Abstract
Angiotensin II (Ang II) type 2 receptor (AT2R) is one of the major components of the renin-angiotensin-aldosterone system. Nevertheless, the physiological role is not well defined compared to the understanding of the Ang II type 1 receptor (AT1R), which is a well characterized G-protein coupled receptor in the cardiovascular system. While the AT2R signaling pathway remains unclear, AT2 receptor interacting protein 1 (ATIP1) has been identified as a candidate molecule for interacting with the C-terminal region of AT2R. In this study, we investigated the ATIP1 dependent AT2R inducible genes in human umbilical vein endothelial cells (HUVECs). CGP42112A, an AT2R specific agonist, resulted in an upregulation of inflammatory genes in HUVECs, which were inhibited by knocking down ATIP1 with siRNA (siATIP1). Among them, we confirmed by quantitative PCR that the induction of COX-2 mRNA expression was significantly downregulated by siATIP1. COX-2 was also upregulated by Ang II stimulation. This upregulation was suppressed by treatment with the AT2R specific antagonist PD123319, which was not replicated by the AT1R antagonist telmisartan.
These findings suggest that ATIP1 plays an important role in AT2R dependent inflammatory responses. This may provide a new approach to the development of cardio-protective drugs.
Keywords: Angiotensin II, AT2R, ATIP1, COX-2, HUVECs, GPCR signaling
Highlights
-
•
Only the AT2 receptor interacting protein 1 (ATIP1) of ATIP isoforms expresses in endothelial cells.
-
•
A novel anti-ATIP monoclonal antibody detected endogenous ATIP1 and revealed ATIP1 localization in endothelial cells.
-
•
AT2 receptor (AT2R) agonist stimulation induced inflammatory gene expression via ATIP1 in endothelial cells.
-
•
An AT2R specific inhibitor blocks the Ang II induction of COX-2 mRNA in endothelial cells.
-
•
There is the AT2R-ATIP1 related pathway of COX-2 induction in endothelial cells.
1. Introduction
The renin-angiotensin system (RAS) is a major cardiovascular regulatory system and is involved in numerous other pathophysiological functions, such as inflammation, aberrant cell growth, fibrosis, and hypertension [1].
Angiotensin II (Ang II) is a vaso-active octa-peptide generated from angiotensinogen by renin and angiotensin converting enzyme in the systemic circulation [1]. There has been increasing attention paid to the fact that Ang II is also generated by other proteases in local tissues, and plays important roles in various pathophysiological phenomena [2].
There are two types of Ang II receptors, the angiotensin II type 1 receptor (AT1R) and type 2 receptor (AT2R). AT1R is a well characterized G-protein coupled receptor (GPCR) involved in cardiovascular regulation [3]. AT2R belongs to the same family of GPCR and has been reported to exhibit opposing effects on the AT1R in terms of cardiovascular function [4]. However, the signaling mechanism of AT2R remains unclear and is considered to be cell, tissue or context dependent [5].
Vascular endothelial cells have been considered to play important roles in vaso-constriction and -dilatation as well as inflammatory responses such as cytokine secretion and prostacyclin production, in which the RAS is also involved [6]. Notably, the endothelium-mediated control of vascular tone has come into new light in terms of the microdomain signaling between the endothelium and smooth muscle cells [7], in particular endothelium-derived hyperpolarizing factor (EDHF) and endothelium-derived contracting factors (EDCF), the latter being the product of cyclooxygenase (COX) [8].
It is known that both AT1R and AT2R are expressed in vascular endothelial cells [9,10]. The physiological functions of these cells depend on their expression levels and signal transduction pathways [1,5]. Variety of ligands have been shown to bind to AT2R with an affinity order, CGP42112A > Ang II ≥ Ang III > compound 21 ≥ PD123319 >> Ang IV > Ang (1–7) [5,11]. The shorter angiotensin peptides are considered to have substantial selectivity to AT2R which can act as endogenous ligands [5,11]. Other than these classical ligands, vasoconstriction-inhibiting factor (VIF) has been reported as a potential endogenous agonist candidate of AT2R [12], and EMA401((S)-2-(diphenylacetyl)-1,2,3,4-tetrahydro-6-methoxy-5-(phenylmethoxy)-3-isoquinolinecarboxylic acid) is developed as highly selective agonist of AT2R for neuropathic pain treatment [13]. Besides, there is a hypothesis that AT2R harbors constitutive activity which does not depend on ligand binding [5].
The C-terminal domain of AT2R, helix8, has been considered to be important in ligand binding and signaling [14], the recent analyses support the notion that AT1R and AT2R are completely different in C-terminus structure [[15], [16], [17]].
To date, ErbB3 [14], PLZF [18], and AT2 receptor interacting protein 1 (ATIP1) [19] have been identified as partners that interact with the C-terminal tail of the AT2R. Interaction between AT2R and ErbB3 regulates cell proliferation and apoptosis [14], and the AT2R-PLZF-p85-p70S6 kinase signaling axis exerts effects in cardiac hypertrophy [18].
The ATIP protein family consists of five transcripts of the MTUS1 gene, as well as ATIP1, ATIP2, ATIP3a, ATIP3b and ATIP4 [19,20]. MTUS1 was first identified as a tumor suppressor gene in various cancers [21]. ATIP1 regulates the transport of AT2R from the Golgi to the membrane [22] and trans-inactivates receptor tyrosine kinases [23]. There have been several reports on ATIP related to cardiovascular function. In the ATIP transgenic mouse, vascular smooth muscle cell senescence is attenuated [24]. ATIP1 knock-down promotes E-selectin production in endothelial cells [25]. The ATIP knockout mouse develops cardiac hypertrophy [26].
The objective of this study is the investigating the role of ATIP in vascular endothelial cells, and we report that the AT2R specific agonist CGP42112A induces COX-2 in endothelial cells ATIP1-dependently. The results that the pathway of Ang II for COX-2 induction goes through AT2R-ATIP1, which will help shed light on a new regulatory mechanism of endothelial mediated vascular tone in inflammation, aging and other aspects of cardiovascular pathophysiology.
2. Materials and Methods
2.1. Reagents
DMEM, PBS and Triton X-100 were purchased from Sigma-Aldrich (UK). Agarose was purchased from nacalai tesque (Japan). Block Ace was purchased from Megmilk Snow Brand (Japan). 4% paraformaldehyde phosphate buffer solution, and bovine serum albumin (BSA) were purchased from the Fuji film Wako Pure Chemical Corporation (Japan).
2.2. Cells
HUVECs were purchased from Lonza (Switzerland). HUVECs were cultured in EBM™-2 Endothelial Cell Growth Basal Medium supplemented with EGM-2 Endothelial SingleQuots Kit (Lonza, Switzerland). HUVECs were used within the first 7 passages. HeLa cells were cultured in DMEM supplemented with 10% FBS and 100U penicillin-streptomycin (Gibco, USA). Cells were grown in a monolayer in dishes at 37 °C in a 5% CO2 incubator.
2.3. RT-PCR for the detection of ATIP isoforms
Total RNA fractions were prepared from HUVECs and HeLa cells using TRIzol Reagent, and cDNA was synthesized from 1 μg samples of each total RNA fraction using SuperScriptII (Invitrogen, USA) following the manufacturer's instructions. RT-PCR was performed with KOD Plus (Toyobo, Japan). The sequences of nine primers, the arbitrary sets of which recognize ATIP variants of ATIP1, ATIP2, ATIP3a, ATIP3b, and ATIP4, are listed below. The recognition sites and primer sets are indicated in Supplemental Fig. 1(A) and (B). The reaction conditions were as follows; initial denaturation for 2 min at 94 °C and 30 cycles of 98 °C for 10 min, annealing at 52 °C for 5 s, and extension at 72 °C for 1 min. Amplified PCR products were run on 1% agarose gel electrophoresis (Supplemental Fig. 1 (C)).
9F; 5’ ATGTATCCAGCCACAGACAG 3’,
12R; 5’ CTCTGAGTGGCTAGCTTCAA 3’,
5F; 5’ TGGTTCAGAGAGGCACTATG 3’,
8F; 5’ CCACCATTCACATACGACTG 3’,
1F; 5’ TCATTACTAGCCAGGCTGTG 3’,
4R; 5’ CCATGACGACTGTGCAGTT 3’,
3R; 5’ GGCAAAACAGAACTGGAC 3’,
6R; 5’ TGATCCTCTGAGGAGATACG 3’,
2R; 5’ GGATCCTGAGGAACCATTCT 3’.
2.4. The detection of AT2R and AT1R in HUVECs
Total RNA fractions were prepared from HUVECs using RNeasy Plus Mini Kit (Qiagen, Germany) and cDNA was synthesized from 1 μg samples of each total RNA fraction using iScript (Bio-Rad, USA) following the manufacturer's instructions. RT-PCR was performed with QuickTaq HS Dye Mix (Toyobo, Japan). The primer sequences which recognize AT2R and AT1R containing restriction enzyme sites are listed below. The primer recognition sites and primer sets are indicated in Supplemental Fig. 2 ((A) and (B)). The reaction conditions were as follows; initial denaturation for 2 min at 94 °C and 34 cycles of 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 68 °C for 1 min. Amplified PCR products were run on 1.5% agarose gel electrophoresis (Supplemental Fig. 2 (C)) and purified using Gel Extraction Kit (Qiagen, Germany). Purified PCR products were digested with KpnI and BamHI restriction enzymes, then ligated into pcDNA4B using DNA Ligation Kit (Takara, Japan). AT2R or AT1R subcloned into the pcDNA4B were confirmed by checking whether the sequence read from T7 or BGH on the vector side matches the human AT2R sequence (NM_000686.5) or AT1R sequence (NM_000685.5).
934F; 5’ CTGGGTACCGCCACCATGTGCGTTAATCCGTTTCTGTATTGTTTTG 3’,
1089R; 5’ CATGGATCCAGACACAAAGGTCTCCATTTCTCTAAGAGAACTGCTT 3’,
964F; 5’ GTATGGTACCGCCACCATGGGAAACCGGTTCCAACAGAAGCTCC 3’,
886F; 5’ GCTCGGGTACCGCCACCATGTGCCTGAATCCTCTTTTTTATG 3’,
1077R; 5’ CATGGATCCCTCAACCTCAAAACATGGTGCAGGCTTCTTGGT 3’,
916F; 5’ GCAGCGGGTACCGCCACCATGGGGAAAAAATTTAAAAGATATTTTC 3’,
T7 primer; 5’ TAATACGACTCACTATAGGG 3’,
BGH primer; 5’ GCTGGCAACTAGAAGGCACAG 3’.
2.5. Generation of monoclonal antibodies against ATIP
The C-terminal region of mouse ATIP1 (327–426 a.a., Supplemental Fig. 1(D) and (E)) expressing budded baculovirus was used to generate the anti-ATIP antibody, as described previously [27]. Briefly, recombinant ATIP-expressing baculovirus was directly immunized to gp64 transgenic mice [27], and the hybridoma was obtained using a common method. The hybridoma clone C1717 was selected by Western blot analysis.
2.6. Gene silencing
siRNAs targeting human ATIP1 were designed and synthesized with BLOCK-iT™ RNAi (Invitrogen). Sequences of siRNA for the N terminus of ATIP1 (18–24 a.a.; siATIP-N), its scrambled control (siControl-N), as well as the C terminus of ATIP1 (414–424 a.a.; siATIP-C) and its scrambled control, are listed below. siRNAs were transfected into HUVECs using Lipofectamin™ RNAiMAX (Invitogen).
siATIP-N; 5’ GCCAAAGGAUUGCUUAGAATT 3’,
siControl-N; 5’ GCCGGAUUAUCGCUAAGAATT 3’
siATIP-C; 5’ AUUCCUUGGUGACUGCAAAGGGAUG 3’,
siControl-C; 5’ AUUGGCCGUUGUGUCAAACGGAAAU 3’.
2.7. Quantitative real-time PCR analysis
Expression of mRNA was quantified by real-time PCR with SYBR Green master mix on an ABI qPCR System (Applied Biosystems, USA) following the manufacturer's instructions. The samples were run in triplicate. The primers for ATIP1, cyclophilin and COX-2 are listed below. A melting point dissociation curve generated by the instrument was used to confirm that only a single product was present. Quantification of the relative gene expression was calculated by the comparative Ct method (2−ΔΔCT). Data were normalized to the cyclophilin mRNA levels.
ATIP1 F; 5’ CCACCATTCACATACGACTGA 3’,
ATIP1 R; 5’ GAGCTGTCTGTGGCTGGATA 3’,
Cyclophilin F; 5’ TTCGTGCTCTGAGCACTGGAGA 3’,
Cyclophilin R; 5’ GGACCCGTATGCTTTAGGATGAAG 3’,
COX-2 F; 5’ TGAGCATCTACGGTTTGCTG 3’,
COX-2 R; 5’ TGCTTGTCTGGAACAACTGC 3’.
2.8. Western blot analysis
Cells were washed with PBS three times and detached with a cell scraper in PBS. After centrifugation at 1000 g for 5 min at 4 °C, cells were lysed with lysis buffer (20 mM Tris-HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% TritonX-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, complete protease cocktail (Roche, Switzerland)) and incubated for 40 min at 4 °C. Cell debris was removed by centrifugation at 18,000 g for 30 min at 4 °C.
An equal amount of protein per well was loaded onto 10% acrylamide gels and the proteins separated by SDS-PAGE under reducing conditions, and subsequently transferred onto a nitrocellulose membrane. For immunoblotting, primary antibodies (an anti-Myc antibody (0.1 μg/ml) (Sigma-Aldrich) and the anti-ATIP antibody C1717 (1.0 μg/ml)) diluted with Tris-buffered saline (TBS) were added onto the membrane and incubated for 1 h after blocking with Block Ace. The membrane was washed with TBS-T (TBS supplemented with 0.05% Tween 20) twice and then incubated with HRP-conjugated anti-mouse antibody for 1 h. After washing twice with TBS-T, the membrane was visualized with SuperSignal West Dura (Thermo Fischer Scientific, USA).
2.9. Microarray analysis
Total RNA was prepared as described in section 2.3. RNA integrity was examined with a bioanalyzer (Agilent Technologies). Preparation of fluorescence labeled RNA and hybridization with a probe in the microarray (GeneChip U133Plus 2.0, Affimetrix, USA) was performed according to the manufacturer's instructions.
2.10. Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed with PBS and then permeabilized with 0.1% Triton X-100 in PBS for 5 min, followed by blocking with 5% BSA for 30 min. For mitochondria staining, 100 nM MitoTracker Red CMXRos (Thermo Fisher Scientific) was added to the cells before they were fixed and incubated for 5 min. Cells were then incubated with primary antibodies (i.e. Fluorescein isothiocyanate (FITC)-labeled C1717 (100 μg/ml) that was labeled with a Fluorescein Labeling Kit (Dojindo, Japan), an anti-VE-cadherin antibody (10 μg/ml) (Abcam) and an anti-golgin 97 antibody (2 μg/ml) (Thermo Fisher Scientific)) for 1 h at room temperature, washed with PBS three times, and subsequently incubated with an Alexa Fluor 594-conjugated anti-mouse antibody or Alexa Fluor 594-conjugated anti-rabbit antibody as the secondary antibody for 1 h at room temperature. After cells were mounted on a slide glass with anti-fade reagent (ProLong Gold, ThermoFisher Scientific), images were taken using confocal microscope (Fluoview FV1000, Olympus).
2.11. Statistical analysis
Data are expressed as the mean ± SEM and statistical significance taken as the differences between the means. Student's t-test was used when comparing two conditions. One-way ANOVA followed by post hoc Tukey's test or 2way ANOVA followed by Sidak's multiple comparisons test were used for multiple comparisons with Prism7.
3. Results
3.1. A novel anti-ATIP monoclonal antibody detects endogenous ATIP1 localization in HUVECs
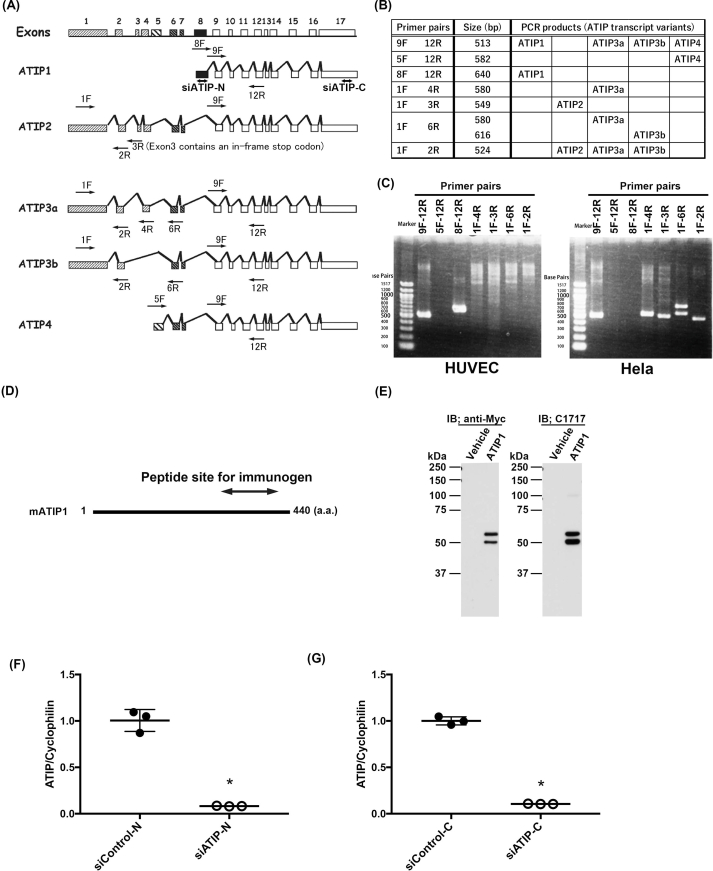
We first validated the ATIP variants expressing in HUVECs. We designed primers specific to exons 1 to 17 constituting ATIP1, ATIP2, ATIP3a, ATIP3b and ATIP4 variants as designated in Supplemental Fig. 1 (A) (B), and checked the expression of ATIP mRNA in HUVECs by RT-PCR. We observed only an ATIP1 corresponding band in HUVECs. In HeLa cells, ATIP1 was not detected. Instead, ATIP2, ATIP3a, and ATIP3b were expressed (Supplemental Fig. 1(B)).
To demonstrate the protein expression and localization of ATIP1 in HUVECs, we next generated an anti-ATIP monoclonal antibody. For the antigen, a C-terminal portion of mouse ATIP1 (327–426 a.a.) was selected (Supplemental Fig. 1(D)). The homology of this portion with human ATIP1 was shown to be 97% (data not shown). This ATIP fragment was fused to baculovirus envelope protein gp64, and the fusion protein expressing baculovirus was directly immunized to gp64 transgenic mice, as previously described [27]. We obtained the hybridoma clone C1717 by a common method. On Western blot analysis, the monoclonal antibody C1717 reacted with the same size bands as the anti-Myc antibody in Myc-tagged ATIP1 transiently expressed HeLa cell lysate (Supplemental Fig. 1 (E)). There was no recognizable band in the vehicle plasmid transfected HeLa cell lysate.
We prepared siRNA for ATIP1 to see the specific reactivity of the ATIP antibody. siRNAs were designed for the N-terminal portion specific for ATIP1 (siATIP-N) and the C-terminal portion containing a region common to all of the ATIP isoforms (siATIP-C), as described in Materials and Methods. Real-time PCR analysis showed that 90% or more of the ATIP1 mRNA was reduced in both siATIP-N and in siATIP-C treated HUVECs at 24 h compared to control siRNA (Supplemental Fig. 1 (F) (G)).
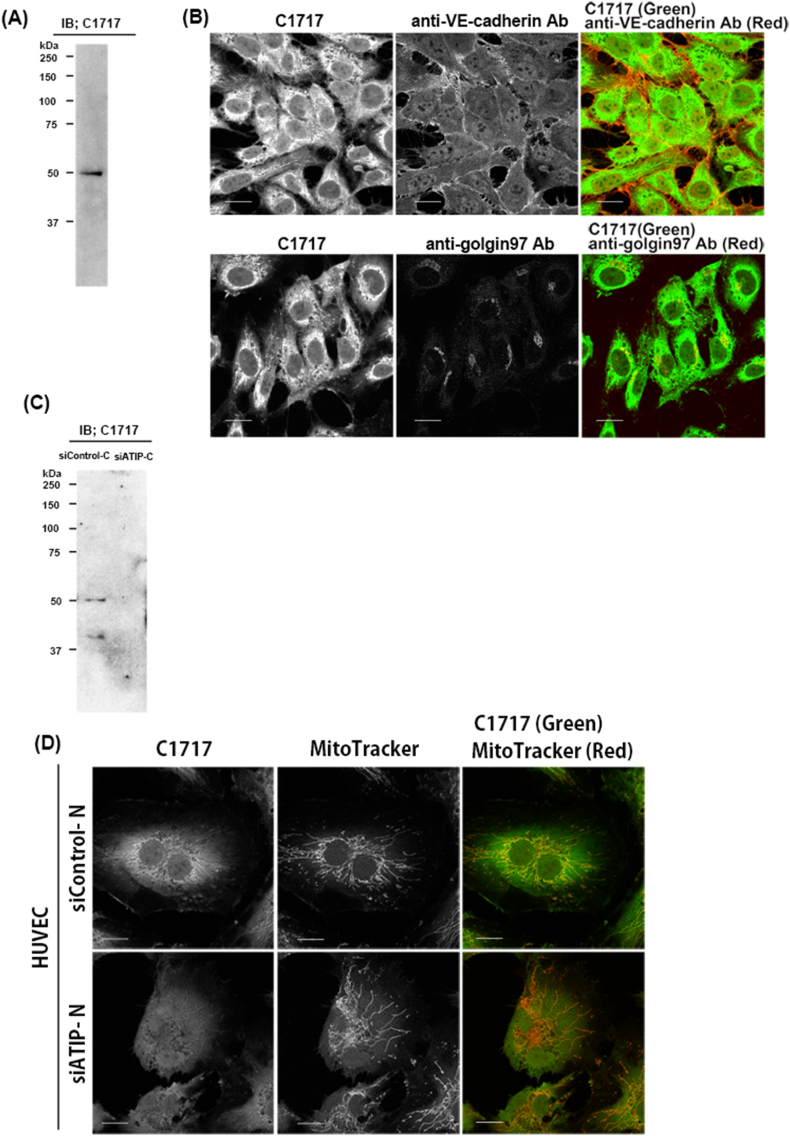
On Western blot analysis, C1717 reacted with an appropriately sized band so as to correspond to endogenous ATIP1 in the HUVEC lysate (Fig. 1(A)), the reactivity of which decreased with siATIP treatment (Fig. 1 (C)).
Fig. 1.
Immunofluorescence analysis of ATIP1 localization in HUVECs using the anti-ATIP monoclonal antibody C1717.
(A) C1717 detected a single 50 kDa band in HUVEC lysate. (IB; immunoblot) (B) HUVECs were stained with C1717, an anti-VE-cadherin antibody (Ab) (Upper panels) and C1717, an anti-golgin 97 Ab (Lower panels). Co-localization of ATIP1 and VE-cadherin (right upper panel) and golgin97 (right bottom panel) are shown in yellow. Scale Bar; 20 μm. (C) The C1717 reactive bands are diminished in siATIP-C treated HUVEC lysate compared to siControl-C. (D) HUVECs were transfected with siControl or siATIP for 48 h. The siControl-N and siATIP-N treated HUVECs were stained with C1717 and MitoTracker. The majority of ATIP1 co-localized to mitochondria, as shown in the upper right panel, an effect which disappeared in HUVECs when ATIP1 was knocked down (right bottom panel). Scale Bar; 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Immunofluorescence staining of HUVECs with C1717 revealed the specific localization of the endogenous ATIP1 which had been reduced by siRNA ATIP (Fig. 1 (D)). A co-localization study with MitoTracker demonstrated that the ATIP1 had localized in both the cytoplasm and mitochondria. We observed the distribution of ATIP1 at the cell membrane upon immunostaining with an anti-VE-cadherin antibody (Fig. 1 (B), the upper-panel). Using an anti-golgin 97 antibody, we also observed that ATIP1 localizes to the Golgi (Fig. 1 (B) bottom-panel).
3.2. AT2R agonist stimulation induces inflammatory gene expression via ATIP1 in HUVECs
We confirmed the expression of AT2R and AT1R in HUVECs used in this study (Supplemental Fig. 2). Then, we analyzed gene expression using a microarray when stimulating HUVECs with the AT2R agonist CGP42112A at the time-points of 4, 8 and 12 h after stimulation. Microarray analysis revealed that the mRNA of more than 30 genes was upregulated 4 h after CGP42112A treatment (>2-fold, Supplemental Table 1). These genes included chemokines, adhesion molecules, and enzymes related to inflammation. The induction of these genes peaked at 4 or 8 h and diminished at 12 h. For reference, the genes down regulated by CGP42112A treatment are listed (Supplemental Table 3).
To see the relationship of ATIP1 in AT2R signaling pathway, we compared the gene induction transcriptome by knocking down ATIP1 with siRNA using siATIP-C and siControl-C, as described in Materials and Methods. We extracted the genes which were upregulated by CGP42112A in siControl-C, but not in siATIP-C (Supplemental Table 2). Among the upregulated genes, we noticed the gene COX-2 (prostaglandin-endoperoxide synthase 2, PTGS2) which has been reported to be an AT2R-related gene in the kidney [28].
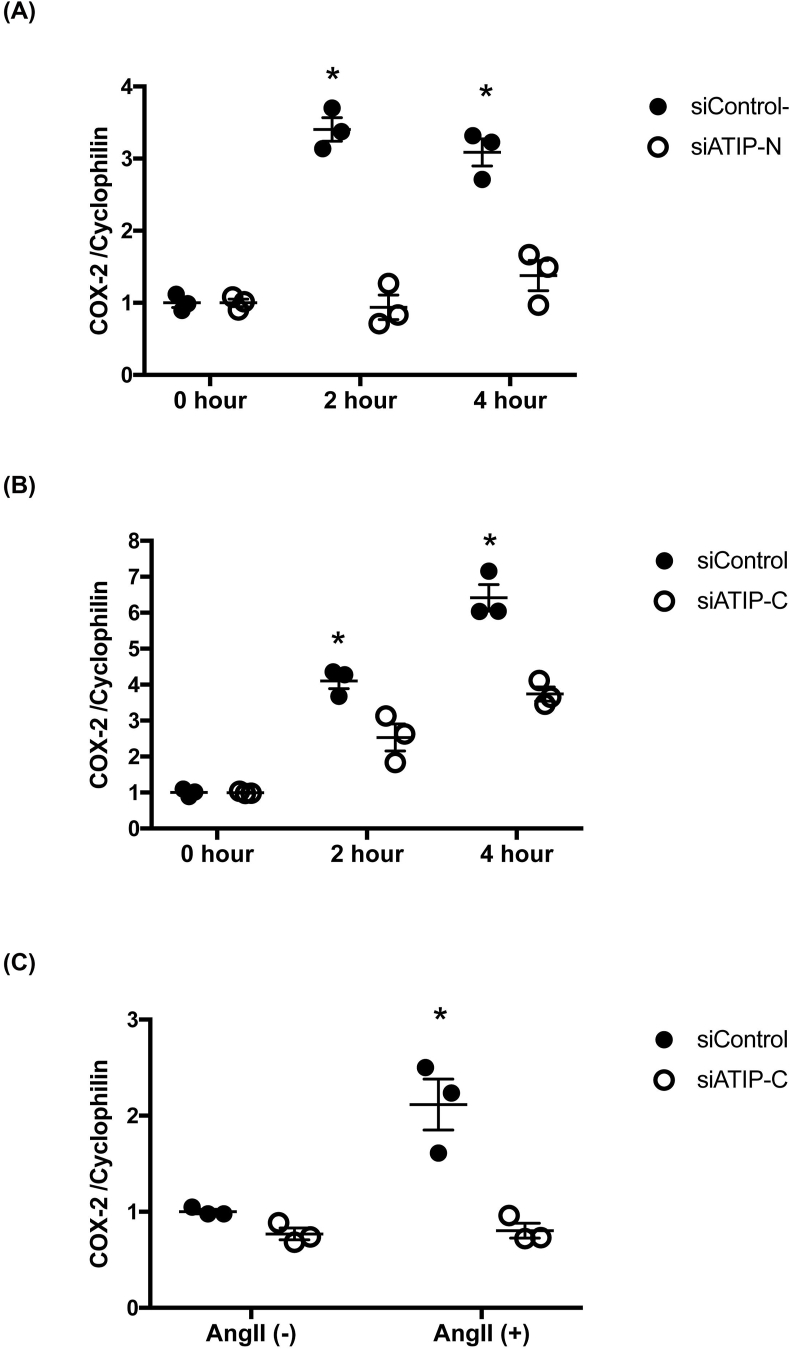
We evaluated COX-2 mRNA expression level quantitatively by qPCR 2 or 4 h after stimulation with CGP42112A in HUVECs. To be certain of the result, we prepared an additional siRNA ATIP1 pair that target the N-terminus of ATIP1 (siATIP-N), along with its control siRNA, siControl-N. The COX-2 mRNA level was upregulated significantly from the time point of 2 h after stimulation, an effect which was significantly suppressed by knocking down ATIP1 with either siATIP-N or siATIP-C (Fig. 2 (A) (B)). Next, we assessed the COX-2 mRNA level by qPCR. when stimulating HUVECs with Ang II. The COX-2 mRNA level was also upregulated by Ang II treatment, an effect which was significantly suppressed by siATIP-C (Fig. 2 (C)).
Fig. 2.
qPCR analysis in HUVECs of COX-2 mRNA induction by CGP42112A or Ang II stimulation.
(A), (B) CGP42112A (10−8 M) upregulated COX-2 mRNA significantly under siControl-N (A) or siControl-C (B) compared to siATIP-N or siATIP-C, respectively (ANOVA, *p < 0.05). The significance between each treatment on the Post Hoc test at the indicated hour is shown (*p < 0.05). (C) Ang II (10−7 M) upregulated COX-2 mRNA, which was suppressed by knocking down siATIP-C (*p < 0.05). Values are the mean ± SEM for all three experiments.
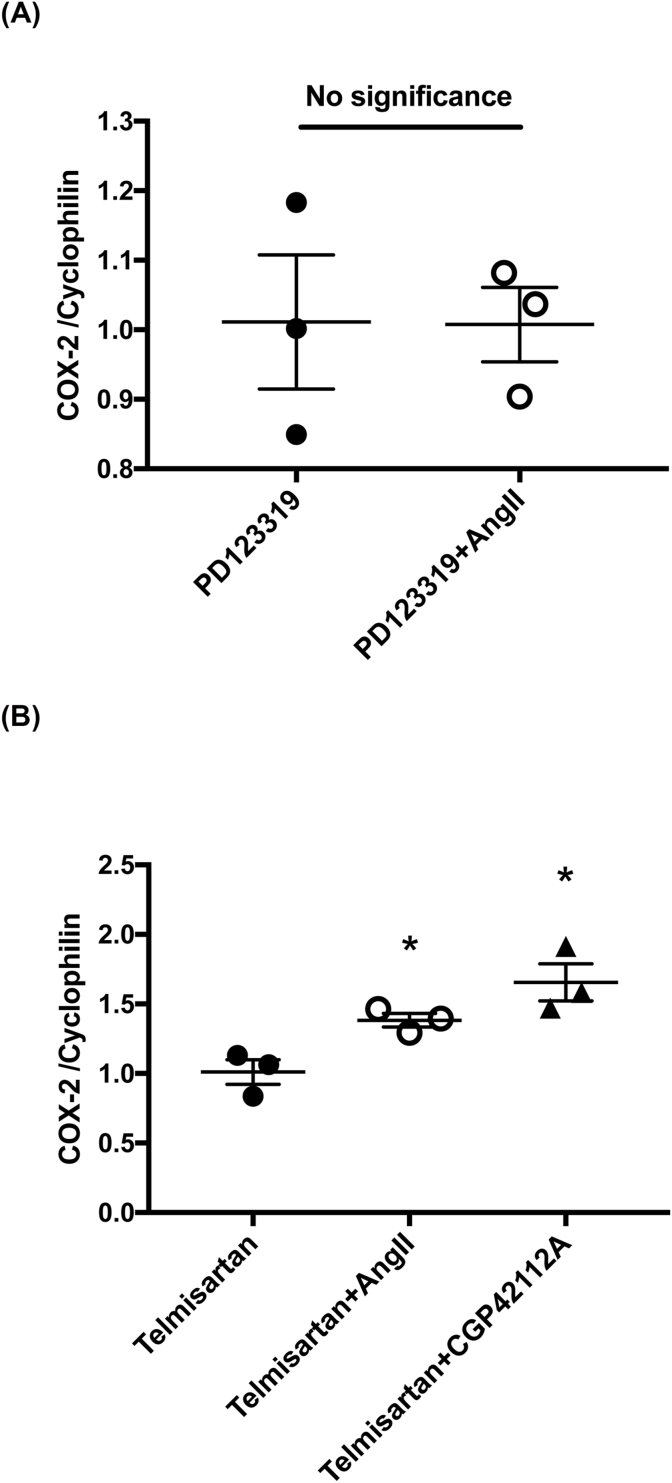
3.3. An AT2R specific inhibitor blocks the Ang II induction of COX-2 mRNA in HUVECs
We verified AT2R-mediated signaling upregulate COX-2 by using an AT2R specific inhibitor or AT1R specific inhibitor. After 16 h of a starvation of HUVECs, we pretreated these HUVECs with the AT2R antagonist PD123319 (10-5 M) for 30 min and stimulated them with Ang II (10-7 M) for 4 h. The COX-2 mRNA level was then analyzed by qPCR. PD123319 treatment suppressed the Ang II induction of COX-2 mRNA in HUVECs (Fig. 3 (A)). In contrast, treatment with the AT1R inhibitor telmisartan (10-5 M) 30 min after 16 h of starvation failed to suppress the induction of COX-2 mRNA by either successive Ang II (10-7 M) or CGP42112A (10-8 M) stimulation for 4 h (Fig. 3 (B)).
Fig. 3.
An AT2R specific inhibitor suppressed COX-2 mRNA induction by Ang II stimulation in HUVECs.
(A) PD123319 (10−5 M) treatment completely suppressed the COX-2 mRNA induction by Ang II (10−7 M) in HUVECs. (B) Treatment with the AT1R blocker telmisartan (10−5 M) suppress Ang II (10-7 M) or CGP42112A (10-8 M) upregulation of COX-2 mRNA in HUVECs. Values are the mean ± SEM for all three experiments. *p < 0.05.
4. Discussion
The relationship between the RAS and the inflammatory response has attracted considerable interest as a potential target of therapeutic intervention [1]. Here, we report that the AT2R-ATIP1 interaction plays an important role in the inflammatory response in endothelial cells.
To investigate ATIP expression in endothelial cells, we designed ATIP primers which distinguish between the ATIP family isoforms (Supplemental Fig. 1(A) and (B)). We observed only the ATIP1 isoform in HUVECs (Supplemental Fig. 1 (C)), which is in accord with previous reports [29,30].
On microarray analysis, treatment of HUVECs with the AT2R agonist CGP42112A induced the inflammatory genes E-selectin (SELE), VCAM1, cyclooxygenase-2 (COX-2), interleukin-8 (IL-8) and ICAM1, whereas those inflammatory genes did not respond in HUVECs in which ATIP1 was knocked down with siATIP-C (Supplemental Tables 1 and 2).
Next, we quantitatively assessed COX-2 upregulation by an AT2R agonist. As shown in Fig. 2, AT2R stimulation with CGP42112A or Ang II induced COX-2 mRNA, an effect which was suppressed by knock down by either siATIP1-N and siATIP-C.
We examined the localization of ATIP1 by raising an anti-ATIP antibody in order to further clarify the relationship between AT2R-ATIP1 (Supplemental Fig. 1(D) and (E)). The hybridoma clone C1717 reacted with ATIP1 in HUVEC cell lysate (Fig. 1 (A), (C)). It was shown by immunofluorescence staining of HUVECs that ATIP1 localizes mainly to the mitochondria and partly to the Golgi apparatus and cell membrane (Fig. 1 (B)).
Seibold et al. used forced expression analysis of ATIP1 [29] and Wang et al. used polyclonal antibody against ATIP [30] to demonstrate that ATIP1 localizes mainly to mitochondria. Wang et al. reported that ATIP1 appeared in two bands in their expression system, which is consistent with our result (Supplemental Fig. 1(E)). On the other hand, Unger et al. showed that GFP-tagged ATIP1 that was transiently expressed in cells distributed to the Golgi, and further demonstrated that ATIP knockdown resulted in a changed pattern of AT2R distribution from the cell membrane to the cytoplasmic area. They suggested that ATIP1 plays an important role in the transport of AT2R from the Golgi to the cell membrane [22]. Using C1717, we confirmed by a colocalization study with immunofluorescence staining that some portion of ATIP1 did localize to the Golgi and cell membrane in HUVECs.
We next checked whether an AT2R antagonist would inhibit AT2R stimulation-dependent COX-2 induction. As shown in Fig. 3, Ang II-induced COX-2 was suppressed by treatment with the AT2R antagonist PD123319, whereas CGP42112A-induced COX-2 displayed little effect as the result of treatment with the AT1R blocker telmisartan. These results are consistent with the data of Zhang et al. that AT2R mediated COX-2 elevation in renal cells [28]. They pointed out that both AT1R and AT2R are involved in COX-2 induction, but the mechanisms are inversely regulated. There is also a report that shows AT1R regulates COX-2 induction in rat endothelial cell using an AT1R blocker [31]. The reason for this discrepancy is not clear, but may depend on the cell type used. Our data simply showed the AT2R and ATIP1 involvement in HUVEC, which does not exclude the possible contribution of AT1R pathway in COX-2 induction by Ang II. It should be noted also that the response may different in cells or tissues used, since the signaling pathway of AT2R is context dependent [5].
COX-2 plays a key role in regulating the biosynthesis of prostaglandins and thromboxane, leading to inflammation in various tissues [32]. To the best of our knowledge, this is the first report that shows COX-2 induction by AT2R via ATIP1. AT2R has been considered to play an anti-inflammatory role as opposing effect against Ang II-AT1R pathway. Our data in this study suggest a possible situation of AT2R-ATIP1 pathway may play a role in pro-inflammatory response. Though it is necessary to show the evidence in vivo, it is important for considering the local RAS role in the control of vascular inflammatory responses [[5], [6], [7], [8], [9]].
The interaction of ATIP1 and the C-terminal portion of AT2R is well documented [22,23], and it is reported that ATIP1 cooperates with AT2R to trans-inactivate receptor tyrosine kinases which is not ligand dependent [23]. Ligand dependent pathway of AT2R-ATIP1 axis is reported by Li JM et al. [33]. Upon ligand binding to AT2R, ATIP trans-locates plasma-membrane to nucleus associating with Src Homology 2 domain-containing protein-tyrosine phosphatase1 (SHP-1) to induce the transcription of ubiquitin-conjugating enzyme MMS2 gene for brain protection [33].
Although the specific effect of ATIP1 to the Ang II signaling is as yet unclear, recent structural analyses revealed that the C-terminal of AT2R is important for the signaling pathway [[15], [16], [17]]. Ang II binding to AT2R induces a structural change in the C-terminus of AT2R that is necessary for activation [17], which is an atypical GPCR signal transduction event that occurs in a G-protein-β-arrestin independent manner [4,[15], [16], [17],34].
On the other hand, Wruck et al. suggested that a dysfunctional interaction between ATIP1 and AT2R may be responsible for certain AT2R-related diseases, such as one form of mental retardation [22]. Considering the localization of ATIP1 in HUVECs, ATIP1 plays an important role in the transfer of AT2R from the Golgi to the cell membrane [22] and may be involved in ROS generation in mitochondria [1].
In conclusion, the findings reported here show that activated AT2R in endothelial cells induces inflammatory genes via ATIP1. This elucidation of the relationship of between ATIP1 and AT2R should open up new targets for the development of anti-inflammatory drugs based on COX-2 regulation.
Author statement
Following the CrediT author statement sample, we declare here the all co-authors are involved in this paper as follows;
Keita Soda: Conceptualization, Investigation, Writing-Original preparation.
Yoshiko Nakada: Preparation of antibody against ATIP.
Hiroko Iwanari: Preparation of antibody against ATIP.
Takao Hamakubo: Conceptualization, Supervision.
Declaration of competing interest
The authors have declared that no conflict of interest exists.
Acknowledgements
We thank Dr. Tadashi Inagami of Vanderbilt University for kindly gift of ATIP1 cDNA and Dr. Kevin Boru of Pacific Edit for review of this article. This study was supported by JSPS Grant-in-Aid for Scientific Research S25220205.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100850.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Supplemental Fig. 1.
ATIP1 variant expression in HUVECs and anti-ATIP monoclonal antibody C1717 generation.
(A) The structural organization of the MTUS1 gene is quoted from the work of Di Benedetto et al. [20]. We designed primers in alternative exons used by the ATIP transcript variants as shown in Material and Methods. siATIP-N in exon 8 and siATIP-C in exon 17 indicates the siRNA region used to knockdown ATIP. (B) The Primer pairs, size (bp) and predicted ATIP transcript variants are listed. (C) The RT-PCR using the primers shown in (B) showed that the ATIP1 variant was expressed in HUVECs but not HeLa cells. (D) We selected the site of the immunogen to generate the anti-ATIP monoclonal antibody in the C-terminal region of mouse ATIP1 (327-426 a.a.), as indicated by both ends of the arrow. (E) C1717 detected Myc-tagged human ATIP1 transiently overexpressed in Hela cells as two bands of approximately 50 kDa. Overexpressed ATIP1 was also detected by anti-Myc antibody at the same size when using C1717. IB; immunoblot. Vehicle; cell lysate of pcDNA4 plasmid transfected to Hela cells. (F), (G) qPCR showed that endogenous ATIP1 was knocked down with siATIP-N or siATIP-C at the mRNA level. Values are the mean ± SEM for all three experiments. *p < 0.05.
Supplemental Fig. 2.
AT2R and AT1R are expressed in HUVECs.
(A) We selected the site of the primers to amplify the C-terminal region of AT2R (Probe 1 and Probe 2) and AT1R (Probe 3 and Probe 4) as indicated by arrows. (B) The Primer pair and predicted PCR-product size (bp) are listed. (C) Amplified PCR products run on 1.5% agarose gel electrophoresis showed that the AT2R and AT1R were expressed in HUVECs.
References
- 1.Forrester S.J., Booz G.W., Sigmund C.D., Coffman T.M., Kawai T., Rizzo V., Scalia R., Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol. Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nehme A., Zouein F.A., Zayeri Z.D., Zibara K. An update on the tissue renin angiotensin system and its role in physiology and pathology. J. Cardiovasc. Dev. Dis. 2019;6 doi: 10.3390/jcdd6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai T., Forrester S.J., O'Brien S., Baggett A., Rizzo V., Eguchi S. AT1 receptor signaling pathways in the cardiovascular system. Pharmacol. Res. 2017;125:4–13. doi: 10.1016/j.phrs.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turu G., Szidonya L., Gaborik Z., Buday L., Spat A., Clark A.J., Hunyady L. Differential beta-arrestin binding of AT1 and AT2 angiotensin receptors. FEBS Lett. 2006;580:41–45. doi: 10.1016/j.febslet.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 5.Karnik S.S., Unal H., Kemp J.R., Tirupula K.C., Eguchi S., Vanderheyden P.M., Thomas W.G. International union of basic and clinical pharmacology. XCIX. Angiotensin receptors: interpreters of pathophysiological angiotensinergic stimuli. Pharmacol. Rev. 2015;67:754–819. doi: 10.1124/pr.114.010454. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flavahan S., Chang F., Flavahan N.A. Local renin-angiotensin system mediates endothelial dilator dysfunction in aging arteries. Am. J. Physiol. Heart Circ. Physiol. 2016;311:H849–H854. doi: 10.1152/ajpheart.00422.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garland C.J., Hiley C.R., Dora K.A. EDHF: spreading the influence of the endothelium. Br. J. Pharmacol. 2011;164:839–852. doi: 10.1111/j.1476-5381.2010.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feletou M., Huang Y., Vanhoutte P.M. Endothelium-mediated control of vascular tone: COX-1 and COX-2 products. Br. J. Pharmacol. 2011;164:894–912. doi: 10.1111/j.1476-5381.2011.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Gasparo M., Catt K.J., Inagami T., Wright J.W., Unger T. International union of pharmacology. XXIII. The angiotensin II receptors. Pharmacol. Rev. 2000;52:415–472. [PubMed] [Google Scholar]
- 10.Sohn H.Y., Raff U., Hoffmann A., Gloe T., Heermeier K., Galle J., Pohl U. Differential role of angiotensin II receptor subtypes on endothelial superoxide formation. Br. J. Pharmacol. 2000;131:667–672. doi: 10.1038/sj.bjp.0703566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosnyak S., Jones E.S., Christopoulos A., Aguilar M.I., Thomas W.G., Widdop R.E. Relative affinity of angiotensin peptides and novel ligands at AT1 and AT2 receptors. Clin. Sci. 1979;121(2011):297–303. doi: 10.1042/CS20110036. London, England. [DOI] [PubMed] [Google Scholar]
- 12.Salem S., Jankowski V., Asare Y., Liehn E., Welker P., Raya-Bermudez A., Pineda-Martos C., Rodriguez M., Muñoz-Castañeda J.R., Bruck H., Marx N., Machado F.B., Staudt M., Heinze G., Zidek W., Jankowski J. Identification of the vasoconstriction-inhibiting factor (VIF), a potent endogenous cofactor of angiotensin II acting on the angiotensin II type 2 receptor. Circulation. 2015;131:1426–1434. doi: 10.1161/CIRCULATIONAHA.114.013168. [DOI] [PubMed] [Google Scholar]
- 13.Rice A.S.C., Dworkin R.H., McCarthy T.D., Anand P., Bountra C., McCloud P.I., Hill J., Cutter G., Kitson G., Desem N., Raff M. EMA401, an orally administered highly selective angiotensin II type 2 receptor antagonist, as a novel treatment for postherpetic neuralgia: a randomised, double-blind, placebo-controlled phase 2 clinical trial. Lancet. 2014;383:1637–1647. doi: 10.1016/S0140-6736(13)62337-5. [DOI] [PubMed] [Google Scholar]
- 14.Pulakat L., Gray A., Johnson J., Knowle D., Burns V., Gavini N. Role of C-terminal cytoplasmic domain of the AT2 receptor in ligand binding and signaling. FEBS Lett. 2002;524:73–78. doi: 10.1016/s0014-5793(02)03005-3. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H., Han G.W., Batyuk A., Ishchenko A., White K.L., Patel N., Sadybekov A., Zamlynny B., Rudd M.T., Hollenstein K., Tolstikova A., White T.A., Hunter M.S., Weierstall U., Liu W., Babaoglu K., Moore E.L., Katz R.D., Shipman J.M., Garcia-Calvo M., Sharma S., Sheth P., Soisson S.M., Stevens R.C., Katritch V., Cherezov V. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asada H., Horita S., Hirata K., Shiroishi M., Shiimura Y., Iwanari H., Hamakubo T., Shimamura T., Nomura N., Kusano-Arai O., Uemura T., Suno C., Kobayashi T., Iwata S. Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog. Nat. Struct. Mol. Biol. 2018;25:570–576. doi: 10.1038/s41594-018-0079-8. [DOI] [PubMed] [Google Scholar]
- 17.Asada H., Inoue A., Ngako Kadji F.M., Hirata K., Shiimura Y., Im D., Shimamura T., Nomura N., Iwanari H., Hamakubo T., Kusano-Arai O., Hisano H., Uemura T., Suno C., Aoki J., Iwata S. The crystal structure of angiotensin II type 2 receptor with endogenous peptide hormone. Structure. 1993;28(2020):418–425. doi: 10.1016/j.str.2019.12.003. e414. [DOI] [PubMed] [Google Scholar]
- 18.Senbonmatsu T., Saito T., Landon E.J., Watanabe O., Price E., Jr., Roberts R.L., Imboden H., Fitzgerald T.G., Gaffney F.A., Inagami T. A novel angiotensin II type 2 receptor signaling pathway: possible role in cardiac hypertrophy. EMBO J. 2003;22:6471–6482. doi: 10.1093/emboj/cdg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodrigues-Ferreira S., Nahmias C. An ATIPical family of angiotensin II AT2 receptor-interacting proteins. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2010;21:684–690. doi: 10.1016/j.tem.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Di Benedetto M., Bieche I., Deshayes F., Vacher S., Nouet S., Collura V., Seitz I., Louis S., Pineau P., Amsellem-Ouazana D., Couraud P.O., Strosberg A.D., Stoppa-Lyonnet D., Lidereau R., Nahmias C. Structural organization and expression of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a family of angiotensin II AT2 receptor-interacting proteins, ATIP. Gene. 2006;380:127–136. doi: 10.1016/j.gene.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Bozgeyik I., Yumrutas O., Bozgeyik E. MTUS1, a gene encoding angiotensin-II type 2 (AT2) receptor-interacting proteins, in health and disease, with special emphasis on its role in carcinogenesis. Gene. 2017;626:54–63. doi: 10.1016/j.gene.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 22.Wruck C.J., Funke-Kaiser H., Pufe T., Kusserow H., Menk M., Schefe J.H., Kruse M.L., Stoll M., Unger T. Regulation of transport of the angiotensin AT2 receptor by a novel membrane-associated Golgi protein. Arterioscler. Thromb. Vasc. Biol. 2005;25:57–64. doi: 10.1161/01.ATV.0000150662.51436.14. [DOI] [PubMed] [Google Scholar]
- 23.Nouet S., Amzallag N., Li J.M., Louis S., Seitz I., Cui T.X., Alleaume A.M., Di Benedetto M., Boden C., Masson M., Strosberg A.D., Horiuchi M., Couraud P.O., Nahmias C. Trans-inactivation of receptor tyrosine kinases by novel angiotensin II AT2 receptor-interacting protein, ATIP. J. Biol. Chem. 2004;279:28989–28997. doi: 10.1074/jbc.M403880200. [DOI] [PubMed] [Google Scholar]
- 24.Min L.J., Mogi M., Iwanami J., Jing F., Tsukuda K., Ohshima K., Horiuchi M. Angiotensin II type 2 receptor-interacting protein prevents vascular senescence. J. Am. Soc. Hypertens. : JASH. 2012;6:179–184. doi: 10.1016/j.jash.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Dai X., Liu Y., Li J., Liu Z., Yin P., Chen J., Wang Y., Wang N., Zhang P. MTUS1 silencing promotes E-selectin production through p38 MAPK-dependent CREB ubiquitination in endothelial cells. J. Mol. Cell. Cardiol. 2016;101:1–10. doi: 10.1016/j.yjmcc.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Zuern C., Krenacs L., Starke S., Heimrich J., Palmetshofer A., Holtmann B., Sendtner M., Fischer T., Galle J., Wanner C., Seibold S. Microtubule associated tumor suppressor 1 deficient mice develop spontaneous heart hypertrophy and SLE-like lymphoproliferative disease. Int. J. Oncol. 2012;40:1079–1088. doi: 10.3892/ijo.2011.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamakubo T., Kusano-Arai O., Iwanari H. Generation of antibodies against membrane proteins. Biochim. Biophys. Acta. 2014;1844:1920–1924. doi: 10.1016/j.bbapap.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M.Z., Yao B., Cheng H.F., Wang S.W., Inagami T., Harris R.C. Renal cortical cyclooxygenase 2 expression is differentially regulated by angiotensin II AT(1) and AT(2) receptors. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16045–16050. doi: 10.1073/pnas.0602176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seibold S., Rudroff C., Weber M., Galle J., Wanner C., Marx M. Identification of a new tumor suppressor gene located at chromosome 8p21.3-22. Faseb. J. : Off. Publ. Feder. Am. Soc. Exp. Biol. 2003;17:1180–1182. doi: 10.1096/fj.02-0934fje. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Huang Y., Liu Y., Li J., Hao Y., Yin P., Liu Z., Chen J., Wang Y., Wang N., Zhang P. Microtubule associated tumor suppressor 1 interacts with mitofusins to regulate mitochondrial morphology in endothelial cells. Faseb. J. : Off. Publ. Feder. Am. Soc. Exp. Biol. 2018;32:4504–4518. doi: 10.1096/fj.201701143RR. [DOI] [PubMed] [Google Scholar]
- 31.Wong S.L., Lau C.W., Wong W.T., Xu A., Au C.L., Ng C.F., Ng S.S., Gollasch M., Yao X., Huang Y. Pivotal role of protein kinase Cdelta in angiotensin II-induced endothelial cyclooxygenase-2 expression: a link to vascular inflammation. Arterioscler. Thromb. Vasc. Biol. 2011;31:1169–1176. doi: 10.1161/ATVBAHA.110.216044. [DOI] [PubMed] [Google Scholar]
- 32.Tsai M.H., Lin Z.C., Liang C.J., Yen F.L., Chiang Y.C., Lee C.W. Eupafolin inhibits PGE2 production and COX2 expression in LPS-stimulated human dermal fibroblasts by blocking JNK/AP-1 and Nox2/p47(phox) pathway. Toxicol. Appl. Pharmacol. 2014;279:240–251. doi: 10.1016/j.taap.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Li J.M., Mogi M., Tsukuda K., Tomochika H., Iwanami J., Min L.J., Nahmias C., Iwai M., Horiuchi M. Angiotensin II-induced neural differentiation via angiotensin II type 2 (AT2) receptor-MMS2 cascade involving interaction between AT2 receptor-interacting protein and Src homology 2 domain-containing protein-tyrosine phosphatase 1. Mol. Endocrinol. 2007;21:499–511. doi: 10.1210/me.2006-0005. [DOI] [PubMed] [Google Scholar]
- 34.Lefkowitz R.J. A brief history of G-protein coupled receptors (Nobel Lecture) Angew. Chem. 2013;52:6366–6378. doi: 10.1002/anie.201301924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.