Abstract
The DNA lesions caused by oxidative damage are principally repaired by the base excision repair (BER) pathway. 8-oxoguanine DNA glycosylase 1 (OGG1) initiates BER through recognizing and cleaving the oxidatively damaged nucleobase 8-oxo-7,8-dihydroguanine (8-oxoG). How the BER machinery detects and accesses lesions within the context of chromatin is largely unknown. Here, we found that the symmetrical dimethylarginine of histone H4 (producing H4R3me2s) serves as a bridge between DNA damage and subsequent repair. Intracellular H4R3me2s was significantly increased after treatment with the DNA oxidant reagent H2O2, and this increase was regulated by OGG1, which could directly interact with the specific arginine methyltransferase, PRMT5. Arginine-methylated H4R3 could associate with flap endonuclease 1 (FEN1) and enhance its nuclease activity and BER efficiency. Furthermore, cells with a decreased level of H4R3me2s were more susceptible to DNA-damaging agents and accumulated more DNA damage lesions in their genome. Taken together, these results demonstrate that H4R3me2s can be recognized as a reader protein that senses DNA damage and a writer protein that promotes DNA repair.
Keywords: H4R3me2s, OGG1, FEN1, BER, Oxidative stress
1. Introduction
Genomic DNA is continuously exposed to oxidative stress arising from various exogenous and endogenous sources, such as chemicals, radiation or normal metabolism, which can cause cells to produce excessive reactive oxygen species (ROS) [1,2]. In cells, ROS can attack DNA, which induces a series of DNA damage effects, including oxidized bases and DNA strand breaks. If not repaired, 8-oxo-7, 8-dihydroguanine (8-oxoG), the most common oxidation products [2], can lead to genetic mutations and subsequent genome instability and cancer initiation [3].
Cells have developed many machineries to respond to oxidative stress. Base excision repair (BER) is the primary repair pathway responsible for repairing oxidative DNA damage [4]. Oxidized guanine can be excised by OGG1, a primary 8-oxoguanine DNA glycosylase, leaving a 5′ deoxyribose phosphate terminus (5′-dRP) and an apurinic/apyrimidinic (AP) site. Then, AP endonuclease 1 (APE1) can recognize the AP site and cleave the backbone to produce a nicked abasic intermediate [5]. This intermediate can be processed through either SP-BER or LP-BER [6]. In SP-BER, only one nucleotide is added to the 3′-end of the nicked AP site, and Pol β then catalyzes the β-elimination of the 5′-sugar phosphate residue with its dRP lyase activity, resulting in a ligatable nick that can be sealed by X-ray repair cross-complementing protein 1 and Ligase IIIα (XRCC1/Ligase IIIα) [7]. In contrast, in LP-BER, Pol β performs strand displacement synthesis, generating a 2–10 nt short DNA flap that is cleaved by flap endonuclease 1 (FEN1). Finally, DNA ligase I seals the nick [[7], [8], [9]].
In mammals, OGG1, the main 8-oxoG glycosylase, is a bifunctional DNA glycosylase with associated AP lyase activity [10,11]. OGG1 also contains targeting signals for both mitochondrial import and nuclear localization and plays important roles in BER. OGG1 was reported to be involved in several oxidative stress-related diseases, including various cancers [10,12], neurological diseases [13] and metabolic syndrome [14,15].
In eukaryotic cells, DNA is packaged into highly compacted chromatin, the basic unit of which is the nucleosome. The nucleosome core particle is composed of an octamer of histone proteins and 146 bp of DNA. The histone N-terminus is located outside the nucleosome structure and is rich in positively charged basic amino acids that can undergo various posttranslational modifications [16]. Histone methylation is an important posttranslational modification, and its function is mainly reflected in heterochromatin formation, gene imprinting, X chromosome inactivation and transcriptional regulation [17]. In recent years, extensive studies have associated histone methylation with both the repression and activation of transcription. A variety of markers have been recognized to activate (H3K4me3 and H3K36me3) and repress (H3K9me2 and H3K27me3) transcription [18]. These specific modifications are regarded as “reader” proteins that regulate subsequent DNA metabolism. Histone arginine modification has recently been regarded as another important posttranslational modification that plays a role in chromatin modeling and the transcriptional state [19].
Although the importance of histone arginine methylation in gene transcription regulation has been addressed in many studies, whether and how histone arginine methylation is involved in the DNA damage response remain unclear. In the current study, we found that H4R3me2s was significantly increased after oxidative reagent treatment, and that symmetrical arginine methylation plays important roles in the DNA damage response. Our results support a novel mechanism of histone modification serving as both the reader and writer protein to sense DNA damage and promote DNA repair.
2. Materials and METHODS
2.1. Antibodies and reagents
The histone peptides were purchased from Chinese Peptide Company, all the primers and DNA substrates used in this paper was synthesized by GenScript, Inc. Four deoxynucleotide triphosphates (dNTPs) were purchased from New England Biolabs (N0446S). [γ-32P]-ATP (BLU002A, 250 μCi) and [α-32P]-dCTP (NEG513H, 250 μCi) were purchased from PerkinElmer. The M2 beads and M-280 beads were purchased from Sigma-Aldrich. The DNA Ligase I was purchased from Abnova. The antibodies used in this paper are as follow: anti-FEN1 (GTX70185, Genetex), anti-PRMT5 (sc-376937, Santa Cruz), anti-8-oxoG (sc-130914, Santa Cruz), anti-H4 (PTM-1003, PTM Bio), anti-H4R3me1 (ab17339, Abcam), anti-H4R3me2s (ab5823, Abcam), anti-OGG1 (ab135940, Abcam), anti-Flag (AP0007MH, Bioword), anti-GAPDH (AP0063, Bioworld), anti-Tubulin (AM031A, Abgent).
2.2. Cell lines and cell culture
HeLa cell line and HEK293T cell line were obtained from ATCC (Manassas, VA, USA) and they were cultured under conditions as directed by the product instructions. MEF WT and Ogg1 null cell lines were kindly supplied by Professor Istvan Boldogh (Sraly Center for Molecular Medicine, Texas). These cell lines are cultured in Dulbecco's modified Eagle's medium (Invitrogen, Shanghai, China) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin–streptomycin (Invitrogen) at 37 °C in a humidified 5% CO2 incubator. We treated the cells with H2O2 under the doses up to 1 mM which were chosen based on sensitivity assay and DNA damage detection for downstream experiments.
2.3. Plasmid construction and purification of recombinant proteins
Human H4 cDNA (NM_175054.2) was cloned from HeLa cells using the following primers: 5′-GATATCGATGTCTGGGCGAGGTAAAGGTGG-3’ (forward) and 5′-CGGGATCCTCAACCGCCGAAACCATAAA-3’ (reverse). Full-length H4 was cloned into pFlag-CMV4 plasmid. The H4 (R3Q) mutant was generated using the QuickChange Site-directed Mutagenesis Kit and the following primers: 5′-CCTTGCCACCTTTACCTTGCCCAGACATCGATATC-3' (forward) and 5′-GATATCGATGTCTGGGCAAGGTAAAGGTGGCAAGG-3' (reverse). Recombinant proteins FEN1, Pol β, APE1, PCNA were expressed in Escherichia coli BL21DE and were purified using Ni2+ affinity chromatography (GE Healthcare). DNA Ligase I (Lig I) was purchased from Abnova.
2.4. FEN1 nuclease activity assay
The nuclease activity reaction was performed in buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl and 5 mM MgCl2. The FAM-labeled DNA substrates, purified FEN1 protein and H4R3me2s or H4R3 peptide were mixed and incubated at 37 °C for 30 min in the dark, stopped by addition of the stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). Finally, the mixture was separated by 15% PAGE containing 8 M urea and analyzed by Odessey FC.
2.5. Electrophoretic mobility shift assay
The same concentration of FEN1 protein and various concentrations of H4R3me2s or H4R3 peptide were incubated with FAM-labeled DNA substrates in a buffer containing 25 mM Tris/HCl (pH 8.0), 1 mM DTT, 5% glycerol, 0.25 mg/mL BSA, 50 mM NaCl and 0.2 mM EDTA. The mixture was assembled on ice and preincubated for 10 min. After further incubation of 10 min at 37 °C, the mixture was separated by a 6% non-denaturing polyacrylamide gel (30:1 acrylamide: bis-acrylamide) in TBE buffer and imaged by Odessey FC.
2.6. Reconstituted LP-BER assay
The reconstituted BER assay was performed in 20 μl of reaction buffer containing 40 mM HEPES–KOH (pH 7.8), 70 mM KCI, 7 mM MgCl2, 1 mM dithiothreitol, 0.5 mM EDTA, 2 mM ATP, 200 U creatine-phosphokinase, 0.5 mM NAD and 5 mM phosphocreatine, 50 μM each of dATP, dTTP and dGTP and 8 μCi [α-32P]-dCTP. For the BER reaction with purified protein, FEN1 (15 ng), APE1 (2 ng), PCNA (2 ng), Ligase I (20 ng), FAM-labeled DNA substrates and various amounts of H4R3me2s or H4R3 peptide (0–2 μg) was mixed on ice and incubated for 30 min at 37 °C. For the BER reaction with the whole cell extract, the dATP, dTTP, dGTP, [α-32P]-dCTP and DNA substrates were incubated with cell extract at 37 °C for 30 min. Reactions were stopped with stop buffer, heated at 95 °C for 10 min, and 10 μl of the reaction mixture were separated by 15% PAGE containing 8 M urea and imaged by Odyssey FC.
2.7. Drug sensitivity assay
Drug sensitivity assay was measured by trypan blue exclusion analysis. Cells were seeded onto a 12-well plate in 1 ml cell culture medium of each well. After transfected with the indicated siRNA or plasmids, the cells were treated with various concentrations of H2O2 for 30 min and continuously cultured in fresh medium for 48 h, the cells were collected and resuspended in cell culture medium. An equal volume of 0.4% trypan blue dye was added and mixed with the cell suspension. The viable cells were measured by Count Star.
2.8. Immunofluorescence
Cells were cultured in 6-well plate which contained an acid-treated slide and incubated overnight. Then the slides were washed with PBS three times, treated with 4% formaldehyde in PBS for 30 min and then washed again with PBS. Triton X-100 (0.05%) was added for 5 min to permeabilize the cells. The slides were blocked by 5% BSA and then incubated with primary antibody at 4 °C overnight. The slides were washed with PBS three times and then incubated with fluorescent secondly antibody conjugated with fluorescein isothiocyanate for 2 h. Then the slides were washed again with PBS and stained with DAPI for 10 min. The mounted slides were viewed with a Nikon 80I 10-1500X microscope, and images were captured with a charge-coupled-device camera.
2.9. Streptavidin pull down assay
The biotin-labeled histone peptides were synthesized by Chinese Peptide Company, HangZhou. Cells were lysed in IP buffer containing PMSF and cocktail inhibitor (Roche) at 4 °C for 3 h. After centrifugation at 12,000 rpm for 10 min at 4 °C, the supernatants were further treated with 100 μg/ml DNase I for 15 min. In vitro pulldown assay was performed by incubating the histone peptides with cell lysates overnight using Streptavidin beads, the next day washed the beads three times with PBS for 5 min each. The proteins that remained bound to the peptides were separated by SDS-PAGE and visualized by silver staining.
3. Results
3.1. H4R3me2s is upregulated under DNA damage stress
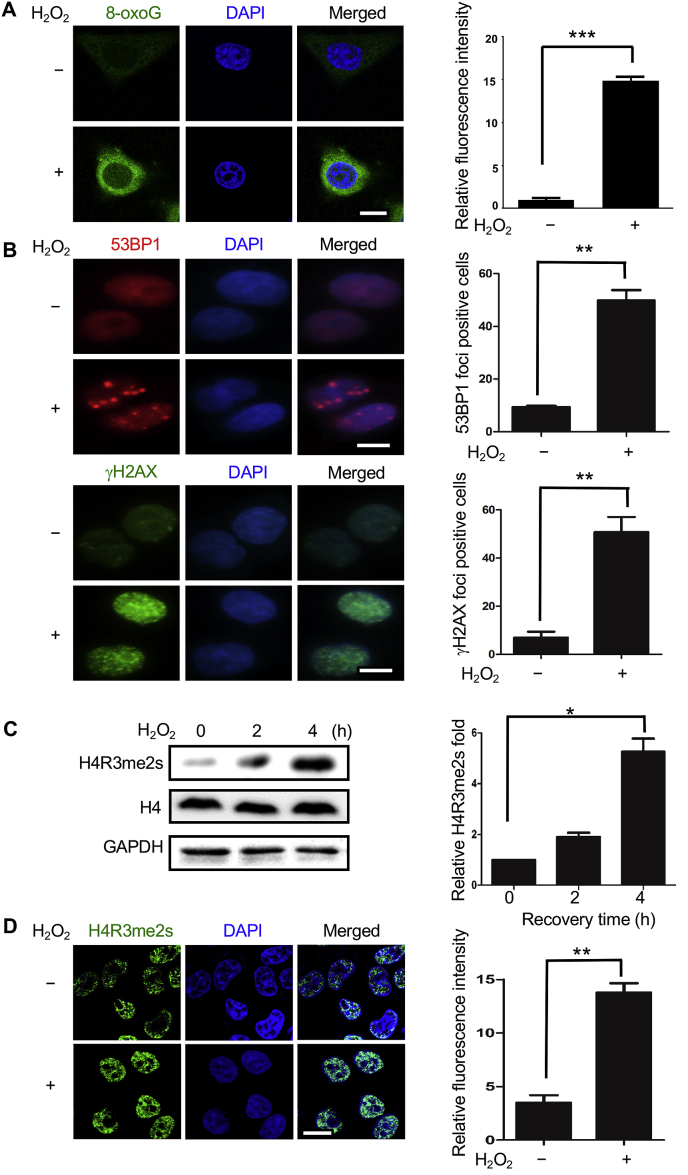
The symmetrical dimethylation of arginine-3 of histone H4 (producing H4R3me2s) is related to the repressed transcription of several genes under different stresses [20]. Oxidative stress is the most common cellular stressor and can lead to oxidatively damaged DNA. To test whether H4R3me2s is a response to this stress, we first chose hydrogen peroxide (H2O2), a well-known oxidizer, to treat HeLa cells. The level of 8-oxoG, the most common oxidation products was significantly increased after H2O2 treatment (Fig. 1A). There was also a dramatic increase in the foci numbers of γH2AX and 53BP1, two established markers of DNA double-strand breaks (DSBs) (Fig. 1B), which indicates that treatment with H2O2 could result in a serious DNA damage in HeLa cells. We then applied Western blotting to examine cellular H4R3me2s level with and without H2O2 treatment and found H2O2 exposure led to a gradual increase in the levels of H4R3me2s (Fig. 1C). This result was further confirmed by immunofluorescence assay (Fig. 1D). Moreover, another oxidant, t-butyl-hydroperoxide also induced the elevated level of H4R3me2s, although such an increase was less than H2O2 (Supplementary Figure S1).
Fig. 1.
Induction of the H4R3me2s modification by H2O2 treatment. (A) HeLa cells were treated with or without 1 mM H2O2 for 30 min, and the media were then changed to fresh media. After 4 h, the cells were fixed and immunostained with antibodies against 8-oxoG (green), DNA was stained with DAPI (blue), and the cells were visualized by laser confocal microscopy. The average fluorescence intensity of three independent experiments is shown on the right. Data are presented as the mean ± SD; ***, P < 0.001, Student's t-test. Scale bars, 10 μm. (B) We examined γH2AX (green) and 53BP1 (red) with the same H2O2 treatment as panel A. The mean numbers of cells with more than ten foci (from >30 fields per group) is plotted in the bar chart (**P < 0.01, Student's t-test). (C) HeLa cells were treated with 1 mM H2O2 for 30 min, and the medium was then changed to fresh medium, followed by continued culture for 0, 2 h, and 4 h. Whole-cell extracts were analyzed by Western blotting with the indicated antibodies. H4R3me2s band intensities were obtained via quantitation with a fluorimager, and the results were normalized to H4. GAPDH served as a loading control. The data represent the mean ± SD of three independent experiments (*P < 0.05, Student's t-test). (D) The cells were immunostained with antibody against H4R3me2s under the same H2O2 treatment as panel A. DNA was stained with DAPI, and the cells were visualized by laser confocal microscopy. The average fluorescence intensity of three independent experiments is shown on the right. Data are presented as the mean ± SD; **, P < 0.01, Student's t-test. Scale bars, 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. OGG1 regulates the level of H4R3me2s modification
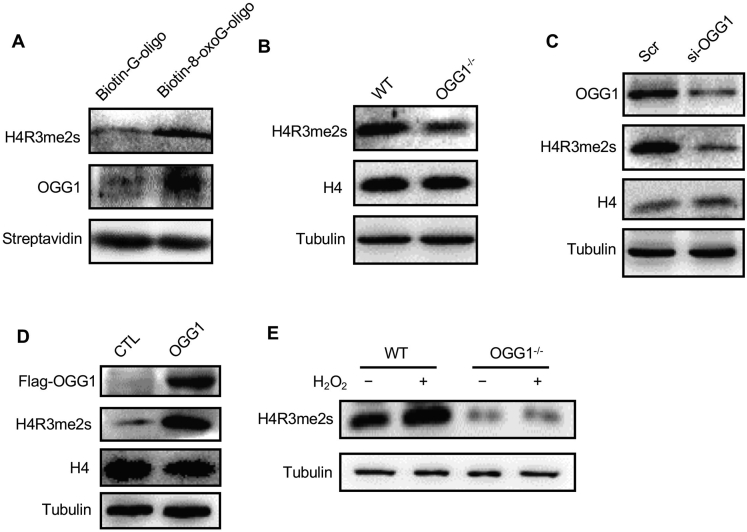
Oxidative stress usually generates oxidatively damaged DNA, among which is the typically easy oxidation of guanine into 8-oxo-7,8-dihydroguanine (8-oxoG). To investigate whether H4R3me2s is preferentially increased at regions of oxidized DNA, we synthesized biotin-labeled nucleic acid probes with or without 8-oxoG (Fig. 2A) and performed pulldown assays with the nuclear lysates from HeLa cells. Our results revealed that more H4R3me2s could be pulled down by probes containing 8-oxoG. OGG1, a specific glycosylase recognizing 8-oxoG, was also detected in the precipitates (Fig. 2A). This result showed that H4R3me2s and OGG1 could be localized at the same DNA damage lesion. Since our previous study has demonstrated that OGG1 was involved in H4R3me2s production [21]. To verify the relationship between histone modification and OGG1, we compared H4R3me2s levels in WT and OGG1−/− mouse embryonic fibroblast (MEF) cells. The H4R3me2s level was noticeably reduced in OGG1−/− MEF cells (Fig. 2B) which was consistent with our previous data. Knockdown of OGG1 with small interfering RNA (siRNA) produced a similar result (Fig. 2C), suggesting that H4R3 arginine modification was downregulated by OGG1 knockdown. To further confirm the effect of OGG1 on H4R3me2s, we overexpressed OGG1 in HEK293T cells, and the H4R3me2s level was clearly increased with OGG1 overexpression (Fig. 2D). Taken together, these results suggest that the H4R3me2s level is regulated by OGG1. Since our data showed that the H4R3me2s level was significantly elevated under H2O2 stimulation, to test whether this elevation was dependent on OGG1, we repeated the H2O2 treatment experiment in WT and OGG1−/− MEF cells. Our data showed that the increase in H2O2-induced H4R3me2s was dramatically reduced in OGG1−/− cells (Fig. 2E). Together, our results demonstrate that OGG1 is indispensable for symmetrical H4R3 arginine methylation under oxidative stress.
Fig. 2.
OGG1 affects the level of H4R3me2s modification. (A) OGG1 and the H4R3me2s′ occupancy of 8-oxoG-containing DNA in NEs. NEs (50 μg per sample) from Hela cells were incubated with G (left lane) or 8-oxoG oligonucleotides (right lane) for 5 min, and protein DNA complexes were pulled down using magnetic streptavidin beads. The washed pellets were subjected to SDS-PAGE and immunoblotting using antibodies against OGG1 or H4R3me2s. (B) The levels of H4R3me2s modification in WT and OGG1−/− MEFs were determined via Western blot analysis. (C) Whole-cell lysates of HEK293T cells transfected with OGG1 siRNA or scrambled siRNA (Scr) were immunoblotted for H4R3me2s and H4. Tubulin was used as a loading control. (D) Whole-cell lysates of HEK293T cells transfected with pcDNA3.1-OGG1 or the control vector were immunoblotted for H4R3me2s and H4. Tubulin was used as a loading control. (E) The level of H4R3me2s modification in WT and OGG1−/− MEFs treated with or without 1 mM H2O2 was detected by Western blotting. Tubulin was used as a loading control.
3.3. OGG1 interacts with PRMT5 to promote H4R3me1 methylation
PRMT5 is the primary methylation transferase responsible for symmetrical arginine methylation of the histone H4R3 [22]. To exclude the possibility that OGG1 regulates H4R3me2s levels because of a change in the protein level of PRMT5, we first examined PRMT5 protein levels in WT and OGG1−/− MEF cells. Western blotting analysis showed that there was no significant difference in the protein level of PRMT5 between the two groups (Fig. 3A), suggesting that OGG1 depletion does not affect PRMT5 expression. We next explored the possibility that OGG1 plays a role in the interaction between PRMT5 and its substrate, H4R3me1. We performed immunoprecipitation experiments in WT and OGG1−/− MEF cells. The data showed that H4R3me1 could be successfully precipitated from WT MEF cells by anti-PRMT5 antibody. Surprisingly, the interaction between PRMT5 and H4R3me1 was nearly abolished in OGG1−/− MEF cells (Fig. 3B), which indicated that OGG1 might play a role in the binding between enzyme and substrate. To investigate how OGG1 is involved in the binding of PRMT5 to H4R3me1, we first applied immunoprecipitation to examine whether OGG1 could bind with PRMT5 or H4R3me1. As shown in Fig. 3C and D, both H4R3me1 and PRMT5 were detected following precipitation by anti-OGG1 antibody (Fig. 3C and D), suggesting that OGG1 can form a complex with these two proteins. A co-IP experiment with anti-PRMT5 antibody confirmed the interaction between OGG1 and PRMT5 (Fig. 3E). To rule out the possibility that binding between OGG1 and PRMT5 is mediated by other proteins, commercial purified proteins were used in the pulldown assay. The results demonstrated that the two purified proteins could directly bind each other (Fig. 3F and G). Taken together, our results showed that the H4R3me2s level was regulated by OGG1 through its interaction with PRMT5.
Fig. 3.
OGG1 interacts with PRMT5 to promote H4R3me1 methylation. (A) The expression of PRMT5 in WT and OGG1−/− MEFs was determined by Western blotting. (B) Whole-cell extracts collected from WT and OGG1−/− MEFs were subjected to IP with anti-PRMT5 antibody or control IgG, followed by IB with anti-H4R3me1 antibody. (C) Whole-cell extracts collected from WT MEFs were subjected to IP with anti-OGG1 antibody or control IgG, followed by IB with anti-H4R3me1 antibody. (D) Co-IP of PRMT5 with OGG1. Whole-cell extracts from WT MEFs were subjected to IP using anti-OGG1 antibody or control IgG. Western blot analysis was performed with anti-PRMT5 antibody. (E) Co-IP of OGG1 with PRMT5. Whole-cell extracts from WT MEFs were subjected to IP using anti-PRMT5 antibody or control IgG. Western blot analysis was performed with anti-OGG1 antibody. (F) Co-IP of purified recombinant OGG1 and PRMT5 proteins using anti-PRMT5 antibody or control IgG. Western blot analysis was performed with anti-OGG1 antibody. (G) Co-IP of purified recombinant PRMT5 and OGG1 proteins using anti-OGG1 antibody or control IgG. Western blot analysis was performed with anti-PRMT5 antibody.
3.4. H4R3me2s interacts with FEN1 in vitro and in vivo
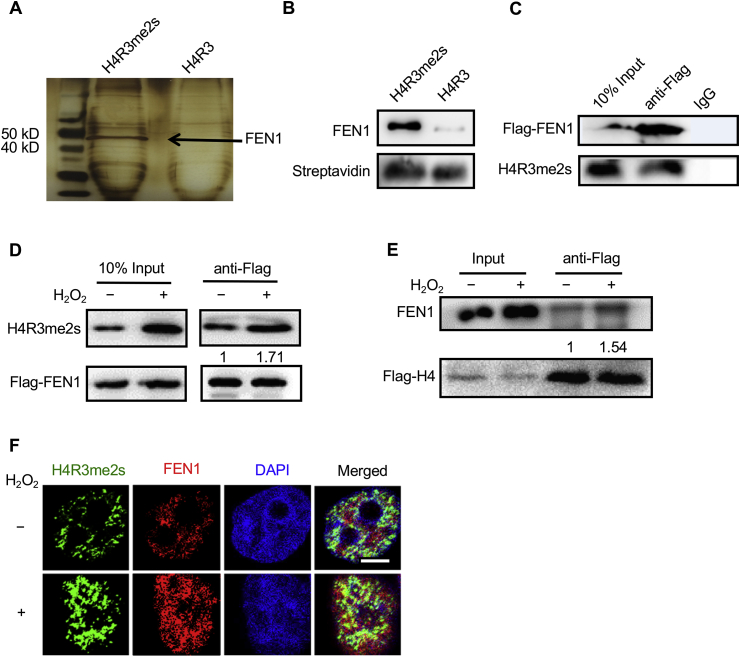
To explore the biological function of the elevated level of H4R3me2s under oxidative stress, we first synthesized two biotin-labeled N-terminal peptides of H4 in which Arg3 was symmetrically modified (H4R3me2s) and H4 with no modification (H4R3). Next, we used the two peptides to perform affinity purification with nuclear extracts from HeLa cells to identify the proteins that specifically interact with H4R3me2s. The components in the precipitant were resolved by 12% SDS-PAGE, and after the gel underwent silver staining, one band with a molecular mass in the range of 40–50 kD was more enriched in the H4R3me2s peptide group than in the H4R3 peptide group (Fig. 4A). Mass spectrometry analysis showed that the FEN1 protein, the core protein of BER, may interact with H4R3me2s (data not shown). Then, we carried out a pulldown assay, and the results confirmed the binding between FEN1 and H4R3me2s (Fig. 4B). Subsequently, immunoprecipitation and Western blot analysis of HeLa cell lysates further confirmed the cellular interaction between H4R3me2s and FEN1 (Fig. 4C). Since FEN1 was required under conditions of H2O2-induced oxidative stress [23], and because our data also showed that H4R3me2s was increased after H2O2 treatment (Fig. 1B and C), we then asked whether the interaction between H4R3me2s and FEN1 is regulated by oxidative stress. To test our hypothesis, we repeated the immunoprecipitation experiment in FEN1-overexpressing HeLa cells under H2O2 treatment for 48 h. The Western blotting data demonstrated that the binding between H4R3me2s and FEN1 was noticeably enhanced under oxidative stress (Fig. 4D). Similar results were obtained in H4-overexpressing HeLa cells (Fig. 4E), which indicated that the FEN1 and H4R3me2s could form a complex in cells. To determine whether the two proteins have the same cellular distribution, immunofluorescence staining was applied to HeLa cells with or without H2O2 treatment. The confocal images showed that more H4R3me2s (green pixels) and FEN1 (red pixels) were colocalized to the chromatin regions (blue pixels) under H2O2 stimulation than without H2O2 stimulation (Fig. 4F).
Fig. 4.
H4R3me2s interacts with FEN1 in vitro and in vivo. (A) Nuclear extracts of HeLa cells were subjected to pull down assays with biotin-labeled H4R3me2s or H4R3 peptides. The eluted protein complex was separated by SDS-PAGE and silver stained. The indicated gel slices were processed for protein identification using mass spectrometry. (B) Recombinant FEN1 was pulled down with biotinylated H4R3me2s or H4R3 N-terminal tail peptides. (C) Whole-cell extracts of HEK293T cells harboring Flag-FEN1 were subjected to co-IP assay with M2 Flag-tagged magnetic beads or control IgG, followed by IB with anti-H4R3me2s antibody. (D) HEK293T cells transfected with Flag-FEN1 were treated with or without 1 mM H2O2 for 30 min, and whole-cell extracts were then collected. A co-IP assay was performed with anti-Flag antibody M2 beads, followed by IB with anti-H4R3me2s antibody. (E) HEK293T cells transfected with Flag-H4 were treated with or without 1 mM H2O2 for 30 min, and whole-cell extracts were then collected. A co-IP assay was performed with anti-Flag M2 beads, followed by IB with anti-FEN1 antibody. The numbers in the figure represent the relative grey values of the bands above (regard control treatment group as 1, quantified with Image J software). (F) HeLa cells were treated with 1 mM H2O2 for 30 min, the medium was then changed to fresh medium. After 4 h, the cells were fixed and immunostained with antibodies against H4R3me2s (green) and FEN1 (red). DNA was stained with DAPI (blue), and cells were visualized by laser confocal microscopy. Scale bars, 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. H4R3me2s increases FEN1 activity through enhancing enzyme-substrate binding
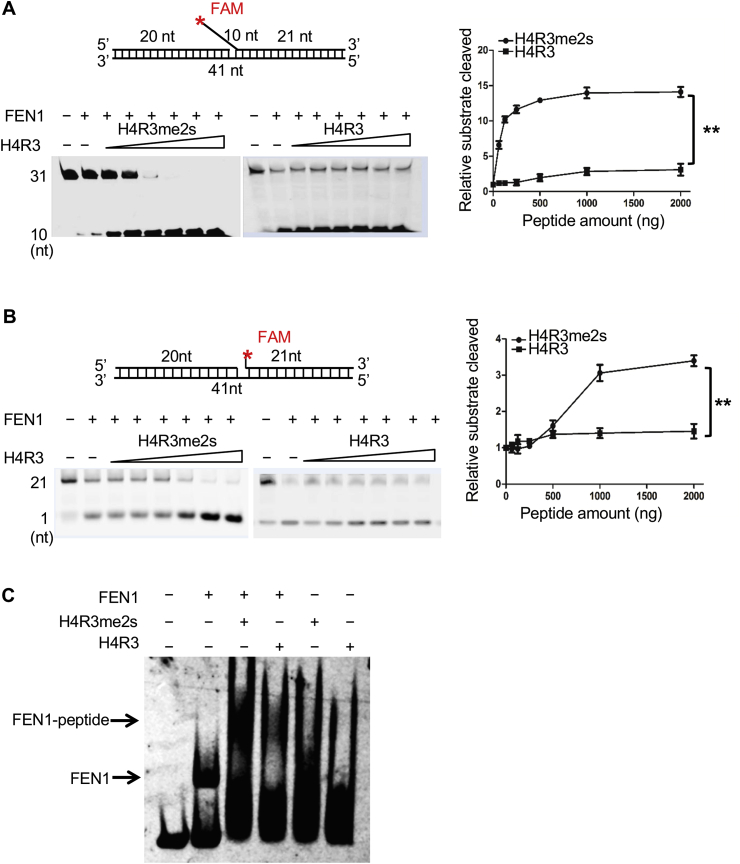
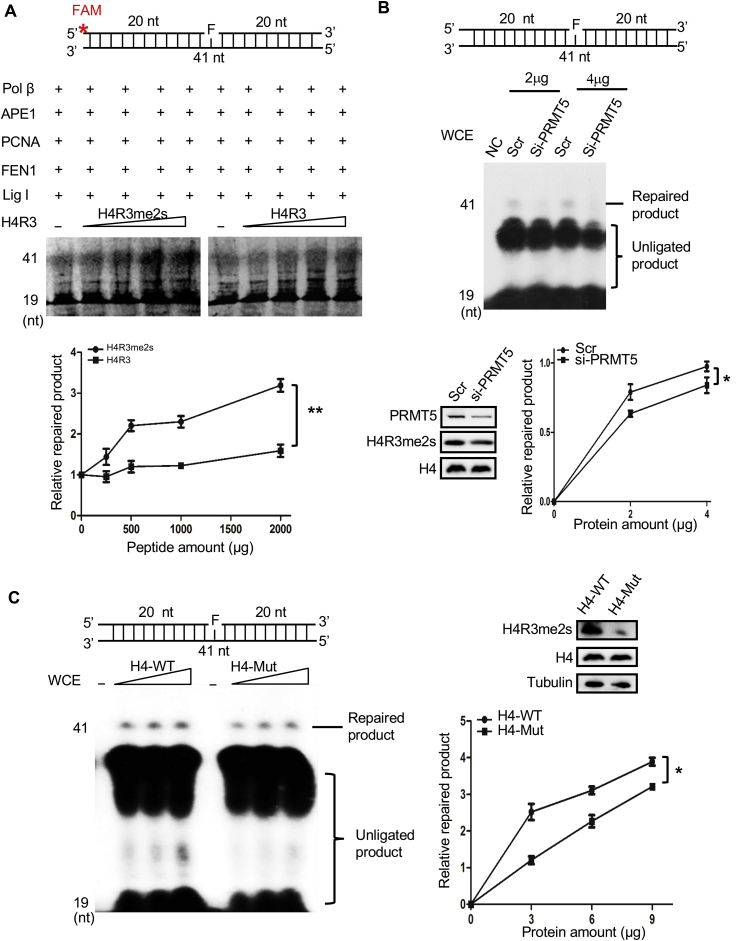
FEN1 is an important nuclease during the DNA repair process. To explore the mechanism and the biological significance of the physical interaction between FEN1 and H4R3me2s, we first investigated whether the methylation of Arg3 of H4 could affect FEN1 activity. To address this question, we mixed recombinant FEN1, DNA substrates and various amounts of each peptide and examined the FEN and EXO activities of FEN1 in the mixture. Fig. 5A shows that the flap-substrates could be cleaved in the presence of FEN1. Moreover, the cleaved products gradually increased with an increasing amount of H4R3me2s peptide in the reaction, while no such effect on the FEN activity of FEN1 was observed with the H4R3 peptide (Fig. 5A). Similarly, H4R3me2s also obviously increased the EXO activity of FEN1 (Fig. 5B). To further understand the exact mechanism by which H4R3me2s affects FEN1 activity, we performed electrophoretic mobility shift assays (EMSAs) to evaluate the binding between FEN1 and its specific DNA substrates. As shown in Fig. 5C, DNA substrates and FEN1 alone could form a DNA-protein complex, and the band for this complex shifted with the addition of H4R3me2s (Fig. 5C). Although H4R3 could also increase the binding of FEN1 to its substrates, this increase was much weaker than that in the presence of H4R3me2s (Fig. 5C). Thus, our results suggested that H4R3me2s can increase FEN1 activity through enhancing the binding of FEN1 to its substrates.
Fig. 5.
H4R3me2s enhances FEN1 nuclease activity through enhancing the substrate–binding affinity of FEN1. Effect of H4R3me2s on FEN1 nuclease activity. The top part of each panel shows schematic structures of the corresponding DNA substrates. (A) Purified FEN1 protein and H4R3me2s and H4R3 peptides were subjected to a FEN activity assay with FAM-labeled flap substrates. Standard nuclease assays were carried out with a fixed amount (10 ng) of FEN1 and increasing amounts (62.5 ng, 125 ng, 250 ng, 500 ng, 1000 ng, 2000 ng) of H4R3me2s or H4R3 peptides. Representative PAGE gels are shown with an arrow denoting the DNA substrates (Sub) and the cleavage products (Prod). The graph represents the quantification of the PAGE results from the image, and the values represent the mean ± SD of three independent assays (**P < 0.01, Student's t-test). (B) Purified FEN1 protein and H4R3me2s and H4R3 peptides were subjected to an EXO activity assay with FAM-labeled nick substrates. Standard assays were carried out with a fixed amount (150 ng) of FEN1 and increasing amounts (62.5 ng, 125 ng, 250 ng, 500 ng, 1000 ng, 2000 ng) of H4R3me2s or H4R3 peptides. The graph represents the quantification of the PAGE results from the image, and the values represent the mean ± SD of three independent assays (**P < 0.01, Student's t-test). (C) Purified FEN1 protein (100 ng) and H4R3me2s or H4R3 peptides (500 ng) were subjected to EMSAs with FAM-labeled flap substrates. The arrow above indicates the position of the FEN1-substrate-peptide complex, and the arrow below indicates the position of the FEN1-substrate complex.
3.6. H4R3me2s enhances LP-BER efficiency
Since FEN1 plays a key role in the removal of flap structures during the LP-BER process, to test whether H4R3me2s can affect LP-BER efficiency through FEN1, an in vitro LP-BER assay was performed using purified BER proteins. A tetrahydrofuran-containing substrate (Pol FEN1-F-FAM, Supplementary Table 1) labeled with a FAM group was used in this assay. As expected, the H4R3me2s peptides dramatically enhanced the efficiency of LP-BER, while the H4R3 peptides failed to do so (Fig. 6A). To further test whether the arginine methylation of histone H4 plays important roles in cellular LP-BER, we performed a reconstituted LP-BER assay with different cell lysates. In this assay, unlabeled tetrahydrofuran-containing (F) substrates were used for LP-BER (Fig. 6B and Supplementary Table 1). Incorporation of [32P]-dCTP into a single nucleotide gap generated by UDG and APE1 resulted in the production of an unligated labeled 21-mer and a fully repaired 41-mer (Fig. 6B). As PRMT5 knockdown is an effective way to downregulate cellular H4R3me2s levels, we first used the extracts from PRMT5 siRNA- or scrambled siRNA-transfected HEK293T cells in the assay. Western blotting data showed that H4R3me2s was successfully decreased in PRMT5 siRNA-transfected cells. Importantly, we observed that the tetrahydrofuran lesions were efficiently repaired in the control group treated with scrambled siRNA but not in the PRMT5 siRNA group (Fig. 6B). Because PRMT5 knockdown could have reduced the arginine modification of other proteins in addition to H4, to exclude the notion that other factors affected LP-BER and to further assess the function of H4R3me2s in this process, we next transfected H4 or H4 R3Q mutant plasmids into HEK293T cells for 48 h. Western blotting data showed that the H4R3me2s level was much higher in H4-overexpressing cells than in H4 R3Q-expressing cells (Fig. 6C). A reconstituted LP-BER assay showed that BER was more efficient in cells overexpressing H4 than in those overexpressing H4 R3Q (Fig. 6C). Altogether, our data suggest that H4R3me2s can improve cellular BER efficiency.
Fig. 6.
H4R3me2s enhances LP-BER efficiency. Effect of H4R3me2s on LP-BER efficiency. The top parts of each panel show schematic structures of the corresponding DNA substrates. (A) Purified BER proteins and increasing amounts (250 ng, 500 ng, 1000 ng, 2000 ng) of H4R3me2s or H4R3 peptides were subjected to a reconstituted LP-BER assay in vitro. The substrates were incubated with proteins at 37 °C for 30 min. Representative PAGE gels are shown with arrows denoting the repaired product and the unligated product. The graph represents the quantification of the PAGE results in the image, and the values represent the mean ± SD of three independent assays (**P < 0.01, Student's t-test). (B) HEK293T cells transfected with PRMT5 siRNA or scrambled siRNA (Scr) were treated with 1 mM H2O2 for 30 min, and the whole-cell extracts were then collected for reconstituted LP-BER and Western blot assays. Representative PAGE gels are shown with arrows denoting the repaired product and the unligated product. Western blot analysis was performed with anti-PRMT5, anti-H4R3me2s and anti-H4 antibodies. The graph represents the quantification of the PAGE results in the image, and the values represent the mean ± SD of three independent assays (*P < 0.05, Student's t-test). (C) HeLa cells transfected with Flag-tagged WT H4 or Flag-tagged mutant H4 R3Q were treated with 1 mM H2O2 for 30min, and whole-cell extracts were then collected for reconstituted LP-BER and Western blot assays. The left panel shows representative PAGE gels. The level of H4R3me2s modification was detected by Western blotting, with Tubulin used as the loading control. The graph represents the quantification of the PAGE results in the image, and the values represent the mean ± SD of three independent experiments (*P < 0.05, Student's t-test).
3.7. The H4R3me2s modification is essential for DNA repair
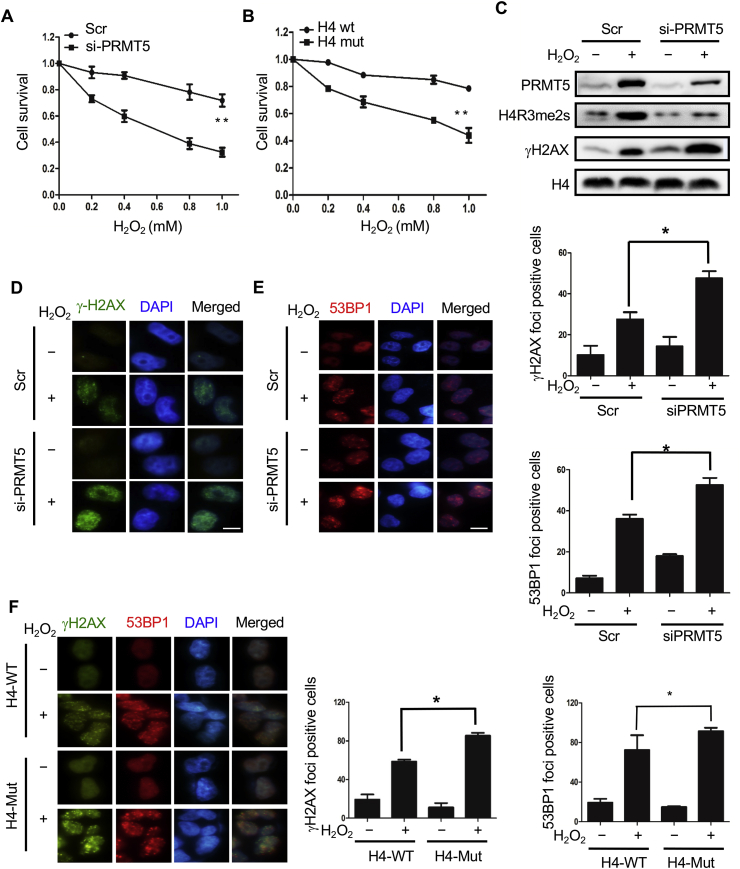
To investigate whether H4R3me2s play an important role in the cellular response to DNA damage, we first downregulated cellular H4R3me2s levels with PRMT5 siRNA and then performed a cell survival assay. As shown in Fig. 7A, PRMT5 siRNA-transfected cells were more sensitive to H2O2 treatment than the control cells (Fig. 7A). Next, we upregulated cellular H4R3me2s levels through overexpressing H4 and carried out the survival assay. Cells overexpressing H4 R3Q were significantly more sensitive to H2O2 than those overexpressing WT H4 (Fig. 7B). Subsequent Western blotting data demonstrated that the cellular γH2AX level was dramatically increased after H2O2 treatment, and this increase was much higher with H4R3me2s reduction (Fig. 7C). Furthermore, immunofluorescence staining of γH2AX and 53BP1 foci was applied to confirm the DNA damage induced by H2O2. More γH2AX and 53BP1 foci were observed in PRMT5 knockdown cells (Fig. 7D and E), which suggested that more DSBs accumulated in the cells. Our data implied that the arginine methylation of H4R3 plays important roles in DNA repair. To further confirm this result, we performed immunofluorescence experiments in HEK293T cells transfected with H4 or H4 R3Q plasmids. The data showed that H4 R3Q-overexpressing cells contained more γH2AX and 53BP1 foci after H2O2 treatment than H4-overexpressing cells (Fig. 7F). Taken together, our results revealed that H4R3me2s is indispensable for the cellular response to DNA damage.
Fig. 7.
The H4R3me2s modification is essential for DNA repair. (A, B) H2O2 sensitivity assays. Cells (1 × 105 per well) were plated in triplicate in six-well plates. Cells transfected with PRMT5 siRNA or scrambled siRNA (A) or Flag-tagged WT H4 or Flag-tagged mutant H4 R3Q (B) were treated with different doses of H2O2 for 30 min, washed with PBS and cultured in fresh media for 2 days. The number of viable cells in every well was determined following the trypsinization of the cells and counting with a cell counter (Countstar IC1000). The control growth ratio was calculated based on the numbers of treated/untreated cells. The data represent the mean ± SD from triplicate wells. Three independent experiments were performed. (C) HeLa cells transfected for 48 h with PRMT5 siRNA or scrambled siRNA were treated with 1 mM H2O2 for 30 min, and cells were then lysed and subject to Western blotting to detect H4R3me2s and γH2AX levels. (D, E) HeLa cells transfected for 48 h with PRMT5 siRNA or scrambled siRNA were treated with 1 mM H2O2 for 30 min γH2AX foci (D) and 53BP1 foci (E) in the cells were detected via immunofluorescence analysis as previously described. Fifty random regions were examined at 400× magnification. Nuclei containing ≥1 foci were counted as positive for focus formation, and the percentage of positive cells was calculated and plotted. The data represent the mean ± SD of three independent experiments. *P < 0.05, Student's t-test. Scale bars, 10 μm. (F) HEK293T cells transfected for 48 h with Flag-tagged WT H4 or Flag-tagged mutant H4 R3Q were treated with 1 mM H2O2 for 30 min. Cells were fixed and stained with antibodies against γH2AX (green) and 53BP1 (red). DNA was stained with DAPI (blue). Left histogram, γH2AX focus analysis; right histogram, 53BP1 focus analysis. The data represent the mean ± SD of three independent experiments. *P < 0.05, Student's t-test. Scale bars, 10 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
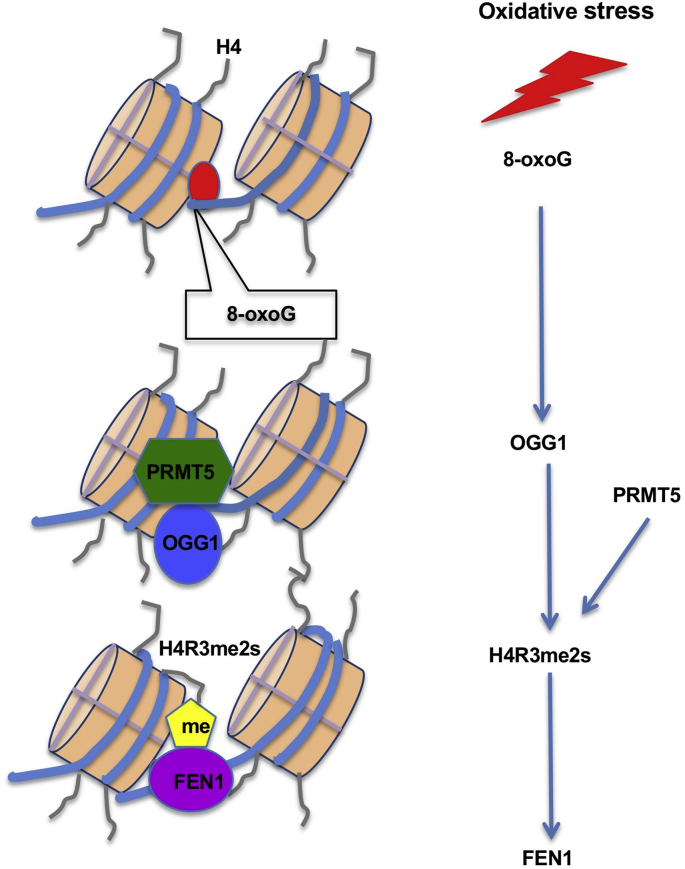
The main finding of this study is that the increase in cellular H4R3me2s upon oxidative stress is mediated by the binding of OGG1 with PRMT5 and that H4R3me2s enhances BER efficiency through directly interacting with FEN1 and enhancing its activity (Fig. 8). Our data showed that H4R3me2s can act as a “reader” that reads oxidatively damaged DNA and can also act as a “writer” to promote DNA damage repair. Altogether, these results suggest that H4R3me2s can serve as a bridge linking DNA damage and repair.
Fig. 8.
Schematic model for histone arginine methylation and DNA repair process. Oxidative stresses induce the conversion of guanine (G) to oxidative guanine (8-oxoG), following which OGG1 can recognize 8-oxoG and recruit PRMT5 to the damaged site. Then, the tail of histone H4 near 8-oxoG is methylated to H4R3me2s by PRMT5. H4R3me2s can increase DNA repair efficiency through binding with FEN1 and enhancing FEN1 activity.
Histone arginine methylation plays important roles in gene transcription and DNA metabolism and can change the structure of chromatin and have downstream functional consequences. H4R3 symmetrical dimethylation (producing H4R3me2s) catalyzed by PRMT5 was shown to be involved in gene silencing and uniquely marked chromatin, mostly at G + C-rich regions in the mouse genome, including imprinting control regions (ICRs) [24]. H4R3me2s can also serve as a direct binding target of DNMT3A, which is required for subsequent DNA methylation [20]. However, little is known about the role of histone arginine methylation in DNA damage repair. In the current work, we present experimental evidence that histone arginine methylation plays important roles in DNA repair.
Various oxidative stresses can induce oxidative DNA damage, and guanine is easily oxidized into 8-oxo-7,8-dihydrogunine due to its low oxidation potential [25]. Once oxidative DNA damage has occurred, DNA glycosylase is actively recruited to regions of open chromatin, where it allows the BER machinery access to these lesions [4]. Here, our data showed that the oxidatively damaged DNA signal can recruit OGG1 to the lesion site, where OGG1 binds with PRMT5 and promotes PRMT5-catalyzed H4R3me2s formation. Our results indicated that OGG1 is indispensable for the symmetrical arginine dimethylation of H4R3 after H2O2 treatment, as OGG1 depletion significantly reduced the H4R3me2s level in MEF cells. This result was also confirmed in HEK293T cells following OGG1 knockdown or overexpression. Previous reports have demonstrated that OGG1 is a damaged base repair enzyme involved in the BER pathway that can specifically recognize and remove the 8-oxoG [10,26]. Girardot et al. reported that H4 symmetrically demethylated at arginine-3 by PRMT5 marks chromatin at G + C-rich regions [24]. These studies further strengthened the possibility that OGG1 and PRMT5 might interact at oxidative G-rich regions. Moreover, our pulldown assay showed that H4R3me2s preferentially bound oxidative guanine, which also supports the possibility that OGG1 regulates H4R3me2s. Depletion of OGG1 did not affect the expression of PRMT5 but nearly abolished the interaction between PRMT5 and H4R3me1, which implied that OGG1 is essential for the binding of PRMT5 to its substrate. Based on these results and those of our study, it seems reasonable to conclude that the oxidative stress-induced increase in H4R3me2s is partially due to the 8-oxoG/OGG1/PRMT5 pathway. In addition to H2O2, other oxidants can also induce the increase in H4R3me2s, and such an increase may be related to the concentration and strength of oxidant.
PRMT5 is an arginine methyltransferase that catalyzes the symmetrical dimethylation of arginine residues in histone and nonhistone proteins [27]. The results of our study of the 8-oxoG/OGG1/PRMT5/H4R3me2s axis suggest that PRMT5 is essential for the DNA damage response. In fact, multiple lines of evidence have shown that PRMT5 can methylate DNA repair proteins and that PRMT5 plays important roles in the cellular response to DNA damage [[28], [29], [30], [31]]. All these studies addressed nonhistone arginine methylation by PRMT5, but the role of histone methylation in DNA repair was not mentioned. Moreover, although there is much evidence suggesting that histone methylation, especially lysine methylation, plays important roles in the DNA damage response [32], histone arginine methylation has not been clearly shown to be involved in this process. Here, our data showed that PRMT5 knockdown decreased BER efficiency and the cell survival ratio. Importantly, cells expressing the H4 R3Q mutant possessed a lower BER efficiency and survival ratio than cells expressing WT H4. Collectively, our results revealed that H4R3me2s can also be an important downstream factor of the function of PRMT5 in DNA repair.
Many mechanisms to address the important roles of histone modification in DNA repair have been proposed. H2AX phosphorylation was the first histone modification involved in the DNA damage response (DDR) to be identified [32]. A reduction in histone H3 and H4 ubiquitylation was shown to impair recruitment of the repair protein XPC to the damaged foci and the repair process [33]. Acetylation at H3K56 and H3K14 in nucleosomes was suggested to promote alternative gap-filling pathways by inhibiting DNA polymerase β activity [34]. Here, we showed that symmetrically dimethylated H4R3 directly interacts with FEN1, promotes FEN1 nuclease activity and increases LP-BER efficiency. These results provide a possible explanation for the function of H4R3me2s in the DNA damage response and are supported by an earlier study by Kwon et al., who discovered that the N-terminal tails of histones possess an intrinsic activity that enhances the nuclease activity of Rad27 (FEN1 in yeast) [35]. Previous studies have revealed that, in response to DNA damage, FEN1 could be post-translationally modified and further affect its functional regulation [36]. It should be noted that it would be possible that H4R3me2s prefers to bind with the post-translationally modified FEN1, although we have no further evidence which form of FEN1 is combined with H4R3me2s in our in vivo and in vitro experiments. In addition, histone posttranslational modification is involved in DNA repair not only because it can be read by DDR factors to promote repair but also because it is a well-known mark that functions in modulating chromatin structure at damage sites [32,37]. H4R3 methylation by PRMT1 was reported to be essential for the establishment or maintenance of a wide range of “active” chromatin modifications [38]. It is reasonable to propose that H4R3me2s is essential for the opening of chromatin to recruit the BER machinery, but this hypothesis requires further investigation.
In summary, this study reveals a novel mechanism by which H4R3me2s serves as a new “histone code” linking oxidative DNA lesions and subsequent base excision repair. Understanding the cellular oxidative response that enables genome stability protection will be helpful.
Author contributions
ZM and FP designed research. ZM and WW performed research. SW, XZ, YM and CW contributed new reagents or analytic tools. WW FP analyzed data. ZM, WW, ZH, LH, and FP discussed and interpreted the data. ZM, WW, SW, FP and ZG wrote the paper.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Professor Istvan Boldogh for kindly supplying MEF WT and Ogg1 null cell lines. This work was supported by National Natural Science Foundation of China (81872284), Changzhou Sci & Tech Program (CE20175035), Jiangsu Key Research and Development Program (Grant No. BE2018714), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (18KJA180006), National Nature Science Foundation (31701179), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101653.
Contributor Information
Feiyan Pan, Email: panfeiyan@njnu.edu.cn.
Zhigang Guo, Email: guozgang@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bystrom L.M., Guzman M.L., Rivella S. Iron and reactive oxygen species: friends or foes of cancer cells? Antioxidants Redox Signal. 2014;20:1917–1924. doi: 10.1089/ars.2012.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marnett L J. Oxyradicals and DNA damage. Carcinogenesis. 2000;21(3):361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 3.Seifermann M., Epe B. Oxidatively generated base modifications in DNA: not only carcinogenic risk factor but also regulatory mark? Free Radic. Biol. Med. 2017;107:258–265. doi: 10.1016/j.freeradbiomed.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 4.Amouroux R., Campalans A., Epe B., Radicella J.P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demple B., Sung J.-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair. 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Frosina G., Fortini P., Rossi O., Carrozzino F., Raspaglio G., Cox L.S., Lane D.P., Abbondandolo A., Dogliotti E. Two pathways for base excision repair in mammalian cells. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 7.Sobol R.W., Horton J.K., Kühn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 8.Andrej Ja Podlutsky I.I.D., Vladimir N.Podust, Vilhelm A.Bohr, Grigory L.Dianov. Human DNA polymerase b initiates DNA synthesis during long-patch repair of reduced AP sites in DNA.pdf. EMBO J. 2001;20:1477–1482. doi: 10.1093/emboj/20.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad R., Dianov G.L., Bohr V.A., Wilson S.H. FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 10.Boiteux S., Radicella J.P. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- 11.Kenichi Nishioka T.O., Oda Hisanobu, Fujiwara Toshiyuki, Kang Dongchon, Sugimachi Keizo, Nakabeppu Yusaku. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevillard S., Radicella J.P., Levalois C., Lebeau J., Poupon M.F., Oudard S., Dutrillaux B., Boiteux S. Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene. 1998;16:3083–3086. doi: 10.1038/sj.onc.1202096. [DOI] [PubMed] [Google Scholar]
- 13.Mao G., Pan X., Zhu B.B., Zhang Y., Yuan F., Huang J., Lovell M.A., Lee M.P., Markesbery W.R., Li G.M. Identification and characterization of OGG1 mutations in patients with Alzheimer's disease. Nucleic Acids Res. 2007;35:2759–2766. doi: 10.1093/nar/gkm189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Björn Tyrberg K.A.A., A Dib Sergio, Rodriguez Jessica Wang, Yoon Kun-Ho, Fred Levine. Islet expression of the DNA repair enzyme 8-oxoguanosine DNA glycosylase (Ogg1) in human type 2 diabetes. BMC Endocr. Disord. 2002;2 doi: 10.1186/1472-6823-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simone S., Gorin Y., Velagapudi C., Abboud H.E., Habib S.L. Mechanism of oxidative DNA damage in diabetes: tuberin inactivation and downregulation of DNA repair enzyme 8-oxo-7,8-dihydro-2'-deoxyguanosine-DNA glycosylase. Diabetes. 2008;57:2626–2636. doi: 10.2337/db07-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Lachner M., O'Sullivan R.J., Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 18.Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 19.Jahan S., Davie J.R. Protein arginine methyltransferases (PRMTs): role in chromatin organization. Adv. Biol. Regul. 2015;57:173–184. doi: 10.1016/j.jbior.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q., Rank G., Tan Y.T., Li H., Moritz R.L., Simpson R.J., Cerruti L., Curtis D.J., Patel D.J., Allis C.D. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Wang W., Du C., Yan F., Yang S., He K., Wang H., Zhao A. OGG1 regulates the level of symmetric dimethylation of histone H4 arginine-3 by interacting with PRMT5. Mol. Cell. Probes. 2018;38:19–24. doi: 10.1016/j.mcp.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Blanc R.S., Richard S. Arginine methylation: the coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki Y., Adachi N., Koyama H. Vertebrate cells lacking FEN-1 endonuclease are viable but hypersensitive to methylating agents and H2O2. Nucleic Acids Res. 2002;30:3273–3277. doi: 10.1093/nar/gkf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girardot M., Hirasawa R., Kacem S., Fritsch L., Pontis J., Kota S.K., Filipponi D., Fabbrizio E., Sardet C., Lohmann F. PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome. Nucleic Acids Res. 2014;42:235–248. doi: 10.1093/nar/gkt884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torkild Visnes ACz-Kr, Wenjing Hao, Olov Wallner, Geoffrey Masuyer, Olga Loseva, Oliver Mortusewicz, Elisée Wiita, Antonio Sarno, Aleksandr Manoilov, Juan Astorga-Wells, Ann-Sofie Jemth, Lang Pan. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.pdf. Science. 2018;362:834–839. doi: 10.1126/science.aar8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L., Wang M., Lv Z., Yang N., Liu Y., Bao S., Gong W., Xu R.M. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20538–20543. doi: 10.1073/pnas.1106946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auclair Y., Richard S. The role of arginine methylation in the DNA damage response. DNA Repair. 2013;12:459–465. doi: 10.1016/j.dnarep.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Hamard P.J., Santiago G.E., Liu F., Karl D.L., Martinez C., Man N., Mookhtiar A.K., Duffort S., Greenblatt S., Verdun R.E. PRMT5 regulates DNA repair by controlling the alternative splicing of histone-modifying enzymes. Cell Rep. 2018;24:2643–2657. doi: 10.1016/j.celrep.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He W., Ma X., Yang X., Zhao Y., Qiu J., Hang H. A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res. 2011;39:4719–4727. doi: 10.1093/nar/gkq1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q., Zhao Y., Yue M., Xue Y., Bao S. The protein arginine methylase 5 (PRMT5/SKB1) gene is required for the maintenance of root stem cells in response to DNA damage. J. Genet. Genom. 2016;43:187–197. doi: 10.1016/j.jgg.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Gong F., Miller K.M. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2017;780:37–47. doi: 10.1016/j.mrrev.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez Y., Hinz J.M., Laughery M.F., Wyrick J.J., Smerdon M.J. Site-specific acetylation of histone H3 decreases polymerase beta activity on nucleosome core particles in vitro. J. Biol. Chem. 2016;291:11434–11445. doi: 10.1074/jbc.M116.725788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon B., Munashingha P.R., Shin Y.K., Lee C.H., Li B., Seo Y.S. Physical and functional interactions between nucleosomes and Rad27, a critical component of DNA processing during DNA metabolism. FEBS J. 2016;283:4247–4262. doi: 10.1111/febs.13934. [DOI] [PubMed] [Google Scholar]
- 36.Xu X., Shi R., Zheng L., Guo Z., Wang L., Zhou M., Zhao Y., Tian B., Truong K., Chen Y. SUMO-1 modification of FEN1 facilitates its interaction with Rad9-Rad1-Hus1 to counteract DNA replication stress. J. Mol. Cell Biol. 2018;10:460–474. doi: 10.1093/jmcb/mjy047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Attikum H., Gasser S.M. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 38.Suming Huang M.L., Felsenfeld Gary. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.