMSO-d6, δ, ppm); 177.2, 161.5(C O), 161.0(C O), 159.8 (C–F), 154.9(C N), 154.8(C N), 137.8, 135.1, 134.1, 131.9, 130.2,128.6, 127.1,126.6,120.7, 118.7,115.1, 114.2, 113.8, 107.0, 65.4(CH2), 39.9(CH2), 36.4(N–CH3), 14.4(CH3); MS: m/z 479 [M+H]+; Anal. Calcd. for C24H19N4O4FS: C, 60.24; H, 4.00; N, 11.71;Found: C, 60.22; H, 3.98; N, 11.68%.
5.4.2. 2-(4-Hydroxy-5-methylpyrimidin-2-ylthio)ethyl 8-methoxy-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5b)
White solid, yield: 72%, m.p. 300–301 °C; Rf, 0.56; FTIR (KBr cm−1): 3162 (O–H), 2954 (N–CH3), 1725, 1659 (C O),1610, 1532(C N), 1153(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.5(s,1H,OH),7.0–8.2 (m, 8H, Ar-H), 4.7 (t, 2H, CH2), 3.9(s, 3H, N–CH3), 3.7(t, 2H, S-CH2), 2.7(t, 3H, CH3), 2.1(t, 3H, OCH3); 13C NMR (DMSO-d6, δ, ppm); 178.2, 167.1(C O), 162.3(C N), 156.2, 155.5(C O), 154.2(C N), 132.5, 132.0, 130.7, 129.9, 129.1,128.3, 128.0,127.2,123.1, 119.2,115.4, 114.2, 111.9, 105.2, 59.1(CH2), 55.7 (OCH3), 37.3(CH2), 35.6(N–CH3), 14.5(CH3); Anal. Calcd. for C25H22N4O5S: C, 61.21; H, 4.52; N, 11.42;Found: C, 61.20; H, 4.50; N, 11.41 %.
5.4.3. 2-(4-Hydroxy-5-methylpyrimidin-2-ylthio)ethyl 8,11-dimethyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5c)
Yellowish solid, yield: 74%, m.p. 297–298 °C; Rf, 0.61; FTIR (KBr cm−1): 3123 (O–H), 2974 (N–CH3), 1710, 1675 (C O),1612, 1518(C N), 1150(C–O);1H NMR (DMSO-d6, δ, ppm): 10.3(s,1H,OH),6.9–8.0 (m, 8H, Ar-H), 5.1 (t, 2H, CH2), 4.4(t, 2H, S–CH2), 4.5(s, 3H, N–CH3), 2.5(t, 3H, CH3), 2.2(t, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 175.1,163.1(C O), 159.2(C N), 154.5, 153.4(C O), 152.6(C N), 132.7, 132.2, 131.3, 130.4, 129.3,128.7, 128.2,127.5, 123.6, 120.1, 119.3, 115.1, 113.5, 101.4, 60.3(CH2), 37.3(CH2), 36.6(N–CH3), 25.7 (CH3), 15.4(CH3); Anal. Calcd. for C25H22N4O4S: C, 63.28; H, 4.67; N, 11.81;Found: C, 63.25; H, 4.62; N, 11.84 %.
5.4.4. 2-(4-Hydroxy-5-methylpyrimidin-2-ylthio)ethyl 11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5d)
Pale yellow crystals, yield: 67%, m.p. 298–299 °C; Rf, 0.65; FTIR (KBr cm−1): 3123 (O–H), 2971 (N–CH3), 1705, 1650 (C O),1624, 1521 (C N), 1132(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.1(s,1H,OH),7.1–8.1 (m, 9H, Ar-H), 5.6 (t, 2H, CH2), 4.3(t, 2H, S-CH2), 3.9(t, 3H, N–CH3), 2.8(t, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 172.7, 165.5(C O), 156.4(C N), 155.5, 154.2(C O), 153.1(C N), 133.6, 133.5, 132.9, 131.2, 130.1,129.3, 128.1,126.6, 122.3, 119.5,118.2, 112.3, 112.1, 104.2, 59.2(CH2), 38.6(CH2), 36.7(N–CH3), 14.5(CH3); Anal. Calcd. for C24H20N4O4S: C, 62.60; H, 4.38; N, 12.17;Found: C, 62.61; H, 4.36; N, 12.15%.
5.4.5. 2-(4-Hydroxypyrimidin-2-ylthio)ethyl-8-fluoro-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5e)
Organ solid, yield: 75%, m.p. 310–311 °C; Rf, 0.71; FTIR (KBr cm−1): 3110 (O–H), 2961 (N–CH3), 1712, 165o (C O),1612, 1540(C N), 1153(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.6(s,1H,OH),7.0–8.0 (m, 9H, Ar-H), 4.8 (t, 2H, CH2), 4.5(t, 2H, S–CH2); 3.6(t, 3H, N–CH3), 13C NMR (DMSO-d6, δ, ppm); 173.4,165.5(C O), 163.4(C N), 156.4 (C–F),155.6(C O), 155.2(C N), 132.3,132.1, 131.4, 130.7, 129.4,128.6, 128.4,127.3,122.2, 117.1,112.4, 112.2, 111.2, 102.5, 59.3(CH2), 38.8(CH2), 36.6(N–CH3); Anal. Calcd. for C23H17N4O4FS: C, 59.48; H, 3.69; N, 12.06; Found: C, 59.45; H, 3.67; N, 12.03%.
5.4.6. 2-(4-Hydroxypyrimidin-2-ylthio)ethyl 8-methoxy-11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5f)
Yellow crystals, yield: 69%, m.p. 312–313 °C; Rf, 0.66; FTIR (KBr cm−1): 3184 (O–H), 2964 (N–CH3), 1713, 1650 (C O),1613, 1530(C N), 1149(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.7(s,1H,OH),7.1–7.9 (m, 9H, Ar-H), 5.1 (t, 2H, CH2), 4.2(t, 2H, S–CH2), 3.6(t, 3H, N–CH3), 2.5(t, 3H, OCH3); 13C NMR (DMSO-d6, δ, ppm); 172.1,165.2(C O), 160.1(C N), 156.3,155.4(C O), 153.5(C N), 132.7, 132.4, 131.8, 130.1, 129.4, 128.2, 128.1, 127.3, 124.2, 119.7,115.2, 112.5, 111.1, 102.3, 56.6(CH2), 54.5(OCH3), 38.8(CH2), 36.6(N–CH3); Anal. Calcd. for C24H20N4O5S: C, 60.49; H, 4.23; N, 11.76;Found: C, 60.50; H, 4.20; N, 11.72%.
5.4.7. 2-(4-Hydroxypyrimidin-2-ylthio)ethyl 8,11-dimethyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5g)
Off-white crystals, yield: 74%, m.p. 297–298 °C; Rf, 0.61; FTIR (KBr cm−1): 3126 (O–H), 2958 (N–CH3), 1732, 1665 (C O),1613, 1519(C N), 1145(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.3(s,1H,OH),6.8–7.9 (m, 9H, Ar-H), 4.9 (t, 2H, CH2), 4.3(t, 2H, S–CH2), 3.6(t, 3H, N–CH3), 2.8(t, 3H, CH3); 13C NMR (DMSO-d6, δ, ppm); 172.4, 165.2(C O), 160.2(C N), 153.5, 152.4(C O), 152.1(C N), 132.4, 132.3, 131.2, 131.0, 129.1,128.4, 128.5, 127.2,124.5, 121.3,120.5, 118.5, 112.7, 109.3, 58.8(CH2), 38.9(CH2), 36.7(N–CH3), 26.1 (CH3); Anal. Calcd. for C24H20N4O4S: C, 62.60; H, 4.38; N, 12.17;Found: C, 62.58; H, 4.36; N, 12.14%.
5.4.8. 2-(4-Hydroxypyrimidin-2-ylthio)ethyl 11-methyl-5-oxo-5H-indolo[3,2-c]isoquinoline-6(11H)-carboxylate (5h)
Greenish, yield: 67%, m.p. 298–299 °C; Rf, 0.65; FTIR (KBr cm−1): 3164 (O–H), 2976 (N–CH3), 1712, 1658 (C O),1611, 1521 (C N), 1130(C–O); 1H NMR (DMSO-d6, δ, ppm): 10.7(s,1H,OH),7.0–8.2 (m, 10H, Ar-H), 5.1 (t, 2H, CH2), 4.6(t, 2H, S–CH2)3.8(t, 3H, N–CH3),; 13C NMR (DMSO-d6, δ, ppm); 172.1,165.3(C O), 157.2(C N), 154.6, 154.2(C O), 152.6(C N), 132.6, 132.5, 132.3, 131.5, 130.7,129.4, 128.6,126.7,122.1, 119.4,118.2, 117.1, 110.4, 102.1, 58.3(CH2), 39.5(CH2), 36.4(N–CH3); Anal. Calcd. for C23H18N4O4S: C, 61.87; H, 4.06; N, 12.55;Found: C, 61.85; H, 4.03; N, 12.52%.
5.5. General procedure for the synthesis of 8-Substitued-6-{[4-(4-nitrophenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-ones (6a-6h)
Compounds (3a-3d) (0.001 mol), hydroxylbenzotriazole (0.012 mol) in dry Tetrahydrofuran (THF) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDCH) (0.012 mol) were mixed and stirred for 30 min. To the reaction mixture, substituted phenylpiperazine (0.001 mol) was included under ice cold temperature and the reaction mixture was further stirred at room temperature for 6 h. After finish of reaction as checked by TLC, Reaction mixture was separated with ethyl acetate, collected organic layer was dried over Na2SO4 and concentrated under vacuum to give compounds 6a-6h. Hexanine as solvent for TLC.
5.5.1. 8-Fluoro-6-{[4-(4- fluoro phenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6a)
Yellow crystals, yield: 68%, m.p. 311–312 °C; Rf, 0.50 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2959 (N–CH3), 1715, 1657 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.1–8.3 (m, 11H, Ar-H), 5.3–4.2(m, 8H, CH2), 3.9(s, 3H, N–CH3);13C NMR (DMSO-d6, δ, ppm); 164.7 (C O), 160.4(C O), 154.8 (C–F), 150.5 (C–F),148.0, 137.1, 134.3, 132.7, 130.5, 129.1, 128.6, 128.2, 127.6, 127.1, 123.9, 123.6, 120.2, 117.3,115.4, 114.3, 113.9, 101.2, 40.5(piperazine ring), 39.8(piperazine ring), 37.6 (N–CH3); MS: m/z 473 [M+H] +; Anal. Calcd. for C27H22N4O4F2: C, 68.63; H, 4.69; N, 11.86; Found: C, 68.60; H, 4.65; N, 11.83 %.
5.5.2. 8-Methoxy-6-{[4-(4- fluorophenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6b)
White crystals, yield: 66%, m.p. 278–279 °C; Rf, 0.47 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2897 (N–CH3), 1687, 1620 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.0 (m, 9H, Ar-H), 5.8–4.1(m, 8H, CH2), 3.9(s, 3H, N–CH3), 2.7 (s,3H OCH3); 13C NMR (DMSO-d6, δ, ppm); 164.2 (C O), 160.1(C O), 151.2(C–F), 143.2, 133.6, 132.4, 131.4, 130.2, 128.5, 128.2, 128.0, 127.2, 122.3, 116.5, 116.2, 115.6, 115.3,113.4, 112.4, 112.2, 105.2, 55.7 (OCH3), 51.3 (piperazine ring), 48.9 (piperazine ring), 36.6 (N–CH3); Anal. Calcd. for C28H25N4O3F: C, 69.41; H, 5.20; N, 11.56; Found: C, 69.43; H, 5.18; N, 11.53 %.
5.5.3. 8-Methyl-6-{[4-(4- fluorophenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one(6c)
White solid, yield: 70%, m.p. 281–282 °C; Rf, 0.38 (Hexanine: EtOAc = 6:4); FTIR (KBr cm−1): 2984 (N–CH3), 1687, 1620 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.1–8.1 (m, 11H, Ar-H), 5.6–40(m, 8H, CH2), 3.8 (s, 3H, N–CH3), 3.2 (s,3H CH3); 13C NMR (DMSO-d6, δ, ppm); 165.1 (C O), 161.2(C O), 152.4 (C–F), 141.1, 132.5, 132.3, 130.9, 129.8, 128.2, 128.5, 127.9, 127.1, 124.2, 117.5, 116.8, 116.2, 115.1, 112.7, 112.1, 112.0, 104.3, 51.3(piperazine ring), 48.9 (piperazine ring), 36.5(N–CH3), 25.7 (CH3); Anal. Calcd. for C28H25N4O2F: C, 71.78; H, 5.38; N, 11.96; Found: C, 71.75; H, 5.37; N, 11.93 %.
5.5.4. 6-{[4-(4- Fluorophenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6d)
White crystals, yield: 74%, m.p. 294–295 °C; Rf, 0.47 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2968 (N–CH3), 1670, 1631 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.2 (m, 12H, Ar-H), 5.5–4.3 (m, 8H, CH2), 3.8 (s, 3H, N–CH3); 13C NMR (DMSO-d6, δ, ppm); 165.1 (C O) 161.2(C O),152.4(C–F), 141.1, 132.5, 132.3, 130.9, 129.8, 128.2, 128.5, 127.9, 127.1, 125.7, 124.2, 117.5, 116.8, 116.2, 115.1, 112.7, 112.1, 112.0, 104.3, 51.3 (piperazine ring), 48.9(piperazine ring), 37.2(N–CH3); Anal. Calcd. for C27H23N4O2F: C, 71.35; H, 5.10; N, 12.33;Found: C, 71.32; H, 5.08; N, 12.30;%.
5.5.5. 8-Fluoro-6-{[4-(4-methoxyphenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6e)
White solid, yield: 68%, m.p. 259–260 °C; Rf, 0.49 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2983 (N–CH3), 1720, 1634 (C O); 1H NMR (DMSO-d6, δ, ppm): 6.8–8.1 (m, 11H, Ar-H), 5.7–4.5(m, 8H, CH2), 3.9(s, 3H, N–CH3), 2.9(s, 3H, OCH3); 13C NMR (DMSO-d6, δ, ppm); 163.5 (C O), 161.3(C O), 154.7 (C–F), 152.7, 142.7, 132.4, 132.1, 131.6, 130.2, 128.4, 128.2, 128.0, 126.3, 122.3, 116.7, 116.2, 115.6, 115.2, 113.7, 112.6, 112.3, 102.4, 55.6 (OCH3), 51.8(piperazine ring), 49.1(piperazine ring), 36.6 (N–CH3); Anal. Calcd. for C28H25N4O3F: C, 69.41; H, 5.20; F, 3.92; N, 11.56; Found: C, 69.39; H, 5.17; F, 3.92; N, 11.47; %.
5.5.6. 8-Methoxy-6-{[4-(4-methoxyphenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one 6f
Yellow solid yield: 67%, m.p. 275–276 °C; Rf, 0.35 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2973 (N–CH3), 1680, 1642 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 11H, Ar-H), 5.6–4.2(m, 8H, CH2), 3.7(s, 3H, N–CH3), 2.7 (s,3H OCH3), 2.3 (s,3H OCH3); 13C NMR (DMSO-d6, δ, ppm); 168.1 (C O), 160.1(C O), 151.2, 143.2, 133.2, 132.3, 131.2, 130.1, 128.3, 128.8, 128.5, 126.7, 122.8, 116.7, 116.4, 115.7, 113.4, 112.4, 112.2, 105.2, 55.7 (OCH3), 55.3 (OCH3), 51.3(piperazine ring), 48.9 (piperazine ring), 34.9 (N–CH3); Anal. Calcd. for C29H28N4O4: C, 70.15; H, 5.68; N, 11.28; Found: C, 70.12; H, 5.64; N, 11.23 %.
5.5.7. 8-Methyl-6-{[4-(4-methoxyphenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6g)
Green solid, yield: 68%, m.p. 264–265 °C; Rf, 0.51 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2985 (N–CH3), 1710, 1640 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.0 (m, 11H, Ar-H), 5.2–4.1 (m, 8H, CH2), 3.9 (s, 3H, N–CH3), 3.1 (s,3H CH3), 2.1 (s,3H OCH3); 13C NMR (DMSO-d6, δ, ppm); 165.1 (C O) 161.2(C O), 152.4 (C–F), 141.1, 132.5, 132.3, 130.9, 129.8, 128.2, 128.5, 127.9, 127.1, 124.2, 117.5, 116.8, 116.2, 115.1, 112.7, 112.1, 112.0, 104.3, 59.7 (OCH3), 50.8(piperazine ring), 49.9 (piperazine ring), 36.9(N–CH3), 28.6 (CH3); Anal. Calcd. for C29H28N4O3: C, 72.48; H, 5.87; N, 11.66; Found: C, 72.47; H, 5.85; N, 11.63%.
5.5.8. 6-{[4-(4-Methoxyphenyl)piperazin-1-yl]oxomethyl}-11-methyl-6H-indolo[3,2-c]isoquinolin-5(11H)-one (6h)
White crystals, yield:71%, m.p. 280–281 °C; Rf, 0.36 (Hexanine:EtOAc = 6:4); FTIR (KBr cm−1): 2964(N–CH3), 1687, 1652 (C O); 1H NMR (DMSO-d6, δ, ppm): 7.0–8.1 (m, 12H, Ar-H), 5.4–4.0 (m, 8H, CH2), 3.7 (s, 3H, N–CH3), 2.4 (s,3H OCH3); 13C NMR (DMSO-d6, δ, ppm); 165.1 (C O) 161.2(C O),152.4(C–F), 141.1, 132.5, 132.3, 130.9, 129.8, 128.2, 128.5, 127.9, 127.1, 125.7,124.2, 117.5, 116.8, 116.2, 115.1,112.7, 112.1, 112.0, 104.3, 55.6 (OCH3); 51.3(piperazine ring),48.9(piperazine ring), 36.6(N–CH3); Anal. Calcd. for C28H26N4O3 C, 72.09; H, 5.62; N, 12.01;Found: C, 72.05; H, 5.64; N, 12.00%.
6. Biological procedure
6.1. Antimicrobial activity
The synthesized compounds were tested against four bacteria and four fungi. The bacterial strains incubated at 106 microbe/mL concentration by the broth dilution method (concentrations 100, 75, 50, 25, 12.5, 6.5 3.12 and 1.5 (µg/mL). The optical density was measured at 655 nm wavelength for each sample and compared with standard drugs streptomycin and fluconazole for antibacterial and antifungal activities, respectively. The lowest concentration of the test compound was considered as precise MIC values. At this concentration, growth of bacteria and fungi were completely inhibited.
6.2. Antioxidant activities
6.2.1. 1, 1-Diphenyl-2-picryl-hydrazil (DPPH) radical scavenging activity (RSA)
The RSA of the test compounds was carried out in methanolic solution at concentrations 25, 50, 75 and 100 µg/mL containing freshly prepared DPPH solution (0.004% w/v) and compared to standards using reported method [57]. All the analysis were performed on averaged of triplicates. The results are expressed as the ratio of absorption decrease in the presence of test compounds and absorption of DPPH solution in the absence of test compounds at 517 nm on ELICO SL171 mini spect spectrometer in percentage. The percentage scavenging activity of the DPPH free radical was determined using the following equation:
where,
Ac is the absorbance of the control reaction and As is the absorbance of the sample or standards.
6.3. Ferrous (Fe2+) ion metal chelating activity
The metal chelating activity of the ferrous ions (Fe2+) towards the synthetic compounds was determined by the Denis method by the ferrous ions and BHA as standards. In this strategy, samples of ethanol solution (0.4 ml) in a solution of ferrous chloride (0.05 ml, 2 ml) were included in 25, 50, 75, and 100 μg / mL at concentrations. The reaction was initiated by the addition of ferrozine (0.02 mL, 5 mM) and the total volume was made up to 4 mL in ethanol. The mixture was shaken vigorously and incubated at room temperature for 10 min and absorbance at 517 nm were estimated using a UV–Visible spectrophotometer. The percentage of inhibition of the ferrozine complex formation was determined using the accompanying equation.
6.4. Anticancer activity
The anticancer activity of synthesized compounds were tested against three human cancer cell lines including MCF-7 (breast carcinoma), A-549 (lung carcinoma) and HeLa (Cervical carcinoma). The synthesized compounds were diluted to various concentrations (10, 5, 2.5 and 1.25 μg ML−1) in Dimethyl Sulfoxide (DMSO) and assessed using 3-(4, 5-dimethyl-2-yl-2, 5-diphenyl tetrazolium bromide (MTT assay). Anticancer activity was determined for cells treated with various concentrations of the titled compounds, the untreated cells (negative control) and Doxorubicin (positive control). A statistical significance was tested between sample and negative control using independent t-test by SPSS 12 program. The concentration of compounds required to kill half of the cell population (IC50) were determined by non-linear regression analysis. Cytotoxic activity was expressed as the mean IC50 of three independent experiments.
6.5. Anti-tuberculosis activity
The anti-TB activity of the synthesized compounds were determined as per reported method. The DMSO dissolved test compounds were further diluted in cation adjusted Mueller–Hinton broth (MHB) to a concentration of 256 mg/L. To each well of 125 μL of MHB, 125 μl of each test solution were added to obtain 128 mg/L starting concentration. A twofold serial dilution of test solutions of range 128–0.25 mg/L was prepared. Columbia blood agar supplemented with 7% defibrinated horse blood was the used media. Bacterial suspension with a turbidity of 0.5 on the MacFarland scale was made in 0.9% NaCl solution and diluted to inoculum density of 5 × 105 cfu/mL. Each well was inoculated with 125 µL of inocula, except the blank. The plates were incubated at 37 °C for 72 h. The lowest concentration of test compounds that completely fail to produce macroscopic growth (MIC) were detected by incubation at 35 °C for 20 min with 20 µL methanolic solution of tetrazolium redox dye (MTT, 3.5 mg/mL). Tests were done in triplicates and ethambutol, isoniazid and rifampicin were used as positive controls.
6.6. Molecular docking protocol
All the synthesized ligands were subsequently prepared using Chemdraw ultra 12 in CDX format. The ligands were converted into three-dimensional structure by Minimize Energy to Minimum RMS Gradient of 0.100 in Chem3D ultra and saved as MDL, MolFiles. The molfile of ligands were then converted to smiles format using Open Babel. The molecular docking studies were performed for PDBs: 6LZE and 6XFN the crystal structure of COVID-19 and SARS-CoV-2 (COVID-19) main proteases downloaded from the Protein Data Bank (https://www.rcsb.org). Protein was prepared using BIOVIA Discovery Studio Visualizer 2020 to define and edit binding site after the preparation of protein and saved as PDB format. Further the ligands’ smiles format and prepared protein (PDB) have been imported into mcule (mcule.com: online drug discovery platform) for molecular docking and the docked protein-ligands interactions poses were downloaded for docking studies.
Declaration of Competing Interest
The authors confirm that there is no conflict of interest in the content of this article.
Acknowledgements
The authors are thankful to the Principal, Sri Prabhu Arts, Science and J. M. Bohra Commerce Degree College, Shorapur-585 224, Yadgir, Karnataka, India for provide laboratory facilities. Authors are grateful to the Directors, IIT Madras, Chennai, India provide spectral data, National Collection of Industrial Microorganisms (NCIM), National Chemical Laboratory (NCL) and National Centre for Cell Science (NCCS), Pune, India to providing test materials.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2020.129829.
Appendix. Supplementary materials
References
- 1.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G., Clercq E.D. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 3.Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.W., Kang Y.M., Lee B., Park S.J. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of Lopinavir/Ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT PCR. J. Korean Med. Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Three drugs fairly effective on novel coronavirus at cellular level-Xinhua | English.news.cn http://www.xinhuanet.com/english/2020-01/30/c_138742163.htm Accessed: 2020-04-11.
- 8.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y., Ea M. Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elfiky A.A., Mahdy S.M., Elshemey W.M. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J. Med. Virol. 2017;89:1040–1047. doi: 10.1002/jmv.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falagas M.E., Kouranos V.D., Athanassa Z., Kopterides P. Tuberculosis and malignancy. Q. J. Med. 2010;103(7):461–487. doi: 10.1093/qjmed/hcq068. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y.I., Goo J.M., Kim H.Y. Coexisting bronchogenic carcinoma and pulmonary tuberculosis in the same lobe: radiologic findings and clinical significance. Korean J. Radiol. 2001;2(3):138–144. doi: 10.3348/kjr.2001.2.3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenner A.V., Wang Z., Kleinerman R.A., Wang L., Zhang S., Metayer C., Chen K., Lei S., Cui H., Lubin J.H. Previous pulmonary diseases and risk of lung cancer in Gansu Province, China. Int. J. Epidemiol. 2001;30:118–124. doi: 10.1093/ije/30.1.118. [DOI] [PubMed] [Google Scholar]
- 15.Storz P. Reactive oxygen species in tumor progression. Front. Biosci. 2005;1(10):1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 16.Szatrowski T.P., Nathan C.F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–798. 1. [PubMed] [Google Scholar]
- 17.Lea A.J. Dietary factors associated with death-rates from certain neoplasms in man. Lancet. 1966;2:332–333. doi: 10.1016/s0140-6736(66)92615-8. [DOI] [PubMed] [Google Scholar]
- 18.Valko M., Leibfritz D., Moncol J., Cronin M.T.D., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell B. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Bulkley G.B. Free radicals and other reactive oxygen metabolites: clinical relevance and the therapeutic efficacy of antioxidant therapy. Surgery. 1993;113(5):479–483. [PubMed] [Google Scholar]
- 20.Halliwell B., Gutteridge M. Oxford Science Publications, Oxford University Press; 1998. Free Radicals in Biology and Medicine. third ed. [Google Scholar]
- 21.Thomson M.J., Frenneaux M.P., Kaski J.C. Antioxidant treatment for heart failure: friend or foe? QJM. 2009;102(5):305–310. doi: 10.1093/qjmed/hcn160. [DOI] [PubMed] [Google Scholar]
- 22.Waris G., Ahsan H.J. Reactive oxygen species: role in the development of cancer and various chronic conditions. J. Carcinog. 2006;5(14):1–8. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen O.H., Spat A., Verkhratsky A., Philos T. Reactive oxygen species in health and disease. R. Soc. B. 2005;360(1464):2197–2199. [Google Scholar]
- 24.Cuzzocrea S., Riley D.P., Caputi A.P., Salvemini D. Antioxidant therapy: a new pharmacological approach in shock, inflammation, and ischemia/reperfusion injury. Pharmacol. Rev. 2001;53(1):135–159. [PubMed] [Google Scholar]
- 25.Moquin-Pattey C., Guyot M. Grossularine-1 and grossularine-2, cytotoxic α-carbolines from the tunicate: Dendrodoa grossularia. Tetrahedron. 1989;45:3445–3450. [Google Scholar]
- 26.Cao R., Chen Q., Hou X., Chen H., Guan H., Ma Y., Peng W., Xu A. Synthesis, acute toxicities, and antitumor effects of novel 9-substituted beta-carboline derivatives. Bioorg. Med. Chem. 2004;12(17):4613–4623. doi: 10.1016/j.bmc.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Dong X., Liu T., Lou J., Jiang C., Huang W., He Q., Yang B., Hu Y. Design, synthesis, and quantitative structure-activity relationship of cytotoxic gamma-carboline derivatives. Bioorg. Med. Chem. 2009;17(9):3324–3331. doi: 10.1016/j.bmc.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 28.Yanagisawa H., Shimoji Y., Hashimoto T. T. Jpn. Kokai Tokkyo Koho JP 05310738, Heisei; Chem. Abstr. 1993;120 [Google Scholar]
- 29.Mazu T.K., Etukala J.R., Jacob M.R., Khan S.I., Walker L.A., Ablordeppey S.Y. δ-Carbolines and their ring-opened analogs: synthesis and evaluation against fungal and bacterial opportunistic pathogens. Eur. J. Med. Chem. 2011;46(6):2378–2385. doi: 10.1016/j.ejmech.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.[a) Arzel E., Rocca P., Grellier P., Labaeid M., Frappier F., Gueritte F., Gaspard C., Marsais F., Godard A., Queguiner G. New synthesis of benzo-delta-carbolines, cryptolepines, and their salts: in vitro cytotoxic, antiplasmodial, and antitrypanosomal activities of delta-carbolines, benzo-delta-carbolines, and cryptolepines. J. Med. Chem. 2001;44:949–960. doi: 10.1021/jm0010419. [DOI] [PubMed] [Google Scholar]; [b) Sayed I.E., der Veken P.V., Steert K., Dhooghe L., Hostyn S., Baelen G.V., Lemière G., Maes B.U.W., Cos P., Maes L., et al. Synthesis and antiplasmodial activity of aminoalkylamino-substituted neocryptolepine derivatives. J. Med. Chem. 2009;52(9):2979–2988. doi: 10.1021/jm801490z. [DOI] [PubMed] [Google Scholar]; [c) Mazu T.K., Etukala J.R., Jacob M.R., Khan S.I., Walker L.A., Ablordeppey S.Y. δ-Carbolines and their ring-opened analogs: synthesis and evaluation against fungal and bacterial opportunistic pathogens. Eur. J. Med. Chem. 2011;46(6):2378–2385. doi: 10.1016/j.ejmech.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorlitzer K., Karmer C., Meyer H., Walte R.D., Jomaa H., Wiesner J. Pyrido [3,2-b]indol-4-yl-amines–synthesis and investigation of activity against malaria. Pharmazie. 2004;59(4):243–250. [PubMed] [Google Scholar]
- 32.[a) Takeuchi Y., Oda T., Chang M.r., Okamoto Y., Ono J., Oda Y., Harada K., Hashigaki K., Yamato M. Synthesis and antitumor activity of fused quinoline derivatives. IV. Novel 11-aminoindolo[3, 2-b]quinolines. Chem. Pharm. Bull. (Tokyo) 1997;45(2):406–411. doi: 10.1248/cpb.45.406. [DOI] [PubMed] [Google Scholar]; [b) Zhou J.M., Zhu X.F., Lu Y.J.R., Deng Z.S., Huang Y.P., Mei Y., Wang W.L., Huang Z.C., Liu L.Q., Gu Y.X., Zeng Senescence and telomere shortening induced by novel potent G-quadruplex interactive agents, quindoline derivatives, in human cancer cell lines. Oncogene. 2006;25(4):503–511. doi: 10.1038/sj.onc.1209067. [DOI] [PubMed] [Google Scholar]; [c) Liu J.N., Deng R., Guo J.F., Zhou J.M., Feng G.K., Huang Z.S., Gu L.Q., Zeng Y.X., Zhu X.F. Inhibition of myc promoter and telomerase activity and induction of delayed apoptosis by SYUIQ-5, a novel G-quadruplex interactive agent in leukemia cells. Leukemia. 2007;21(6):1300–1302. doi: 10.1038/sj.leu.2404652. [DOI] [PubMed] [Google Scholar]
- 33.Pongnan F., Saucier J.M., Paoletti C., Kaczmarek L., Nantka-Namirski P., Mordarski M., Peczynska-Czoch W. A carboline derivative as a novel mammalian DNA topoisomerase II targeting agent. Biochem. Pharmacol. 1992;44(11):2149–2155. doi: 10.1016/0006-2952(92)90341-f. [DOI] [PubMed] [Google Scholar]
- 34.Kaczmarek L., Peczynska-Czoch W., Osiadacz J., Mordarski M., Sokalski W.A., Boratynski J., Marcinkowska E., Glazman-Kusnierczyk H., Radzikowski C. Synthesis, and cytotoxic activity of some novel indolo[2,3-b]quinoline derivatives: DNA topoisomerase II inhibitors. Bioorg. Med. Chem. 1999;7(11):2457–2464. doi: 10.1016/s0968-0896(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 35.Katarzyna S., Marta S., Joanna W., Anna J., Pietka-Ottlik M., Piotr C., Joanna Z., Wojciech S., Lukasz K., Peczynska-Czoch W. Synthesis and biological evaluation of new amino acid and dipeptide derivatives of neocryptolepine as anticancer agents. J. Med. Chem. 2012;55:5077–5087. doi: 10.1021/jm300468t. [DOI] [PubMed] [Google Scholar]
- 36.Peczynska-Czoch W., Pognan F., Kaczmarek L., Boratynski J. Synthesis and structure–activity relationship of methyl-substituted indolo[2,3-b]quinolines: novel cytotoxic, DNA topoisomerase II inhibitors. J. Med. Chem. 1994;37:3503–3510. doi: 10.1021/jm00047a008. [DOI] [PubMed] [Google Scholar]
- 37.Winters G., DiMola N., Berti M., Arioli V. Synthesis and biological activities of some indolo[2,3-c]isoquinoline derivatives. Farmaco. Sci. 1979;34(6):507–517. doi: 10.1002/chin.197949235. Chem. Abstr, 1979, 91, 175235m. [DOI] [PubMed] [Google Scholar]
- 38.Ishizumi K., Katsube J. Indolo isoquinolines and processes for producing them Brit. Pat, 2, 025, 932. Chem. Abstr. 1980;93 [Google Scholar]
- 39.Ishizumi K., Katsube J. 10-Chloro-7-methyl-7H-indolo[2,3-c]isoquinolin-5(6H)-one Japan Kokai, 7,879,899. Chem. Abstr. 1978;89 [Google Scholar]
- 40.Sumitomo Chemical Co.Ltd Jpn Kokai Tokkyo Koho, Jap.Pant. Indolo isoquinolines 5869, 882 (8369, 882) Chem. Abstr. 1983;99:88182p. [Google Scholar]
- 41.Hiremath S.P., Saundane A.R., Mruthyunjayaswamy B.H.M. Synthesis and biological evaluation of some substituted 5H,6H,7H,-indolo[2,3-c]isoquinoline-5-thiones and their derivatives. Indian J. Heterocycl. Chem. 1993;3:37–42. [Google Scholar]
- 42.Hiremath S.P., Saundane A.R., Mruthyunjayaswamy B.H.M. Synthesis of [10-substituted 6H,7H-indolo[2,3-c]isoquinolin-5-one-6-yl]acetyl-3,5-disubstituted-pyrazoles/ pyrazolones and 5-[10-substituted 6H,7H-indolo[2,3-c]isoquinolin-5-one-6-yl]methyl-1,3,4-oxadiazol-2-thiones. J. Indian Chem. Soc. 1995;72(10):735–738. [Google Scholar]
- 43.Hiremath S.P., Rudresh K., Saundane A.R. Syntesis and biological activities of new 5-hydrazino-10-substituted-7H-indolo[2,3-c]isoquinolines and 1-(10-substituted-7H-indolo[2,3-c]isoquinoline-5-yl)-3,5-disubstituted pyrazoles, -3-methyl, pyrazol-5-ones and -3,5-disubstituted pyrazolines. Indian J. Chem. 2002;41B:394–399. [Google Scholar]
- 44.Saundane A.R., Vaijinath A.V., Katkar V. Synthesis, antimicrobial and antioxidant activities of some new 1′-(10-substituted 5H,6H,7H,-indolo[2,3-c]isoquinoline-5-ylthio)formyl-3′,5′-disubstituted pyrazoles, -3′-methylpyrazole-5′-ones and -1′,3′,4′-oxadiazole-2′-thiones. Heterocycl. Lett. 2012;(3):333–348. [Google Scholar]
- 45.Saundane A.R., Vaijinath A.V., Katkar V. Synthesis of some new indolo[2,3-c]isoquinolinyl pyrazoles, -1,3,4-oxadizoles and their biological activities. Med. Chem. Res. 2013;22(8):3787–3793. [Google Scholar]
- 46.Vaijinath A.V. Synthesis, antimicrobial, and antioxidant studies of some new indolo[3,2-c]isoquinoline derivatives. Russ. J. Gen. Chem. 2018;88(12):2628–2645. [Google Scholar]
- 47.Vaijinath A.V., Saundane A.R. Synthesis of some novel 5-(8-substituted-11h-indolo[3,2-c]isoquinolin-5-ylthio)-1′,3′,4′-oxadiazol-2-amines bearing thiazolidinones and azetidinones as potential antimicrobial, antioxidant, antituberculosis, and anticancer agents. Polycycl. Aromat. Compd. 2019 doi: 10.1080/10406638.2019.1628782. [DOI] [Google Scholar]
- 48.Toharo K.A., Toharo A. Vol. 3. W.C. Brown Publisher; Dubuque: 1993. p. 326. (Foundation of Microbiology). edition. [Google Scholar]
- 49.Deepa S.M., Beverley D.G., Santy D. Melatonin: new places in therapy. Biosci. Rep. 2007;27(6):299–320. doi: 10.1007/s10540-007-9052-1. Dec. [DOI] [PubMed] [Google Scholar]
- 50.National Committee for Clinical Laboratory Standards (NCCLS) NCCLS; Villanova, PA: 1997. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard M27-A. [Google Scholar]
- 51.National Committee for Clinical Laboratory Standards (NCCLS), Standard methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 2nd ed. Approved standard M7-A2 (Villanova, PA: National Committee for Clinical Laboratory Standards, 1990).
- 52.National Committee for Clinical Laboratory Standards (NCCLS) National Committee for Clinical Laboratory Standards; Villanova, PA: 1999. Performance Standards for Antimicrobial Susceptibility Testing; Ninth Informational Supplement. M100-S9. [Google Scholar]
- 53.Garmroodi F.G., Omidi M., Saeedi M., Sarrafzadeh F., Rafinejad A., Mahdavi M., Bardajee G.R., Akbarzadeh T., Firoozpour L., Shafiee A., Foroumadi A. Simple and efficient syntheses of novel benzo[4,5]imidazo[1,2-a]pyridine derivatives. Tetrahedron. Lett. 2015;56:743–746. [Google Scholar]
- 54.Lerner A., Adler A., Abu-Hanna J., Cohen-Percia S., Kazma-Matalon M., Carmeli Y. Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin. Microbiol. Infect. 2015;21(5):470.e1. doi: 10.1016/j.cmi.2014.12.015. 7. [DOI] [PubMed] [Google Scholar]
- 55.Biswal A., Venkataraghavan R., Pazhamalai V., Romauld S.I. Molecular docking of various bioactive compounds from essential oil of Trachyaspermum ammi against the fungal enzyme Candidapepsin-1. J. Appl. Pharm. Sci. 2019;9(05):021–032. [Google Scholar]
- 56.Hatano T., Kagawa H., Yasuhara T., Okuda T. Two new flavonoids and other constituents in licorice root: their relative astringency and radical scavenging effects. Chem. Pharm. Bull. (Tokyo) 1988;36(6):2090–2097. doi: 10.1248/cpb.36.2090. [DOI] [PubMed] [Google Scholar]
- 57.Dinis T.C., Maderia V.M., Almeida L.M. Action of phenolic derivatives (acetaminophen, salicylate, and 5-aminosalicylate) as inhibitors of membrane lipid peroxidation and as peroxyl radical scavengers. Arch. Biochem. Biophys. 1994;315(1):161–169. doi: 10.1006/abbi.1994.1485. [DOI] [PubMed] [Google Scholar]
- 58.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 59.Schinkovitz A., Gibbons S., Stavri M., Cocksedge M.J., Bucar F. Ostruthin: an antimycobacterial coumarin from the roots of Peucedanum ostruthium. Planta Med. 2003;69(4):369–371. doi: 10.1055/s-2003-38876. [DOI] [PubMed] [Google Scholar]
- 60.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.