Abstract
The mitochondrial electron transport chain utilizes a series of electron transfer reactions to generate cellular ATP through oxidative phosphorylation. A consequence of electron transfer is the generation of reactive oxygen species (ROS), which contributes to both homeostatic signaling as well as oxidative stress during pathology. In this graphical review we provide an overview of oxidative phosphorylation and its inter-relationship with ROS production by the electron transport chain. We also outline traditional and novel translational methodology for assessing mitochondrial energetics in health and disease.
Keywords: Electron transport chain, Mitochondrial reactive oxygen species, Mitochondria, Oxidative phosphorylation
1. Introduction
In 1957, Peter Siekevitz branded the mitochondrion the “powerhouse” of the cell [1]. Less than a decade later came the first reports that the organelle generated reactive oxygen species (ROS) as a byproduct of cellular respiration [2]. Since then, it has become apparent that mitochondria are highly dynamic organelles that contribute to cellular homeostasis not only through maintaining adenosine triphosphate (ATP) levels, but also through the generation of low levels of ROS for cell signaling, and that dysfunction in either of these processes can propagate pathology. Notably, the importance of each of these functions varies by cell type. For example, cardiomyocytes rely on mitochondria to supply >95% of the energy required for their function [3]. In contrast, endothelial cells rely more heavily on glycolysis than mitochondria for ATP, but mitochondrial ROS production is essential for endothelial homeostatic signaling [4].
Despite this variation between cell types, mitochondrial ATP generation and ROS production are intimately linked through function of the electron transport chain (ETC), and thus efficient measurement of ETC function can provide insight into mechanisms of physiology and disease pathogenesis. In this review, we will provide an overview of the function of the ETC, focusing on oxidative phosphorylation and its relationship to ROS production. Further, we will compare current methodologies to measure bioenergetic function and mitochondrial ROS production.
2. Overview of oxidative phosphorylation
Cellular metabolism comprises the utilization of carbohydrates, fats, and proteins, to synthesize energy. The processes for the catabolism of glucose (via glycolysis and subsequent pyruvate oxidation), fatty acids (via fatty acid β-oxidation), and amino acids (via oxidative deamination and transamination) are reviewed in detail elsewhere [5]. However, the molecules derived from these processes are used in the tricarboxylic acid (TCA) cycle to generate substrates that enter the ETC for oxidative phosphorylation. Here we summarize the reactions that occur in the ETC to produce energy, but for a more detailed review, refer to Zhao et al. [6].
The ETC is embedded within the extensive inner membrane of the mitochondrion, in close proximity to the mitochondrial matrix in which the TCA cycle is localized (Fig. 1). NADH and FADH2 generated by the TCA cycle donate electrons to the ETC at either Complex I (NADH:ubiquinone oxidoreductase) or Complex II (succinate dehydrogenase), respectively (Fig. 1). The electrons from NADH are passed to ubiquinone (CoQ) through a chain of co-factors including a flavin mononucleotide (FMN) followed by seven low to high potential iron-sulfur (FeS) clusters in Complex I to enter the Q cycle, where CoQ is reduced to ubiquinol (QH2). This electron transfer induces the pumping of protons by Complex I from the matrix into the intermembrane space. Though the mechanism linking electron transfer to proton pumping remains unclear, one hypothesis speculates an indirect pumping of two protons via a conformation-coupled manner and the direct pumping of the other two protons via the ubiquinone redox reaction, while another hypothesis suggests that changes in the conformation and density of water in Complex I dictates the proton translocation [6]. Regardless of the exact mechanism, the transfer of two electrons from NADH results in the pumping of four protons.
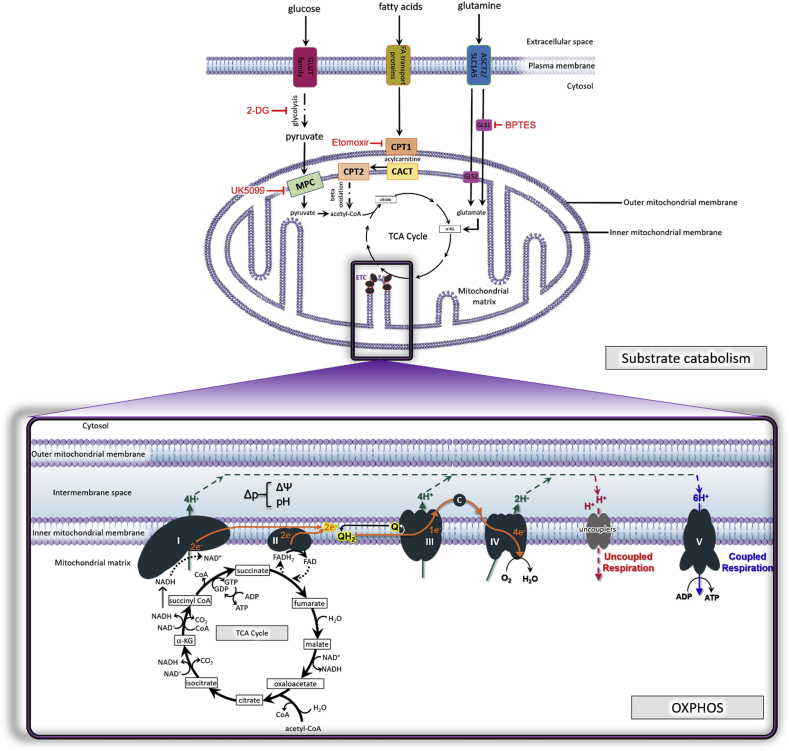
Fig. 1.
(A) Substrate supply for mitochondrial respiration. Glucose, fatty acids, and amino acids (glutamine shown here) undergo catabolism to feed into the tricarboxylic acid cycle (TCA cycle), which generates substrates for the electron transport chain (ETC). Glucose is metabolized by glycolysis. Red inhibitory signs denote pharmacologic agents utilized to inhibit specific sources of substrate in order to delineate substrate supply to the ETC, particularly in in vitro respirometric assays as outlined in the text. 2-deoxy-d-glucose (2-DG) inhibits glycolysis. Etomoxir inhibits fatty acid entry into the mitochondria through carnitine palmitoyltransferase 1 (CPT1). Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)-ethyl sulfide (BPTES) inhibits glutaminase (GLS) to inhibit glutamine metabolism to glutamate. 2-Cyano-3-(1-phenyl-1H-indol-3-yl)-2-propenoic acid (UK5099) inhibits the mitochondrial pyruvate carrier (MPC) to prevent pyruvate entry into the organelle. Within the mitochondrial matrix, acetyl-CoA is produced from pyruvate or from the beta-oxidation of fatty acids and serves as a point of entry to the TCA cycle. Citrate synthase converts acetyl-CoA and oxaloacetate to citrate. Citrate is converted to isocitrate by aconitase, and isocitrate is in turn oxidized into α-ketoglutarate, which reduces NAD+ to NADH. Next, α-ketoglutarate which is derived from the hydrolysis of extracellular amino acid glutamine to glutamate via the glutaminase (GLS) I and II pathways, enters the TCA cycle. α-ketoglutarate undergoes oxidative decarboxylation with NAD+ and CoA-SH (CoA not attached to an acyl group) to irreversibly form succinyl-CoA, NADH, CO2 and H+. The succinyl-CoA is then hydrolyzed to form succinate, CoA-SH and energy in the form of GTP. Succinate is oxidized to fumarate by Complex II/succinate dehydrogenase while converting FAD to FADH2. Hydration of fumarate by fumarase results in the formation of l-malate that then becomes oxidized to form oxaloacetate, NADH and H+. At this point the cycle is complete and the aldol condensation of oxaloacetate with acetyl CoA and water can restart the TCA cycle. Throughout the TCA cycle, the reduction of NAD+ to NADH is coupled to the release of CO2.
(B) Mitochondrial oxidative phosphorylation (OXPHOS). The mitochondrial electron transport chain (ETC) consists of five protein complexes integrated into the inner mitochondrial membrane. The TCA cycle in the mitochondrial matrix supplies NADH and FADH2 to the ETC, each of which donates a pair of electrons to the ETC via Complexes I and II respectively. The transfer of electrons from Complex I to the Q cycle results in a net pumping of 4 protons across the inner membrane into the intermembrane space (IMS). Of note, Complex II does not span the inner membrane and does not participate in proton translocation. The electrons from either Complex I (2 electrons) or Complex II (2 electrons one at a time) are donated to ubiquinone (Q) which is reduced to ubiquinol (QH2). Ubiquinol is oxidized by Complex III allowing one electron at a time to continue the journey through cytochrome c (c). For every electron transferred to cytochrome c, 2 protons (H+) are pumped into the IMS, resulting in 4H+ pumped into the IMS for every electron pair moved through the cycle. Cytochrome c transports electrons to Complex IV where molecular oxygen acts as a terminal electron acceptor and is reduced to water. The reduction of one molecule of O2 requires 4 electrons. The reduction of O2 to H2O results in the pumping of 4 protons to the IMS, but 2 protons are consumed in the process, netting a total of 2 H+ pumped into the IMS at Complex IV. The movement of protons from the mitochondrial matrix into the intermembrane space in response to electron transfer creates a protonmotive force (Δp), which is the proton concentration (pH) combined with the electrochemical proton gradient known as the mitochondrial membrane potential (ΔΨ). The membrane potential is dissipated by the re-entry of H+ back into the matrix through Complex V, which is coupled to the production of ATP from ADP. In contrast, uncoupled respiration due to proton leak is facilitated by adenine nucleotide translocase (ANT) and uncoupling proteins (UCP) and dissipates membrane potential at the expense of ATP production.
Abbreviations: protonmotive force, Δp; oxaloacetic acid, OAA, alpha-ketoglutarate, α-KG; carbon dioxide, CO2; reduced flavin adenine dinucleotide reduced FADH2; glutaminase, GLS; carnitine palmityl transferase, CPT; carnitine-acylcarnitine translocase, CACT; mitochondrial pyruvate carrier, MPT. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Electrons also enter the ETC through Complex II, which is a component of both the TCA cycle and the ETC. Electrons donated from FADH2 are transferred sequentially to CoQ via the FeS cluster of Complex II, in a similar manner as at Complex I. Unlike Complex I, electron transport at Complex II is not accompanied by proton translocation from the matrix to the intermembrane space [6].
Once in the Q-cycle, electrons are transferred to Complex III (coenzyme Q:cytochrome c reductase) and then to cytochrome c. First, QH2 binds to the cytoplasmic side of Complex III and releases two protons into the intermembrane space. One electron is transferred from QH2 to the catalytically active and high-potential iron-sulfur cluster in the Rieske center of Complex III, while the second electron is transferred to cytochrome b within the complex. This electron is then transferred from cytochrome b to a second molecule of Q that is bound to the matrix side of the complex to generate ubisemiquinone (Q·-). Concomitantly, the electron at the 2Fe–2S cluster is transferred to cytochrome c1, from where it is transferred to, and reduces the mobile carrier, cytochrome c. At this point, a second QH2 molecule binds to the membrane side of the complex and undergoes the same oxidation process to reduce Q·- back to QH2, complete the Q-cycle, and pump two more protons into the intermembrane space [6].
Once the mobile electron carrier cytochrome c is reduced, it ferries single electrons from Complex III to Complex IV (cytochrome c oxidase), where molecular oxygen binds and is reduced to water. Complex IV consists of three core subunits, namely, the central subunit I that contains the redox-active metal centers heme a (Fea) and a binuclear center composed of heme a3 (Fea3) and CuB, and flanking subunit II, that contains the other redox-active metal center, namely, CuA and subunit III that is involved in proton pumping. Reduced cytochrome c can simultaneously interact with subunit II of Complex IV, and transfer electrons to the CuA site of subunit II to then pass these electrons from heme a to the binuclear center of subunit I, where O2 binds and is reduced to H2O. Therefore, at Complex IV, a total of eight protons are pumped from the matrix, of which, four are used to form two water molecules, and the other four are transferred into the intermembrane space. This process of oxygen consumption is known as mitochondrial respiration [6].
In response to electron transport, a total of ten protons (H+) (two from Complex III, and four from each Complex I and Complex IV) are pumped from the matrix into the intermembrane space, where they accumulate to generate an electrochemical proton gradient known as the mitochondrial membrane potential (ΔΨ). ΔΨ combined with proton concentration (pH) generates a protonmotive force (Δp) which is an essential component in the process of energy storage during OXPHOS since it couples electron transport (complexes I-IV) (and oxygen consumption) to the activity of Complex V (ATP synthase), where protons re-enter the matrix to dissipate the proton gradient. Complex V is a multi-subunit complex comprised of two distinct domains, extra-membranous (termed F1) and transmembrane (termed FO), and functions under a rotational motor mechanism to allow for ATP production. Proton movement through FO from the intermembrane space is coupled to the rotation that results in the addition of a phosphate to adenosine diphosphate (ADP) to synthesize adenosine triphosphate (ATP) at sites in F1 (Fig. 1) [6].
3. Generation of superoxide from the ETC
It is now well established that the mitochondrion is a significant source of cellular ROS (Fig. 2). While multiple sites of ROS production have been identified in the organelle and reviewed in detail elsewhere [7], this section will focus on generation of ROS by the complexes of the ETC and its interplay with cellular respiration and energetics. The predominant route of ROS production by the ETC is the premature leak of electrons from complexes I, II, and III to mediate the one electron reduction of oxygen to superoxide (O2•−), which can then be dismutated to hydrogen peroxide (H2O2) [6]. The rate of O2•− production from the ETC is dependent on the concentration of the one-electron donor at a particular site and the rate at which this redox active donor reacts with oxygen O2 [8]. Importantly, factors such as local O2 tension, Δp, electron flux, and ATPase activity can change these factors to dynamically modulate O2•− from the ETC, and these factors will be considered along with a brief explanation of the mechanisms of ETC O2•− generation at each complex.
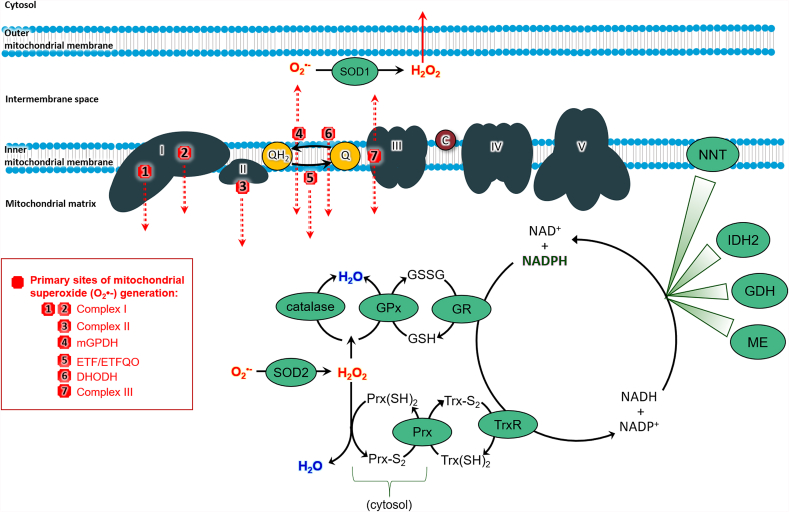
Fig. 2.
Sites of mitochondrial ROS production and antioxidant systems. Seven major ETC sites of ROS generation are shown here, with red lines indicating on which side of the membrane the ROS are formed. Superoxide (O2•–) is the primary ROS generated by the ETC. O2•− is dismutated to H2O2 by MnSOD in the matrix and Cu/Zn SOD in the intermembrane space. Also within the matrix is the glutathione peroxidase (GPx)/glutathione reductase (GR) system and catalase, found in liver and cardiac tissues. The peroxiredoxin (Prx)/thioredoxin (TrxR) system overlaps between the cytosol and the matrix. NADPH/NADP + renews these antioxidant systems with its reducing potential. Matrix enzymes (malic enzyme (ME), glutamate dehydrogenase (GDH), and isocitrate dehydrogenase (IDH2)) and the inner membrane-associated nicotinamide nucleotide transhydrogenase (NNT) regenerate the pool of NADPH.
Abbreviations: mitochondrial glycerol-3-phosphate dehydrogenase, mGPDH; dihydroorotate dehydrogenase, DHODH, electron transfer flavoprotein oxidoreductase, ETFQO; superoxide, O2•-; hydrogen peroxide, H2O2; water, H2O; glutathione, GSH; glutathione disulfide, GSSG; glutathione peroxidase, GPx; GR; thioredoxin reductase,TR; reduced thioredoxin, Trx (SH)2; oxidized thioredoxin Trx-S2; peroxiredoxin, Prx; reduced peroxiredoxin, Prx (SH)2; oxidized peroxiredoxin, Prx-S2; malic enzyme, ME; glutamate dehydrogenase, GDH; NADP+-dependent isocitrate dehydrogenase, IDH2, and nicotinamide nucleotide transhydrogenase, NNT. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Two sites of O2•− generation have been identified at Complex I - 1) the FMN cofactor which accepts electrons from NADH and 2) the Q binding site at which two electrons are transferred the terminal Fe–S to Q. With forward electron transport, electrons from NADH fully reduce the FMN center, which reacts with oxygen to generate O2•− [8]. Thus, O2•− production at this center is regulated by the ratio of NADH to NAD+ and in physiological conditions is relatively low. However, conditions leading to the accumulation of NADH, such as low ATP demand resulting in decreased respiration or damage to the ETC, lead to greater reduction of the FMN and increased O2•− production at this site. Notably, the Complex I inhibitor rotenone, which binds to the Q binding site, stimulates O2•− production by potentiating electron accumulation and reduction of the FMN site. A second mechanism by which Complex I generates O2•− is dependent on reverse electron transport (RET) [9]. RET occurs when the Q pool is highly reduced and Δp is high. In these conditions, the energy is high enough to drive electron transport against the redox potential gradient of the ETC and electrons are driven back from QH2 into Complex I. Superoxide generated by RET is thought to occur at the Q binding site of Complex I. This is supported by the fact that RET-dependent O2•− production is abolished by rotenone. Further, this mechanism of ROS production is highly sensitive to minimal changes in pH. Given that proton pumping is linked with the Q binding site, it is likely that Q•− is formed during proton pumping and RET, leading to the O2•− generation observed. While RET-induced ROS production was once thought to be a strictly in vitro phenomenon, it is now known to contribute to the pathogenesis of bacterial sepsis, as well as physiological hypoxia-sensing in the carotid body [10].
It is important to note that in physiological conditions, when respiration and ATP production are high (and Δp is low), Complex I ROS production is low. While early studies suggested that physiological ROS production by Complex I was high, it is now recognized that much of the ROS measured in those studies may have originated from flavin-dependent dehydrogenases that utilize the NADH/NAD+ pool and operate at a similar redox potential as Complex I. These enzymes, which include α-ketoglutarate dehydrogenase, pyruvate dehydrogenase, and branched chain 2-oxoacid dehydrogenase are now known to be significant generators of ROS and generate O2•− in the matrix of the mitochondrion (similarly to both sites of the Complex I) (Fig. 2) [11]. However, under conditions of high NADH/NAD+ or high QH2/Q and elevated Δp, O2•− production is significantly enhanced by FMN reduction and RET respectively at Complex I, significantly out-producing these dehydrogenase complexes.
Complex III, specifically the site of QH2 oxidation, is also a generator of O2•− within the ETC. As described above, the two electrons carried by QH2 when it binds Complex III are transferred from QH2 to the Rieske Fe–S center and then to cytochrome c in sequence. The transfer of one electron with the other remaining on Q favors the formation of the unstable Q•− species, which can react with oxygen to produce O2•−. The rate of ROS production at this site is physiologically low, and favored when the site is only partially reduced, which occurs when substrate is at submaximal levels. Alternatively, inhibition of the complex downstream of Q•− also potentiates ROS production [11]. For example, treatment with the Complex III inhibitor Antimycin A binds the Qi site of the complex, stabilizing the semiquinone radical in the Qo site. Addition of Qo inhibitors such as myxathiazol completely inhibit ROS production by Complex III [12]. Similarly, to dehydrogenases that contribute to the NADH/NAD+ pool, several enzymes responsible for substrate catabolism and operate at the same redox potential as Complex III are generators of O2•−. These enzymes include mitochondrial glycerol-3-phosphate dehydrogenase (mGPDH), the electron transferring flavoprotein/ETF:ubiquinone oxidoreductase system, and dihydroorotate dehydrogenase (DHODH), all of which donate electrons to the CoQ pool. Of note, ROS generated by these enzymes as well as complex III are released on both the matrix and intermembrane side of the mitochondrion (Fig. 2) [11].
While Complex II is not generally considered a major source of ROS in comparison to Complexes I and III, several reports describe O2•− production by the complex. While there is some debate surrounding the exact site of ROS production in the complex, Brand and colleagues have demonstrated that the flavin site, at which FAD binds the active site of the enzyme, is responsible for the production. These studies demonstrate that in isolated mitochondria ROS generation by Complex II is highly regulated by succinate concentration, with a bell-shaped response in which ROS production is optimized when succinate concentration is not too high or too low. Moreover, the Q pool must be highly reduced such as what is observed with Complex III inhibition [11]. While the exact mechanism by which Complex II generates ROS is being elucidated, its contribution to overall mitochondrial ROS production in physiological conditions remains unknown.
Early studies estimated that 1–2% of electrons entering the ETC contributed to O2•− production. It is now appreciated that the rate of mitochondrial ROS generation is likely lower than this value in vivo [13] and fluctuates depending on ETC function. As outlined above, each site of mitochondrial ROS production is differentially sensitive to factors such as substrate supply, rate of ATP production and Δp depending on how these conditions affect the reduction of particular site of ROS production. Importantly, discussion of the impact of these factors assumes high efficiency of oxidative phosphorylation in which respiration is “coupled” to ATP generation by Δp and the membrane potential (Δψm) is maintained within a homeostatic range (140–160 mV) [14]. Physiologically, oxidative phosphorylation is not completely coupled, and low levels of proton leak dissipate Δp and decrease Δψ, which can attenuate ROS production. The level of basal proton leak is determined by inner membrane structure and composition as well as proteins such as the adenine nucleotide translocase (ANT), which regulates the Δψm– driven exchange of ATP for ADP across the inner membrane, but also is responsible for over half of basal proton leak [6]. In addition to ANT, the mitochondrial ATP-sensitive potassium channel (mitoKATP) also mediates uncoupling in response to high concentrations of superoxide and H2O2. Free fatty acids can also act as uncouplers, though this function appears to be distinct from their ability to act as substrate to fuel respiration [15]. Beyond these proteins and substrates that are constitutive to the mitochondrion, expression of specific uncoupling proteins (UCP) within the inner mitochondrial membrane facilitate the leak of protons from the intermembrane space back to the matrix without involvement of the ATP synthase. Expression of these UCP proteins is induced by many stimuli, including inner membrane hyperpolarization and high levels of ROS. Expression of UCP1 was first identified in brown adipose tissue, in which uncoupling of oxidative phosphorylation generates heat [16]. However, it is now known that at least five different UCP isoforms exist and expression of these proteins are implicated in a variety of physiological signaling pathways ranging from T-cell maturation [17] and neuroprotection [18], to mediating the protective effects of caloric restriction [19]. For an in-depth review of UCP function see Ref. [15].
A final consideration in the factors regulating O2•− production from the ETC is the availability of oxygen as an electron acceptor. While decreased availability of oxygen should attenuate O2•− production, a number of studies in cell systems demonstrate that mitochondrial ROS production is paradoxically increased in hypoxic conditions [12,20]. Several hypotheses have been proposed to explain this effect including the notion that oxygen limitation at Complex IV causes accumulation of electrons and increased reduction of Complexes I-III. However, given the low Km of Complex IV for O2 (<1 μM), limitation at Complex IV should also limit oxygen available for reduction to O2•− [21]. More recent studies have suggested that hypoxia induces a conformational change in Complex III such that Q•− is stabilized or that there is more access of O2 to the Q•− [12,20]. Notably, Brookes and colleagues showed in controlled experiments utilizing isolated mitochondria and fixed substrate concentration that unlike in cells, ROS production by the ETC decreased as O2 concentrations were lowered below ~1 μM O2 [21]. These data demonstrate a discrepancy between the effect of hypoxia in isolated mitochondria and cell systems, and suggest that cellular factors outside the mitochondrion may regulate ETC ROS production during hypoxia in cells. These factors may include nitric oxide (NO) production, which regulates the ETC at Complex V [22], changes in substrate entry into the ETC or utilization, or cellular kinase activity that potentially regulates ETC activity through phosphorylation [23].
4. Mitochondrial antioxidant systems and ROS-mediated signaling
While the mitochondrion is a site for ROS production, antioxidant systems contribute to the regulation of the concentration and redox species in the organelle. Though the proximal ROS generated by the ETC is O2•−, this species is rapidly dismutated to H2O2 by manganese superoxide dismutase (MnSOD) localized in the mitochondrial matrix and low concentrations of copper/zinc SOD located in the intermembrane space [6]. Notably, unlike O2•−, H2O2 is stable and uncharged, and thus able to leave the mitochondrion to mediate cytosolic cell signaling. This signaling occurs predominantly through the oxidation of either metal cofactors or reduced thiols on cytosolic proteins, changing their function. It is now well recognized that mitochondrial H2O2 regulates a number of signaling pathways by this mechanism. While discussion of each of these specific pathways is outside the scope of this article, we direct the reader to reviews of the essential role of mitochondrial ROS signaling in hypoxic adaptation [20,24], apoptosis [25], regulation of phosphorylation signaling [26,27], and cell growth and differentiation [28,29].
The concentration of H2O2 that leaves the mitochondrion is regulated by two main antioxidant systems in the mitochondrial matrix: the glutathione and thioredoxin/peroxiredoxin systems (Fig. 2). Glutathione (GSH) is oxidized by H2O2 to form the glutathione disulfide (GSSG), a reaction catalyzed by glutathione peroxidase (GPx). GSSG is then reduced back to GSH by Glutathione Reductase (GR). Similarly, H2O2 oxidizes a cysteine residue in the catalytic site of peroxiredoxin (PRx), which forms a disulfide bridge with a neighboring cysteine. The reductive power of thioredoxin (TRx) reduces PRx through a disulfide exchange reaction and TRx is then reduced by Thioredoxin reductase (TR). Importantly, both GR and TR require NADPH for their reductive activity. The pool of reduced NADPH is maintained by several enzymes in the mitochondrial matrix including malic enzyme (ME), glutamate dehydrogenase (GDH), and isocitrate dehydrogenase (IDH2) [30]. Additionally, membrane-associated nicotinamide nucleotide transhydrogenase (NNT) is particularly important in maintaining NADPH pools through its function of pumping protons into the matrix to regenerate NADPH by coupling oxidation of NADH to the reduction of NADP+. Importantly, the ability of NNT to renew the pool of available NADPH is dependent on ΔΨ. States of low ΔΨ decrease the amount of available reduced NADPH which then decreases the amount of reduced glutathione and thioredoxin to buffer H2O2.
5. Measurement of mitochondrial bioenergetics in human disease
In the previous sections we have outlined the mechanisms by which the ETC functions to generate ATP and produce ROS, highlighting the interplay between these two processes. While physiological function of the ETC generates ROS at levels required for homeostatic signaling, ETC dysfunction, particularly that which decreases mitochondrial energy production and enhances ROS generation, has been linked to the onset and development of a number of biological changes including obesity and aging, and pathology in all organ systems including cardiovascular diseases. Thus, accurate and efficient assessment of energetic function and mitochondrial ROS production in humans could potentially enable the diagnosis of disease and the mechanistic understanding of pathogenesis. In this section, we focus on a translational approach to the measurement of ETC function. Currently, no standard methodologies exist to directly measure mitochondrial ROS production in humans. Thus, we will give an overview of methodology to assess mitochondrial energetic function in humans.
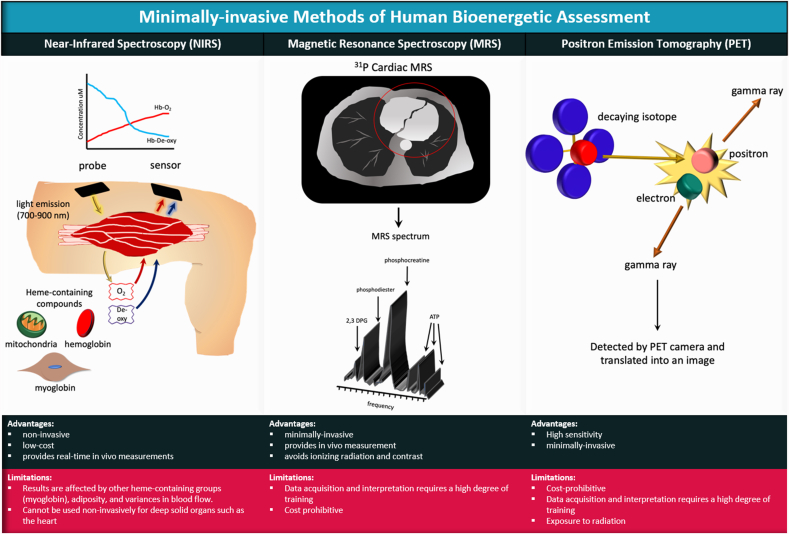
A number of non- and minimally-invasive technologies such as near-infrared spectroscopy (NIRS), magnetic resonance spectroscopy (MRS), and positron emission tomography (PET) can be utilized to indirectly assess mitochondrial function in humans (Fig. 3). Near-infrared spectroscopy capitalizes on the principle that near-infrared light is absorbed at differential wavelengths by the oxygenated and deoxygenated heme groups of hemoglobin and myoglobin, allowing the calculation of tissue oxygen consumption, which is proportional to mitochondrial respiration. While NIRS is relatively inexpensive, one major disadvantage of this method is that near infrared light applied non-invasively cannot reach solid organs such as the heart, and thus is best utilized for exercising large muscles closer to the skin [31]. Magnetic resonance spectroscopy is a non-invasive and ionizing-radiation-free method that can be used in the heart and other solid organs to measure the presence of metabolites such as choline-containing compounds, creatine, glucose, alanine and lactate by measuring the resonance of MR-visible isotopes [32]. 31Phosphorous (31P) is most frequently utilized to visualize concentrations of ATP, phosphocreatine, and inorganic phosphate, which can provide a dynamic measure of ATP production in the tissue. 13Carbon (13C) and proton NMR (1H) can also be used to measure intermediates in the TCA cycle [33,34]. In contrast to NMR, PET imaging employs the detection of injected positron-emitting radionuclide tracers to measure the accumulation or consumption of metabolic intermediates [35,36]. While PET and MRS provide a solid assessment of energetics, both techniques require costly equipment and a high degree of training for data acquisition and analysis. In addition, PET has the added obstacle of radiopharmaceutical production and the exposure of patients to radiation. Further, these technologies are highly specialized and not suited for high throughput clinical application, limiting their mainstream use.
Fig. 3.
Minimally-invasive methods of human bioenergetic assessment. Near Infrared Spectroscopy (NIRS) operates on the principle that near-infrared light (700–900 nm) penetrates tissue with little scatter and is absorbed by heme-containing groups (e.g., hemoglobin, myoglobin, and the heme-containing prosthetic groups within the mitochondrial electron chain complexes) in an oxygen-dependent manner. Measurement of changes in absorbance can be used to reflect changes in tissue oxygenation and mitochondrial oxidative capacity. Magnetic Resonance Spectroscopy (MRS) utilizes magnetic resonant-visible isotopes (eg, 1H, 31P and 13C), with each resonating at characteristic frequencies within a magnetic field. The distinct resonance of known isotopes creates a signature for identification of compounds within living tissues. MRS can distinguish products of glycolysis, creatine metabolism, choline metabolism, and amino acid metabolism. Positron Emission Tomography (PET) utilizes a radioactive isotope tracer such as glucose analogue 18-fluorodeoxyglucose ([18F]FDG) or other radio-labeled metabolites (acetate, choline, methionine or glutamine). The tracer is administered intravenously and becomes trapped within metabolically active cells. The nucleus of the isotope emits positrons as it decays. The positrons make contact with electrons to generate high-energy photons (gamma rays) that are detected by the PET camera and translated into an electrical signal to produce an image. The image that is produced is dependent on the metabolism of the cells/tissues that take up the isotope.
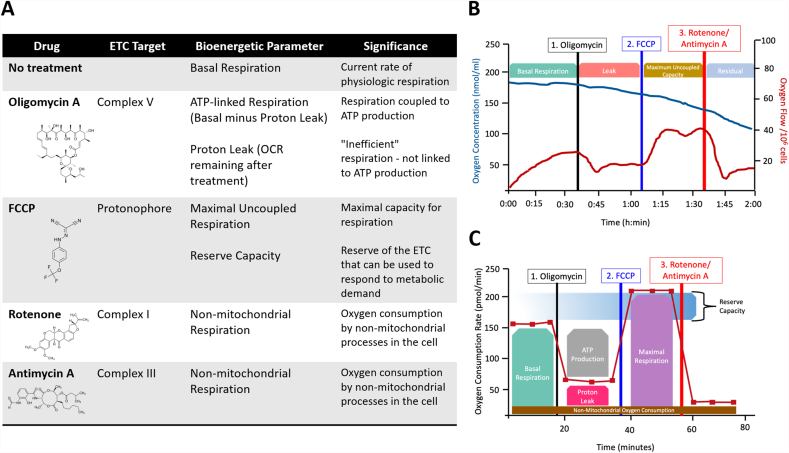
The gold standard for the direct assessment of mitochondrial ETC function in humans is the measurement of mitochondrial function in small biopsies of tissue [37,38]. Once obtained, biopsies are most frequently subjected to measurement of mitochondrial oxygen consumption, which provides information on multiple facets of ETC function. The Clark-type oxygen electrode, developed in the 1960's, enabled the polarographic measurement of oxygen consumption rate (OCR) by isolated mitochondria or cells in a sealed chamber. This technology was later utilized by the Oroboros O2k system to provide amperometric measurements of O2 consumption with maximal sensitivity and precision. In addition, the Oroboros O2k contains closed, air-tight reaction chambers that minimize O2 back-diffusion, thus limiting over estimations of OCR. Further, it allows the measurement of OCR at fixed oxygen tensions and the potential for simultaneous measurement of multiple parameters including pH, Δψm, [Ca2+], and NO in the same chamber. More recently, development of the Seahorse extracellular flux (XF) analyzer, has enabled the measurement of OCR in an intact monolayer of cells in a high throughput multi-well format. The Seahorse analyzer simultaneously measures extracellular pH to enable the estimation of glycolytic rate. Despite key differences between the two systems, such as a greater sample size required for the Oroboros and the need for a monolayer for the Seahorse versus sample in suspension for the Oroboros, both systems can be used with a series of pharmacologic mitochondrial modulators to provide a profile of ETC function (Fig. 4). In a typical bioenergetic profile, basal OCR is measured to determine the current turnover of the ETC. This is followed by the addition of oligomycin A, an inhibitor of the ATP synthase, in order to measure OCR that is not contributing to ATP production (proton leak). A protonophore, such as trifluoro-methoxy -carbonyl cyanide-4-phenylhydrazone (FCCP) is then added to uncouple respiration, yielding the maximal rate of OCR. Finally, a mitochondrial inhibitor is added such that the rate of non-mitochondrial OCR is revealed [39]. Notably, if intact cells are being utilized, pharmacologic inhibitors of glucose, fatty acid, or amino acid oxidation/entry into the mitochondrion can be utilized (as shown in Fig. 1) to determine the source of substrate for the ETC.
Fig. 4.
Respirometric methods of bioenergetic assessment. Panel (A) shows the common pharmacologic modulators of the respiratory chain utilized to generate a bioenergetic profile (along with their chemical structures), the target of these modulators, the bioenergetic parameter measured in their presence, and the significance of each parameter. (B) A typical bioenergetic profile generated in the Oroboros system showing oxygen concentration in the chamber over time (blue trace) and the calculated oxygen flux (red trace). (C) A typical bioenergetic profile generated by the Seahorse XF analyzer in which oxygen consumption rate (OCR) over time is shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
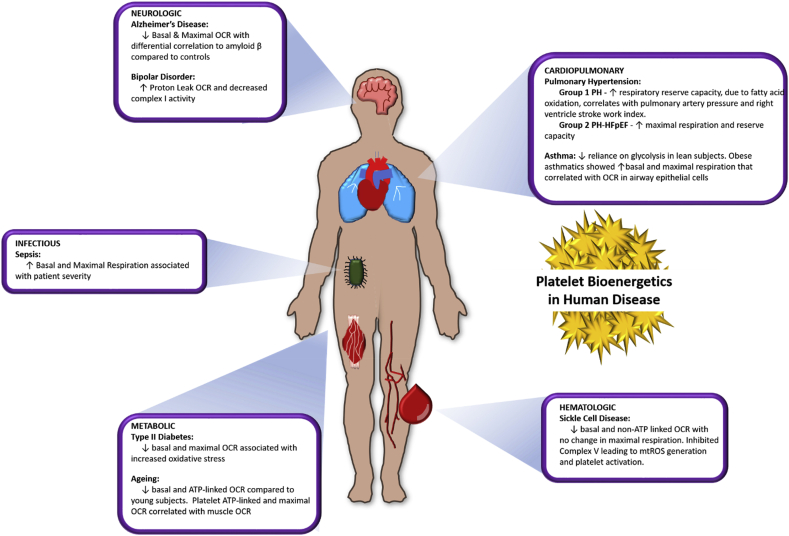
While the respirometric techniques described above can be applied to any cell type, traditionally for human studies, skeletal muscle biopsies or cells cultured from skin grafts have been most frequently used as a source of viable tissue/mitochondria. However, the invasiveness of obtaining these samples is prohibitive of their widespread use for routine mitochondrial measurement. Recently, a number of labs have advanced the concept of performing respirometric measures in circulating blood cells, including platelets and peripheral blood mononuclear cells, as an assessment of systemic bioenergetic function [[40], [41], [42]]. Blood cells contain fully functional mitochondria, and are abundant, self-renewing, and minimally invasive to obtain. Our group and others have successfully employed Seahorse XF and/or Oroboros to circulating cells to demonstrate that this methodology is feasible [43,44], reproducible in human populations [40], and that bioenergetics of circulating cells reflect bioenergetics in solid tissues such as the heart [42], skeletal muscle [42,45], brain [46], and lungs [47]. Further, these studies demonstrate alterations in blood cell bioenergetics in a number of natural biological conditions (e.g aging) or pathologies, and correlate with physical or clinical parameters of these conditions [45,[47], [48], [49]]. Altered platelet bioenergetics have been measured in neurologic [46], metabolic [46], hematologic [44], cardiopulmonary [47,49,50], and infectious diseases [51] (Fig. 5).
Fig. 5.
Changes in platelet bioenergetics in human disease. The bioenergetic profiles of platelets reflect alterations that occur at a systemic level during human disease states. Bioenergetic profiles have been measured in platelets isolated from patients with a wide array of diagnosed diseases. This figure demonstrates published platelet bioenergetic alterations in neurologic disease including Alzheimer's disease [53] and bipolar disorder [54], metabolic changes including type II diabetes [55] and advanced age [45], cardiopulmonary diseases including pulmonary hypertension [50,56] and asthma [47,57], sickle cell disease as a hematologic disease [44], and infectious disease such as sepsis [58].
One example of platelet alterations in cardiovascular disease is in sickle cell disease in which a bioenergetic “screen” of platelets from a cohort of sickle cell patients demonstrated that basal OCR was decreased but maximal uncoupled OCR was unchanged compared to age and race matched healthy subjects [44]. This pattern of alteration potentially suggested that while the capacity for respiration was unchanged in sickle cell patients, Δψm was higher (leading to decreased basal OCR). Direct measurement of Δψm confirmed this hypothesis, and further measurement of individual complex activities demonstrated the increased Δψm was due to Complex V inhibition and resulted in increased O2•− production. This production of mitochondrial O2•− is linked to platelet activation in sickle cell patients (REF 44) and the measurement of Complex V activity and markers of ROS in platelets from these patients can now be utilized to not only elucidate the pathogenesis of the disease but also test the ability of potential therapeutics to attenuate this mitochondrial-driven oxidative pathogenesis [52]. These data demonstrate the potential for the measurement of ETC function to reveal changes in both mitochondrial energetic and redox function and highlight the potential use of blood bioenergetic measurements as a translational and clinical tool.
In summary, the mitochondrial ETC is a redox hub that regulates cellular homeostasis through the production of ATP and the generation of ROS, two processes that are intimately linked. Given that alterations in both of these processes have been associated with the pathogenesis of a myriad of diseases spanning all organ systems, the ability to accurately and efficiently measure ETC function in humans may provide useful diagnostic and mechanistic information.
Funding sources
This work was supported by NIH RO1 HL133003-01A1, funds from the Hemophilia Center of Western Pennsylvania to SS and AHA 19POST34380956 to ACB, and funds from UPMC Children’s Trust Young Investigator Foundation Fund 020284 to DND
Declaration of competing interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101674.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.P S. Powerhouse of the cell. Sci. Am. 1957;197(1):131–140. [Google Scholar]
- 2.Jensen P.K. Antimycin-insensitive oxidation of succinate and reduced nicotinamide-adenine dinucleotide in electron-transport particles. II. Steroid effects. Biochim. Biophys. Acta. 1966;122(2):167–174. doi: 10.1016/0926-6593(66)90058-0. [DOI] [PubMed] [Google Scholar]
- 3.Ventura-Clapier R. Bioenergetics of the failing heart. Biochim. Biophys. Acta. 2011;1813(7):1360–1372. doi: 10.1016/j.bbamcr.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Widlansky M.E., Gutterman D.D. Regulation of endothelial function by mitochondrial reactive oxygen species. Antioxidants Redox Signal. 2011;15(6):1517–1530. doi: 10.1089/ars.2010.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stryer L. fourth ed. W.H. Freeman; New York: 1995. Biochemistry; p. 1064. xxxiv. [Google Scholar]
- 6.Zhao R.Z. Mitochondrial electron transport chain, ROS generation and uncoupling (Review) Int. J. Mol. Med. 2019;44(1):3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kowaltowski A.J. Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 2009;47(4):333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robb E.L. Control of mitochondrial superoxide production by reverse electron transport at complex I. J. Biol. Chem. 2018;293(25):9869–9879. doi: 10.1074/jbc.RA118.003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scialo F., Fernandez-Ayala D.J., Sanz A. Role of mitochondrial reverse electron transport in ROS signaling: potential roles in health and disease. Front. Physiol. 2017;8:428. doi: 10.3389/fphys.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong H.S. Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J. Biol. Chem. 2017;292(41):16804–16809. doi: 10.1074/jbc.R117.789271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzy R.D., Schumacker P.T. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp. Physiol. 2006;91(5):807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 13.Boveris A., Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem. J. 1973;134(3):707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dzbek J., Korzeniewski B. Control over the contribution of the mitochondrial membrane potential (DeltaPsi) and proton gradient (DeltapH) to the protonmotive force (Deltap). In silico studies. J. Biol. Chem. 2008;283(48):33232–33239. doi: 10.1074/jbc.M802404200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demine S., Renard P., Arnould T. Mitochondrial uncoupling: a key controller of biological processes in physiology and diseases. Cells. 2019;8(8) doi: 10.3390/cells8080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri L. Uncoupling protein 2 regulates metabolic reprogramming and fate of antigen-stimulated CD8+ T cells. Cancer Immunol. Immunother. 2016;65(7):869–874. doi: 10.1007/s00262-016-1851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normoyle K.P. The emerging neuroprotective role of mitochondrial uncoupling protein-2 in traumatic brain injury. Transl. Neurosci. 2015;6(1):179–186. doi: 10.1515/tnsci-2015-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asami D.K. Effect of aging, caloric restriction, and uncoupling protein 3 (UCP3) on mitochondrial proton leak in mice. Exp. Gerontol. 2008;43(12):1069–1076. doi: 10.1016/j.exger.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waypa G.B., Schumacker P.T. Oxygen sensing in hypoxic pulmonary vasoconstriction: using new tools to answer an age-old question. Exp. Physiol. 2008;93(1):133–138. doi: 10.1113/expphysiol.2007.041236. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman D.L., Salter J.D., Brookes P.S. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: implications for hypoxic cell signaling. Am. J. Physiol. Heart Circ. Physiol. 2007;292(1):H101–H108. doi: 10.1152/ajpheart.00699.2006. [DOI] [PubMed] [Google Scholar]
- 22.Mateo J. Regulation of hypoxia-inducible factor-1 alpha by nitric oxide through mitochondria-dependent and -independent pathways. Biochem. J. 2003;376(Pt 2):537–544. doi: 10.1042/BJ20031155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L., Lemasters J.J. Dephosphorylation of the Rieske iron-sulfur protein after induction of the mitochondrial permeability transition. Biochem. Biophys. Res. Commun. 2005;334(3):829–837. doi: 10.1016/j.bbrc.2005.06.170. [DOI] [PubMed] [Google Scholar]
- 24.Taylor C.T., Moncada S. Nitric oxide, cytochrome C oxidase, and the cellular response to hypoxia. Arterioscler. Thromb. Vasc. Biol. 2010;30(4):643–647. doi: 10.1161/ATVBAHA.108.181628. [DOI] [PubMed] [Google Scholar]
- 25.Sabharwal S.S., Schumacker P.T. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat. Rev. Canc. 2014;14(11):709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamanaka R.B., Chandel N.S. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 2010;35(9):505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163(3):560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer H., Wartenberg M., Hescheler J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell. Physiol. Biochem. 2001;11(4):173–186. doi: 10.1159/000047804. [DOI] [PubMed] [Google Scholar]
- 29.Kim K.H. Potential mechanisms for the inhibition of tumor cell growth by manganese superoxide dismutase. Antioxidants Redox Signal. 2001;3(3):361–373. doi: 10.1089/15230860152409013. [DOI] [PubMed] [Google Scholar]
- 30.Mailloux R.J. Mitochondrial antioxidants and the maintenance of cellular hydrogen peroxide levels. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/7857251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willingham T.B., McCully K.K. In vivo assessment of mitochondrial dysfunction in clinical populations using near-infrared spectroscopy. Front. Physiol. 2017;8:689. doi: 10.3389/fphys.2017.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layec G. Reproducibility assessment of metabolic variables characterizing muscle energetics in vivo: a 31P-MRS study. Magn. Reson. Med. 2009;62(4):840–854. doi: 10.1002/mrm.22085. [DOI] [PubMed] [Google Scholar]
- 33.Befroy D.E. Assessment of in vivo mitochondrial metabolism by magnetic resonance spectroscopy. Methods Enzymol. 2009;457:373–393. doi: 10.1016/S0076-6879(09)05021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagana Gowda G.A., Abell L., Tian R. Extending the scope of (1)H NMR spectroscopy for the analysis of cellular coenzyme A and acetyl coenzyme A. Anal. Chem. 2019;91(3):2464–2471. doi: 10.1021/acs.analchem.8b05286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen I.Y., Wu J.C. Cardiovascular molecular imaging: focus on clinical translation. Circulation. 2011;123(4):425–443. doi: 10.1161/CIRCULATIONAHA.109.916338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al Badarin F.J., Malhotra S. Diagnosis and prognosis of coronary artery disease with SPECT and PET. Curr. Cardiol. Rep. 2019;21(7):57. doi: 10.1007/s11886-019-1146-4. [DOI] [PubMed] [Google Scholar]
- 37.Bharadwaj M.S. Preparation and respirometric assessment of mitochondria isolated from skeletal muscle tissue obtained by percutaneous needle biopsy. JoVE. 2015;(96) doi: 10.3791/52350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doerrier C. High-resolution FluoRespirometry and OXPHOS protocols for human cells, permeabilized fibers from small biopsies of muscle, and isolated mitochondria. Methods Mol. Biol. 2018;1782:31–70. doi: 10.1007/978-1-4939-7831-1_3. [DOI] [PubMed] [Google Scholar]
- 39.Salabei J.K., Gibb A.A., Hill B.G. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 2014;9(2):421–438. doi: 10.1038/nprot.2014.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braganza A., Annarapu G.K., Shiva S. Blood-based bioenergetics: an emerging translational and clinical tool. Mol. Aspect. Med. 2020;71 doi: 10.1016/j.mam.2019.100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chacko B.K. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clin. Sci. (Lond.) 2014;127(6):367–373. doi: 10.1042/CS20140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyrrell D.J. Blood cell respirometry is associated with skeletal and cardiac muscle bioenergetics: implications for a minimally invasive biomarker of mitochondrial health. Redox Biol. 2016;10:65–77. doi: 10.1016/j.redox.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chacko B.K. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Lab. Invest. 2013;93(6):690–700. doi: 10.1038/labinvest.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardenes N. Platelet bioenergetic screen in sickle cell patients reveals mitochondrial complex V inhibition, which contributes to platelet activation. Blood. 2014;123(18):2864–2872. doi: 10.1182/blood-2013-09-529420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braganza A.C. Platelet bioenergetics correlate with muscle energetics and are altered in older adults. JCI Insight. 2019;5 doi: 10.1172/jci.insight.128248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyrrell D.J. Blood-Based bioenergetic profiling reflects differences in brain bioenergetics and metabolism. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/7317251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winnica D. Bioenergetic differences in the airway epithelium of lean versus obese asthmatics are driven by nitric oxide and reflected in circulating platelets. Antioxidants Redox Signal. 2019;31(10):673–686. doi: 10.1089/ars.2018.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyrrell D.J. Respirometric profiling of muscle mitochondria and blood cells are associated with differences in gait speed among community-dwelling older adults. J Gerontol A Biol Sci Med Sci. 2015;70(11):1394–1399. doi: 10.1093/gerona/glu096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen Q.L. Platelets from pulmonary hypertension patients show increased mitochondrial reserve capacity. Jci Insight. 2017;2(5) doi: 10.1172/jci.insight.91415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen Q.L. Alterations in platelet bioenergetics in Group 2 PH-HFpEF patients. PLoS One. 2019;14(7) doi: 10.1371/journal.pone.0220490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternfeld T. Mitochondrial membrane potential and apoptosis of blood mononuclear cells in untreated HIV-1 infected patients. HIV Med. 2009;10(8):512–519. doi: 10.1111/j.1468-1293.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 52.Morris C.R. Impact of arginine therapy on mitochondrial function in children with sickle cell disease during vaso-occlusive pain. Blood. 2020 doi: 10.1182/blood.2019003672. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fisar Z. Plasma amyloid beta levels and platelet mitochondrial respiration in patients with Alzheimer's disease. Clin. Biochem. 2019;72:71–80. doi: 10.1016/j.clinbiochem.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Zverova M. Disturbances of mitochondrial parameters to distinguish patients with depressive episode of bipolar disorder and major depressive disorder. Neuropsychiatric Dis. Treat. 2019;15:233–240. doi: 10.2147/NDT.S188964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avila C. Platelet mitochondrial dysfunction is evident in type 2 diabetes in association with modifications of mitochondrial anti-oxidant stress proteins. Exp. Clin. Endocrinol. Diabetes. 2012;120(4):248–251. doi: 10.1055/s-0031-1285833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen Q.L. Platelets from pulmonary hypertension patients show increased mitochondrial reserve capacity. JCI Insight. 2017;2(5) doi: 10.1172/jci.insight.91415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu W. Platelets from asthmatic individuals show less reliance on glycolysis. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sjovall F. Temporal increase of platelet mitochondrial respiration is negatively associated with clinical outcome in patients with sepsis. Crit. Care. 2010;14(6) doi: 10.1186/cc9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.