Abstract
Reactive oxygen species (ROS) are critical for the progression of cardiovascular diseases, inflammations and tumors. However, the mechanisms of how ROS sense metabolic stress, regulate metabolic pathways and initiate proliferation, inflammation and cell death responses remain poorly characterized. In this analytic review, we concluded that: 1) Based on different features and functions, eleven types of ROS can be classified into seven functional groups: metabolic stress-sensing, chemical connecting, organelle communication, stress branch-out, inflammasome-activating, dual functions and triple functions ROS. 2) Among the ROS generation systems, mitochondria consume the most amount of oxygen; and nine types of ROS are generated; thus, mitochondrial ROS systems serve as the central hub for connecting ROS with inflammasome activation, trained immunity and immunometabolic pathways. 3) Increased nuclear ROS production significantly promotes cell death in comparison to that in other organelles. Nuclear ROS systems serve as a convergent hub and decision-makers to connect unbearable and alarming metabolic stresses to inflammation and cell death. 4) Balanced ROS levels indicate physiological homeostasis of various metabolic processes in subcellular organelles and cytosol, while imbalanced ROS levels present alarms for pathological organelle stresses in metabolic processes. Based on these analyses, we propose a working model that ROS systems are a new integrated network for sensing homeostasis and alarming stress in metabolic processes in various subcellular organelles. Our model provides novel insights on the roles of the ROS systems in bridging metabolic stress to inflammation, cell death and tumorigenesis; and provide novel therapeutic targets for treating those diseases. (Word count: 246).
Keywords: Reactive oxygen species (ROS), A sensing network for metabolic stress, Inflammation, Trained immunity, Nuclear signaling
1. Introduction
Reactive oxygen species (ROS) are defined as oxygen-containing reactive species. It is well established that increased ROS play a vital role in promoting cardiovascular disease [[1], [2], [3]] such as hypertension, aortic aneurysm, hypercholesterolemia, atherosclerosis [4], diabetic vascular complication, cardiac ischemia reperfusion injury, myocardial infarction [5], heart failure, and cardiac arrhythmias [6], chronic kidney disease [[7], [8], [9], [10], [11]], hyperhomocysteinemia [[12], [13], [14]], metabolic syndrome [15], induction of regulatory T cells (Treg) and T cell-mediated inflammation [16,17], cigarette smoking [18], metabolically healthy obesity [19,20] and obesity [21], and tumorigenesis [22]. Additionally, ROS have been studied as the therapeutic targets for these disease progression and complications [23]. However, two important issues remain unknown, whether and how the ROS system senses danger associated molecular pattern (DAMP) [24] and conditional DAMP [25,26] signals in metabolic diseases [27] and connects danger signals to proliferative, inflammatory and cell death-related responses.
ROS are generated by oxidant enzymes and scavenged by scavenging systems, which include enzymatic or non-enzymatic reactions. Imbalances between generation and scavenging ROS systems increase ROS levels and represent alarming stresses. ROS are generated in almost every subcellular organelle in cell [28], including plasma membrane [29], cytosol [30], mitochondria [31], nucleus [31], peroxisome [32], endoplasmic reticulum (ER) [33], Golgi [29], and others. Of note, 41 human enzymes involved in generating H2O2 and O2− listed in the Supplemental Table 1 demonstrated the proof of principle that various metabolic processes contribute to generation of ROS [34]. Studies found that mitochondrial ROS production promotes endothelial cell activation and monocyte migration [[35], [36], [37], [38], [39], [40], [41], [42]]; and cytosolic ROS production via nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX) activation contributes to innate immunity. These findings suggest that while each organelle can generate ROS, they differ in sensing danger signals [43,44]. Current studies have found that 2801 metabolic pathways(https://metacyc.org/) exist in various organelles [45] with efficient metabolic requirements for space and compartmentalization [46,47], as we discussed in our recent paper [48].
However, several important questions remain: 1) are there sensing and monitoring systems for each of 2801 metabolic pathways [49], in one-to-one basis, or an integrated sensing system for all the metabolic pathways compartmentalized in all the subcellular organelles; 2) it is non-evolutionally economic and efficient to have one-to-one sensing system for each of 2801 metabolic pathways. An example of an evolutionally efficient sensing system is the humoral immune system, where only four groups of IgG subtypes [50] as backbone structures are needed in order to recognize millions of antigens. So, how can it be possible for danger-associated molecular pattern receptors (DAMP-receptors) [24,27,36] and conditional DAMP receptors [25], including plasma membrane-localized Toll-like receptors, and cytosolically localized caspase-1 [4,5,[51], [52], [53], [54], [55], [56], [57]]/canonical and caspase-4 (humans)/caspase-11 (mice) non-canonical inflammasome pathways [58,59], to monitor the two cellular status, physiological homeostasis and pathologic abnormality of metabolic processes in all the organelles, in an evolutionarily efficient manner; and 3) is there any currently known sensing system capable of: i) localizing in all the subcellular organelles; ii) trafficking across the membranes of organelles; iii) utilizing a universal process shared by all the metabolic pathways, such as being sensitive to any abnormalities of electron donation and electron acceptance as elucidated in our previous reviews [[37], [38], [39], [40]] and reports [41,42,60,61]; and iv) converging and activating inflammasomes [62]; and v) regulating gene expression, damaging DNAs and determining cell proliferation and cell death [44].
To address those significant issues, we analyzed recent progress in the ROS field. ROS levels are tired to generally every reaction via reception and donation of electrons and participate in various intra-organelle signaling pathways. Thus, ROS might act as an evolutionarily efficient monitoring system for cellular metabolic status. However, whether the functions of ROS in organelles other than mitochondria are connected still needs to be elucidated. Superoxide (O2▪−) [30], nitric oxide radical (NO−) [63] and hydrogen peroxide (H2O2) [64] are the most studied ROS subtypes, but other subtypes of ROS are not mentioned in many studies. Increased ROS are damaging to lipids, proteins and DNA; and some of these ROS can cross lipid membranes. However, the functional differences between ROS subtypes remain unknown. Furthermore, studies found that ROS are not only by-products of metabolic reactions, but also participate in physiological signaling [[65], [66], [67], [68], [69], [70], [71]], while increased ROS levels contribute to various metabolic outcomes via regulating metabolic processes and pathways [43,[72], [73], [74], [75]]. Therefore, some antioxidant therapies fail because of the undesirable disruption of physiological functions caused by ROS. Therefore, the therapeutic window is important in order to maintain the ROS in normal levels. In addition to the regulatory mechanisms of gene transcription, mRNA splicing, mRNA stability with microRNA modulation, translation efficiency, and post-translation modifications, activations of different metabolic enzymes are also affected by metabolic processes. These enzymes alter the ROS production and scavenging, which qualify the ROS system for their metabolic sensing functions.
In this review, we analyzed ROS literature extensively regarding ROS generation, scavenging, subcellular localization, physiological and pathological signaling and outcomes. Since ROS are functionally communicated each other between organelles, we propose a new working model that ROS is an integrated network for sensing homeostasis and alarming stress in metabolic processes in various subcellular organelles. Based on increased ROS levels, ROS participated in various metabolic pathways that can relay the metabolic stress to inflammasomes or branch out and cross-talk to other cell stress and stress-sensing pathways (also see section 5) to initiate corresponded reactions such as proliferation, inflammation and cell death [76].
2. ROS serve as the new sensing network for homeostasis and stress of metabolic processes in various subcellular organelles
ROS are generated by oxidant enzymes localized in different subcellular organelles; and ROS scavengers also contribute to maintaining redox homeostasis through enzymatic or non-enzymatic mechanisms. Oxidative stress, as a concept formulated in 1985, is induced when ROS generation is increased, or ROS scavengers are dysregulated [77]. Since oxidative stress is strongly correlated with inflammation and cell death [74,78,79], the imbalance of ROS generation and scavenging systems is an alarming signal for cellular metabolisms. In addition, mitochondrial ROS convey information to the nucleus and modulate the expression of nuclear genes accordingly in both physiological [80] and pathological states [66]. Based on the studies we mentioned above, ROS work as a sensor of organelles’ metabolic homeostasis and alarming stress, additionally, ROS transduce signals among organelles and contribute to the determination of cellular responses [34,81]. To better understand the functions of ROS in physiological and pathological conditions, we classify eleven of ROS based on their chemical structures. However, only three types of ROS, including superoxide (O2▪−), nitric oxide radical (NO▪) and hydrogen peroxide (H2O2), are mostly studied and characterized. In addition, even though the important signaling transducing roles of ROS have been found, the subcellular localizations and functions of different types of ROS are less studied and discussed.
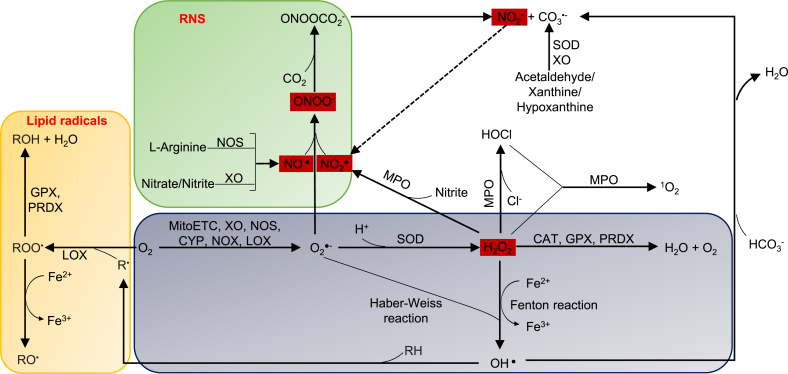
As we mentioned above, eleven types of ROS are classified into two groups, radical ROS and non-radical ROS. Radical ROS include seven types such as superoxide (O2▪−), nitric oxide radical (NO▪), nitrogen dioxide (NO2▪), hydroxyl radical (OH▪), peroxyl radical (ROO▪), alkoxyl radical (RO▪) and carbonate radical (CO3▪-). Non-radical ROS includes four types such as hydrogen peroxide (H2O2), peroxynitrite (ONOO−), singlet oxygen (1O2) and hypochlorous acid (HOCl). In addition, three other types of ROS are less studied, such as hydroperoxyl radical (HO2▪), nitrocarbonate (ONOOCO2−), and ozone (O3), and are not discussed in this review. Reactive oxygen and nitrogen reactive species (RONS) are generally discussed as ROS in this review including NO▪, NO2▪ and ONOO− [82]. As showed in Fig. 1, among the eleven types of ROS, four types of ROS such as NO▪, NO2▪, H2O2 and ONOO− have the ability to cross lipid membrane and transduce signaling across organelles and cells, which we classify as organelle communication ROS in our new working model based on their functions in new integrated sensing and alarming system for metabolic stress. For the other types of ROS, they can hardly pass through lipid membrane due to limited half-life or its physical property, but they contribute to the production of other aggressive ROS or biological molecule damages [83], which contribute to activation of other stress pathways [44]. Nitrogen-containing ROS mostly are communication ROS. Lipid radicals are dominant in peroxisome. The ROS, which are generated by hydrogen and oxygen, such as HO▪ and H2O2, are connected with different types of ROS systems, and are functionally classified as the chemical connecting ROS in our new working model.
Fig. 1.
Four types of ROS (red box) that are generated by nitrogen and oxygen are communication ROS, which are, connected with different types of ROS systems. NO▪, NO2 ▪, H2O2 and ONOO− have the ability to cross lipid membrane and are classified as communication ROS. Three types of nitrogen-containing ROS are communication ROS.
Abbreviations: O2▪−—superoxide, NO▪—nitric oxide radical, NO2 ▪—nitrogen dioxide, OH▪—hydroxyl radical, ROO▪—peroxyl radical, RO▪— alkoxyl radical, CO3▪-—carbonate radical, H2O2—hydrogen peroxide, ONOO−—peroxynitrite, 1O2—singlet oxygen, HOCl—hypochlorous acid, ONOOCO2 −—nitrocarbonate, MitoETC: Mitochondrial electron transport chain, XO—xanthine oxidase, NOS—nitric oxide synthase, CYP— cytochrome P450, NOX—NADPH oxidase, LOX—lipoxygenase, SOD—superoxide dismutase, MPO—myeloperoxidase, MAO— monoaminoxidase. GPX—glutathione peroxidase, PRDX—peroxiredoxin, CAT—catalase. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The enzymes generated and subcellular localization of eleven types of ROS are listed in Table 1A. The following results in Table 1A are highlighted: first, among seven radical ROS, O2▪− and NO▪ are the dominant radicals, which are generated by 10 enzymes and 4 enzymes, respectively. Second, among non-radical ROS, H2O2 and 1O2 are major ROS, which are generated by 11 and 8 enzymes or reactions, respectively. Notably, cell type information related to those ROS are under-investigated. Enzymatic and non-enzymatic scavenging systems and organelle localization of eleven types of ROS are listed in Table 1B. Among the 11 types of ROS, five types of ROS such as O2▪−, ROO▪, CO3▪-, H2O2 and ONOO− can be scavenged by enzymes; and the rest are found to be scavenged by non-enzymatic scavengers but not enzymatic scavengers, suggesting that they are modulated indirectly by gene product regulations. We integrated the subcellular localization of each type of ROS in Table 1C, and the following results are easily found: 1) more than five types of ROS are found to be generated and functioned in the metabolic intensively active organelles, including mitochondria, peroxisome, plasma membrane, cytosol and nucleus. 2) the generation and scavenging enzymes of ROS are both found in some organelles. However, for some types of ROS only generation or scavenging system is found in the organelle. For example, O2▪− was found to be generated in Golgi and ER, but the corresponded scavenging enzymes have not been reported in these organelles, indicating that O2▪− related metabolic stress can either relay to other types of ROS or branch out to other stress pathways [84]. ROO▪ scavenging enzymes are found to be expressed in the nucleus, whereas it is currently unknown if ROO▪ is generated in the nucleus. Further studies are needed to determine the subcellular location of ROO▪ synthesis.
Table 1A.
Reactive oxygen species (ROS) include seven radical- and four non-radical ROS; the generation enzymes and subcellular locations are presented. Among radical ROS, superoxide and nitric oxide are the dominant radicals which are generated by 10 enzymes and 4 enzymes, respectively. Among non-radical ROS, hydrogen peroxide, singlet oxygen and peroxynitrite are major ROS which are generated by 11 and 8 reactions, respectively.
| Category | Name | Formula | Classification | Generating system | Intracellular organelle | PMID | Ref # |
|---|---|---|---|---|---|---|---|
| Radicals | Superoxide | O2▪− | ROS | Complex I | Mt | 24481843 | [122] |
| Complex II | Mt | 24481843 | [122] | ||||
| Complex III | Mt | 24481843 | [122] | ||||
| XO | Mt, Px, Cyto | 15475499 | [30] | ||||
| eNOS | Golgi, PM | 24180388 | [229] | ||||
| iNOS | Px | 24180388 | [229] | ||||
| nNOS | SR | 24180388 | [229] | ||||
| CYP | ER, Mt | 12605691 | [33] | ||||
| NOX | NOX1 (Caveolae), NOX2 (Phagosome, PM), NOX3/5 and Duox1/2 (PM), NOX4 (Mt, ER, Nuc) | 15475499 | [30] | ||||
| LOX | Cyto, Nuc | 15475499 | [30] | ||||
| Radicals | Nitric oxide | NO▪ | RONS | eNOS | Golgi, PM, Mt | 22178243, 20388537, 11729179 | [29,63,230] |
| iNOS | Px | 22178243, 12085352 | [29,88] | ||||
| nNOS | SR | 22178243 | [29] | ||||
| XO | Px | 22178243 | [29] | ||||
| Radicals | Nitrogen dioxide | NO2▪ | RONS | MPO | Nuc, Cyto, Mt | 19298861 | [89] |
| Radicals | hydroxyl radical | OH▪ | ROS | Fenton reaction | Mt, Cyto, Nuc, Px | 24987008 | [31] |
| Haber-Weiss Reaction | Mt, Cyto | 24987008 | [31] | ||||
| Radicals | peroxyl radical | ROO▪ | ROS | LOX | Microsome, Mt, Px, PM | 15728540 | [96] |
| Radicals | alkoxyl radical | RO▪ | ROS | LOX | Microsome, Mt, Px, PM | 15728540 | [96] |
| Radicals | Carbonate radical | CO3▪- | ROS | SOD1 | Px | 17505962 | [97] |
| XO | Px | 17505962 | [97] | ||||
| Nonradicals | hydrogen peroxide | H2O2∗ | ROS | SOD | SOD1 (Cyto, Mt, Nuc, Px), SOD2 (Mt matrix), | 17505962 | [97] |
| MAO | Mt | 24987008 | [31] | ||||
| XO | Mt, Px, Cyto | 25678748 | [64] | ||||
| p66shc | Cyto, Mt | 27925481 | [39] | ||||
| acyl-CoA oxidases | Px | 25906193 | [32] | ||||
| urate oxidase | Px | 25906193 | [32] | ||||
| d-amino acid oxidase | Px | 25906193 | [32] | ||||
| d-aspartate oxidase | Px | 25906193 | [32] | ||||
| l-pipecolic acid oxidase | Px | 25906193 | [32] | ||||
| l-α-hydroxyacid oxidase | Px | 25906193 | [32] | ||||
| polyamine oxidase | Px | 25906193 | [32] | ||||
| Nonradicals | peroxynitrite | ONOO− | RONS | – | – | 17505962 | [97] |
| Nonradicals | singlet oxygen | 1O2 | ROS | Photooxygenation | Mt, Cyto | 22266568 | [106] |
| MPO | Cyto | 27042259 | [105] | ||||
| LOX | Px | 169247 | [104] | ||||
| dioxygenase | Px | 169247 | [104] | ||||
| lactoperoxidase | Px | 169247 | [104] | ||||
| CYP | ER | 29081894 | [107] | ||||
| Cytochrome C | Mt | 29081894 | [107] | ||||
| Fenton reaction | Px | 26070643 | [108] | ||||
| Nonradicals | hypochlorous acid | HOCl | ROS | MPO | Mt, Cyto, Nuc | 27042259, 26632272 | [105,111] |
Abbreviations: XO—xanthine oxidase, NOS—nitric oxide synthase, CYP—cytochrome P450, NOX—NADPH oxidase, LOX—lipoxygenase, SOD—superoxide dismutase, eNOS—endothelial NOS, iNOS—inducible NOS, nNOS—neuronal NOS, MPO—myeloperoxidase, MAO—monoaminoxidase, Mt—mitochondria, Px—peroxisome, Cyto-cytosol, PM—plasma membrane, ER--endoplasmic reticulum, Nuc—Nucleus, SR--Sarcoplasmic reticulum, ECM—extracellular matrix.
41 specific generation enzymes for hydrogen peroxide and superoxide are also studied in Suppl. Table 1.
Table 1B.
ROS scavengers and subcellular localization are indicated, and the scavengers include enzymatic- and non-enzymatic- scavengers. Among the 11 types of ROS, superoxide, peroxyl radical, carbonate radical, hydrogen peroxide and peroxynitrite can be scavenged by enzymes; and the rest six types of ROS are found to be scavenged by non-enzymatic scavengers but not enzymatic scavengers.
| Formula | Enzymatic scavenger | Localization (enzymatic-scavenger) | PMID | Ref # | Non-enzymatic scavenger | PMID | Ref# |
|---|---|---|---|---|---|---|---|
| O2▪− | SOD | SOD1 (Cyto, Mt, Nuc, Px), SOD2 (Mt matrix), SOD3 (ECM) | 23442817 | [83] | Bilirubin | 31400697 | [131] |
| NO▪ | |||||||
| NO2▪ | Urate | 22101009 | [231] | ||||
| Ferrocyanide | 22101009 | [231] | |||||
| OH▪ | Vitamin E | 31400697 | [131] | ||||
| Uric acid | 31400697 | [131] | |||||
| NDGA | 20415502 | [232] | |||||
| Carotenoids | 23015774 | [233] | |||||
| NAC | 21118657 | [234] | |||||
| Vitamin C | 9090754 | [235] | |||||
| ROO▪ | GPX4/5 | Cyto, Nuc, Mt | 22178243 | [29] | Carotenoids | 23015774 | [233] |
| PRDX1/2/3/4/5 | Cyto, Nuc, Mt, Px | 31400697 | [131] | Vitamin E | 31400697 | [131] | |
| Uric acid | 31400697 | [131] | |||||
| RO▪ | Vitamin C | 31400697 | [131] | ||||
| Edaravone | 17115906 | [236] | |||||
| CO3▪- | CAT | Px, Mt, Cyto, Nuc | 17505962 | [97] | Melatonin | 16153306 | [237] |
| AMK | 16153306 | [237] | |||||
| H2O2 | CAT | Px, Mt, Cyto, Nuc | 31467634, 22178243, 25646037 | [29,93,98] | Bilirubin | 31400697 | [131] |
| GPX1/2/3/6/7/8 | Cyto, ECM, Mt, ER | 31819197 | [238] | ||||
| PRDX1/2/3/4/5/6 | Cyto, Nuc, Mt, Px | 22178243 | [29] | ||||
| ONOO− | GPX | Cyto, ECM | Bilirubin | 31400697 | [131] | ||
| PRDX5 | Cyto, Nuc, Mt, Px | 22178243 | [29] | Uric acid | 9435251 | [239] | |
| 1O2 | Sodium azide | 21491580 | [240] | ||||
| Vitamin E | 9119263 | [241] | |||||
| Carotenoids | 23015774 | [233] | |||||
| Edaravone | 21871447 | [242] | |||||
| HOCl | Uric acid | 31400697 | [131] | ||||
| Carotenoids | 23015774 | [233] | |||||
| Bilirubin | 2542140 | [243] | |||||
| NAC | 21118657 | [234] | |||||
| 5-HT/Serotonin | 26699077 | [244] |
Abbreviations: GPX—glutathione peroxidase, PRDX—peroxiredoxin, CAT—catalase, NDGA-- nordihydroguaiaretic acid, AMK-- N1-acetyl-5-methoxykynuramine, NAC-- N-acetylcysteine, 5-HT/Serotonin-- 5-Hydroxytryptamine.
Table 1C.
The subcellular organelles of ROS generation and scavenging system are organized as follows. Among the twelve subcellular localizations, mitochondria, peroxisome, plasma membrane, cytosol and nucleus cause more than five types of ROS.
| Organelle | Mt | Px | PM | Cyto | Nuc | Golgi | Caveolae | ER | Phagosome | Microsome | SR | ECM | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O2▪− | + | – | + | – | + | + | – | + | – | + | + | + | + | – | ||||||||||
| NO▪ | + | + | + | + | + | |||||||||||||||||||
| NO2▪ | + | + | + | |||||||||||||||||||||
| OH▪ | + | + | + | + | ||||||||||||||||||||
| ROO▪ | + | – | + | – | + | – | – | + | ||||||||||||||||
| RO▪ | + | + | + | + | ||||||||||||||||||||
| CO3▪ | – | + | – | – | – | |||||||||||||||||||
| H2O2 | + | – | + | – | + | – | + | – | – | – | ||||||||||||||
| ONOO− | – | – | – | – | – | |||||||||||||||||||
| 1O2 | + | + | + | + | ||||||||||||||||||||
| HOCl | + | + | + | |||||||||||||||||||||
Indicates relative ROS generation enzyme subcellular localization and - indicates relative ROS scavenging enzyme subcellular localization.
To clarify, the ROS generated by enzymes in the plasma membrane are released into the cytosol and are scavenged by intracellular systems. For example, ROO▪ is generated by enzymes located on the plasma membrane, and the scavenge enzymes are localized in the cytosol. To better illustrate the functions of these ROS, the biochemical methods and specific inhibitors are indicated in supplemental (S.) Supplemental table 2, Supplemental table 3. Based on the roles of different types of ROS in relaying the metabolic stress signals from 2801 metabolic pathways in organelles to a final end in activating inflammasomes, we classify various types of ROS into seven functional groups in our new working model: 1) metabolic stress-sensing (initiation), 2) chemical connecting (one type ROS converted/dismutated into another type of ROS), 3) organelle communication (organelle membrane crossing RNAs and capable of relaying metabolic stress to mitochondrial ROS central hub and/or nuclear ROS converging hub), 4) stress branch-out (non-inflammasome activating and crosstalk to other stress pathways (also see section 5) such as misfolded proteins for endoplasmic reticulum (ER) stress [84], DNA damage for nuclear stress [44], RNA damage for RNA degradation [85]), 5) inflammasome-activating (final stop for this sensing system), 6) dual functions, and 7) triple functions. We will explain the rationale for this functional categorization of each type of ROS in the following paragraphs.
2.1. Superoxide (O2▪−)
Up to 1–4% of oxygen is reduced into O2▪−, which is the first formed ROS. We classify O2▪− as the dual function ROS with metabolic stress-sensing and chemical connecting. O2▪− is generated in most organelles such as mitochondria, peroxisome, cytosol, plasma membrane, ER and nucleus. It could be generated by eight types of oxidant enzymes, including mitochondrial electron transport chain complex I, complex II, complex III, as well as xanthine oxidase (XO), NO synthases (NOS), cytochrome p 450 enzymes (CYP), NADPH oxidases (NOX) and lipoxygenase (LOX). O2▪− is unstable in aqueous solutions because of its short half-life. O2▪− can be scavenged or converted in three pathways: 1) O2▪− is quickly dismutated to H2O2 after generation; 2) if the production level of O2▪− is low (picomolar range), O2▪− reacts with NO▪ to form ONOO−, which process is even faster than dismutation into H2O2 [30]; and 3) at high levels of O2▪−, it can react with H2O2 to produce highly reactive radical OH▪ via protein iron-sulfur center reaction and iron release [30]. Of note, it has been reported that endogenously produced O2▪− and H2O2 primarily contribute to NLRP3 inflammasome formation and activation [86] in mouse glomeruli resulting in glomerular injury or consequent sclerosis during hyperhomocysteinemia [87]. Therefore, O2▪− and H2O2 are also functionally classified as the inflammasome activating ROS.
2.2. Nitric oxide (NO▪)/nitrogen dioxide (NO2▪)
NO▪ is also the first formed ROS and the triple-function ROS with metabolic stress-sensing, chemical connecting and organelle communication, which is produced by a family of NO synthases (NOS), including neuronal NOS (NOS1, nNOS), inducible NOS (NOS2, iNOS) and endothelial NOS (NOS3, eNOS). In addition to NO▪, studies found that NOS also produces O2▪− when NOS is not coupled with its cofactors or substrate; and this process is called NOS uncoupling [88]. However, other publications also reported that NOS also generates O2▪− in coupled conditions [88]. Besides, NO▪ is also generated by xanthine oxidase (XO) via reducing nitrates and nitrites [29]. Nitrites reacts with H2O2 to generate NO2▪ via the function of enzyme myeloperoxidase (MPO). NO2▪ also reacts with O2▪− and produces ONOO− [89]. They both can cross the lipid membrane, which are responsible for communication between organelles and cells.
2.3. Hydroxyl radical (OH▪)
As mentioned above, OH▪ is generated from H2O2 and O2▪− through the Haber-Weiss reaction or Fe3+-mediated decomposition of H2O2 through Fenton reaction [31]. However, no enzymes have been identified for OH▪ generation [90]. In addition, there are no existing enzymatic systems to scavenge OH▪ [90]. These findings suggest that OH▪ generation and conversion overcome no energy barriers and do not require the enzymes. Furthermore, OH▪ has the capability to damage different cellular components via lipid peroxidation, protein damage, DNA bases damage, and membrane destruction [31]. Therefore, the generation and accumulation of OH▪ lead to cell death. However, based on recent understanding, the organelles that contain O2▪− and H2O2 generation enzymes can produce OH▪. Since OH▪ does not convert into other types of ROS, and does not activate nucleotide-binding domain, leucine-rich-repeat containing family, pyrin domain-containing 3 (NLRP3) inflammasome [87] but damage and “branch out oxidative stress” to other molecules and pathways, therefore, OH▪ is functionally classified as the stress-branch out ROS.
2.4. Peroxyl radical (ROO▪)/Alkoxyl radical (RO▪)
ROO▪ is generated as intermediates in the lipid peroxidation reactions [91]. Lipid peroxidation is a chain reaction process resulting in the generation of ROO▪, which is characterized by repetitive hydrogen abstraction [92] and addition of oxygen to R▪. 0.3% of cytosolic O2▪− is in the protonated form as HOO▪, which is the simplest form of ROO▪ [93]. ROO▪ nearly attacks all major classes of biomolecules, mainly the polyunsaturated fatty acids (PUFA) of cell membranes [94] and DNA bases [95]. RO▪ is generated form ROO▪ via a tetroxide. RO▪ is a less aggressive ROS than HO▪, but more reactive than ROO▪ [29]. Studies reported that LOX contributes to lipid peroxidation and is localized in cytoplasm and nucleus [96], which indicating the subcellular localization of ROO▪. They are the most dominant ROS in peroxisome, which may have critical sensing functions in peroxisome. Therefore, ROO▪ is functionally classified as the triple function ROS with metabolic stress sensing, chemical connecting and ROS-converting and stress-branch-out.
2.5. Carbonate radical (CO3▪-)
CO3▪- is a negatively charged radical in physiological and pathological conditions, and is formed in the following four reactions: first, superoxide dismutase 1 (SOD1) peroxidase activity. Second, XO-mediated oxidation of acetaldehyde, xanthine and hypoxanthine [97]. Third, hydrogen abstraction from bicarbonate by OH▪. Fourth, nitrocarbonate is converted into NO2▪ and CO3▪- [97]. CO3▪- is less oxidizing than OH▪, but it has larger ranges of oxidative actions because of its longer half-life in comparison to OH▪ [97]. CO3▪- could rapidly oxidizes DNA guanine residues and protein residues [97]. However, CO3▪- is unable to produce stable adducts, which makes it difficult to prove its production compared with other radicals. CO3▪- is functionally classified as the stress-branch out ROS.
2.6. Hydrogen peroxide (H2O2)
H2O2 is generated via SOD-mediated dismutation of O2▪− [31]. There are three isoforms of SOD. First, Cu, Zn-dependent SOD (SOD1) converts O2▪− into H2O2 in mitochondrial intermembrane space (IMS) and cytosol. Second, Mn-dependent SOD (SOD2) converts O2▪− into H2O2 in the mitochondrial matrix (MM). Third, Cu, Zn-dependent SOD (SOD3) converts O2▪− into H2O2 in extracellular space [83]. Monoaminoxidase (MAO) resides in the outer mitochondrial membrane and generates H2O2 [31,98]. XO is one of the most studied ROS generation enzymes in the cytosol [83,99] and mediates H2O2 production through the process of hypoxanthine and xanthine catalyzation [64]. H2O2 is also generated in peroxisomes via various enzymes from the normal catalytic cycle [100]. These enzymes are mainly flavoproteins, including acyl-CoA oxidases, urate oxidase, d-amino acid oxidase, d-aspartate oxidase, l-pipecolic acid oxidase, l-α-hydroxyacid oxidase, polyamine oxidase, and XO [32]. The adaptor and ROS-regulating protein P66shc normally resides in the cytosol and translocates into the mitochondria under oxidative stress via a protein kinase C (PKC)-dependent manner. After translocation, p66shc oxidizes cytochrome c, and generates H2O2 [101]. H2O2 is more stable than O2▪−, and the concentrations of H2O2 are also 100 times higher than O2▪− in mitochondria. These properties render H2O2 as an ideal signaling molecule in cells [83]. H2O2 is released via aquaporin 8 formed channel from cell membrane. As discussed above, H2O2 is functionally classified as the dual function ROS with organelle communication and inflammasome activating.
To demonstrate the proof of principle that how ROS generating enzymes are connected to metabolic pathways, we also list the metabolic functions of 41H2O2-generating enzymes and O2- generating enzymes, which suggests that many metabolic pathways contribute H2O2 generation and O2- generation, which are further connected to inflammasome activation and other cellular stress pathways [102]. The enzymes participate in various metabolic processes and pathways as shown in Supplemental Table 1. For example, by searching the metabolomics database (https://hmdb.ca/metabolites/HMDB0004062) using the method that we reported [11], we can find that the enzyme aldehyde oxidase 1 is functional in amino acid valine, leucine and isoleucine degradation, tyrosine metabolism, tryptophan metabolism, vitamin B6 metabolism and nicotinate and nicotinamide metabolism.
2.7. Peroxynitrite (ONOO−)
As mentioned above, the powerful oxidant ONOO− is produced from the rapid reaction between O2▪− and NO▪ [97]. ONOO− causes DNA single-strand breakage and activates poly-ADP-ribose polymerase (PARP) [103]. ONOO− also induces nitration and S-nitrosylation of proteins and lipids [103]. It is capable to cross lipid membranes and thus has the potential to communicate cellular status between organelles. ONOO− is functionally classified as the dual function ROS with organelle communication and stress branch-out.
2.8. Singlet oxygen (1O2)
1O2 is formed via photooxygenation and enzymatic reactions. The enzymatic reactions for 1O2 production is mediated by several peroxidases in lipid peroxidation, including lipoxygenase (LOX), dioxygenase and lactoperoxidase [92,104]. Oxidation of H2O2 and HOCl mediated by MPO [105] contributes to the production of 1O2 in a chemical system [104,106]. In addition, 1O2 is also formed by CYP 2E1 and cytochrome c in the ER and mitochondria, respectively [107]. Fenton-type reactions may also give rise to the production in peroxisomes of 1O2. Thus, 1O2 could be a new player in the peroxisome-derived signaling network [108]. Furthermore, studies also reported that 1O2 causes mitochondrial ETC proteins oxidation. In addition, studies found that 1O2 generation after light stimulation significantly affects the activities of mitochondrial ETC complex I, complex III, complex IV but except complex II [109]. Studies found that the secondary generation of O2▪− and H2O2 are induced in 1O2 caused dysfunctional mitochondrial respiration [109,110]. Thus, we classify 1O2 as the triple function ROS with metabolic sensing, chemical connecting, and stress branch-out.
2.9. Hypochlorous acid (HOCl)
MPO catalyzes the formation of HOCl from H2O2 and chloride ion [105]. MPO is found to localize to the nucleus, mitochondria and cytoplasm, which may explain the subcellular localization of HOCl seen in neutrophil and monocytes [111]. However, it remains unclear whether HOCl roles in our integrated sensing and alarming system for metabolic stress.
3. Mitochondrial ROS systems serve as the central hub for connecting ROS systems with inflammasomes, trained immunity and immunometabolic pathways
Mitochondria consume the greatest amount of oxygen (80–95%) to allow oxidative phosphorylation (OXPHOS) for energy generation [112]. OXPHOS produces 95% of cell energy by coupling tricarboxylic acid (TCA) cycle and mitochondrial electron transport chain (ETC) in most cell types [113], except for endothelial cells (both tip cells and non-tip cells) [114], which mostly take use of glycolysis for energy generation [115]. During OXPHOS, mitochondrial ROS is generated at mitochondria ETC [38]. In addition, studies found that mitochondrial ROS production [38] accounts for 2% of total oxygen consumed in mitochondria in physiological condition, and fluctuates from 0.25% to 11% depending on the animal species and respiration rates in pathophysiological conditions [31]. Studies found that mitochondrial ROS (mtROS) are connected with ROS systems in other organelles, and initiated inflammatory [78,116] or cell death signals [117]. Therefore, as the most oxygen consuming and ROS generation organelle, mitochondria serves as the central hub for connecting ROS systems in other organelles (upstream) with the downstream DAMP sensing pathways such as inflammasomes [27], trained immunity [42,48,118] and immunometabolic pathways [119]. At the central hub, the metabolic homeostasis, and alarming stress in each organelle are sensed and bridged to DAMP sensing pathways in the downstream into the cytosol (for example, inflammasomes).
MtROS are generated at mitochondrial ETC and by other mitochondrial oxidant enzyme systems. Mitochondrial ETC includes five complexes from complex I to complex V [120]. The complexes except complex II are assembled crossing both inner and outer mitochondrial membranes, while complex II expresses on mitochondrial inner membrane. TCA cycle products such as nicotine adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2) donating electrons at complex I and complex II, respectively, and during which process, O2▪− is generated and released into mitochondrial matrix from complex I and complex II. The electrons then pass through complex III and ultimately reduce O2 to water at complex IV (cytochrome C); and O2▪− is also generated and released into both mitochondrial intermembrane space (IMS) and mitochondrial matrix (MM) through this process at complex III. Meanwhile, positively charged protons (H+) from complex I, III and IV, but not complex II, are actively being pumped from the mitochondrial matrix into the intermembrane space, and thus creating a mitochondrial membrane potential (Δψm) across the inner mitochondrial membrane. This proton-motive force allows complex V-- ATP synthase (ATPase) to generate adenosine triphosphate (ATP) when protons re-enter the mitochondrial matrix via the complex V enzyme [83]. However, increased mtROS generation is uncoupled from ATP production and promoted by proton leaks, which means the reactions between complex I to complex IV is accelerated and uncoupled from ATP generation as we reported [37,39,40,60,121]. Interestingly, due to the low levels of complex II substrate—succinate in physiological conditions, the mtROS generation of complex II draws less attention [31]. However, recent studies also found that complex II also contributes to O2▪− generation in pathological conditions [122]. For example, tumor necrosis factor- α (TNF-α) induced mtROS production is mediated by complex II but not complex I and complex III in retinal pigment epithelium (RPE) cells [78]. mtROS production rates are higher by using complex II substrate—succinate in comparison to pyruvate, glutamate and malate [101]. Additionally, complex II mediated O2▪− release is uncoupled with proton, which is unlike complex I and III as mentioned above.
MtROS is mostly produced by the mitochondrial ETC; however, several other oxidant enzymes also contribute to mtROS generation, including four ETC associated enzymes (glycerol 3-phosphate dehydrogenase, Q oxidoreductase, pyruvate dehydrogenase, and 2-oxoglutarate dehydrogenase) [83], XO, CYP, NOX4, eNOS, MPO, LOX and MAO (Table 1A). O2▪− is the first formed ROS, and subsequently, O2▪− is quickly dismutated to H2O2 by SOD2 in the mitochondrial matrix (MM) and SOD1 in intermembrane space (IMS) (Table 1B). Therefore, the most studied mtROS are O2▪− and H2O2; however, nine of the eleven types of ROS are found to exist in mitochondria except CO3▪- and ONOO−. But ONOO− may also be localized in mitochondria because of the rapid interactions of O2▪− and NO▪. Even though other types of ROS are harmful to lipid, protein and DNA in mitochondria and cells (stress branch-out function), there has not been much published on the determination of their functions and signals. One of the reasons may be the limited detection methods of different types of ROS. Furthermore, mitochondrial DNA (mtDNA) encodes seven subunits of complex I, one subunit of complex III, three subunits of complex IV and two subunits of complex V [123]. However, mtDNA is not protected by histones as nuclear DNA, and the rates of mtDNA damage are higher than that of nuclear DNA [123]. MtDNA damage includes mtDNA mutations and decreased mtDNA copy numbers [124]. Studies reported that mtDNA damage and dysfunction are susceptible to increased cytosolic and mitochondrial ROS levels in metabolic disease and age-related disease [[124], [125], [126]]. Although mtRNA is one of mitochondrial DAMPs [127], however, the relationship between mtDNA damages and nuclear ROS levels is not clear.
MtROS is connected to ROS systems of other organelles; and the signals are transduced between organelle. Studies found that extra-mitochondria ROS could transduce the signals to mitochondria and induce the production of mtROS (Table 2). Extra-mitochondria O2▪− induce opening of inner membrane anion channel (IMAC) and depolarize Δψm; and O2▪− is produced at complex III [128,129]. In addition, extra-mitochondrial H2O2 activate mitochondrial ATP sensitive potassium channel via protein kinase C (PKC)-ε pathway, thus potassium influx and mtROS generation are increased [130,131]. Moreover, extra-mitochondrial ROS-- HOCl, H2O2 and 1O2, all triggers mtROS production via affecting ETC enzyme activities [132,133] or mitochondrial permeability transition pore (MPTP) opening [134,135]. On contrast, mitochondrial generated ONOO− and H2O2 cause eNOS uncoupling and activation of NOX, which lead to extra-mitochondrial ROS generation [131,[136], [137], [138], [139], [140]]. After the communications between mitochondria and other organelles, imbalanced ROS participates and regulates various metabolic pathways to respond, especially mtROS via a feedback mechanism. MtROS are important signal transducer and regulates downstream inflammatory and cell death pathways (Fig. 2A–a). For examples, mtROS promotes inflammation via NLRP3- inflammasome/caspase-1 activation [78,116,141], promotes cell proliferation or migration via the key inflammatory transcription factor NF-kB [142] or Rac1 (a small G-protein in the Rho family) pathway [143] in inflammasome-dependent or independent manners, and promotes apoptosis via caspase-3 activation [117]. In addition, a new conditional DAMP [25], pro-atherogenic stimuli lysophosphatidylcholine (LPC) [144]-induced increased mtROS production contributes to trained immunity via histone 3 lysine 14 acetylation [145]. Furthermore, studies found that anti-inflammatory cytokine interleukin 35 (IL-35) inhibits endothelial cell activation via inhibiting the increase of mtROS [146]. In addition, we analyzed the target enzymes of various treatments in Fig. 4 and genes of their downstream pathways by circus plot (Fig. 2A and b; also see the Supplemental Table 4 for the details). Various stimuli have different ROS generation enzymes and downstream signaling, however, their signaling pathways crosstalk with each other.
Table 2.
Extra-mitochondrial ROS modulate mtROS production, and two types of mtROS modulate extra-mitochondrial ROS production.
| Category | Triggering ROS | Pivotal proteins | Findings | PMID | Ref # |
|---|---|---|---|---|---|
| Extra-mt ROS induced Mt ROS | O2•- | IMAC | Extra-Mt O2•- induced opening of IMAC which depolarized ΔΨm; O2•- was produced in ETC complex III. |
12930841 24657720 |
[128,129] |
| H2O2/NO• | Mt-KATP | Extra-Mt H2O2 activated Mt-KATP via PKC-ε leading to the increase of K+ influx and production of O2•- |
18586884 31400697 |
[130,131] | |
| HOCl | ETC enzymes | HOCl–oxLDL leads to Mt ROS generation via decreasing activities of ETC dehydrogenase and reductases |
19843872 18282575 |
[132,133] | |
| 1O2/H2O2 | MPTP | Mt ROS was triggered by external ROS (possibly 1O2) and linked to MPTP opening and calcium sparks. |
11015441 28279675 |
[134,135] | |
| Mt ROS induced extra-mt ROS | ONOO− | Zinc cluster or BH4 | O2•- reacts with NO• to form ONOO− which causes uncoupling of eNOS via oxidation of zinc cluster or BH4 |
11901190 14656731 31400697 |
[131,136,137] |
| Cysteine of XDH | O2•- reacts with NO• to form ONOO− which induces disulfide formation between two cysteines of XDH |
22657349 31400697 |
[131,138] | ||
| H2O2 | c-Src | Activation of c-Src by mitochondrial H2O2 can activate cytoplasmic NOX or increasing its mRNA levels, increasing cytosolic O2•- production |
24053613 24759683 31400697 |
[131,139,140] |
Abbreviation: ΔΨm, Mitochondrial membrane potential; BH4, Tetrahydrobiopterin; c-Src, Proto-oncogene tyrosine-protein kinase Src; eNOS, Endothelial nitric oxide synthase; ETC, Electron transport chain; H2O2, Hydrogen peroxide; HOCl, Hypochlorous acid; IMAC, Inner membrane anion channel; Mt, Mitochondrion; Mt-KATP, Mitochondrial adenosine triphosphate (ATP)-sensitive potassium channel; MPTP, mitochondrial permeability transition pore; NO, Nitric oxide; NOX NADPH oxidase; O2•-, Superoxide radical anion; 1O2, Singlet oxygen; ONOO−, Peroxynitrite; oxLDL, Oxidized low-density lipoprotein; PKC-ε, Protein kinase C-epsilon; ROS, Reactive oxygen species; XDH, xanthine dehydrogenase.
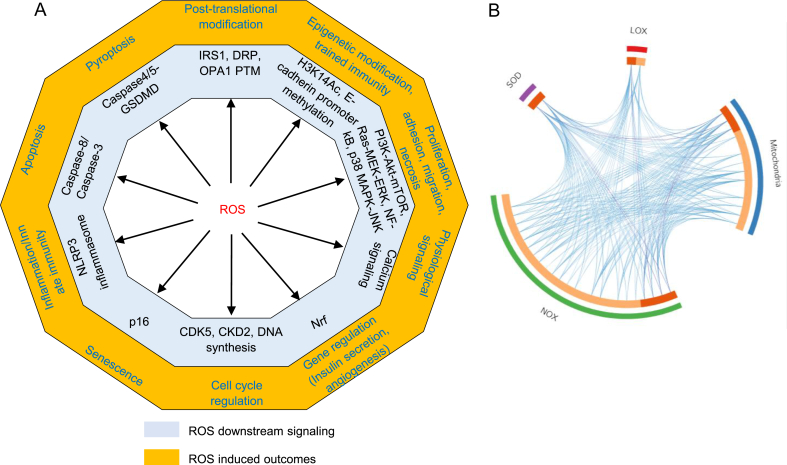
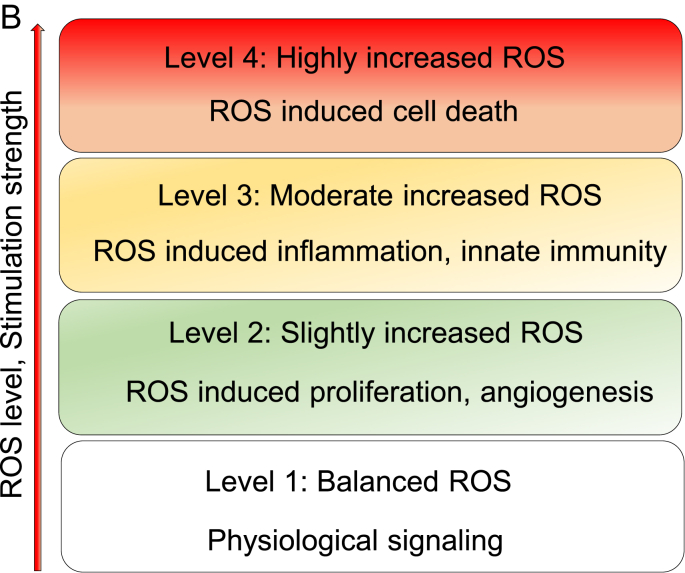
Fig. 2.
A. Left panel (a): ROS induces ten types of cell functions (yellow) via various specific downstream pathways (grey). The ten cell functions include: 1) physiological signaling, 2) proliferation, 3) gene regulation, 4) cell cycle regulation, 5) epigenetic modification, 6) post-translational modification, 7) inflammation/innate immunity, 8) pyroptosis, 9) apoptosis, and 10) senescence. Right panel (b): The circus plot indicates that four major ROS generating and anti-ROS regulatory systems have both shared genes and unique genes. On the outside, each arc represents the identity of each gene list。On the inside, each arc represent a gene list, where each gene has a spot on the arc. Dark orange color represents the genes that appear in multiple lists and light orange color represents genes that are unique to that gene list. Purple lines link the same gene that are shared by multiple gene lists. Blue lines link the different genes where they fall into the same ontology them. (also see the detailed information in a supplemental table). Detailed information of Fig. 2A-b see supplemental Table 4.
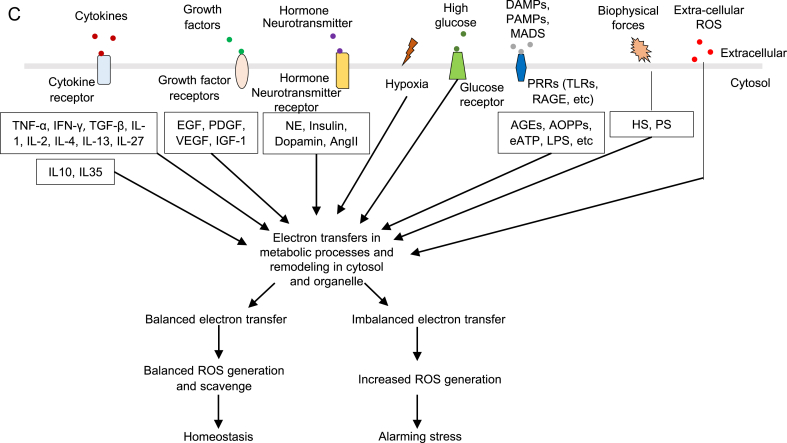
Abbreviations: IRS1—insulin receptor substrate 1, DRP--dynamin-related protein, OPA1--optic atrophy 1, PTM—post-translational modification, H3K14Ac—histone 3 lysine 14 acetylation, PI3K--phosphatidylinositol-3-kinase, Akt--protein kinase B, mTOR--mammalian target of rapamycin, Ras—GTPase, MEK--MAPK kinase, ERK--extracellular signals-regulated kinase, NF-kB--nuclear factor kappa-lightchain- enhancer of activated B cells, MAPK-- mitogen-activated protein kinases, JNK-- MAPK-Jun N-terminal kinase, Nrf-- Nuclear factor erythroid2-related factor, CDK5--cyclin dependent kinase 5, CKD2-- cyclin dependent kinase 2, NLRP3-- NOD-, LRR- and pyrin domain containing protein 3, GSDMD—gasdermin D, SOD-- superoxide dismutase, LOX--lipoxygenase, NOX--NADPH oxidase. B. The ladder indicates different levels of ROS contribute to different cellular functions. In redox homeostasis state, ROS contributes to physiological signaling. Low ROS increase contributes to proliferation and angiogenesis. Moderated ROS increase contributes to inflammatory response and innate immunity. High ROS increase contributes to various types of cell death. C. Seven types of stimulations promote ROS production, including cytokines, growth factors, hormone/neurotransmitters, hypoxia, high glucose, DAMPs and extracellular ROS.
Abbreviations: TNF-α--tumor necrosis factor- α, IFN-γ-- interferon gamma, IL-1--interleukin 1, IL-2--interleukin 2, IL-4--interleukin 4, IL-13-- interleukin 13, IL-27--interleukin 27, IL-10--interleukin 10, IL-35--interleukin 35, EGF-- epidermal growth factor, PDGF--Platelet derived growth factor, VEGF-- vascular endothelial growth factor, IGF-1--Insulin-like growth factor 1, NE—Norepinephrine, AngII—angiotensin II, AGE-- AGEsadvanced glycation end products, AOPP-- advanced oxidation protein products, ATP-- adenosine triphosphate, LPS—lipopolysaccharide, HS-- Hemodynamic strain, PS-- Pulmonary stretch.
Nucleotide-binding oligomerization domain (NOD)-like receptor-containing pyrin domain 3 (NLRP3) inflammasome is a multiprotein complex, which is composed of NLRP3, apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD) (ASC, PYCARD) and pro-caspase 1 [27]. Increased ROS levels contribute to NLRP3 inflammasome activation. Among various subtypes of ROS, ONOO−, O2▪−, and H2O2 are reported to be involved in inflammasome activation [147]. Upon activation by ROS, NLRP3 inflammasome promotes secretion of pro-inflammatory cytokines such as IL-1β and IL-18 and an inflammatory form of cell death [4]. Inhibition of NOX activity, p22phox subunits and ONOO− production significantly prevented caspase-1 activation and IL-1β production [147]. The structure of disulfide bond that connects the PYD domain and the nucleotide-binding site domain in NLRP3 is highly sensitive to ROS [147]. The inactive form of NLRP3 resides on ER and translocates on mitochondrial outer membrane upon activation [[148], [149], [150], [151]]. NLRP3 located in mitochondria interacts with mitochondrial ASC to assemble inflammasome or with mitochondrial antiviral signaling protein (MAVS) [152]. MAVS is a mitochondrial adaptor protein which mediates the translocation and association of NLRP3 to mitochondria. MAVS facilitates NLRP3 binding with ASC and pro-caspase 1 [147,153]. In addition, ER and mitochondria associated membrane are found to be the potential site for NLRP3 assembly [154]. Reports of subcellular localization of ASC varies significantly [53], including mitochondria [155], cytosol [150] and nucleus [156]. Upon inflammasome activation, ASC is especially observed in cytosol, which is correlated with the subcellular localization of assembled NLRP3 inflammasome. Furthermore, ROS activate NLRP3 inflammasome via direct and indirect ways. Studies found that increased ROS level induced mitochondrial DNA damage is also an indirect pathway for NLRP3 inflammasome activation by ROS [152,154]. In high level of ROS, thioredoxin-interacting protein (TXNIP) is dissociated with TRX and activates NLRP3 inflammasome [157]. Therefore, mitochondrial ROS systems serve as the central hub for connecting ROS systems in other organelles (upstream) with the downstream DAMP sensing pathways such as inflammasomes, trained immunity, immunometabolic pathways, cell proliferation, migration and cell death.
4. Nuclear ROS systems serve as a convergent hub and decision-makers to connect unbearable and alarming metabolic stresses from intracellular organelles to inflammation and cell death
Our previous reports showed that proatherogenic lipids (lysophosphatidylcholines, LPC)-induced mitochondrial ETC-generated ROS upregulate endothelial adhesion molecule intercellular adhesion molecule-1 (ICAM-1) expression and human aortic endothelial cell (HAEC) activation [42,60] via increasing histone 3 lysine 14 acetylation (H3K14ac) and increasing transcription factor AP-1 binding to ICAM-1 promoter in the nucleus [41]. In addition, we also found that LPC activates HAEC by increasing the enzyme expression of trained immunity (innate immune memory) [118]-related three metabolic pathways such as glycolysis, acetyl-CoA generation and mevalonate pathways via H3K14ac-mediated epigenetic innate immune memory mechanisms [48]. Our results have demonstrated that mitoROS and metabolic pathways are connected to an epigenetic histone modification and gene expression in the nucleus via so-called mito-nuclear communication [158], which is functional as a form of newly characterized innate immune memory (also termed trained immunity) [118,159]. Moreover, we found that intracellular organelle-generated stress can be converged into the nucleus and regulate DNA damage and DNA repair pathways [44]; and presumably cytosol-located inflammasome-caspase-1 activation [5,55,56,160] can regulate gene expression in caspase-1 processed IL-1β-, IL-18-, and sirtuin-1-, independent manners [52]. Taken together, our reports have demonstrated that in addition to mitochondria, cytosol stress and organelle stress are also converged into the nucleus in inflammasomes-dependent or independent manners [53].
The nucleus is responsible for epigenetic and genomic regulation of gene expression, cell differentiation, proliferation, senescence, and cell death [161]. Studies found that the nuclear membrane also contains cytochrome oxidases and electron transport systems, and may contribute to nuclear ROS production [162]. The basal levels of ROS in the nucleus are higher than those in the cytosol at resting state [163], but the potential role of nuclear ROS have rarely been studied or discussed. On the other hand, an important question remains unknown whether nuclear ROS are generated in the nucleus or due to the trafficking from increased cytosolic ROS. In Table 1A, ROS generation enzymes are found to be expressed in the nucleus, which presents strong evidence for ROS production in the nucleus. In addition, comparing increased ROS levels in various organelles including cytosol, mitochondria, endosome, Golgi and nucleus [164], increased nuclear ROS significantly promote cell death compared to that of other organelles; in contrast, increased ROS in the ER does not always affect cell viability [164]. Therefore, nuclear ROS serves as a convergent hub and decision-makers to connect unbearable and alarming metabolic stresses from intracellular organelles to inflammation or cell death [165]. In addition, nucleus ROS connected with ROS systems from extracellular space or extra-nucleus also regulate inflammatory or cell death responses [76]. Theoretically, there are at least two means for microenvironmental ROS to affect cellular homeostasis via an impact on the cell nucleus [165]. For example, extracellular H2O2 (50 μmol/L) stimulation induces sustained increase of nuclear ROS and regulates nuclear calcium signaling (Table 3); and the antioxidant glutathione (GSH) [166] could reverse the increase of nuclear ROS [163]. In addition, the nuclear envelope membranes contain several G protein-coupled receptors, including prostaglandin E2 (EP3R) and endothelin-1 (ET-1) receptors. Activation of EP3R increases endothelial nitric oxide synthase (eNOS) RNA expression in nuclei. eNOS and inducible NOS (iNOS) are reported to also be present at the nuclear levels. Moreover, stimulation with NO donor sodium nitroprusside [167] results in an increase of intranuclear calcium that is dependent on guanylate cyclase activation, but independent of mitogen-activated protein kinases (MAPK) [168]. H2O2 and ET-1 increase both cytosolic and nuclear ROS in human endocardial endothelial cells and in human aortic vascular smooth muscle cells [163]. Furthermore, pro-hypertension hormone angiotensin II (Ang II) (1 nM) stimulation activates Ang II type 1 receptor (AT1R), which is highly expressed in nuclei, and increases ROS production via NADPH oxidase 4 (NOX4) activation, which has 65% expression in nuclei [169]. Based on these studies, nuclear ROS senses the status of cell conditions. The imbalanced ROS levels contribute to cell inflammation and cell death. Studies on nuclear ROS remain less than that of other organelles, and further studies are needed to clearly uncover the mechanisms underlying death signaling and nuclear ROS generation and scavenging. One of the reasons for this may be due to the fact there is not a simple, convenient and specific method for nuclear ROS detection except for ROS staining with fluorescent ROS probe carboxy H2DCFDA (DCF) and co-stained with live nucleic acid stain Syto-11 [163].
Table 3.
Extracellular ROS induce intracellular ROS generation and promote different signaling and functions.
| ROS | Target | Concentration | Function | PMID | Ref # | |
|---|---|---|---|---|---|---|
| ONOO- | Increase | 100uM | Angiogenesis | 18309287 | [192] | |
| Increase | Apoptosis | 9020024 | [218] | |||
| H2O2 | Increase | 1-4uM | Insulin secretion | 17400930 | [70] | |
| Increase | NOX4 | 10 uM | Proliferation, Migration | 25315297 | [191] | |
| Increase | 10-20uM | Proliferation | 15109912 | [193] | ||
| Increase | Cyto/Nuc ROS | 50uM | Nuclear Calcium signaling | 20393594 | [71] | |
| Increase | 100uM | 26461342 | [142] | |||
| Increase | 200uM | Apoptosis | 29061494 | [187] | ||
| Increase | 300uM | Tumor progression | 18801366 | [198] | ||
| Increase | 1 mM | Caspase-independent apoptosis | 20084055 | [69] |
In our new model proposed here, we emphasize the mitochondria/organelle-to-nucleus retrograde communications [41]. Actually, recent progresses include nucleus-to-mitochondria (anterograde) and mitochondria-to-nucleus (retrograde) communication, mito-nuclear feedback signaling and proteostasis (pathways that control the biogenesis, folding, trafficking and degradation of proteins) regulation, the integrated stress response and non-cell-autonomous communication [170].
5. Balanced ROS levels indicate a physiological homeostasis of various metabolic processes in subcellular organelles while imbalanced ROS levels present alarms for pathological organelle stresses in metabolic processes
ROS systems are found to play important roles in promoting the progression in metabolic diseases, such as cardiovascular disease (CVD), chronic kidney disease, inflammations, diabetes and tumors. However, the detailed mechanisms of how ROS play the critical roles remain poorly characterized. We propose that ROS system serves as the sensing network for cellular homeostasis and alarming stress in metabolic processes. Therefore, ROS levels are increased in metabolic disorders and participate in disease progression. Balanced ROS contribute to physiological signals and functions after sensing metabolic homeostasis, while imbalanced ROS participate in metabolic processes regulating proliferation, inflammation or cell death in pathological conditions. Studies found that lightly increased ROS (1.1 fold changes) contribute to cell proliferation, angiogenesis and metastasis, moderate increased ROS (1.1–20 fold change) reach the maximum inflammatory responses, and highly increased ROS (more than 20 fold change) promote apoptosis, necroptosis, autophagy, pyroptosis and ferroptosis [171,172] (Fig. 2B). However, cell types and stimuli cause variations for fold differences in ROS change effects. Thus, the criteria of increased ROS levels may be used in the same stimulation conditions and cell types, and it is not suitable for comparing across different experiments. Importantly, lightly increased ROS can be recognized as a complimentary signal to rescue the imbalanced ROS system and alarming stress and promote cell survival. Therefore, ROS system is a network connecting various DAMPs/conditional DAMPs, including cytokines, growth factors, hormones/neurotransmitters, hypoxia, and high glucose, and other disease risk factors, to the initiation of metabolic processes (Fig. 2C). In Table 4A, even though the fold changes do not match the criteria mentioned above, the response severity and scales are dose-dependently increased according to the stimulation strength, such as interleukin 1 (IL-1) [78,173], IL-13 [174,175] and Ang II [169,[176], [177], [178], [179], [180], [181]]. Here, we attempted to collect the evidences to determine whether ROS systems are functionally qualified in sensing metabolic stress in organelles. One requirement for this function is the ROS system must have the capacity to sense the strengths of stress with different functional consequences to activate inflammasomes and cross-talk to different stress pathways in inflammasomes-dependent, or independent manners.
Table 4A.
Eight cytokines, four growth factors, three out of four hormone/neurotransmitters promote ROS production, and anti-inflammatory cytokines IL-10 and Il-35 inhibit ROS production. IGF-1, NE and Dopamin also decrease ROS production in certain condition.
| ROS | Sensor | Target | Concentration | Function | PMID | Ref# | ||
|---|---|---|---|---|---|---|---|---|
| Cytokine | TNF-α | Increase | TNFR | 5-lipoxygenase | 10 ng/ml | Inflammation | 10934206 | [72] |
| Increase | TNFR1 | Complex II, ΔΨm | 20 ng/ml | Inflammation, Apoptosis | 17765224, 20203691 | [78,117] | ||
| IFN-γ | Increase | IFN-γR | Complex II/NOX | 20 U/ml | Inflammation | 17765224 | [78] | |
| Increase | IFN-γR | Duox2 | 100 U/mL | Innate immunity | 16111680 | [174] | ||
| TGF-β | Increase | TGFBR | NOX, Complex I | 10 ng/ml | Fibronectin secretion/EMT | 15677311 | [245] | |
| Increase | TGFBR | NOX4 | 50 ng/ml | Angiogenesis | 25315297 | [191] | ||
| IL-1 | Increase | IL1R | 10 ng/ml | Angiogenesis | 26811540 | [173] | ||
| Increase | IL1R | NOX | 20 ng/ml | Inflammation | 17765224 | [78] | ||
| IL-2 | Increase | IL2R | 100 U/ml | Angiogenesis | 18309287 | [192] | ||
| IL-4 | Increase | IL4R | Duox1 | 10 ng/ml | Inflammation, Innate immunity | 16249002, 16111680 | [174,206] | |
| IL-13 | Increase | IL13R | Duox1 | 10 ng/ml | Innate immunity | 16111680 | [174] | |
| Increase | IL13R | NOX | 1 μg/μl | Cell death/damage | 19752235 | [175] | ||
| IL-27 | Enhance | IL27R | NOX2 | 100 ng/ml | Inflammation | 28240310 | [79] | |
| IL-10 | Decrease | IL10R | 10 ng/ml | Inhibit endothelial cell activation | 31731100 | [146] | ||
| IL-35 | Decrease | IL12R/IL27R | Mt | 10 ng/ml | Inhibit endothelial cell activation | 29371247, 31731100 | [146,208] | |
| Growth factor | EGF | Increase | EGFR | 5 ng/ml | Adhesion, Migration, Proliferation | 19635476 | [209] | |
| Increase | EGFR | NOX | 10 ng/ml | Proliferation | 25122478 | [188] | ||
| Increase | EGFR | 40 ng/ml | Angiogenesis | 17045920 | [194] | |||
| Increase | EGFR | 100 ng/ml | Necrosis | 10854274 | [73] | |||
| PDGF | Increase | PDGFR | 1 ng/ml | Proliferation | 15109912 | [193] | ||
| Increase | PDGFR | 15 ng/ml | Senescence | 16081426 | [220] | |||
| Increase | PDGFR | 20 ng/ml | Inflammation | 23774581 | [246] | |||
| VEGF | Increase | VEGFR | mtROS | 50 ng/ml | Migration | 31653897 | [143] | |
| IGF-I | Decrease | IGF-1R | 100 ng/ml | Survival | 20084055 | [69] | ||
| Increase | IGF-1R | NOX4 | 200 ng/ml | Migration | 18567639 | [210] | ||
| Hormone/Neurotransmitter | NE | Decrease | 10uM | Protect DNA damage | 26167254 | [185] | ||
| Insulin | Enhanced | IR | 10–100 nM | Enhance insulin sensitivity | 19808019 | [183] | ||
| Dopamine | Increase | MAO | 0-50uM | Physiological signaling | 20547771 | [67] | ||
| Increase | mtROS (SOD) | 1uM | Physiological signaling | 23994527 | [68] | |||
| Increase | MAO | 100μM, 500uM | Cell death | 20547771 | [67] | |||
| Decrease | D5R | NOX | Protect from hypertension | 23425954 | [186] | |||
| Serotonin | Increase | 5-HT1BR | NOX1 | 1uM | Post-translational oxidative modification of protein, proliferation | 28473438 | [247] | |
| 10-100uM | Caspase-3 dependent apoptosis | 16286591 | – | |||||
| AngII | Increase | AT1R | NOX4 (Nuc/Cyto) | 1 nM | Gene regulation, Protect from cell damage | 19409874, 19948986 | [169,176] | |
| Increase | AT1R | NOX | 100 nM | maintain arterial tone | 29472601 | [177] | ||
| Increase | ET-1AR | NOX | 100 nM | Proliferation | 14642698 | [178] | ||
| Increase | AT1/2R | NOX | 100 nM | Apoptosis, Necrosis, Senescence | 12606818, 21270817 | [179,180] | ||
| Increase | AT1R | NOX | 1uM | Impair insulin signaling | 16982630 | [181] |
Balanced ROS levels are critical for cell signaling and homeostasis in physiological condition. In physiological homeostasis conditions, ROS initiate diverse cellular responses including triggering signaling pathways and regulating cell protection besides to activate inflammasome pathways. In addition, ROS participates in coordinating activation of mitochondrial fission fusion and mitophagy to optimize clearance of abnormal mitochondria and cell [31]. These process prevent the damage signals to spread to neighboring mitochondria or cells [31]. In addition, ROS regulate various metabolic processes in cell cycle and survival via calcium signaling in physiological conditions [67,68,177,182]. Moreover, lightly increased ROS complementarily rescue the imbalanced ROS system and alarming stress via regulating metabolic processes. For example, slightly increased ROS enhance insulin sensitivity via phosphatidylinositol-3-kinase (PI3K)-protein kinase B (Akt)-mammalian target of rapamycin (mTOR) pathway [183] and protein synthesis via Ras (a small GTPase)-MAPK kinase (MEK)-extracellular signals-regulated kinase (ERK) pathway [184], protect from DNA damage [185] and increase cell survival [69,186,187]. Additionally, ROS are slightly increased to induce cell proliferation [178,[188], [189], [190], [191]] and angiogenesis [173,191,192] via regulating p38 MAPK-Jun N-terminal kinase (JNK) pathway [193], hypoxia induced factors alpha (HIF-α) stabilization [194] and signal transducer and activator of transcription 3 (Stat3) activation [195]. Studies found that NLRP3 inflammasome could be activated via PI3K-Akt pathway [196] and p38 MAPK pathway [197]. ROS serve as a bridge to connect metabolic stress and inflammasome activation and regulate metabolic pathways in a feedback mechanism. On the other hand, ROS contribute chronically to tumorigenesis or metastasis [198,199]. In addition to ROS regulation of certain signaling pathways above discussed, it remains poorly defined whether ROS in physiological conditions regulate a functional status of vascular cell type-endothelial cells, which we recently classified as innate immune cells [36,200]. To address the issue, we examined human aortic endothelial cells (HAECs). Both mtROS and ATP are produced as a result of electron transport chain activity [38], but it remained enigmatic whether mitochondrial ROS (mtROS) could be generated independently from ATP synthesis. Our report shed light on this important question and found that, during endothelial cell (EC) activation, mtROS could be upregulated in a proton leak-coupled, but ATP synthesis-uncoupled manner [60]. As a result, EC could upregulate low dose mtROS production for physiological EC activation without compromising mitochondrial membrane potential and ATP generation, and consequently without causing mitochondrial damage and EC death. Thus, a novel pathophysiological role of proton leak [37,40] in driving mtROS production is uncovered for low grade EC activation, patrolling immunosurveillance cell (for example, non-classical monocytes [201]) trans-endothelial migration and other signaling events without compromising cellular survival. This new working model explains how mtROS could be increasingly generated independently from ATP synthesis and endothelial damage or death. Mapping the connections among mitochondrial metabolism, physiological EC activation, patrolling cell migration, and pathological inflammation is significant towards the development of novel therapies for inflammatory diseases and cancers [39].
Moderate increased ROS promote cytokine secretion [202], inflammatory cell adhesion [203,204] and migration [205]. Studies found that six proinflammatory cytokines cause increased cytosolic and mitochondrial ROS generation, including TNF-α [72,78,117], interferon gamma (IFN-γ) [78,174], interleukin 1 (IL-1) [78,173], IL-4 [174,206], IL-13 [174,175] and IL-27 [79] (Table 4A). The increased ROS generation promote inflammatory responses and innate immunity via regulating cyclin dependent kinase 5 (CDK5)-related cell cycle [207], histone 3 lysine 14 acetylation [208], caspase-1 inflammasome activation [116], NF-kB pathway [142] and expressions monocyte chemoattractant protein-1 (MCP1) [206]. In addition, anti-inflammatory cytokines, IL-10 [146] and IL-35 [146,208], are found to inhibit endothelial cell activation via decreasing mtROS production in the presence of pro-inflammatory stimulation. Growth factors, epidermal growth factor (EGF) [209] and vascular endothelial growth factor (VEGF) [143,210], induced cell migration via increasing ROS production and matrix metalloproteinases (MMP)2/9 expression [143] (Table 4A). DAMPs stimulation, such as advanced oxidation protein products (AOPPs) [211], AGEs-advanced glycation end products (AGEs) [212], respiratory syncytial virus (RSV) [141], lipopolysaccharide (LPS) [213] and integrin [43], induce ROS generation via NOX activation, therefore, promote cytokine secretion and innate immunity (Table 4B). Furthermore, cells are exposed to several types of biophysical forces (hemodynamic strain and pulmonary stretch) in physiological and pathological condition [[214], [215], [216], [217]]. Studies found that these biophysical forces also induce higher level of cytosolic and mitochondrial ROS which promote the inflammatory signaling and disease progression (Table 4B).
Table 4B.
Hypoxia, hyperglycemia and eighteen types of DAMPs promote ROS production, and hypoxia can also decrease ROS generation in certain conditions.
| ROS | Sensor | Target | Concentration | Function | PMID | Ref # | ||
|---|---|---|---|---|---|---|---|---|
| Hypoxia | Increase | NOX4 | 1% O2 | Angiogenesis, Migration | 26297045, 29123322 | [195,205] | ||
| Increase | SOD | 5% O2 | Proliferation | 16624953 | [189] | |||
| Decrease | 1% O2 | Protect from apoptosis | 29061494 | [187] | ||||
| Hyperglycemia | High glucose | Increase | GLUT | H2O2 | 20 mM | Insulin secretion | 17400930 | [70] |
| Increase | GLUT | NOX | 33 mM | Angiogenesis | 23274526 | [248] | ||
| DAMPs | LPA | Increase | Edg | NOX | 10 nM | Proliferation | 20934509 | [190] |
| Increase | LPAR5 | 1uM | pro-inflammatory cytokine and chemokine | 29258556 | [202] | |||
| LPC | Increase | mtROS | 40uM | Trained immunity | 31153039 | [48] | ||
| AGEs | Increase | RAGE | 300μg/ml | Cytokine secretion, Caspase-dependent apoptosis | 31587010, 22944044 | [212,223] | ||
| Increase | Galectin-3, CD36, SR-AI and RAGE | NOX | 400μg/ml | Liver fibrosis | 20133001 | [249] | ||
| AOPPs | Increase | NOX4 | 50uM | Cytokine secretion | 31539804 | [211] | ||
| Increase | NOX1/4 | 150uM | S-phase arrest | 29032312 | [250] | |||
| Increase | RAGE | NOX2 | 50μg/ml | Apoptosis | 23453926 | [224] | ||
| Increase | 200μg/ml | Apoptosis | 28000869 | [225] | ||||
| HSA | Increase | 10 mg/ml | Cell death | 25713411 | [74] | |||
| Fatty acid | Increase | Mt | Mitochondrial fission | 29092894 | [251] | |||
| eATP | Increase | P2X7 | 130uM | Cell death | 23431238 | [252] | ||
| Increase | DORN1 | NOX | 200uM | Bacterial defense | 29273780 | [253] | ||
| Zymosan | Increase | TLR2 | NOX | 24121038 | [254] | |||
| Concanavalin A | Increase | TLR2/4/9 | NOX | 24121038 | [254] | |||
| dsDNA (NCS) | Increase | NOX1 | 0.5ug/ml | Apoptosis | 22237206 | [75] | ||
| Thrombin | Increase | GPIbα, PAR4 | FAK/NOX1 | 1–2U/ml | 29569550 | [255] | ||
| Increase | NOX | 20U/4ul | Neurodegeneration | 15843610 | [256] | |||
| Integrin | Increase | PRR (β2 integrin Mac-1) | NOX2 | Innate immunity (Phagocytosis) | 29544096 | [43] | ||
| Increase | RTKR | LOX, NOX | Cell adhesion | 12796479 | [204] | |||
| ET-1 | Increase | ET1R | eNOS | 1 nM | Nuclear Calcium homoeostasis | 20393594 | [71] | |
| Increase | ET1R | NOX | 10 nM | Protein synthesis | 15203192 | [184] | ||
| Increase | ETAR | NOX | Cardiac hypertrophy | 16107552 | [257] | |||
| Methionine | Increase | Mt | 29679893 | [258] | ||||
| Homocysteine | Increase | Mt | 500uM | Caspase-1 inflammation activation | 27006445 | [259] | ||
| PAMPs | RSV | Increase | TLR2 | Cytokine secretion | 22295065 | [141] | ||
| DENV | Increase | TLR9 | Mt | Innate immunity | 29880709 | [116] | ||
| Shiga toxin | Increase | CD77 | Caspase-4/GSDMD | 200 ng/ml | Pyroptosis | 30404007 | [221] | |
| LPS | Increase | CD14/TLR4 | Caspase-5 | 10 ng/ml | Inflammation | 26508369 | [213] | |
| Increase | TLR4 | NOX | 10μg/ml | Monocyte adhesion | 23153039, 24121038 | [203,254] | ||
| Biophysical forces | Hemodynamic strain | Increase | N/A | N/A | Average strain (12%) | Adhesion molecule expression, Monocyte adhesion | 9449403, 19186986 | [214,215] |
| Increase | Integrins | Mito complex I | Uniaxial cyclic stretch (20%) | Membrane spreading | 23738008 | [216] | ||
| Pulmonary strech | Increase | Integrin, EC receptor and ion channel | NOX, XO, eNOS | N/A | Pulmonary hypertension and lung injury | 30623676 | [217] |
Abbreviations: EGF-Epidermal growth factor; PDGF-platelet-derived growth factor; VEGF-vesicular epithelial growth factor; IGF-I-insulin-like growth factor; LPA-lysophosphatidic acid; LPC--lysophosphatidylcholine; LPS-lipopolysaccharide; AGEs-advanced glycation end products; AOPPs-advanced oxidation protein products; HAS-human serum albumin; eATP-extracellular ATP; RSV-respiratory syncytial virus; DENV-dengue RNA virus; dsDNA-double-stranded DNA; endothelin-1—ET1; RIRR-ROS-induced ROS release; TBE cell-Tracheobronchial epithelial cells; EC-Endothelial cell; EPC-Endothelial progenitor cell; HCC-hepatocellular carcinoma cell; PPP-pattern recognition receptors; EMT-epithelial mesenchymal transition; ΔΨm, Mitochondrial membrane potential; NE-Norepinephrine; LPC-lysophosphatidylcholine.
Highly increased ROS significantly contributes to different types of cell death, including apoptosis, pyroptosis (inflammatory cell death), necrosis and senescence [76]. TNF-α induced ROS generation by affecting mitochondrial membrane potential contributes to apoptosis [117]. IL-13 leads to cell death via NOX activation and ROS generation in microglia [175]. A high concentration of extracellular ROS [187], such as ONOO− and H2O2, promotes apoptosis via DNA fragmentation [218], potentially NLRP3 inflammasome-DNA damage pathway [219], and caspase-independent pathway [69]. Epidermal growth factor (EGF) induced H2O2 generation promotes cell necrosis and senescence via regulating Ras-MEK-ERK pathway [73,220]. In addition, Shiga toxin 2 increases ROS generation via caspase-4 activation/presumably noncanonical inflammasome pathway [59] and then promotes cell pyroptosis [221]. Moreover, mitochondrial membrane potential (MMP) collapse and ROS generation induce NLRP3 inflammasome activation. The elimination of ROS alleviates the cleavage of Gasdermin D (GSDMD) carried out by activated caspase-1/caspases-4/caspase-11 and non-canonical cytokine secretory pathways mediated by N-terminal Gasdermin D-protein pores on the plasma membrane [59]. Hydrogen peroxide treatment augments the cleavage of GSDMD by caspase-1. Four amino acid residues of GSDMD are oxidized under oxidative stress in macrophages, suggesting that GSDMD oxidation serves as a de novo mechanism, by which mitochondrial ROS promote NLRP3 inflammasome-dependent pyroptosis [222]. Other DAMPs, including AOPPs, serotonin and AGEs [223], accelerate cell death [75] via increased ER stress [74], p38 MAPK-JNK pathway [224,225] and Gasdermin family [59] (Table 4B).
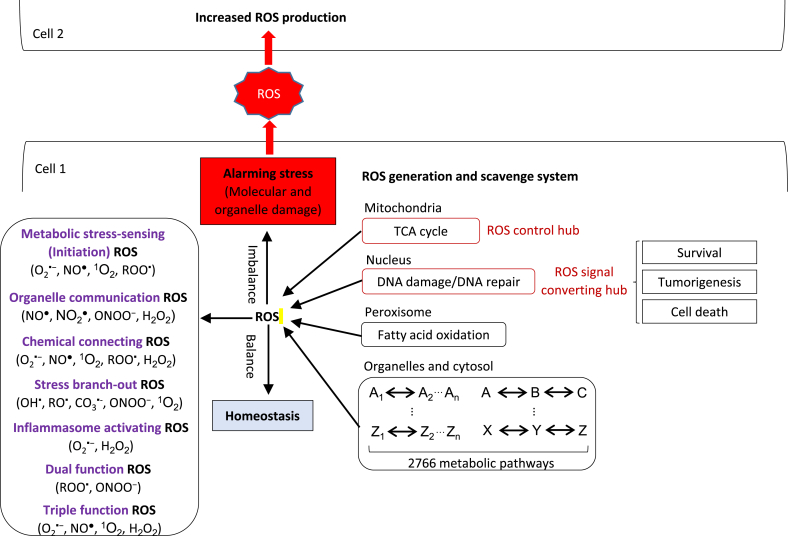
6. Our new working model: ROS systems are a new integrated network for sensing homeostasis and alarming stress in metabolic processes in various subcellular organelles
The key knowledge gap that this analytic review attempted to address is how an integrated system can sense homeostasis and stress in 2801 metabolic pathways localized in various intracellular organelles, cytosol and nucleus. We proposed that ROS are a new integrated network for sensing homeostasis and alarming stress in metabolic processes in various subcellular organelles. The following evidence are found: first, ROS are generated in most organelles during metabolic processes and are participating in cellular signaling; second, the electron transfer process existed in most metabolic reactions is the most convenient cue, which could be sensed and monitored in the 2801 complex metabolic pathways. Indeed, we classify a few types of ROS that are functional in sensing metabolic stress; third, ROS systems make use of the most general chemical resources in cytosol and organelles—oxygen, which is also generally used in metabolic processes; Fourth, four types of ROS are our newly classified organelle communication ROS that are able to cross lipid membrane which is responsible for the communication of cellular metabolic status between organelles and cells, including NO●, NO2●, H2O2 and ONOO− (Fig. 3). In addition, increased levels of ROS can activate inflammasome pathways in cytosol and are harmful to DNA, protein and lipid, that are newly classified stress branch-out (cross-talking) ROS. Moreover, some ROS can be converted into another ROS, which we classify as chemical converting ROS as mentioned in Section 2 in details. Fifth, after sensing the imbalance of metabolism in metabolic disease in the form of electron transfer imbalance, ROS activate inflammation initiation pathways such as capase-1 inflammasomes in the cytosol to initiate inflammation, which we classify as inflammasome-activating ROS. Furthermore, mitochondria consume 80–95% oxygen and generates mtROS, thus mitochondria are the ROS control hub for connecting cell status and metabolic processes. Increased nuclear ROS significantly increase cell death compared to other organelles. Therefore, the nucleus is the ROS signal converting hub in retrograde, which determines the functions of survival, tumorigenesis and various types of cell death. In homeostasis status, balanced ROS initiate physiological signaling, while imbalanced ROS in organelles as an integrated network promote pathological signals after sensing alarming stress [226]. The increased ROS cross organelle lipid membrane to share signals between organelles or plasma membrane to spread danger signals to neighboring cells via exosomes [227,228]. Our model provides novel insights on the roles of ROS system in bridging metabolic stress to inflammation, cell death and tumorigenesis; and provide novel therapeutic targets for treating those diseases.
Fig. 3.
A novel working model: 1) ROS are a novel integrated network for sensing homeostasis and alarming stress in organelle- or cytosolic-metabolic stresses; and 2) ROS also serve as cellular communication signaling to increase neighboring cell ROS production.
Abbreviations: O2▪−—superoxide, NO▪—nitric oxide radical, NO2 ▪—nitrogen dioxide, OH▪—hydroxyl radical, ROO▪—peroxyl radical, RO▪— alkoxyl radical, CO3▪-—carbonate radical, H2O2—hydrogen peroxide, ONOO−—peroxynitrite, 1O2—singlet oxygen, HOCl—hypochlorous acid, TCA: tricarboxylic acid cycle.
Declaration of competing interest
None.
Acknowledgements
YS carried out the primary literature search and drafted the manuscript. Others provided material input and helped revise the manuscript. XY supervised the study, data analysis, and manuscript writing. The authors are very grateful to Dr. Christ Kevil at Louisiana State Univ. Health Sciences Center Shreveport, Dr. Yabing Chen and Dr. Rakesh P. Patel at Univ. of Alabama at Birmingham for invitation and advices. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101696.
Sources of funding
Our research activities are supported by grants from National Institutes of Health (NIH) grants. The content in this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplemental table 1.
The 41 enzymes which are regulating superoxide and hydrogen peroxide are actively functioned and regulated in metabolic processes.
| Namec | Abbreviation | Function |
|---|---|---|
| Aldehyde oxidase | AOX1 | Valine, leucine and isoleucine degradation, Tyrosine metabolism, Tryptophan metabolism, Vitamin B6 metabolism and Nicotinate and nicotinamide metabolism |
| Amine oxidase (flavin- containing) A | AOFA | Deamination of 5-hydroxytryptamine, norepinephrine and epinephrine. |
| Amine oxidase (flavin- containing) B | AOFB | Deamination of benzylamine and phenylethylamine |
| d- Amino acid oxidase | OXDA | Regulate the level of neuromodulator d-serine and dopamine synthesis |
| l- Amino acid oxidase | OXLA | Lysosomal antigen processing and presentation |
| d- Aspartate oxidase | OXDD | Deamination of d-aspartate and its N-methylated derivative, N-methyl d-aspartate |
| Amiloride- sensitive amino oxidase (copper containing) | AOC1 | Degradation of putrescine, histamine, spermine and spermidine |
| Cytochrome P450 3A4 | CP3A4 | Metabolism of sterols, steroid hormones, retinoids and fatty acids |
| Cytochrome P450 2D6 | CP2D6 | Metabolism of fatty acids, steroid and retinoids |
| Cytochrome P450 2E1 | CP2E1 | Metabolism of saturated fatty acids and xenobiotics |
| Cytochrome P450 4A11 | CP4AB | Metabolism of fatty acids and their oxygenated derivatives (oxylipins) |
| ERO1- like protein- α | ERO1A | Involved in disulfide bond formation in ER and reoxidizes P4HB/PDI |
| ERO1- like protein- β | ERO1B | Involved in disulfide bond formation in ER and reoxidizes P4HB/PDI |
| FAD- linked sulfhydryl oxidase ALR | ALR | Regenerate the redox-active disulfide bonds in CHCHD4/MIA40 |
| Hydroxyacid oxidase 1 | HAOX1 | Active on 2-carbon substrate glycolate and 2-hydroxy fatty acids |
| Hydroxyacid oxidase 2 | HAOX2 | Oxidation of L-alpha-hydroxy acids and L-alpha-amino acids |
| Membrane primary amine oxidase | AOC3 | Semicarbazide-sensitive (SSAO) monoamine oxidase activity |
| Peroxisomal N1- acetylspermine/spermidine oxidase | PAOX | Oxidation of N1-acetylspermine to spermidine and putrescine, and polyamine back-conversion |
| Peroxisomal acyl- CoA oxidase 1 | ACOX1 | Desaturation of acyl-CoAs to 2-trans-enoyl-CoAs |
| Peroxisomal acyl- CoA oxidase 3 | ACOX3 | Oxidizes the CoA-esters of 2-methyl-branched fatty acids |
| Peroxisomal sarcosine oxidase | SOX | Metabolizes sarcosine, l-pipecolic acid and l-proline |
| Prenylcysteine oxidase 1 | PCYOX | Degradation of prenylated proteins and cleavage of thioether bond of prenyl-l-cysteines |
| Prenylcysteine oxidase- like | PCYXL | Prenlcysteine oxidase activity |
| Protein- lysine 6- oxidase | LYOX | Post-translational oxidative deamination of peptidyl lysine residues and regulate Ras expression |
| Pyridoxine 5′- phosphate oxidase | PNPO | Oxidation of pyridoxine 5′-phosphate and pyridoxamine 5′-phosphate |
| Retina- specific copper amine oxidase | AOC2 | Oxidation of 2-phenylethylamine and tryptamine |
| Spermine oxidase | SMOX | Oxidation of spermine and metabolism of N1-acetylspermine and spermidine |
| Sulfhydryl oxidase 1 | QSOX1 | Oxidation of sulfhydryl groups in peptide and protein thiols |
| Sulfhydryl oxidase 2 | QSOX2 | Oxidation of sulfhydryl groups in peptide and protein thiols |
| Sulfite oxidase, mitochondrial | SUOX | Metabolism of sulfur |
| Xanthine dehydrogenase/oxidase | XDH | Purine degradation; oxidation of hypoxanthine and xanthine |
| NADPH oxidase 1 | NOX1 | Regulate cellular pH and mediate proton currents |
| NADPH oxidase 2 | NOX2 (CY24B) | Regulate cellular pH and mediate proton currents |
| NADPH oxidase 3 | NOX3 | Biogenesis of otoconia/otolith |
| NADPH oxidase 4 | NOX4 | Inhibit phosphatase and regulate KCNK3/TASK-1 patassium channel |
| NADPH oxidase 5 | NOX5 | calcium-dependent proton channel and may regulate redox-dependent processes in lymphocytes and spermatozoa |
| Dual oxidase 1 | DUOX1 | Synthesis thyroid hormone, contribute to thyroid peroxidase/TPO and lactoperoxidase/LPO activity |
| Dual oxidase 2 | DUOX2 | Synthesis thyroid hormone, contribute to thyroid peroxidase/TPO and lactoperoxidase/LPO activity |
| Superoxide dismutase [Cu–Zn] | SOD1 | Form fibrillar aggregates in the absence of intramolecular disulfide bond |
| Superoxide dismutase [Mn], mitochondrial | SOD2 | Transcription of RNA polymerase Ii and release of cytochrome C |
| Extracellular superoxide dismutase [Cu–Zn] | SOD3 | Response to copper ion and hypoxia |
The 41 enzymes are generated from PMID:32231263 [102]. The function of enzymes are collected from Human metabolome database (https://hmdb.ca/) and Uniport database (https://www.uniprot.org/).
Supplemental table 2.
MitoSOX Red and Amplex Red are used for mtROS detection in vitro; and mitoB is used for mtROS detection in vivo.
| Category | Method | Reagent | Off-target response | Comment | PMID | Ref # |
|---|---|---|---|---|---|---|
| ROS, RNS | ESR | Specific, sensitive, expensive | 24862270 | [260] | ||
| Extracellular O2▪− | Fluorescent probe | HPr+ | Membrane impermeable | 29790367 | [261] | |
| General ROS | Fluorescent probe | DCFH-DA | 24862270, 16297980, 27909341 | [260,262,263] | ||
| General ROS | Fluorescent probe | H2DCF | 23665586 | [264] | ||
| General ROS | Fluorescent probe | CM-H2DCFDA | 24862270, 27302663 | [260,265] | ||
| O2▪− | Fluorescent probe | HE/DHE | OH▪, H2O2, OONO- | Cell permeable, intensity quantifiable, low specificity | 24862270, 16297980, 27909341 | [260,262,263] |
| O2▪− | Fluorescent probe | DPBF | 16297980 | [262] | ||
| O2▪− | Fluorescent probe | 2-(2-Pyridil)-benzothiazoline | 16297980, 27909341 | [262,263] | ||
| O2▪− | Chemiluminescent probe | L-012 | 24862270 | [260] | ||
| O2▪− | Chemiluminescent probe | Lucigenin | ONOO- | Cell permeable, low selectivity/sensitivity | 24862270 | [260] |
| O2▪− | Spectrophotometry | NBT | 24862270, 20387054 | [260,266] | ||
| O2▪− | Fluorescent probe | cpYFP | 24862270, 27288722 | [260,267] | ||
| OH▪ | Fluorescent probe | CHD | 16297980, 27909341 | [262,263] | ||
| OH▪ | Fluorescent probe | 3-CCA | 16297980, 26516887 | [262,268] | ||
| OH▪ | Fluorescent probe | APF | HOCl | 16297980, 26516887 | [262,268] | |
| OH▪ | Fluorescent probe | HPF | HOCl | 16297980, 16168507 | [262,269] | |
| OH▪ | Fluorescent probe | FL | 16297980, 27909341 | [262,263] | ||
| ROO▪ | Fluorescent probe | C11-BODIPY581/591 | HO•, RO•, ONOO− | Lipid peroxidation sensor | 16297980, 21396693 | [262,270] |
| ROO▪ | Fluorescent probe | DPPP | Lipid peroxidation sensor | 16297980 | [262] | |
| H2O2 | Histochemical staining | DAB | In Situ imaging of ROS | 24862270, 20387054 | [260,266] | |
| H2O2 | Fluorescent probe | DCFH | CO3•−, NO2•, HO• | 16297980, 27909341 | [262,263] | |
| H2O2 | Fluorescent probe | HyPer | 24862270, 27302663 | [260,265] | ||
| ONOO- | Fluorescent probe | DHR | HOCl | 16297980 | [262] | |
| 1O2 | Fluorescent probe | DMA | 16297980 | [262] | ||
| 1O2 | Fluorescent probe | DPAX | 16297980, 19794917 | [262,271] | ||
| 1O2 | Fluorescent probe | DMAX | 16297980, 19794917 | [262,271] | ||
| HOCl | Fluorescent probe | BODIPY-based | 26043093 | [272] | ||
| Mt O2▪− | Fluorescent probe | MitoHE | 23668959 | [273] | ||
| Mt O2▪− | Fluorescent probe | MitoSox Red | 24862270 | [260] | ||
| Mt H2O2 | MitoB | Analyzed by mass spectrometry ex vivo | 23726990 | [274] | ||
| O2▪−, H2O2 | Fluorescent probe | Amplex Red | High sensitivity, low background fluorescence | 24862270, 16297980, 27909341 | [260,262,263] | |
| O2▪−, H2O2 | Chemiluminescent probe | Luminol | OH▪, ONOO- | 24862270, 29780884 | [260,275] | |
| ONOO-, H2O2 | Spectrophotometry | Boronates | 24862270, 23665586 | [260,264] | ||
| Redox status changes | Fluorescent probe | roGFP | Slow in reaction, non-sensitive, real-time, cell friendly but restriction in receptor cells | 24862270, 27306519 | [260,276] | |
| Redox status changes | Fluorescent probe | rxYFPs | 24862270, 27306519 | [260,276] | ||
| Redox status changes | Fluorescent probe | rxRFP | 24862270. 27208426 | [260,277] |
ESR: electron spin resonance; DCFH-DA: 2,7-Dichlorodihydrofluorescein diacetate; H2DCF: 2′,7′-dichloro-dihydrofluorescein; DHE/HE:dihydroethidium; DPBF: 1,3-Diphenylisobenzofuran; L-012: luminol analogue 8-amino-5-chloro-2,3-dihydro-7-phenylpyrido; NBT: Nitroblue tetrazolium; CHD: 1,3-Cyclohexanedione; 3-CCA: Coumarin, coumarin-3-carboxylic acid; SECCA:N-succinimidyl ester of coumarin-3-carboxylic acid; HPF: 2-[6-(4′-Hydroxy)phenoxy-3H-xanthen-3-on-9-yl]benzoic acid; APF: 2-[6-(4′-amino)phenoxy-3H-xanthen-3-on-9-yl] benzoic acid; FL: Fluorescein; DMA: 9,10-Dimethylanthracene; DPAXs: 9-[2-(3-Carboxy-9,10-diphenyl)anthryl]-6-hydroxy-3H-xanthen-3-ones; DMAX: [2-(3-Carboxy-9,10-dimethyl)anthryl]-6-hydroxy-3H-xanthen-3-one; CM-H2DCFDA: 5-(and 6)-chloromethyl-2′,7′-dichlorohydrofluorescein diacetate; scopoletin: 7-hydroxy-6-methoxy-coumarin; HVA: Homovanillic acid (4-hydroxy-3-methoxy-phenylacetic acid; DHR: Dihydrorhodamine 123; Amplex Red:10-acetyl-3,7-dihydroxyphenoxazineDAB: Diaminobenzidine; C11-BODIPY581/591: 4,4-Difluoro-5-(4-phenyl-1,3-butadienyl)-4-bora-3a,4a-diaza-s-indacene-3-undecanoic acid; DPPP: Diphenyl-1-pyrenylphosphine; mitoHE: mito-hydroethidine; HPr+: hydropropidine.
Supplemental table 3.
ROS generation Inhibitors.
| Target | Drug | Cell type | Concentration | PMID | Ref # | |
|---|---|---|---|---|---|---|
| General ROS | NAC | RPE cell | 1 mM | 17765224 | [78] | |
| Epithelial cell | 30 mM | 15109912 | [193] | |||
| Fibroblast | 20 mM | 16081426 | [220] | |||
| INS-1 cell | 0.4 mM | 17400930 | [70] | |||
| PDTC | RPE cell | 50uM | 17765224 | [78] | ||
| Vitamin C | Endothelial cell | 10uM | 10828488 | [278] | ||
| Vitamin E | Endothelial cell | 10uM | 10828488 | [278] | ||
| Tempol | Astrocyte | 3 nM | 21270817 | [180] | ||
| MnTBAP | Astrocyte | 50uM | 20547771 | [67] | ||
| Mitochondrial ROS | complex I | rotenone | RPE cell | 2.5uM | 17765224 | [78] |
| NIH-3T3 fibroblast | 5uM | 12796479 | [204] | |||
| complex II | TTFA | RPE cell | 10uM | 17765224 | [78] | |
| Atpenin A5 | Cardiomyocyte | 100 nM-1uM | 19242645 | [279] | ||
| Malonate | Heart Mt | 25-100uM | 18433712 | [280] | ||
| Complex III | Myxothizol | Liver Mt | 0.56 mg/kg | 24661880 | [281] | |
| Antimycin A | hepa1-6,Huh-7 cell | 1ug/ml | 26461342 | [142] | ||
| RPE cell | 100 ng/ml | 17765224 | [78] | |||
| Compled IV | Sodium azide | Motoneurone | 2 mM | 14660707 | [282] | |
| ATPase | Oligomycin | 20107899 | [283] | |||
| Non-selective | Mito-Tempo | HAEC | 1 mM | 27127201 | [60] | |
| Non-selective | Mito-Vit-E | HUVEC | 1uM | 31653897 | [143] | |
| NOS | Non-selective | l-NAME | L6 myotube | 1 mM | 16982630 | [181] |
| nNOS | NOSIP | 19298861 | [89] | |||
| eNOS | NMMA | 28904082 | [284] | |||
| NOX | Non-selective | apocynin | SMC | 50uM | 29472601 | [177] |
| VSMC | 100uM | 23774581 | [246] | |||
| DPI | RPE cell | 5uM | 17765224 | [78] | ||
| Endothelial cell | 15uM | 16249002 | [206] | |||
| HT-29 cell | 10uM | 25122478 | [188] | |||
| Fibroblast | 20uM | 16081426 | [220] | |||
| NIH-3T3 fibroblast | 5uM | 12796479 | [204] | |||
| AEBSF | 0.9uM | 23251382 | [285] | |||
| NOX1 | ML171 | SMC | 1uM | 29472601 | [177] | |
| NOX1/4 | GKT136901 | 24319690 | [286] | |||
| LOX | NDGA | Epithelial cell | 19635476 | [209] | ||
| NIH-3T3 fibroblast | 10uM | 12796479 | [204] | |||
| MPO | ABAH | 23251382 | [285] | |||
| MAO | Selegiline | Astrocyte | 20uM | 20547771 | [67] |
NAC– N-acetyl-l-cysteine, PDTC– pyrrolidene-dithiocarbamate, MnTBAP– Manganese III tetrakis, TTFA– 2-thenoyltrifluoroacetone,l-NAME—N omega-Nitro-l-arginine methyl ester hydrochloride, NMMA– NG-monomethyl-arginine, NOSIP– nitric oxide synthase interacting protein, DPI– diphenylene iodonium, SEBSF– 4-(2-aminoethyl)-benzenesulfonyl fluoride, NDGA– nordihydroguaiaretic acid, ABAH– 4-aminobenzoic acid hydrazide.
Supplemental Table 4.
The targets in various treatments in Figure 4 (MitoETC and membrane potential, NOX, SOD and LOX) and genes of their downstream pathways are analyzed by circus plot (see details in Fig. 2A and b).
| Target | Mitochondria ETC and membrane potential | NOX | SOD | LOX |
|---|---|---|---|---|
| Downstream signaling genes | ROMO1 | CDK5 | MAPK3 | PTK2 |
| BCL2L1 | MAPK1 | MAPK1 | PLA2G4A | |
| CASP3 | MAPK2 | |||
| CASP8 | MAPK3 | |||
| NFKB1 | NOS3 | |||
| MAPK1 | PIK3CB | |||
| MAPK2 | AKT1 | |||
| MAPK3 | HMOX1 | |||
| RAC1 | NFKB1 | |||
| AKAP1 | MMP2 | |||
| UTRN | MMP9 | |||
| OPA1 | PRRT2 | |||
| NLRP3 | CDKN2A | |||
| PYCARD | IRS1 | |||
| CASP1 | NFE2L2 | |||
| NOS2 | ||||
| MAP1LC3A | ||||
| PTK2 | ||||
| STAT3 | ||||
| VEGFA | ||||
| MAPK3 | ||||
| MAPK1 | ||||
| CTNNB1 | ||||
| CDK2 | ||||
| MAPK8 | ||||
| MAP2K7 | ||||
| VCAM1 | ||||
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Panth N., Paudel K.R., Parajuli K. Reactive oxygen species: a key hallmark of cardiovascular disease. Advances in medicine. 2016;2016 doi: 10.1155/2016/9152732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He F., Zuo L. Redox roles of reactive oxygen species in cardiovascular diseases. Int. J. Mol. Sci. 2015;16(11):27770–27780. doi: 10.3390/ijms161126059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senoner T., Dichtl W. Oxidative stress in cardiovascular diseases: still a therapeutic target? Nutrients. 2019;11(9):2090. doi: 10.3390/nu11092090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin Y. Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arterioscler. Thromb. Vasc. Biol. 2015;35(4):804–816. doi: 10.1161/ATVBAHA.115.305282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y.F. Caspase-1 mediates hyperlipidemia-weakened progenitor cell vessel repair. Front. Biosci. 2016;21:178–191. doi: 10.2741/4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y. NADPH oxidases and oxidase crosstalk in cardiovascular diseases: novel therapeutic targets. Nat. Rev. Cardiol. 2020;17(3):170–194. doi: 10.1038/s41569-019-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modaresi A., Nafar M., Sahraei Z. Oxidative stress in chronic kidney disease. Iranian journal of kidney diseases. 2015;9(3):165. [PubMed] [Google Scholar]
- 8.Ratliff B.B. Oxidant mechanisms in renal injury and disease. Antioxidants Redox Signal. 2016;25(3):119–146. doi: 10.1089/ars.2016.6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y. Uremic toxins are conditional danger-or homeostasis-associated molecular patterns. Front. Biosci. 2018;23:348. doi: 10.2741/4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R. End-stage renal disease is different from chronic kidney disease in upregulating ROS-modulated proinflammatory secretome in PBMCs - a novel multiple-hit model for disease progression. Redox Biol. 2020:101460. doi: 10.1016/j.redox.2020.101460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y. Uremic toxins are conditional danger- or homeostasis-associated molecular patterns. Front. Biosci. 2018;23:348–387. doi: 10.2741/4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumder A. Restoration of skeletal muscle homeostasis by hydrogen sulfide during hyperhomocysteinemia-mediated oxidative/ER stress condition (1) Can. J. Physiol. Pharmacol. 2019;97(6):441–456. doi: 10.1139/cjpp-2018-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai J. Metabolism-associated danger signal-induced immune response and reverse immune checkpoint-activated CD40(+) monocyte differentiation. J. Hematol. Oncol. 2017;10(1):141. doi: 10.1186/s13045-017-0504-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen W. Homocysteine-methionine cycle is a metabolic sensor system controlling methylation-regulated pathological signaling. Redox Biol. 2020;28:101322. doi: 10.1016/j.redox.2019.101322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester S.J. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018;122(6):877–902. doi: 10.1161/CIRCRESAHA.117.311401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraaij M.D. Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 2010;107(41):17686–17691. doi: 10.1073/pnas.1012016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kempkes R.W.M. Metabolic pathways involved in regulatory T cell functionality. Front. Immunol. 2019;10:2839. doi: 10.3389/fimmu.2019.02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamceva G. Cigarette smoking and oxidative stress in patients with coronary artery disease. Open Access Maced J Med Sci. 2016;4(4):636–640. doi: 10.3889/oamjms.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virtue A. MicroRNA-155 deficiency leads to decreased atherosclerosis, increased white adipose tissue obesity, and non-alcoholic fatty liver disease: a novel mouse model OF obesity paradox. J. Biol. Chem. 2017;292(4):1267–1287. doi: 10.1074/jbc.M116.739839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson C. Increased expression of resistin in MicroRNA-155-deficient white adipose tissues may Be a possible driver of metabolically healthy obesity transition to classical obesity. Front. Physiol. 2018;9:1297. doi: 10.3389/fphys.2018.01297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith G.I., Mittendorfer B., Klein S. Metabolically healthy obesity: facts and fantasies. J. Clin. Invest. 2019;129(10):3978–3989. doi: 10.1172/JCI129186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumari S., Badana A.K., Malla R. Reactive oxygen species: a key constituent in cancer survival. Biomark. Insights. 2018;13 doi: 10.1177/1177271918755391. 1177271918755391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perillo B. ROS in cancer therapy: the bright side of the moon. Exp. Mol. Med. 2020:1–12. doi: 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X.F., Yin Y., Wang H. VASCULAR INFLAMMATION AND ATHEROGENESIS ARE ACTIVATED VIA RECEPTORS FOR PAMPs AND SUPPRESSED BY REGULATORY T CELLS. Drug Discov. Today Ther. Strat. 2008;5(2):125–142. doi: 10.1016/j.ddstr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X. Lysophospholipid receptors, as novel conditional danger receptors and homeostatic receptors modulate inflammation-novel paradigm and therapeutic potential. J Cardiovasc Transl Res. 2016;9(4):343–359. doi: 10.1007/s12265-016-9700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao Y. Lysophospholipids and their receptors serve as conditional DAMPs and DAMP receptors in tissue oxidative and inflammatory injury. Antioxidants Redox Signal. 2017 doi: 10.1089/ars.2017.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y. Inflammasomes: sensors of metabolic stresses for vascular inflammation. Front. Biosci. 2013;18:638–649. doi: 10.2741/4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corpas F.J., Gupta D.K., Palma J.M. Reactive Oxygen Species and Oxidative Damage in Plants under Stress. Springer; 2015. Production sites of reactive oxygen species (ROS) in organelles from plant cells; pp. 1–22. [Google Scholar]
- 29.Fransen M. Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2012;1822(9):1363–1373. doi: 10.1016/j.bbadis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Li J.-M., Shah A.M. Endothelial cell superoxide generation: regulation and relevance for cardiovascular pathophysiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287(5):R1014–R1030. doi: 10.1152/ajpregu.00124.2004. [DOI] [PubMed] [Google Scholar]
- 31.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94(3):909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinnerthaler M. Oxidative stress in aging human skin. Biomolecules. 2015;5(2):545–589. doi: 10.3390/biom5020545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahsan H., Ali A., Ali R. Oxygen free radicals and systemic autoimmunity. Clin. Exp. Immunol. 2003;131(3):398–404. doi: 10.1046/j.1365-2249.2003.02104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020;21(7):363–383. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 35.Li X. Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler. Thromb. Vasc. Biol. 2016;36(6):1090–1100. doi: 10.1161/ATVBAHA.115.306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao Y. Vascular endothelial cells and innate immunity. Arterioscler. Thromb. Vasc. Biol. 2020;40(6):e138–e152. doi: 10.1161/ATVBAHA.120.314330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng J. Mitochondrial proton leak plays a critical role in pathogenesis of cardiovascular diseases. Adv. Exp. Med. Biol. 2017;982:359–370. doi: 10.1007/978-3-319-55330-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X. Mitochondrial ROS, uncoupled from ATP synthesis, determine endothelial activation for both physiological recruitment of patrolling cells and pathological recruitment of inflammatory cells. Can. J. Physiol. Pharmacol. 2017;95(3):247–252. doi: 10.1139/cjpp-2016-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanayakkara G.K., Wang H., Yang X. Proton leak regulates mitochondrial reactive oxygen species generation in endothelial cell activation and inflammation - a novel concept. Arch. Biochem. Biophys. 2019;662:68–74. doi: 10.1016/j.abb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X. IL-35 (Interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14) Arterioscler. Thromb. Vasc. Biol. 2018;38(3):599–609. doi: 10.1161/ATVBAHA.117.310626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X. Anti-inflammatory cytokines IL-35 and IL-10 block atherogenic lysophosphatidylcholine-induced, mitochondrial ROS-mediated innate immune activation, but spare innate immune memory signature in endothelial cells. Redox Biol. 2020;28:101373. doi: 10.1016/j.redox.2019.101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gluschko A. The β2 integrin Mac-1 induces protective LC3-associated phagocytosis of Listeria monocytogenes. Cell Host Microbe. 2018;23(3):324–337. doi: 10.1016/j.chom.2018.01.018. e5. [DOI] [PubMed] [Google Scholar]
- 44.Zeng H. DNA checkpoint and repair factors are nuclear sensors for intracellular organelle stresses-inflammations and cancers can have high genomic risks. Front. Physiol. 2018;9:516. doi: 10.3389/fphys.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caspi R. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alam-Nazki A., Krishnan J. Spatial control of biochemical modification cascades and pathways. Biophys. J. 2015;108(12):2912–2924. doi: 10.1016/j.bpj.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinzpeter F., Gerland U., Tostevin F. Optimal compartmentalization strategies for metabolic microcompartments. Biophys. J. 2017;112(4):767–779. doi: 10.1016/j.bpj.2016.11.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y. Increased acetylation of H3K14 in the genomic regions that encode trained immunity enzymes in lysophosphatidylcholine-activated human aortic endothelial cells - novel qualification markers for chronic disease risk factors and conditional DAMPs. Redox Biol. 2019;24:101221. doi: 10.1016/j.redox.2019.101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L. Metabolic reprogramming in immune response and tissue inflammation. Arterioscler. Thromb. Vasc. Biol. 2020 doi: 10.1161/ATVBAHA.120.314037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin Y. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int. J. Immunopathol. Pharmacol. 2009;22(2):311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y.F. Analyses of caspase-1-regulated transcriptomes in various tissues lead to identification of novel IL-1beta-, IL-18- and sirtuin-1-independent pathways. J. Hematol. Oncol. 2017;10(1):40. doi: 10.1186/s13045-017-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L. Novel extracellular and nuclear caspase-1 and inflammasomes propagate inflammation and regulate gene expression: a comprehensive database mining study. J. Hematol. Oncol. 2016;9(1):122. doi: 10.1186/s13045-016-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J. Caspase-1 recognizes extended cleavage sites in its natural substrates. Atherosclerosis. 2010;210(2):422–429. doi: 10.1016/j.atherosclerosis.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Pastrana J. Inhibition of caspase-1 activation in endothelial cells improves angiogenesis: a novel therapeutic potential for ischemia. J. Biol. Chem. 2015;290(28):17485–17494. doi: 10.1074/jbc.M115.641191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrer L.M. Caspase-1 plays a critical role in accelerating chronic kidney disease-promoted neointimal hyperplasia in the carotid artery. J Cardiovasc Transl Res. 2016;9(2):135–144. doi: 10.1007/s12265-016-9683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xi H. Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ. Res. 2016;118(10):1525–1539. doi: 10.1161/CIRCRESAHA.116.308501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017;42(4):245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Broz P., Pelegrin P., Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020;20(3):143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 60.Li X. Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler. Thromb. Vasc. Biol. 2016;36(6):1090–1100. doi: 10.1161/ATVBAHA.115.306964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li X. Lysophospholipids induce innate immune transdifferentiation of endothelial cells, resulting in prolonged endothelial activation. J. Biol. Chem. 2018 doi: 10.1074/jbc.RA118.002752. jbc.RA118.002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathur A., Hayward J.A., Man S.M. Molecular mechanisms of inflammasome signaling. J. Leukoc. Biol. 2018;103(2):233–257. doi: 10.1189/jlb.3MR0617-250R. [DOI] [PubMed] [Google Scholar]
- 63.Villanueva C., Giulivi C. Subcellular and cellular locations of nitric oxide synthase isoforms as determinants of health and disease. Free Radic. Biol. Med. 2010;49(3):307–316. doi: 10.1016/j.freeradbiomed.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakuma S. Hydrogen peroxide generated by xanthine/xanthine oxidase system represses the proliferation of colorectal cancer cell line Caco-2. J. Clin. Biochem. Nutr. 2015;56(1):15–19. doi: 10.3164/jcbn.14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr. Biol. 2014;24(10):R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaarmann A., Gandhi S., Abramov A.Y. Dopamine induces Ca2+ signaling in astrocytes through reactive oxygen species generated by monoamine oxidase. J. Biol. Chem. 2010;285(32):25018–25023. doi: 10.1074/jbc.M110.111450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Acquier A.B. Reactive oxygen species mediate dopamine-induced signaling in renal proximal tubule cells. FEBS Lett. 2013;587(19):3254–3260. doi: 10.1016/j.febslet.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 69.Yang S.Y. Pretreatment with insulin-like growth factor I protects skeletal muscle cells against oxidative damage via PI3K/Akt and ERK1/2 MAPK pathways. Lab. Invest. 2010;90(3):391–401. doi: 10.1038/labinvest.2009.139. [DOI] [PubMed] [Google Scholar]
- 70.Pi J. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56(7):1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 71.Provost C. Nitric oxide and reactive oxygen species in the nucleus revisited. Can. J. Physiol. Pharmacol. 2010;88(3):296–304. doi: 10.1139/Y10-011. [DOI] [PubMed] [Google Scholar]
- 72.Woo C.-H. Tumor necrosis factor-α generates reactive oxygen species via a cytosolic phospholipase A2-linked cascade. J. Biol. Chem. 2000;275(41):32357–32362. doi: 10.1074/jbc.M005638200. [DOI] [PubMed] [Google Scholar]
- 73.Cha Y.K. Epidermal growth factor induces oxidative neuronal injury in cortical culture. J. Neurochem. 2000;75(1):298–303. doi: 10.1046/j.1471-4159.2000.0750298.x. [DOI] [PubMed] [Google Scholar]
- 74.Cheng Y.-C. Albumin overload down-regulates integrin-β1 through reactive oxygen species-endoplasmic reticulum stress pathway in podocytes. J. Biochem. 2015;158(2):101–108. doi: 10.1093/jb/mvv020. [DOI] [PubMed] [Google Scholar]
- 75.Kang M. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012;3(1) doi: 10.1038/cddis.2011.134. e249-e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J., Lai B., Nanayakkara G., Yang Q., Sun Y., Lu Y., Shao Y., Yu D., Yang W.Y., Cueto R., Fu H., Zeng H., Shen W., Wu S., Zhang C., Liu Y., Choi, Wang H., Yang X. Experimental data-mining analyses reveal new roles of low-intensity ultrasound in differentiating cell death regulatome in cancer and non-cancer cells via potential modulation of chromatin long-range interactions. Frontiers in Oncology. 2019;2019 doi: 10.3389/fonc.2019.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang D. Pro-inflammatory cytokines increase reactive oxygen species through mitochondria and NADPH oxidase in cultured RPE cells. Exp. Eye Res. 2007;85(4):462–472. doi: 10.1016/j.exer.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sowrirajan B. Interleukin-27 enhances the potential of reactive oxygen species generation from monocyte-derived macrophages and dendritic cells by induction of p47 phox. Sci. Rep. 2017;7:43441. doi: 10.1038/srep43441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kleine T., Leister D. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta. 2016;1857(8):1313–1325. doi: 10.1016/j.bbabio.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 81.Zeng H. DNA checkpoint and repair factors are nuclear sensors for intracellular organelle stresses—inflammations and cancers can have high genomic risks. Front. Physiol. 2018;9:516. doi: 10.3389/fphys.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ozcan A., Ogun M. Biochemistry of reactive oxygen and nitrogen species. Basic principles and clinical significance of oxidative stress. 2015;3:37–58. [Google Scholar]
- 83.Li X. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6(1):19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saaoud F. Bone marrow deficiency of mRNA decaying protein Tristetraprolin increases inflammation and mitochondrial ROS but reduces hepatic lipoprotein production in LDLR knockout mice. Redox Biol. 2020:101609. doi: 10.1016/j.redox.2020.101609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abderrazak A. NLRP3 inflammasome: from a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases. Redox Biol. 2015;4:296–307. doi: 10.1016/j.redox.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abais J.M. Contribution of endogenously produced reactive oxygen species to the activation of podocyte NLRP3 inflammasomes in hyperhomocysteinemia. Free Radic. Biol. Med. 2014;67:211–220. doi: 10.1016/j.freeradbiomed.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stolz D.B. Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology. 2002;36(1):81–93. doi: 10.1053/jhep.2002.33716. [DOI] [PubMed] [Google Scholar]
- 89.Zhou L., Zhu D.-Y. Neuronal nitric oxide synthase: structure, subcellular localization, regulation, and clinical implications. Nitric Oxide. 2009;20(4):223–230. doi: 10.1016/j.niox.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 90.Das K., Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Frontiers in Environmental Science. 2014;2:53. [Google Scholar]
- 91.Spiteller G. Peroxyl radicals: inductors of neurodegenerative and other inflammatory diseases. Their origin and how they transform cholesterol, phospholipids, plasmalogens, polyunsaturated fatty acids, sugars, and proteins into deleterious products. Free Radic. Biol. Med. 2006;41(3):362–387. doi: 10.1016/j.freeradbiomed.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 92.Repetto M., Semprine J., Boveris A. vol. 1. 2012. (Lipid Peroxidation: Chemical Mechanism, Biological Implications and Analytical Determination). chapter. [Google Scholar]
- 93.Phaniendra A., Jestadi D.B., Periyasamy L. Free radicals: properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015;30(1):11–26. doi: 10.1007/s12291-014-0446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nimse S.B., Pal D. Free radicals, natural antioxidants, and their reaction mechanisms. RSC Adv. 2015;5(35):27986–28006. [Google Scholar]
- 95.Lim P. Peroxyl radical mediated oxidative DNA base damage: implications for lipid peroxidation induced mutagenesis. Biochemistry. 2004;43(49):15339–15348. doi: 10.1021/bi048276x. [DOI] [PubMed] [Google Scholar]
- 96.Chang M.S. Detection and subcellular localization of two 15S-lipoxygenases in human cornea. Invest. Ophthalmol. Vis. Sci. 2005;46(3):849–856. doi: 10.1167/iovs.04-1166. [DOI] [PubMed] [Google Scholar]
- 97.Medinas D.B. The carbonate radical and related oxidants derived from bicarbonate buffer. IUBMB Life. 2007;59(4‐5):255–262. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]
- 98.Snezhkina A.V. Oxidative medicine and cellular longevity; 2019. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ives A. Xanthine oxidoreductase regulates macrophage IL1β secretion upon NLRP3 inflammasome activation. Nat. Commun. 2015;6(1):1–11. doi: 10.1038/ncomms7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Del Río L.A., López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016;57(7):1364–1376. doi: 10.1093/pcp/pcw076. [DOI] [PubMed] [Google Scholar]
- 101.Lambert A.J., Brand M.D. Mitochondrial DNA. Springer; 2009. Reactive oxygen species production by mitochondria; pp. 165–181. [DOI] [PubMed] [Google Scholar]
- 102.Sies H., Jones D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020:1–21. doi: 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- 103.Forstermann U., Sessa W.C. Nitric oxide synthases: regulation and function. Eur. Heart J. 2012;33(7):829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.King M.M., Lai E.K., McCay P.B. Singlet oxygen production associated with enzyme-catalyzed lipid peroxidation in liver microsomes. J. Biol. Chem. 1975;250(16):6496–6502. [PubMed] [Google Scholar]
- 105.Onyango A.N. Oxidative medicine and cellular longevity; 2016. Endogenous Generation of Singlet Oxygen and Ozone in Human and Animal Tissues: Mechanisms, Biological Significance, and Influence of Dietary Components. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Agnez-Lima L.F. DNA damage by singlet oxygen and cellular protective mechanisms. Mutat. Res. Rev. Mutat. Res. 2012;751(1):15–28. doi: 10.1016/j.mrrev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 107.Onyango A.N. Oxidative medicine and cellular longevity; 2017. The Contribution of Singlet Oxygen to Insulin Resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sandalio L., Romero-Puertas M. Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 2015;116(4):475–485. doi: 10.1093/aob/mcv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qian W. Chemoptogenetic damage to mitochondria causes rapid telomere dysfunction. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116(37):18435–18444. doi: 10.1073/pnas.1910574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou X. Laser controlled singlet oxygen generation in mitochondria to promote mitochondrial DNA replication in vitro. Sci. Rep. 2015;5:16925. doi: 10.1038/srep16925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Okada S.S. Myeloperoxidase in human peripheral blood lymphocytes: production and subcellular localization. Cell. Immunol. 2016;300:18–25. doi: 10.1016/j.cellimm.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 112.Zhang L. Biochemical basis and metabolic interplay of redox regulation. Redox Biol. 2019;26:101284. doi: 10.1016/j.redox.2019.101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhalla K. Role of hypoxia in Diffuse Large B-cell Lymphoma: metabolic repression and selective translation of HK2 facilitates development of DLBCL. Sci. Rep. 2018;8(1):1–15. doi: 10.1038/s41598-018-19182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fu H. Thrombus leukocytes exhibit more endothelial cell-specific angiogenic markers than peripheral blood leukocytes do in acute coronary syndrome patients, suggesting a possibility of trans-differentiation: a comprehensive database mining study. J. Hematol. Oncol. 2017;10(1):74. doi: 10.1186/s13045-017-0440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yetkin-Arik B. Endothelial tip cells in vitro are less glycolytic and have a more flexible response to metabolic stress than non-tip cells. Sci. Rep. 2019;9(1):10414. doi: 10.1038/s41598-019-46503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lai J.H. Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO Rep. 2018;19(8) doi: 10.15252/embr.201846182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim J. TNF-α-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X L. Cell Death Differ. 2010;17(9):1420–1434. doi: 10.1038/cdd.2010.19. [DOI] [PubMed] [Google Scholar]
- 118.Zhong C. Trained immunity: an underlying driver of inflammatory atherosclerosis. Front. Immunol. 2020;11:284. doi: 10.3389/fimmu.2020.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lai B. Twenty novel disease group-specific and 12 new shared macrophage pathways in eight groups of 34 diseases including 24 inflammatory organ diseases and 10 types of tumors. Front. Immunol. 2019;10:2612. doi: 10.3389/fimmu.2019.02612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cueto R. Identification of homocysteine-suppressive mitochondrial ETC complex genes and tissue expression profile - novel hypothesis establishment. Redox Biol. 2018;17:70–88. doi: 10.1016/j.redox.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nanayakkara G.K., Wang H., Yang X. Proton leak regulates mitochondrial reactive oxygen species generation in endothelial cell activation and inflammation-A novel concept. Arch. Biochem. Biophys. 2019;662:68–74. doi: 10.1016/j.abb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Y.-R., Zweier J.L. Cardiac mitochondria and reactive oxygen species generation. Circ. Res. 2014;114(3):524–537. doi: 10.1161/CIRCRESAHA.114.300559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sabharwal S.S., Schumacker P.T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat. Rev. Canc. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quan Y. Mitochondrial ROS-modulated mtDNA: a potential target for cardiac aging. Oxidative Medicine and Cellular Longevity. 2020:2020. doi: 10.1155/2020/9423593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kaarniranta K. Role of mitochondrial DNA damage in ROS-mediated pathogenesis of age-related macular degeneration (AMD) Int. J. Mol. Sci. 2019;20(10):2374. doi: 10.3390/ijms20102374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ide T. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ. Res. 2001;88(5):529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 127.Grazioli S., Pugin J. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front. Immunol. 2018;9:832. doi: 10.3389/fimmu.2018.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aon M.A. Synchronized whole cell oscillations in mitochondrial metabolism triggered by a local release of reactive oxygen species in cardiac myocytes. J. Biol. Chem. 2003;278(45):44735–44744. doi: 10.1074/jbc.M302673200. [DOI] [PubMed] [Google Scholar]
- 129.Nickel A., Kohlhaas M., Maack C. Mitochondrial reactive oxygen species production and elimination. J. Mol. Cell. Cardiol. 2014;73:26–33. doi: 10.1016/j.yjmcc.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 130.Costa A.D., Garlid K.D. Intramitochondrial signaling: interactions among mitoKATP, PKCε, ROS, and MPT. Am. J. Physiol. Heart Circ. Physiol. 2008;295(2):H874–H882. doi: 10.1152/ajpheart.01189.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang L. Biochemical basis and metabolic interplay of redox regulation. Redox biology. 2019:101284. doi: 10.1016/j.redox.2019.101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Roy Chowdhury S.K. Effects of extensively oxidized low-density lipoprotein on mitochondrial function and reactive oxygen species in porcine aortic endothelial cells. Am. J. Physiol. Endocrinol. Metabol. 2010;298(1):E89–E98. doi: 10.1152/ajpendo.00433.2009. [DOI] [PubMed] [Google Scholar]
- 133.Ermak N. Role of reactive oxygen species and Bax in oxidized low density lipoprotein-induced apoptosis of human monocytes. Atherosclerosis. 2008;200(2):247–256. doi: 10.1016/j.atherosclerosis.2007.12.052. [DOI] [PubMed] [Google Scholar]
- 134.Zorov D.B. Reactive oxygen species (Ros-Induced) Ros release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med. 2000;192(7):1001–1014. doi: 10.1084/jem.192.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuznetsov A.V. Synchronism in mitochondrial ROS flashes, membrane depolarization and calcium sparks in human carcinoma cells. Biochim. Biophys. Acta Bioenerg. 2017;1858(6):418–431. doi: 10.1016/j.bbabio.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zou M.-H., Shi C., Cohen R.A. Oxidation of the zinc-thiolate complex and uncoupling of endothelial nitric oxide synthase by peroxynitrite. J. Clin. Invest. 2002;109(6):817–826. doi: 10.1172/JCI14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alp N.J., Channon K.M. Regulation of endothelial nitric oxide synthase by tetrahydrobiopterin in vascular disease. Arterioscler. Thromb. Vasc. Biol. 2004;24(3):413–420. doi: 10.1161/01.ATV.0000110785.96039.f6. [DOI] [PubMed] [Google Scholar]
- 138.Schulz E. Mitochondrial redox signaling: interaction of mitochondrial reactive oxygen species with other sources of oxidative stress. Antioxidants Redox Signal. 2014;20(2):308–324. doi: 10.1089/ars.2012.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dikalov S.I. Nox2-induced production of mitochondrial superoxide in angiotensin II-mediated endothelial oxidative stress and hypertension. Antioxidants Redox Signal. 2014;20(2):281–294. doi: 10.1089/ars.2012.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang R. Mitochondrial regulation of NADPH oxidase in hindlimb unweighting rat cerebral arteries. PloS One. 2014;9(4) doi: 10.1371/journal.pone.0095916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Segovia J. TLR2/MyD88/NF-κB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PloS One. 2012;7(1) doi: 10.1371/journal.pone.0029695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kastl L. TNF-a stimulation enhances ROS-dependent cell migration via NF-? B activation in liver cells. Free Radic. Biol. Med. 2014;75:S32. doi: 10.1016/j.freeradbiomed.2014.10.765. [DOI] [PubMed] [Google Scholar]
- 143.Somvanshi P.R., Tomar M., Kareenhalli V. Computational analysis of insulin-glucagon signalling network: implications of bistability to metabolic homeostasis and disease states. Sci. Rep. 2019;9(1):1–11. doi: 10.1038/s41598-019-50889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li Y.F. Lysophospholipids and their G protein-coupled receptors in atherosclerosis. Front. Biosci. 2016;21:70–88. doi: 10.2741/4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lu Y. Increased acetylation of H3K14 in the genomic regions that encode trained immunity enzymes in lysophosphatidylcholine-activated human aortic endothelial cells–Novel qualification markers for chronic disease risk factors and conditional DAMPs. Redox biology. 2019;24:101221. doi: 10.1016/j.redox.2019.101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li X. Anti-inflammatory cytokines IL-35 and IL-10 block atherogenic lysophosphatidylcholine-induced, mitochondrial ROS-mediated innate immune activation, but spare innate immune memory signature in endothelial cells. Redox biology. 2020;28:101373. doi: 10.1016/j.redox.2019.101373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Abais J.M. Redox regulation of NLRP3 inflammasomes: ROS as trigger or effector? Antioxidants Redox Signal. 2015;22(13):1111–1129. doi: 10.1089/ars.2014.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee J.-W. A translocator protein 18 kDa ligand, Ro5-4864, inhibits ATP-induced NLRP3 inflammasome activation. Biochem. Biophys. Res. Commun. 2016;474(3):587–593. doi: 10.1016/j.bbrc.2016.04.080. [DOI] [PubMed] [Google Scholar]
- 149.Zhang Z. Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J. Exp. Med. 2017;214(9):2671–2693. doi: 10.1084/jem.20162040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhou R. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 151.Liu Q. The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol. 2018;103:115–124. doi: 10.1016/j.molimm.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 152.Moretti J., Blander J.M. Increasing complexity of NLRP3 inflammasome regulation. J. Leukoc. Biol. 2020 doi: 10.1002/JLB.3MR0520-104RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gurung P., Lukens J.R., Kanneganti T.-D. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol. Med. 2015;21(3):193–201. doi: 10.1016/j.molmed.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Holley C.L., Schroder K. The rOX‐stars of inflammation: links between the inflammasome and mitochondrial meltdown. Clinical & translational immunology. 2020;9(2) doi: 10.1002/cti2.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Misawa T. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 2013;14(5):454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 156.Sagulenko V. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20(9):1149–1160. doi: 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Castejón-Vega B., Giampieri F., Alvarez-Suarez J.M. Nutraceutical compounds targeting inflammasomes in human diseases. Int. J. Mol. Sci. 2020;21(14):4829. doi: 10.3390/ijms21144829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Janssen J.J.E. Mito-nuclear communication by mitochondrial metabolites and its regulation by B-vitamins. Front. Physiol. 2019;10:78. doi: 10.3389/fphys.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shao Y., Saredy J., Yang W., Sun Y., Lu Y., Saaoud F., Drummer IV C., Johnson C., Xu K., Jiang X., Wang H., Yang X. Vascular endothelial cells and innate immunity. Arterioscler. Thromb. Vasc. Biol. 2020;40(6) doi: 10.1161/ATVBAHA.120.314330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yin Y. Early hyperlipidemia promotes endothelial activation via a caspase-1-sirtuin 1 pathway. Arterioscler. Thromb. Vasc. Biol. 2015;35(4):804–816. doi: 10.1161/ATVBAHA.115.305282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wiese M., Bannister A.J. Two genomes, one cell: mitochondrial-nuclear coordination via epigenetic pathways. Mol Metab. 2020 doi: 10.1016/j.molmet.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Thannickal V.J., Fanburg B.L. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 2000;279(6):L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 163.Provost C. Nitric oxide and reactive oxygen species in the nucleus revisited. Can. J. Physiol. Pharmacol. 2010;88(3):296–304. doi: 10.1139/Y10-011. [DOI] [PubMed] [Google Scholar]
- 164.Paardekooper L.M. Radical stress is more cytotoxic in the nucleus than in other organelles. Int. J. Mol. Sci. 2019;20(17):4147. doi: 10.3390/ijms20174147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chittiboyina S., Bai Y., Lelievre S.A. Microenvironment-cell nucleus relationship in the context of oxidative stress. Front Cell Dev Biol. 2018;6:23. doi: 10.3389/fcell.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Espinosa-Diez C. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shao Y. Immunosuppressive/anti-inflammatory cytokines directly and indirectly inhibit endothelial dysfunction-a novel mechanism for maintaining vascular function. J. Hematol. Oncol. 2014;7(1):80. doi: 10.1186/s13045-014-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sha X. Interleukin-35 inhibits endothelial cell activation by suppressing MAPK-AP-1 pathway. J. Biol. Chem. 2015;290(31):19307–19318. doi: 10.1074/jbc.M115.663286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pendergrass K.D. The Angiotensin II–AT1 Receptor stimulates reactive oxygen species within the cell nucleus. Biochem. Biophys. Res. Commun. 2009;384(2):149–154. doi: 10.1016/j.bbrc.2009.04.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17(4):213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 171.Tuet W.Y. Dose-dependent intracellular reactive oxygen and nitrogen species (ROS/RNS) production from particulate matter exposure: comparison to oxidative potential and chemical composition. Atmos. Environ. 2016;144:335–344. [Google Scholar]
- 172.Galadari S. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic. Biol. Med. 2017;104:144–164. doi: 10.1016/j.freeradbiomed.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 173.Chien S.-Y. Interleukin-1β induces fibroblast growth factor 2 expression and subsequently promotes endothelial progenitor cell angiogenesis in chondrocytes. Clin. Sci. 2016;130(9):667–681. doi: 10.1042/CS20150622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harper R.W. Differential regulation of dual NADPH oxidases/peroxidases, Duox1 and Duox2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett. 2005;579(21):4911–4917. doi: 10.1016/j.febslet.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 175.Park K.W., Baik H.H., Jin B.K. IL-13-induced oxidative stress via microglial NADPH oxidase contributes to death of hippocampal neurons in vivo. J. Immunol. 2009;183(7):4666–4674. doi: 10.4049/jimmunol.0803392. [DOI] [PubMed] [Google Scholar]
- 176.Gwathmey T.M. Angiotensin-(1-7)-angiotensin-converting enzyme 2 attenuates reactive oxygen species formation to angiotensin II within the cell nucleus. Hypertension. 2010;55(1):166–171. doi: 10.1161/HYPERTENSIONAHA.109.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hashad A.M. Reactive oxygen species mediate the suppression of arterial smooth muscle T-type Ca 2+ channels by angiotensin II. Sci. Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-21899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cheng T.-H. Involvement of reactive oxygen species in angiotensin II-induced endothelin-1 gene expression in rat cardiac fibroblasts. J. Am. Coll. Cardiol. 2003;42(10):1845–1854. doi: 10.1016/j.jacc.2003.06.010. [DOI] [PubMed] [Google Scholar]
- 179.Lodha S. Angiotensin II-induced mesangial cell apoptosis: role of oxidative stress. Mol. Med. 2002;8(12):830–840. [PMC free article] [PubMed] [Google Scholar]
- 180.Liu G. Angiotensin II induces human astrocyte senescence through reactive oxygen species production. Hypertens. Res. 2011;34(4):479–483. doi: 10.1038/hr.2010.269. [DOI] [PubMed] [Google Scholar]
- 181.Wei Y. Angiotensin II-induced NADPH oxidase activation impairs insulin signaling in skeletal muscle cells. J. Biol. Chem. 2006;281(46):35137–35146. doi: 10.1074/jbc.M601320200. [DOI] [PubMed] [Google Scholar]
- 182.Bae Y.S. Regulation of reactive oxygen species generation in cell signaling. Mol. Cell. 2011;32(6):491–509. doi: 10.1007/s10059-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Loh K. Reactive oxygen species enhance insulin sensitivity. Cell Metabol. 2009;10(4):260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Daou G.B., Srivastava A.K. Reactive oxygen species mediate Endothelin-1-induced activation of ERK1/2, PKB, and Pyk2 signaling, as well as protein synthesis, in vascular smooth muscle cells. Free Radic. Biol. Med. 2004;37(2):208–215. doi: 10.1016/j.freeradbiomed.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 185.Patel P.R. Norepinephrine reduces reactive oxygen species (ROS) and DNA damage in ovarian surface epithelial cells. J. Bioanal. Biomed. 2015;7(3):75. doi: 10.4172/1948-593X.1000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lu Q. D 5 dopamine receptor decreases NADPH oxidase, reactive oxygen species and blood pressure via heme oxygenase-1. Hypertens. Res. 2013;36(8):684–690. doi: 10.1038/hr.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang L.-B. TIGAR mediates the inhibitory role of hypoxia on ROS production and apoptosis in rat nucleus pulposus cells. Osteoarthritis Cartilage. 2018;26(1):138–148. doi: 10.1016/j.joca.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 188.Lien G.-S. Epidermal growth factor stimulates nuclear factor-κB activation and heme oxygenase-1 expression via c-Src, NADPH oxidase, PI3K, and Akt in human colon cancer cells. PloS One. 2014;9(8) doi: 10.1371/journal.pone.0104891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu J. Mild hypoxia promotes survival and proliferation of SOD2-deficient astrocytes via c-Myc activation. J. Neurosci. 2006;26(16):4329–4337. doi: 10.1523/JNEUROSCI.0382-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Saunders J.A. Reactive oxygen species mediate lysophosphatidic acid induced signaling in ovarian cancer cells. Free Radic. Biol. Med. 2010;49(12):2058–2067. doi: 10.1016/j.freeradbiomed.2010.10.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chen L. Both hydrogen peroxide and transforming growth factor beta 1 contribute to endothelial Nox4 mediated angiogenesis in endothelial Nox4 transgenic mouse lines. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2014;1842(12):2489–2499. doi: 10.1016/j.bbadis.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 192.Bae J. Interleukin-2 promotes angiogenesis by activation of Akt and increase of ROS. J. Microbiol. Biotechnol. 2008;18(2):377–382. [PubMed] [Google Scholar]
- 193.Chen K.C.-W. Platelet derived growth factor (PDGF)-induced reactive oxygen species in the lens epithelial cells: the redox signaling. Exp. Eye Res. 2004;78(6):1057–1067. doi: 10.1016/j.exer.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 194.Liu L.-Z. Reactive oxygen species regulate epidermal growth factor-induced vascular endothelial growth factor and hypoxia-inducible factor-1α expression through activation of AKT and P70S6K1 in human ovarian cancer cells. Free Radic. Biol. Med. 2006;41(10):1521–1533. doi: 10.1016/j.freeradbiomed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 195.Yu M.O. Reactive oxygen species production has a critical role in hypoxia-induced Stat3 activation and angiogenesis in human glioblastoma. Journal of neuro-oncology. 2015;125(1):55–63. doi: 10.1007/s11060-015-1889-8. [DOI] [PubMed] [Google Scholar]
- 196.Lee Y.C. Eur Respiratory Soc; 2015. NLRP3 Inflammasome Is Activated via Phosphoinositide 3-kinase δ Pathway in Aspergillus Fumigatus-Induced Allergic Airway Inflammation. [Google Scholar]
- 197.Li D. Regulation of the NLRP3 inflammasome and macrophage pyroptosis by the p38 MAPK signaling pathway in a mouse model of acute lung injury. Mol. Med. Rep. 2018;18(5):4399–4409. doi: 10.3892/mmr.2018.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lim S.-O. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135(6):2128–2140. doi: 10.1053/j.gastro.2008.07.027. e8. [DOI] [PubMed] [Google Scholar]
- 199.Aggarwal V. Role of reactive oxygen species in cancer progression: molecular mechanisms and recent advancements. Biomolecules. 2019;9(11):735. doi: 10.3390/biom9110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mai J. An evolving new paradigm: endothelial cells--conditional innate immune cells. J. Hematol. Oncol. 2013;6:61. doi: 10.1186/1756-8722-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Buscher K. Patrolling mechanics of non-classical monocytes in vascular inflammation. Front Cardiovasc Med. 2017;4:80. doi: 10.3389/fcvm.2017.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Plastira I. Lysophosphatidic acid via LPA-receptor 5/protein kinase D-dependent pathways induces a motile and pro-inflammatory microglial phenotype. J. Neuroinflammation. 2017;14(1):253. doi: 10.1186/s12974-017-1024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lee I.-T. Role of TLR4/NADPH oxidase/ROS-activated p38 MAPK in VCAM-1 expression induced by lipopolysaccharide in human renal mesangial cells. Cell Commun. Signal. 2012;10(1):33. doi: 10.1186/1478-811X-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chiarugi P. Reactive oxygen species as essential mediators of cell adhesion: the oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J. Cell Biol. 2003;161(5):933–944. doi: 10.1083/jcb.200211118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Azimi I. Hypoxia-induced reactive oxygen species mediate N-cadherin and SERPINE1 expression, EGFR signalling and motility in MDA-MB-468 breast cancer cells. Sci. Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-15474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Walch L. Pro-atherogenic effect of interleukin-4 in endothelial cells: modulation of oxidative stress, nitric oxide and monocyte chemoattractant protein-1 expression. Atherosclerosis. 2006;187(2):285–291. doi: 10.1016/j.atherosclerosis.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 207.Sandoval R. TNF-α increases production of reactive oxygen species through Cdk5 activation in nociceptive neurons. Front. Physiol. 2018;9:65. doi: 10.3389/fphys.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li X. IL-35 (interleukin-35) suppresses endothelial cell activation by inhibiting mitochondrial reactive oxygen species-mediated site-specific acetylation of H3K14 (histone 3 lysine 14) Arterioscler. Thromb. Vasc. Biol. 2018;38(3):599–609. doi: 10.1161/ATVBAHA.117.310626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Huo Y. Reactive oxygen species (ROS) are essential mediators in epidermal growth factor (EGF)-stimulated corneal epithelial cell proliferation, adhesion, migration, and wound healing. Exp. Eye Res. 2009;89(6):876–886. doi: 10.1016/j.exer.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 210.Meng D., Lv D.-D., Fang J. Insulin-like growth factor-I induces reactive oxygen species production and cell migration through Nox4 and Rac1 in vascular smooth muscle cells. Cardiovasc. Res. 2008;80(2):299–308. doi: 10.1093/cvr/cvn173. [DOI] [PubMed] [Google Scholar]
- 211.Liao C.-R. Advanced oxidation protein products increase TNF-α and IL-1β expression in chondrocytes via NADPH oxidase 4 and accelerate cartilage degeneration in osteoarthritis progression. Redox biology. 2020;28:101306. doi: 10.1016/j.redox.2019.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Morita M. Advanced glycation end products-induced reactive oxygen species generation is partly through NF-kappa B activation in human aortic endothelial cells. J. Diabetes Complicat. 2013;27(1):11–15. doi: 10.1016/j.jdiacomp.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 213.Viganò E. Human caspase-4 and caspase-5 regulate the one-step non-canonical inflammasome activation in monocytes. Nat. Commun. 2015;6:8761. doi: 10.1038/ncomms9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cheng J.-J. Cyclic strain–induced reactive oxygen species involved in ICAM-1 gene induction in endothelial cells. Hypertension. 1998;31(1):125–130. doi: 10.1161/01.hyp.31.1.125. [DOI] [PubMed] [Google Scholar]
- 215.Birukov K.G. Cyclic stretch, reactive oxygen species, and vascular remodeling. Antioxidants Redox Signal. 2009;11(7):1651–1667. doi: 10.1089/ars.2008.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zeller K.S. The role of mechanical force and ROS in integrin-dependent signals. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0064897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zemskov E.A. Biomechanical forces and oxidative stress: implications for pulmonary vascular disease. Antioxidants Redox Signal. 2019;31(12):819–842. doi: 10.1089/ars.2018.7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lin K.-T. Reactive oxygen species participate in peroxynitrite-induced apoptosis in HL-60 cells. Biochem. Biophys. Res. Commun. 1997;230(1):115–119. doi: 10.1006/bbrc.1996.5897. [DOI] [PubMed] [Google Scholar]
- 219.Licandro G. The NLRP3 inflammasome affects DNA damage responses after oxidative and genotoxic stress in dendritic cells. Eur. J. Immunol. 2013;43(8):2126–2137. doi: 10.1002/eji.201242918. [DOI] [PubMed] [Google Scholar]
- 220.Svegliati S. Platelet-derived growth factor and reactive oxygen species (ROS) regulate Ras protein levels in primary human fibroblasts via ERK1/2 amplification OF ROS and Ras IN systemic sclerosis fibroblasts. J. Biol. Chem. 2005;280(43):36474–36482. doi: 10.1074/jbc.M502851200. [DOI] [PubMed] [Google Scholar]
- 221.Platnich J.M. Shiga toxin/lipopolysaccharide activates caspase-4 and gasdermin D to trigger mitochondrial reactive oxygen species upstream of the NLRP3 inflammasome. Cell Rep. 2018;25(6):1525–1536. doi: 10.1016/j.celrep.2018.09.071. e7. [DOI] [PubMed] [Google Scholar]
- 222.Wang Y. Mitochondrial ROS promote macrophage pyroptosis by inducing GSDMD oxidation. J. Mol. Cell Biol. 2019;11(12):1069–1082. doi: 10.1093/jmcb/mjz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dai J., Chen H., Chai Y. Advanced glycation end products (AGEs) induce apoptosis of fibroblasts by activation of NLRP3 inflammasome via reactive oxygen species (ROS) signaling pathway. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res.: international medical journal of experimental and clinical research. 2019;25:7499. doi: 10.12659/MSM.915806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Valente A.J. Advanced oxidation protein products induce cardiomyocyte death via Nox2/Rac1/superoxide-dependent TRAF3IP2/JNK signaling. Free Radic. Biol. Med. 2013;60:125–135. doi: 10.1016/j.freeradbiomed.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yu C. Advanced oxidation protein products induce apoptosis, and upregulate sclerostin and RANKL expression, in osteocytic MLO-Y4 cells via JNK/p38 MAPK activation. Mol. Med. Rep. 2017;15(2):543–550. doi: 10.3892/mmr.2016.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yang Z., Han, Oppenheim J.J. Alarmins and immunity. Immunol. Rev. 2017;280(1):41–56. doi: 10.1111/imr.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kramer-Albers E.M. Exosomes deliver ROS for regeneration. Nat. Cell Biol. 2018;20(3):225–226. doi: 10.1038/s41556-018-0048-9. [DOI] [PubMed] [Google Scholar]
- 228.Yang Q. Low-Intensity ultrasound-induced anti-inflammatory effects are mediated by several new mechanisms including gene induction, immunosuppressor cell promotion, and enhancement of exosome biogenesis and docking. Front. Physiol. 2017;8:818. doi: 10.3389/fphys.2017.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Luo S. Molecular mechanisms of endothelial NO synthase uncoupling. Curr. Pharmaceut. Des. 2014;20(22):3548–3553. doi: 10.2174/13816128113196660746. [DOI] [PubMed] [Google Scholar]
- 230.Fulton D. Localization of endothelial nitric-oxide synthase phosphorylated on serine 1179 and nitric oxide in Golgi and plasma membrane defines the existence of two pools of active enzyme. J. Biol. Chem. 2002;277(6):4277–4284. doi: 10.1074/jbc.M106302200. [DOI] [PubMed] [Google Scholar]
- 231.Silkstone R.S. Nitrogen dioxide oxidizes mitochondrial cytochrome c. Free Radic. Biol. Med. 2012;52(1):80–87. doi: 10.1016/j.freeradbiomed.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Galano A. Mechanism of the OH radical scavenging activity of nordihydroguaiaretic acid: a combined theoretical and experimental study. J. Phys. Chem. B. 2010;114(19):6625–6635. doi: 10.1021/jp912001c. [DOI] [PubMed] [Google Scholar]
- 233.Rodrigues E., Mariutti L.R., Mercadante A.Z. Scavenging capacity of marine carotenoids against reactive oxygen and nitrogen species in a membrane-mimicking system. Mar. Drugs. 2012;10(8):1784–1798. doi: 10.3390/md10081784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dean O., Giorlando F., Berk M. N-acetylcysteine in psychiatry: current therapeutic evidence and potential mechanisms of action. J. Psychiatry Neurosci.: JPN. 2011;36(2):78. doi: 10.1503/jpn.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bagchi D. Oxygen free radical scavenging abilities of vitamins C and E, and a grape seed proanthocyanidin extract in vitro. Res. Commun. Mol. Pathol. Pharmacol. 1997;95(2):179–189. [PubMed] [Google Scholar]
- 236.Dohi K. Alkoxyl radical-scavenging activity of edaravone in patients with traumatic brain injury. J. Neurotrauma. 2006;23(11):1591–1599. doi: 10.1089/neu.2006.23.1591. [DOI] [PubMed] [Google Scholar]
- 237.Hardeland R., Pandi-Perumal S. Melatonin, a potent agent in antioxidative defense: actions as a natural food constituent, gastrointestinal factor, drug and prodrug. Nutr. Metabol. 2005;2(1):22. doi: 10.1186/1743-7075-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Raimondi V., Ciccarese F., Ciminale V. Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br. J. Canc. 2019:1–14. doi: 10.1038/s41416-019-0651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Hooper D. Uric acid, a natural scavenger of peroxynitrite, in experimental allergic encephalomyelitis and multiple sclerosis. Proc. Natl. Acad. Sci. Unit. States Am. 1998;95(2):675–680. doi: 10.1073/pnas.95.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bancirova M. Sodium azide as a specific quencher of singlet oxygen during chemiluminescent detection by luminol and Cypridina luciferin analogues. Luminescence. 2011;26(6):685–688. doi: 10.1002/bio.1296. [DOI] [PubMed] [Google Scholar]
- 241.Fukuzawa K. Kinetics and dynamics of singlet oxygen scavenging by α-tocopherol in phospholipid model membranes. Free Radic. Biol. Med. 1997;22(5):923–930. doi: 10.1016/s0891-5849(96)00485-6. [DOI] [PubMed] [Google Scholar]
- 242.Nishinaka Y. Singlet oxygen is essential for neutrophil extracellular trap formation. Biochem. Biophys. Res. Commun. 2011;413(1):75–79. doi: 10.1016/j.bbrc.2011.08.052. [DOI] [PubMed] [Google Scholar]
- 243.Stocker R., Peterhans E. Antioxidant properties of conjugated bilirubin and biliverdin: biologically relevant scavenging of hypochlorous acid. Free Radic. Res. Commun. 1989;6(1):57–66. doi: 10.3109/10715768909073428. [DOI] [PubMed] [Google Scholar]
- 244.Kalogiannis M., Delikatny E.J., Jeitner T.M. Serotonin as a putative scavenger of hypohalous acid in the brain. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2016;1862(4):651–661. doi: 10.1016/j.bbadis.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rhyu D.Y. Role of reactive oxygen species in TGF-β1-induced mitogen-activated protein kinase activation and epithelial-mesenchymal transition in renal tubular epithelial cells. J. Am. Soc. Nephrol. 2005;16(3):667–675. doi: 10.1681/ASN.2004050425. [DOI] [PubMed] [Google Scholar]
- 246.Pi Y. Inhibition of reactive oxygen species generation attenuates TLR4-mediated proinflammatory and proliferative phenotype of vascular smooth muscle cells. Lab. Invest. 2013;93(8):880. doi: 10.1038/labinvest.2013.79. [DOI] [PubMed] [Google Scholar]
- 247.Hood K.Y. Serotonin signaling through the 5-HT1B receptor and NADPH oxidase 1 in pulmonary arterial hypertension. Arterioscler. Thromb. Vasc. Biol. 2017;37(7):1361–1370. doi: 10.1161/ATVBAHA.116.308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tsai C.-H. High glucose induces vascular endothelial growth factor production in human synovial fibroblasts through reactive oxygen species generation. Biochim. Biophys. Acta Gen. Subj. 2013;1830(3):2649–2658. doi: 10.1016/j.bbagen.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 249.Guimarães E.L. Advanced glycation end products induce production of reactive oxygen species via the activation of NADPH oxidase in murine hepatic stellate cells. J. Hepatol. 2010;52(3):389–397. doi: 10.1016/j.jhep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 250.Sun S. Advanced oxidation protein products induce S-phase arrest of hepatocytes via the ROS-dependent, β-catenin-CDK2-mediated pathway. Redox biology. 2018;14:338–353. doi: 10.1016/j.redox.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tsushima K. Mitochondrial reactive oxygen species in lipotoxic hearts induce post-translational modifications of AKAP121, DRP1, and OPA1 that promote mitochondrial fission. Circ. Res. 2018;122(1):58–73. doi: 10.1161/CIRCRESAHA.117.311307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bartlett R., Yerbury J.J., Sluyter R. P2X7 receptor activation induces reactive oxygen species formation and cell death in murine EOC13 microglia. Mediat. Inflamm. 2013:2013. doi: 10.1155/2013/271813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chen D. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-02340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fagundes-Netto F. The production of reactive oxygen species in TLR-stimulated granulocytes is not enhanced by hyperglycemia in diabetes. Int. Immunopharm. 2013;17(3):924–929. doi: 10.1016/j.intimp.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 255.Arbab-Zavar M.H. Electrochemical hydride generation atomic absorption spectrometry for determination of cadmium. Anal. Chim. Acta. 2005;546(1):126–132. doi: 10.1016/j.aca.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 256.Choi S.-H. Thrombin-induced oxidative stress contributes to the death of hippocampal neurons in vivo: role of microglial NADPH oxidase. J. Neurosci. 2005;25(16):4082–4090. doi: 10.1523/JNEUROSCI.4306-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lund A.K. Endothelin-1–Mediated increase in reactive oxygen species and NADPH oxidase activity in hearts of aryl hydrocarbon receptor (AhR) null mice. Toxicol. Sci. 2005;88(1):265–273. doi: 10.1093/toxsci/kfi284. [DOI] [PubMed] [Google Scholar]
- 258.Cueto R. Identification of homocysteine-suppressive mitochondrial ETC complex genes and tissue expression profile–Novel hypothesis establishment. Redox biology. 2018;17:70–88. doi: 10.1016/j.redox.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xi H. Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ. Res. 2016;118(10):1525–1539. doi: 10.1161/CIRCRESAHA.116.308501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wojtala A. Methods in Enzymology. Elsevier; 2014. Methods to monitor ROS production by fluorescence microscopy and fluorometry; pp. 243–262. [DOI] [PubMed] [Google Scholar]
- 261.Xiao Y., Meierhofer D. Mary Ann Liebert, Inc.; 2019. Are Hydroethidine-Based Probes Reliable for Reactive Oxygen Species Detection? publishers 140 Huguenot Street, 3rd Floor New …. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gomes A., Fernandes E., Lima J.L. Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods. 2005;65(2–3):45–80. doi: 10.1016/j.jbbm.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 263.Castro-Alférez M., Polo-López M.I., Fernández-Ibáñez P. Intracellular mechanisms of solar water disinfection. Sci. Rep. 2016;6:38145. doi: 10.1038/srep38145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Winterbourn C.C. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochim. Biophys. Acta Gen. Subj. 2014;1840(2):730–738. doi: 10.1016/j.bbagen.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 265.Oparka M. Quantifying ROS levels using CM-H2DCFDA and HyPer. Methods. 2016;109:3–11. doi: 10.1016/j.ymeth.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 266.Jambunathan N. Plant Stress Tolerance. Springer; 2010. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants; pp. 291–297. [DOI] [PubMed] [Google Scholar]
- 267.Wang W., Zhang H., Cheng H. Mitochondrial flashes: from indicator characterization to in vivo imaging. Methods. 2016;109:12–20. doi: 10.1016/j.ymeth.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Son Y. A novel high-throughput approach to measure hydroxyl radicals induced by airborne particulate matter. Int. J. Environ. Res. Publ. Health. 2015;12(11):13678–13695. doi: 10.3390/ijerph121113678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Tomizawa S. The detection and quantification of highly reactive oxygen species using the novel HPF fluorescence probe in a rat model of focal cerebral ischemia. Neurosci. Res. 2005;53(3):304–313. doi: 10.1016/j.neures.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 270.Partyka A. Detection of lipid peroxidation in frozen-thawed avian spermatozoa using C11-BODIPY581/591. Theriogenology. 2011;75(9):1623–1629. doi: 10.1016/j.theriogenology.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 271.Nagano T. Bioimaging probes for reactive oxygen species and reactive nitrogen species. J. Clin. Biochem. Nutr. 2009;45(2):111–124. doi: 10.3164/jcbn.R09-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chen W.-C., Venkatesan P., Wu S.-P. A highly selective turn-on fluorescent probe for hypochlorous acid based on hypochlorous acid-induced oxidative intramolecular cyclization of boron dipyrromethene-hydrazone. Anal. Chim. Acta. 2015;882:68–75. doi: 10.1016/j.aca.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 273.Kalyanaraman B. HPLC-based monitoring of products formed from hydroethidine-based fluorogenic probes—the ultimate approach for intra-and extracellular superoxide detection. Biochim. Biophys. Acta Gen. Subj. 2014;1840(2):739–744. doi: 10.1016/j.bbagen.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Logan A. Using exomarkers to assess mitochondrial reactive species in vivo. Biochim. Biophys. Acta Gen. Subj. 2014;1840(2):923–930. doi: 10.1016/j.bbagen.2013.05.026. [DOI] [PubMed] [Google Scholar]
- 275.Zhu H. A Highly sensitive chemiluminometric assay for real-time detection of biological hydrogen peroxide formation. Reactive oxygen species (Apex, NC) 2016;1(3):216. doi: 10.20455/ros.2016.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Braeckman B.P. In vivo detection of reactive oxygen species and redox status in Caenorhabditis elegans. Antioxidants Redox Signal. 2016;25(10):577–592. doi: 10.1089/ars.2016.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wages P.A. Live-cell imaging approaches for the investigation of xenobiotic-induced oxidant stress. Biochim. Biophys. Acta Gen. Subj. 2016;1860(12):2802–2815. doi: 10.1016/j.bbagen.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Dimmeler S., Zeiher A.M. Reactive oxygen species and vascular cell apoptosis in response to angiotensin II and pro-atherosclerotic factors. Regul. Pept. 2000;90(1–3):19–25. doi: 10.1016/s0167-0115(00)00105-1. [DOI] [PubMed] [Google Scholar]
- 279.Wojtovich A.P., Brookes P.S. The complex II inhibitor atpenin A5 protects against cardiac ischemia-reperfusion injury via activation of mitochondrial K ATP channels. Basic Res. Cardiol. 2009;104(2):121–129. doi: 10.1007/s00395-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wojtovich A.P., Brookes P.S. The endogenous mitochondrial complex II inhibitor malonate regulates mitochondrial ATP-sensitive potassium channels: implications for ischemic preconditioning. Biochim. Biophys. Acta Bioenerg. 2008;1777(7–8):882–889. doi: 10.1016/j.bbabio.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Davoudi M. A mouse model of mitochondrial complex III dysfunction induced by myxothiazol. Biochem. Biophys. Res. Commun. 2014;446(4):1079–1084. doi: 10.1016/j.bbrc.2014.03.058. [DOI] [PubMed] [Google Scholar]
- 282.Bergmann F., Keller B.U. Impact of mitochondrial inhibition on excitability and cytosolic Ca2+ levels in brainstem motoneurones from mouse. J. Physiol. 2004;555(1):45–59. doi: 10.1113/jphysiol.2003.053900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Veenman L. Potential involvement of F 0 F 1-ATP (synth) ase and reactive oxygen species in apoptosis induction by the antineoplastic agent erucylphosphohomocholine in glioblastoma cell lines. Apoptosis. 2010;15(7):753–768. doi: 10.1007/s10495-010-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Klawitter J. A relative L‐arginine deficiency contributes to endothelial dysfunction across the stages of the menopausal transition. Physiological reports. 2017;5(17) doi: 10.14814/phy2.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Liu C. Effects of a novel pharmacologic inhibitor of myeloperoxidase in a mouse atherosclerosis model. PloS One. 2012;7(12) doi: 10.1371/journal.pone.0050767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Chen S., Meng X.-F., Zhang C. Role of NADPH oxidase-mediated reactive oxygen species in podocyte injury. BioMed Res. Int. 2013;2013 doi: 10.1155/2013/839761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.