Abstract
Breast cancer ranked second among most frequent cancer in the world playing a significant role in mortality rate. Having prior knowledge on differentially expressed genes in breast cell carcinoma elucidated important indications to understand the molecular mechanism underneath breast carcinogenesis. In this study we have investigated the distinguished CSN1S1 expression in human breast cancer. We have analyzed CSN1S1 mRNA expression between cancer and normal tissues using TCGA datasets. Moreover, analysis including promoter methylation, mutations, prognosis, co-expression, gene ontology, and pathways of CSN1S1 were performed by the TCGA Wanderer, UCSC Xena, cBioPortal, PrognoScan, UALCAN, and Enricher server. We have observed low mRNA expression and high promoter methylation of CSN1S1 in cancer tissues compared to normal tissues. Furthermore, we have also identified low mRNA expression in clinicopathological patients, as well as 9 deleterious mutations with highly co-expressed protein MRC1, and significantly related signaling pathways. We have found a positive correlation between the lower expression of CSN1S1 and patients surviving with breast cancer. Here we have concluded that CSN1S1 acts as a biomarker for the surveillance and prognosis of breast cancer, and also works as a novel therapeutic target at the molecular and pathway levels.
Keywords: CSN1S1, Breast cancer, Mutational analysis, Biomarker, Transcriptional expression
Highlights
-
•
Low transcriptional expression and low survival rate of CSN1S1 in breast cancer.
-
•
The investigation of clinical profiles and mutational positions of CSN1S1 in breast cancer.
-
•
The investigation of gene ontology and signaling pathway of CSN1S1 and their co-expressed genes.
-
•
We identified CSN1S1 and also their co-expressed proteins are the potential biomarkers in breast cancer.
1. Introduction
Being the most prevalent and frequent tumor types, breast cancer affects around 2.1 million women per year around the world. It is estimated that approximately 0.6 million women have succumbed to death due to breast cancer in 2018. This is an apprehensive figure as 15% cancer death has been brought among women only by breast cancer stated by the World Health Organization (WHO). Rates of breast cancer have been more recurrent in the full-fledged areas but these percentages are substantially burgeoning and can be a global timid in every corner of the world [1,2].
Though significant progress achieved in plausible diagnosis, early observations, and easy screening method, several effective treatment options open a new door to treat breast cancer. Despite attaining hopeful advancement in recent decades, poor prognosis is a remarkable global burden [3,4]. Prognostic establishment and outcome prediction did use clinical, pathological and molecular features. Finding accurate biomarkers and establishing condign prognosis can improve the productivity of current interventions [4]. Thus finding more sensitive and specific biomarkers is the running trend in cancer research replacing these previous features [[4], [5], [6], [7], [8], [9]].
Human milk has a great deal of noteworthy aspects including various neonatal defense mechanisms interacting proteins [10,11]. Caseins are the fundamental milk proteins which are a heterogeneous group of phosphoproteins, shaping micelles with calcium phosphate and different parts [12,13]. The imminent function of caseins is to provide the trace elements along with essential amino acids [13]. Owing to natural evolution, ‘romiscuous nature’ of some proteins do fortuitous additional tasks besides their proposed initial functionality [14]. Cognizant research corroborates caseins as such proteins as it can modulate immune response of intestinal cells. Recent studies discovered the immunomodulation nature of human α-S1-casein. By toll like receptor 4 signaling in monocytic cells, proinflammatory cytokine expression is induced CSN1S1 [15]. Moreover, contemporary research substantiates that α-casein functions as a ‘tumor suppressor’ with effective prevention of breast cancer, tumor growth and metastasis by activating STAT1 signaling [16]. For this reason, the down-regulation of α-casein may participate in tumor growth and metastasis. Even so, the characteristics of CSN1S1 in breast cancer and the potential molecular mechanisms has not yet been perused and need more research attention.
In our study, CSN1S1 gene expression evaluated by publicly available bioinformatics databases related to human breast cancer. Down regulation of mRNA expression on breast cancer explained by the Oncomine data set and two other data sets e.g. UALCAN and GENT2 also evaluate the CSN1S1 mRNA expression profile. CSN1S1 mRNAexpression shown by the Human Protein Atlas website where it indicated that CSN1S1 mRNA expression is less than the cancerous cells. We also mined prognostic value from ProgoScan where a significant mortal probability related to a low-expression profile. Subsequently relation among different types of receptors, DNA methylation, mutations, co-alteration, gene ontology, and pathways also shown by different currently available databases.
2. Methods and materials
2.1. Human cancer expression analysis
We investigated the differences between normal and cancer tissues for finding the variable CSN1S1 expression in distinct types of cancers. For the following purpose, we used three bioinformatics websites including Oncmine (https://www.oncomine.org/resource/login.html) [17,18], UALCAN webpage (http://ualcan.path.uab.edu/index.html) [19], GENT2 server (GPL570 HG-U133_Plus_2 platform) (http://gent2.appex.kr/gent2/) [20].
2.2. Breast cancer expression exploration
We analyzed the expression of CSN1S1 in various datasets, each dataset was taken from the Oncomine database. Expression of CSN1S1 was observed in the datasets of various breast cancer subtypes such as invasive breast carcinoma, invasive lobular breast carcinoma, intraductal cribriform breast adenocarcinoma, breast carcinoma, invasive ductal, lobular breast carcinoma, mixed lobular and ductal breast carcinoma, male breast carcinoma, mucinous breast carcinoma, and medullary breast carcinoma. From GEPIA 2 database, we got the expression of CSN1S1 gene in breast cancer compared to their normal embodiments [21]. TCGA dataset was observed for the expression of CSN1S1 via UALCAN tool. The level of protein was investigated between normal breast and breast carcinoma cells by using the Human Protein Atlas database (https://www.proteinatlas.org/) [22].
2.3. Evaluation of relationship between mRNA expression and clinicopathological parameters in breast cancer
We evaluated the expression of CSN1S1 in human breast cancer using bcGenExMiner v4.2server [23]. The mRNA expression of CSN1S1in hormonal receptors, histology subtype, nodal status and various factors have been focused to count the foretelling impact of this postulant gene in breast cancer and to identify the possible foretelling markers for human breast cancer.
2.4. Exploration of CSN1S1 expression, promoter methylation and mutations
DNA methylation in breast invasive carcinoma and normal tissue embodiments were investigated by TCGA mplab tool called Wanderer (http://maplab.imppc.org/wanderer/) [24]. Using the UCSC Xena browser (https://xe nabrowser.net/heatmap/), we generated a heat map that included BREAST INVASIVE CARCINOMA PAM50 subtypes and breast invasive carcinoma [25]. Mutations and genomics alteration frequencies of CSN1S1 in breast cancer were presented in cBioPortal web (http://www.cbioportal.org/) [26].
2.5. CSN1S1 in breast cancer of co-expressed exploration
The co-expression of CSN1S1 with variant genes has been utilized by UALCAN server. The MRC1 was the top positive co-expressed gene in breast invasive carcinoma with CSN1S1. We used UCSC Xena website for the exploration of heatmap of CSN1S1 and MRC1 transcriptional expression analysis. Positive correlation was found between CSN1S1 and MRC1 transcript levels by the exploration of breast invasive carcinoma patients data using UCSC Xena. Also the correlation between CSN1S1 and MRC1 transcriptional expression was studied by GEPIA2 web.
2.6. Pathway exploration of breast cancer for correlated genes
We analyzed the gene ontology and pathway using the co-expressed gene from the UALCAN website.The gene ontology and pathway analysis by the Enricher website (https://amp.pharm. mssm.edu/Enrichr) [27].
2.7. Prognostic analysis of CSN1S1 gene
We checked the prognostic value by using PrognoScan as it gives important data about general tumor growth and danger of backsliding (http://dna00.bio.kyutech.ac.jp/PrognoScan/). We analyzed the expression plot, expression histogram plot, p-value plot, survival time plot, attribute plot and Kaplan Meir plot using PrognoScan server [28].
3. Results
3.1. Investigation of mRNA expression in human cancer
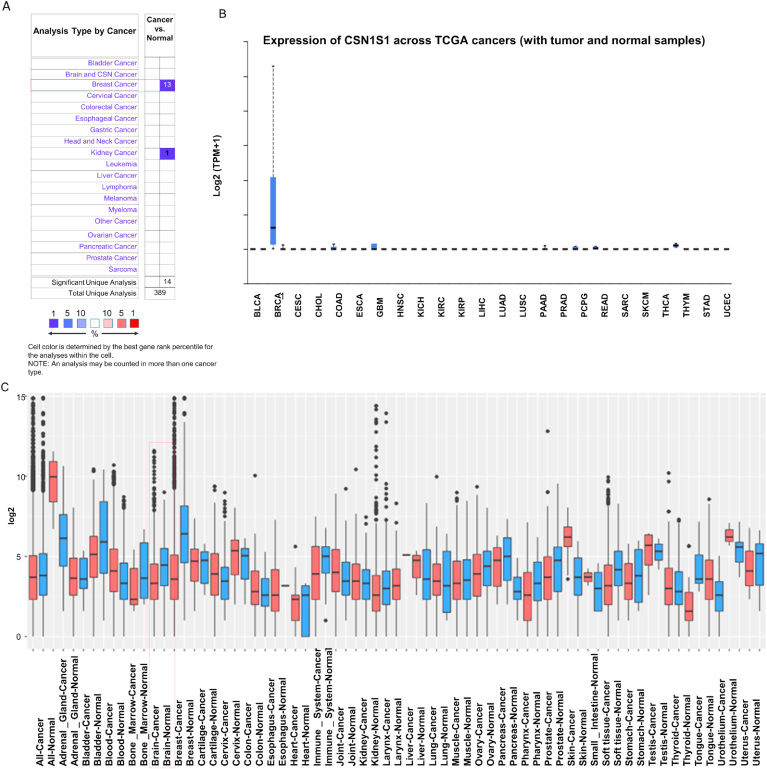
We explored the CSN1S1 mRNA expression level between their normal and cancer tissues using the Oncomine, UALCAN and GENT2. In Oncomine online platform, it showed the differences of mRNA expression between the normal tissue and cancer tissue which was explored with the specification of fold-change > 2 and p-value is less than 0.00001. The down regulation of CSN1S1 in almost all cancers was found by comparing the expression pattern within each type of cancer vs. normal counterpart (Fig. 1A). The decrease of CSN1S1 expression in breast cancer was highest (Fig. 1A). Expression of CSN1S1 across TCGA cancers (with tumor and normal samples) were observed using the UALCAN database (Fig. 1B). In the GENT2 database, gene expression was analyzed using the GPL570 HG-U133_Plus_2 platforms. The CSN1S1 transcription was down expressed in some specific cancer types, together with bladder, cervix, breast, lung, pancreatic, stomach and ovarian cancers (Fig. 1C). CSN1S1 was also up regulated in several cancer types like blood, colon, adipose, liver, small intestine. The result showed remarkably decreased expression of CSN1S1 in the human cancers. All database showed that the expressions of CSN1S1 in breast cancer were remarkably lower compared to their normal tissue expression patterns of CSN1S1 (Fig. 1).
Fig. 1.
The analysis of mRNA expression of CSN1S1 in human breast cancer in various cancer types (A) This dataset indicate CSN1S1 mRNA under regulation (blue colour) and overexpression (red colour) in cancer along with normal tissues utilizing from Oncomine server. The threshold values were defaults. (B) The breast invasive carcinoma expression of CSN1S1 across TCGA cancers (with tumor and normal samples) has been obtained from ‘Pan-cancer view’ in UALCAN webpage. (C) The CSN1S1 in human breast cancer expression patterns of cancer were utilizing from the GENT2 database. In this figure, blue colour represents healthy cells and red colour represents cancer cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Breast cancer expression exploration
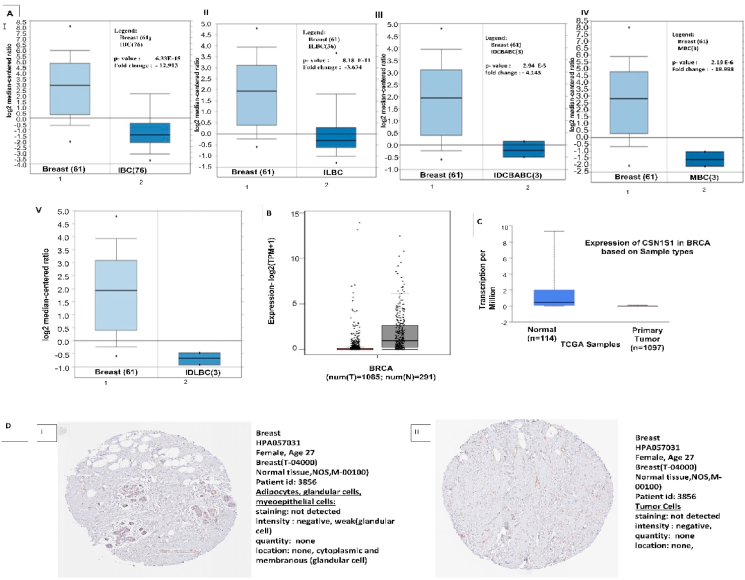
We identified mRNA expression derived from the Oncomine online portal to examine the expression of CSN1S1 in various subtypes of breast cancer (Fig. 2A). Down-regulation of CSN1S1 was noticed with invasive lobular breast carcinoma, invasive breast carcinoma, invasive ductal and lobular breast carcinoma, intra ductal cribriform breast adenocarcinoma breast carcinoma, ductal breast carcinoma, benign breast carcinoma, mixed lobular and ductal breast carcinoma, male breast carcinoma, mucinous breast carcinoma, breast phyllodes tumor, mucinous breast carcinoma, medullary breast carcinoma (Table 1). The data acquired from GEPIA 2 server showed the significant decrease of CSN1S1 expression in breast cancer compared to healthy cells (Fig. 2B). Decreased expression of CSN1S1 in the TCGA dataset of breast cancer also confirmed using the UALCAN tool (Fig. 2C). Later, we observed the protein levels in breast cells (healthy tissue) as well as in breast carcinoma (cancer tissue). The immunohistochemical images downloaded from human protein atlas of patient samples showed high staining whereas the total normal breast cells in healthy breast tissue showed detectable CSN1S1 expression (Fig. 2D). In general, transcription levels were suggested that CSN1S1 expression could be less in breast cancer compared to non-infected tissues.
Fig. 2.
CSN1S1 mRNA expression in breast cancer: (A) Decreased expression of CSN1S1 was observed in normal and cancer tissues datasets with (I) invasive breast carcinoma, (II) invasive lobular breast carcinoma, (III) intra ductal cribriform breast adenocarcinoma breast carcinoma, and (IV) male breast carcinoma, by using Oncomine web portal. (B) Graph shows the CSN1S1 expression in breast invasive carcinoma and non-infected tissues based on Sample types which was derived from UALCAN platform. (C) The given proteins of CSN1S1 of human breast cancer were analyzed from the webpage of Human Protein Atlas.
Table 1.
The CSN1S1 expression levels between breast cancer and healthy tissues using TCGA dataset.
| Subtype of breast cancer | p- value | Fold change | Rank % | Sample | Dataset type |
|---|---|---|---|---|---|
| Invasive Breast Carcinoma | 6.33 E-15 | −12.913 | 7 | 76 | TCGA |
| Invasive Lobular Breast Carcinoma | 8.18 E-11 | −3.634 | 6 | 36 | TCGA |
| Intraductal Cribriform Breast Adenocarcinoma Breast Carcinoma | 2.94 E-5 | −4.143 | 3 | 3 | TCGA |
| Male Breast Carcinoma | 2.19 E-6 | −19.998 | 3 | 3 | TCGA |
| Invasive Ductal And Lobular Breast Carcinoma | 1.2 E-9 | −5.847 | 2 | 3 | TCGA |
| Mixed Lobular And Ductal Breast Carcinoma | 6.96 E-8 | −4.078 | 1 | 7 | TCGA |
| Mucinous Breast Carcinoma | 2.08 E-9 | −4.854 | 10 | 4 | TCGA |
| Ductal Breast Carcinoma | 2.34 E-12 | −2.404 | 1 | 4 | Curtis |
| Benign Breast Carcinoma | 1.05 E-12 | −2.561 | 1 | 4 | Curtis |
| Breast Phyllodes Tumor | 2.72 E-13 | −2.583 | 1 | 4 | Curtis |
| Mucinous Breast Carcinoma | 2.38 E-13 | −2.548 | 1 | 4 | Curtis |
| Medullary Breast Carcinoma | 2.42 E-13 | −2.452 | 4 | 4 | Curtis |
| Breast Carcinoma | 2.38 E-13 | −2.548 | 1 | 14 | Curtis |
3.3. CSN1S1 expression with different clinical parameters
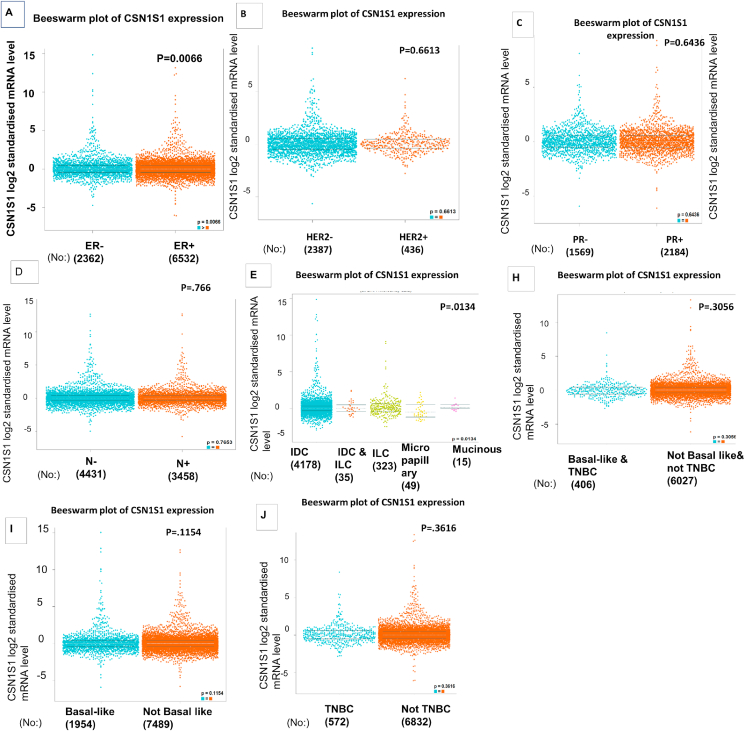
The bc-GenExMiner was used to indicate that CSN1S1 expression remarkably varied in different receptor status, nodal status, histology subtype, basal-like or TNBC. Expression of mRNA was remarkably different among patients with different receptor status. CSN1S1 expression was upregulated in ER−, HER2-, PR + breast cancer compared to ER+, HER2+, PR-breast cancer (Fig. 3A–C). mRNA expression was also different in patients with different histology subtype and basal like or TNBC. Considerable variation in the CSN1S1 mRNA expression between patients with basal-like and/or TNBC were observed. The expression of CSN1S1 mRNA was down regulated in patients with basal-like and/or TNBC than in patients without these subtypes (Fig. 3F–H). Expression levels of CSN1S1 mRNA were higher in patients with invasive ductal carcinoma and invasive lobular carcinoma than any other subtypes (Fig. 3E). CSN1S1 mRNA expression in patients with lymph node positive was lower than lymph node negative (Fig. 3D). CSN1S1 mRNA expression was also downregulated in clinicopathological parameters including cancer stages, age, tumor grade, race, gender, menopause status, histological subtype and cancer subclass using UALCAN serever (Table 2).
Fig. 3.
Clinic pathological analysis of CSN1S1 in breast cancer: in figure A, B, C, D, F, G and H the red colour represents negative and blue represents positive result. A) Estrogen receptor status (ER) (B) HER2 status and (C) PR status, (D) Nodal status, (E) Histological subtype, and (F–H) basal like or TNBC. These were obtained from bc-GenExMiner online portal. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
The relationship between the CSN1S1 and the clinicopathological characteristics of breast cancer using TCGA.
| Parameters | mRNA expression | n | p-value |
|---|---|---|---|
| SampleType | |||
| Normal | ↑ | 114 | |
| Primary tumor | ↓ | 1097 | 0.51 |
| PatientsRace | |||
| Normal | ↑ | 114 | |
| Caucasian | ↓ | 745 | 0.83 |
| African -american | ↓ | 179 | 0.05 |
| Asian | ↓ | 61 | 0.05 |
| Cancer Stages | |||
| Normal | ↑ | 114 | |
| Stage 1 | ↓ | 153 | 0.87 |
| Stage 2 | ↓ | 615 | 0.68 |
| Stage 3 | ↓ | 247 | 0.04 |
| Stage 4 | ↓ | 20 | 0.04 |
| Cancer Subclasses | |||
| Normal | ↑ | 114 | |
| Luminal | ↓ | 566 | 0.74 |
| HER2 positive | ↓ | 37 | 0.04 |
| Triple negative | ↓ | 116 | 0.65 |
| Patient's Gender | |||
| Normal | ↑ | 114 | |
| Male | ↓ | 12 | 0.04 |
| Female | ↓ | 1075 | 0.52 |
| Patient's Age | |||
| Normal | ↑ | 114 | |
| 21–40 | ↓ | 97 | 0.58 |
| 41–60 | ↓ | 505 | 0.83 |
| 61–80 | ↓ | 431 | 0.05 |
| 81–100 | ↓ | 54 | 0.04 |
| Menopause Status | |||
| Normal | ↑ | 114 | |
| Pre-menopause | ↓ | 230 | 4.04E-01 |
| Peri-menopause | ↓ | 37 | 4.33E-02 |
| Post- menopause | ↓ | 700 | 4.44E-02 |
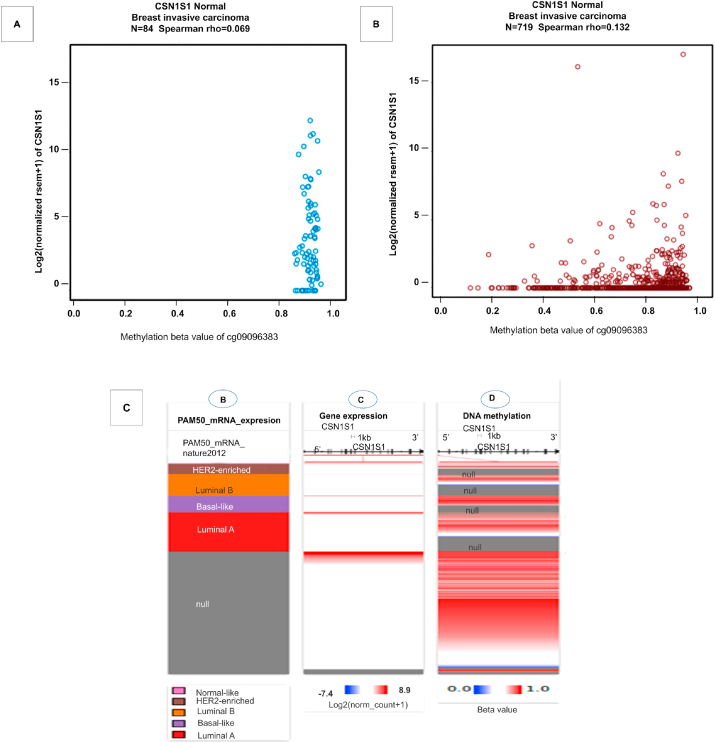
3.4. Methylation status and relationship between CSN1S1 expression of breast cancer patients
DNA methylation is an epigenetic mechanism in which methyl (CH3) group is added to DNA. In consequence, it frequently changes the function of the genes and influences the gene expression. DNA methylation was remarkably increased in breast invasive carcinoma tumors compared to healthy tissue counterparts in the TCGA Wanderer analysis of TCGA data (Fig. 4A and B). It was confirmed by examining data of methylation using an UCSC Xena database (Fig. 4C). The comparison between the CSN1S1 mRNA expression heatmap and the DNA methylation status confirmed that CSN1S1 expression gradually decreased with increasing DNA methylation, which resolved that the transcript expression of CSN1S1 could be negatively associated with a number of CpG sites. Overall, these data suggest the decreased mRNA expression of CSN1S1 and increased DNA methylation in breast invasive carcinoma (Fig. 4).
Fig. 4.
The methylation status of CSN1S1 in human breast cancer: The methylation status of probe cg09096383 (A) indicates healthy tissues and (B) tumor tissues, datasets generated from TCGA Wanderer (http://maplab.imppc.org/wanderer/). (C) The CSN1S1 heat map expression and its DNA methylation status were obtained from UCSC Xena.
3.5. CSN1S1 in human breast cancer mutation and copy number alteration
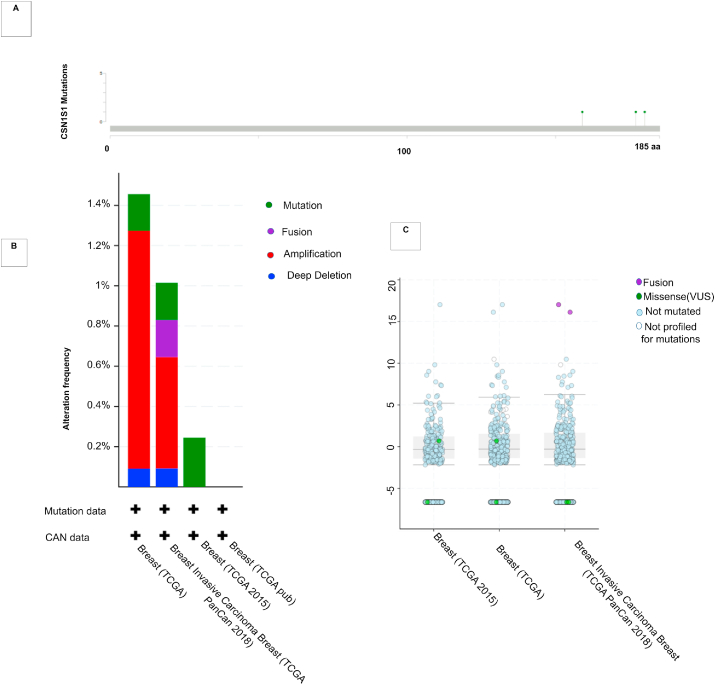
Mutation and CNAs of CSN1S1 in breast cancer patients was observed using cBioPortal. We found nine mutations including three duplicate mutations and 0.2% somatic mutation frequency. There are six missense mutations, and three other mutations. Total nine mutation spots were detected from amino acids 0 and 185 of the CSN1S1 (Fig. 5A). Moreover, mutations and genomics alteration frequencies of CSN1S1 in breast cancer were presented as bar diagrams. This diagram was obtained from a cancer type summary from cBioPortal (Fig. 5B). The mutation frequencies were respectively, nearly 1.4% breast TCGA and 0.2% breast TCGA pub in the TCGA datasets (Fig. 5C). TCGA data shows the most CNAs of CSN1S1. This dataset is also obtained from cBioPortal (Fig. 5C).
Fig. 5.
CSN1S1 in human breast cancer mutation and copy number alteration datasets were derived from cBioPortal: (A) Graph depicts nine mutations including three duplicate mutations in patients with multiple samples. (B) The bar diagram represents CSN1S1 mutation frequencies in breast cancer. Green represents mutation, purple represents fusion, red represents amplification and blue represents deep deletion. (C) The graph shows the correlation between expression of CSN1S1 and copy number alterations in breast cancer of TCGA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Correlation of CSN1S1 in human breast cancer
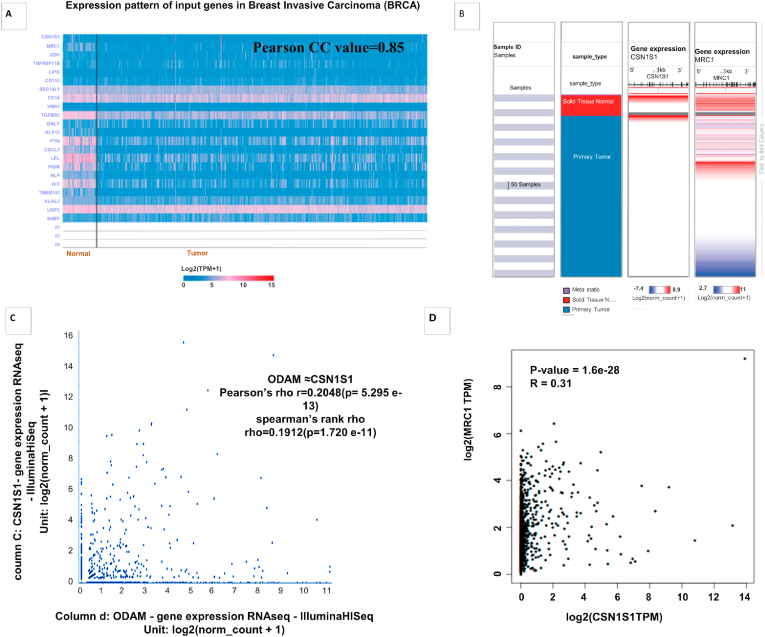
We marked a group of genes that were positively correlated with CSN1S1 in breast cancer using the UALCAN database. Among these genes, expression of MRC1 was highly co-expressed (R = 0.85CC) (Fig. 6A) [20]. TCGA data was analyzed with GEPIA2 web which confirmed the notable positive correlation between CSN1S1 and MRC1 expression (Fig. 6D). Finally, it was justified that there is positive correlation between CSN1S1 and MRC1 by using spearman (r = 0.1912)) and Pearson (r = 0.2048) correlation analyses with TCGA data of breast invasive carcinoma patient from UCSC Xena web (Fig. 6C). We found the co-expression of CSN1S1 with MRC1 through heat map analysis using the UCSC database (Fig. 6B). Our outcomes suggest that expression of CSN1S1 and MRC1 might be closely correlated and may have an effective role in a signaling pathway in breast invasive carcinoma.
Fig. 6.
Co-expression and correlation of CSN1S1 with various genes in breast cancer: (A) the expression of CSN1S1 with the pattern of input genes in breast invasive carcinoma obtained from UALCAN database. (B) Co-expression of CSN1S1 with MRC1 were found with heat map analysis using the UCSC Xena database. (C) Co-expression of mRNA between CSN1S1 and MRC1 in breast cancer generated from UCSC Xena web. (D) Correlation analysis between CSN1S1 and MRC1 obtained from GEPIA 2 web.
3.7. Pathway and gene ontology analysis with CSN1S1 in breast cancer
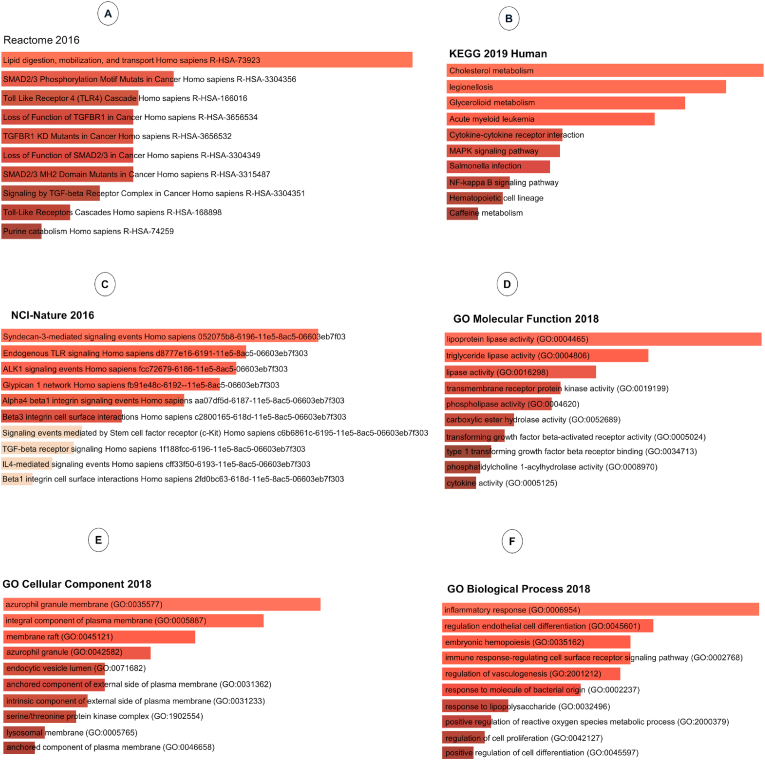
To identify the possible signaling pathways from the set of co-altered genes with CSN1S1 in breast invasive carcinoma, we carry out ontology analysis with 24 positively co-altered genes with CSN1S1 obtained from the UALCAN database (Fig. 7). The top 10 REACTOME pathways obtained from CSN1S1 correlated genes were mainly related to the Lipid digestion, mobilization, and transport, SMAD2/3 phosphorylation motif mutants in cancer, loss of function of TGFBR1 in cancer, toll-like receptor 4 (TLR4) cascade, TGFBR1 KD mutants in cancer, loss of function of SMAD2/3 in cancer, SMAD2/3 MH2 domain mutants in cancer, signaling by TGF-beta receptor complex in cancer, toll-like receptors cascades, purine catabolism (Fig. 7A). Furthermore, the top 10 KEGG for CSN1S1 and its correlated genes were mainly related to cholesterol metabolism, legionellosis, Glycerolipid metabolism, acute myeloid leukemia, cytokine-cytokine receptor interaction, MAPK signaling pathway, salmonella infection, NF-kappa B signaling pathway, hematopoietic cell lineage, caffeine metabolism. Changes in these pathways might be involved in breast invasive carcinoma tumorigenesis (Fig. 7B). Moreover, the top 10 national cancer institute-nature pathways (NCI-Nature) obtained from CSN1S1 correlated genes were mainly related to syndecan-3-mediated signaling events, endogenous TLR signaling, ALK1 signaling events, glypican 1 network, Alpha4 beta1 integrin signaling events, beta3 integrin cell surface interactions, signaling events mediated by stem cell factor receptor (c-Kit), TGF-beta receptor signaling, IL4-mediated signaling event, beta 1 integrin cell surface interactions (Fig. 7C). Then, gene ontology analysis revealed positively correlated genes with CSN1S1. Enrichr was used to find out functions in cellular components, molecular mechanism and biological processes of these correlated genes. These genes were mostly related to the transcription regulation from the RNA components (Fig. 7D–F).
Fig. 7.
We used the Enricher web to profile the co-expressed genes with the CSN1S1 in breast cancer. (A) The bar graphs illustrate pathway analysis (REACTOME pathways 2016). (B) This graph shows the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways 2019. (C) This represents NCI-Nature 2016 which terms in proteomic analysis. (D) The data represents the GO molecular function (2018) terms in proteomic analysis. (E) Illustration of GO cellular component (2018) shows proteomic analysis. (F) The graph of GO biological process (2018) terms in proteomic analysis.
3.8. Analysis of the prognostic value of CSN1S1
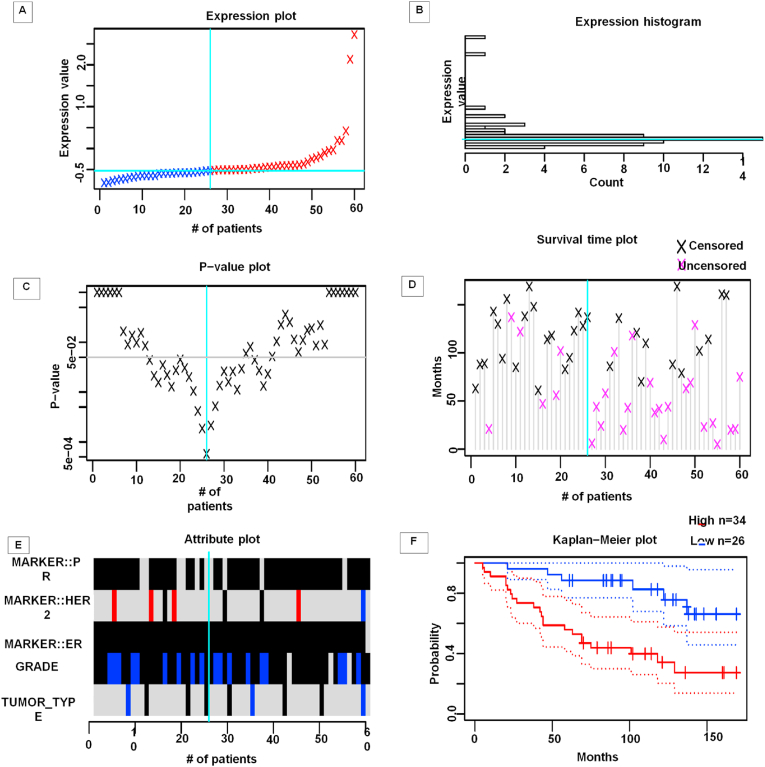
We checked prognostic value using Prognoscan as it gives valuable information about overall cancer outcome and risk of relapse. Here the Figure dichotomizes 60 patients into two groups (i.e. low and high) with a cyan cut-point according to their expression value (Fig. 8A and B). It is observed that more than 14 patients have the expression value on which cut-point has been set off and this value is close to the base-line for which it is considered as “low expression value” (Fig. 8B). Patients with low-expression value accumulate above and around the cut-point and few patients seem to have high expression value. It seems there is no significant difference between survival and high-low expression as the curve is the same in either side of the cut-off point under 5% significance level (gray line) (Fig. 8C). Most of the patients were followed-up (Fig. 8D). Attribute plot shows a various grade and receptor expression among the patients (Fig. 8E). Defining another optimal cut-point, patients undergo survival analysis and it is observed that probability of survival is reduced in patients with low-expression than who have high-expression according to Figure (Fig. 8F).
Fig. 8.
Different plots have been analyzed by using prognoscan server: (a) Expression plot: Patients are ordered in terms of CSN1S1 expression values. The cumulative number of patients are plotted against the expression value. The optimal cut points are shown by the straight lines (cyan) in which patients are separated into high (red) and low (blue) expression groups. (b) Expression histogram: The distribution of the expression value is presented where the X-axis represents the number of patients and the Y-axis represents the expression value on the same scale as the expression plot. The optimal cutpoint line (cyan) is also present. (c) p-value plot: Calculations are done on patients regarding survival difference between high and low expression groups. The X-axis indicates the cumulative number of patients on the same scale as the expression plot where the raw P-values (log scale) is presented in Y - axis. The cutpoint to minimize the P-value which is determined and indicated by the cyan line. The gray line indicates the 5% significance level. (d) Survival Plot: The events (relapse, progression, death) may not be observed for some individuals within the study time period, producing censored observations. The X-axis represents the cumulative number of patients while Y-axis represents the study time. The cut point is indicated by the cyan line. (e) Attribute plot: Patients are separated based on grade and Receptor Expression. The X-axis represents the cumulative number of patients on the same scale as the expression plot whereas the Y-axis represents parameters based on which we mentioned. The cut point is indicated by the cyan line. (f) Kaplan-Meier plot: Survival curves for high (red) and low (blue) expression groups are separated at the optimal cut point. Times are plotted against the survival rate. 95% confidence intervals for each group are also indicated by dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
The number of new cases of breast cancer globally is estimated to be 1.38 million each year. It is noticeable that developed countries have the highest incidence rates than developing countries. The highest incidence rates are seen in Western Europe, with 89.7 new cases per 100,000 people (world standard), the lowest rates in Africa with about 20 cases per 100,000 people. Geographically, maternal cancer mortality rates are significantly lower (somewhere within the range between 6 and 19 deaths per 100,000) than because patients in high-level countries even have strong prognosis. However, breast cancer appears to be the leading common cause of death in women, both in developing countries and developed countries [29]. Death rates have recently decreased in most Western nations, particularly in more young age groups, as a result of increased treatment and prior diagnosis [30]. Advanced methodology including focused on treatments dependent on molecular profiling of breast cancer growth have improved endurance in a few instances of progressive breast cancer [30,31]. The current findings indicate that the decreased expression of CSN1S1 is not positively associated with the survival of patients with breast cancer, indicating that CSN1S1 may be a valuable therapeutic target for breast cancer development.
The present deliberate study utilizing bioinformatics analyses of open datasets shows the prognostic estimation of CSN1S1 in breast cancer growth and explicit changes according to breast cancer at the mRNA expression, DNA methylation, and protein (expression and subcellular localization) levels. The TCGA datasets analyses uncovered the noteworthy down regulation of CSN1S1 in breast cancer tissues and the levels of its expression are negatively correlated with the various clinicopathological characteristics of breast cancer patients. In mutation from cBioPortal nine mutations including three duplicate mutations and 0.2% somatic mutation frequency, mutations and genomics alteration frequencies of CSN1S1 in BC were presented as bar diagrams, expression dataset nearly 1.4% breast TCGA and 0.2% breast TCGA pub in the TCGA datasets were found. TCGA data were analyzed for methylation via oncomine website.
In addition to CSN1S1 which investigates the modified pathway of CSN1S1, some genes have been co-altered. Oncomine database are analyzed among positively correlated genes that express MRC1 as mostly related to co-alter with CSN1S1. MRC1 and CSN1S1 co-alteration was observed by cBioPortal online platforms. MRC1 gene belongs to the mannose receptor family. Potential uses of the mannose receptor family are specified for health and disease. There is likewise the capacity to utilize the mannose receptor as a target to improve macrophage activation and antigen introduction [32,33]. Many genes were analyzed to observe CSN1S1 pathway co-expression and 24 genes were identified as co-expressed along with CSN1S1 Additionally, we utilized Enricher web tools to appoint pathways related with normally correlated genes of CSN1S1 in breast cancer. In addition, for functional viewpoint, GO enrichment analyses were performed by CSN1S1 and co-altered genes as cellular components, biological process and molecular functions. Moreover, the REACTOME pathway shows the activation of lipid digestion, mobilization and transportation to tumor cells which were related to highly expressed in CSN1S1 correlated pathways. These pathways play a role in controlling the cancer micro-environment and are proposed as an immunotherapy target for cancer [34,35].
Several studies have shown that mass cholesterol ester (CE) promotes breast cancer cell proliferation. These results highlight intra tumoral CE accumulation as a potential indicator in the diagnosis of human breast cancer. Moreover, increased free fatty acids (FFAs) not only induce the expression of plasminogen activator inhibitor-1 in breast cancer cells but also inhibit the degradation of fibrin, facilitating breast cancer invasion and metastasis [36]. CSN1S1 co-alteration and a group of genes that are involved in activation of the lipid digestion, mobilization and transport system implicit a potential role of CSN1S1 expression regulating cancer microenvironment. In the next, the most correlated pathway was the cholesterol metabolism and Syndecan-3-mediated signaling events that have an important role in various cancers suppression [37]. The present analysis demonstrated that CSN1S1 was down regulated in breast cancer and positively associated with survival time.
5. Conclusion
To analyze the mRNA expression of CSN1S1, we used bioinformatics web tools in human breast cancer. This study revealed that CSN1S1 is down regulated exemplarily and this expression showed negative correlation with clinical outcomes in breast carcinoma. Lower expression of CSN1S1 could be monitored due to promoter methylation, mutations, and CNAs. These findings also acknowledge the significance of CSN1S1 expression and possible CSN1S1-related pathways regarding tumor progression. All these results may facilitate further therapeutic studies in breast cancer treatment with a breakthrough multiomics approach. We hope that further knowledge development based on gene analyzes will help to improve the prevalence of breast cancer in the future.
Authors contribution
AS and FA designed the project; MAM, DMJH, MSAH, NAK, MMI, and RA performed the analysis of data; MAM, DMJH, MSAH, NAK, MMI, and RA evaluated and interpreted the results; MAM, DMJH, MSAH, NAK, MMI, SR, and RA prepared the draft manuscript; AS and FA critically reviewed and finalized the manuscript. Finally, all authors approved the final version for journal submission.
Funding
Not applicable.
Declaration of competing interest
Authors declare no conflict of interest.
Acknowledgments
The authors like to thank the Biological Solution Centre (BioSol Centre, www.biosolcentre.org) for their technical support.
Contributor Information
Mohsina Akter Mou, Email: mou.mohsinaakter@gmail.com.
Nawshin Atia Keya, Email: nawshin.atia@gmail.com.
Majharul Islam, Email: majharulislam1318@gmail.com.
Md. Jahid Hossain, Email: Jahid.hossain@icddrb.org.
Md. Syeed Al Habib, Email: Syeedhabib141@gmail.com.
Rahat Alam, Email: rahatalam1643@gmail.com.
Sohel Rana, Email: sohelrana3940@gmail.com.
Abdus Samad, Email: kazisamad50@gmail.com.
Foysal Ahammad, Email: foysalgebt@gmail.com.
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Canc. Apr 15, 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Canc J Clin. Nov, 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X., Rice M., Tworoger S.S., Rosner B.A., Eliassen A.H., Tamimi R.M., Joshi A.D., Lindstrom S., Qian J., Colditz G.A., Willett W.C., Kraft P., Hankinson S.E. Addition of a polygenic risk score, mammographic density, and endogenous hormones to existing breast cancer risk prediction models: a nested case-control study. PLoS Med. Sep, 2018;15(9):e1002644. doi: 10.1371/journal.pmed.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy M.J., Walsh S., McDermott E.W., Crown J. Biomarkers in breast cancer: where are we and where are we going? Adv. Clin. Chem. 2015;71:1–23. doi: 10.1016/bs.acc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Samad A., Jafar T., Rafi J.H. Identification of angiotensin-converting enzyme 2 (ACE2) protein as the potential biomarker in SARS-CoV-2 infection-related lung cancer using computational analyses. Genomics. 2020;112(6):4912–4923. doi: 10.1016/j.ygeno.2020.09.002. 11/01/, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha S.K., Kader M.A., Samad K.A., Biswas K.C., Rahman M.A., Parvez M.A.K., Rahman M.S. Prognostic and clinico-pathological significance of BIN1 in breast cancer. Infomat. Med. Unlocked. 2020;19:100327. 01/01/, 2020. [Google Scholar]
- 7.Samad A., Haque F., Nain Z., Alam R., Al Noman M.A., Rahman Molla M.H., Hossen M.S., Islam M.R., Khan M.I., Ahammad F. Computational assessment of MCM2 transcriptional expression and identification of the prognostic biomarker for human breast cancer. Heliyon. 2020;6(10) doi: 10.1016/j.heliyon.2020.e05087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barman U.D., Saha S.K., Kader M.A., Jamal M.A.H.M., Sharma S.P., Samad A., Rahman M.S. Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Network Modeling Analysis in Health Informat. Bioinformat. 2020;9(1):24. 04/05, 2020. [Google Scholar]
- 9.Karim M.A., Samad A., Adhikari U.K., Kader M.A., Kabir M.M., Islam M.A., Hasan M.N. A multi-omics analysis of bone morphogenetic protein 5 (BMP5) mRNA expression and clinical prognostic outcomes in different cancers using bioinformatics approaches. Biomedicines. Jan 21, 2020;8(2) doi: 10.3390/biomedicines8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lönnerdal B. Biochemistry and physiological function of human milk proteins. Am. J. Clin. Nutr. Dec, 1985;42(6):1299–1317. doi: 10.1093/ajcn/42.6.1299. [DOI] [PubMed] [Google Scholar]
- 11.Lönnerdal B., Forsum E. Casein content of human milk. Am. J. Clin. Nutr. Jan, 1985;41(1):113–120. doi: 10.1093/ajcn/41.1.113. [DOI] [PubMed] [Google Scholar]
- 12.Jenness R., Holt C. Casein and lactose concentrations in milk of 31 species are negatively correlated. Experientia. Sep 15, 1987;43(9):1015–1018. doi: 10.1007/BF01952224. [DOI] [PubMed] [Google Scholar]
- 13.Petermann K., Vordenbäumen S., Maas R., Braukmann A., Bleck E., Saenger T., Schneider M., Jose J. Autoantibodies to αS1-casein are induced by breast-feeding. PloS One. 2012;7(4) doi: 10.1371/journal.pone.0032716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco O.L. Peptide promiscuity: an evolutionary concept for plant defense. FEBS Lett. Apr 6, 2011;585(7):995–1000. doi: 10.1016/j.febslet.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Vordenbäumen S., Saenger T., Braukmann A., Tahan T., Bleck E., Jose J., Schneider M. Human casein alpha s1 induces proinflammatory cytokine expression in monocytic cells by TLR4 signaling. Mol. Nutr. Food Res. May, 2016;60(5):1079–1089. doi: 10.1002/mnfr.201500792. [DOI] [PubMed] [Google Scholar]
- 16.Bonuccelli G., Castello-Cros R., Capozza F., Martinez-Outschoorn U.E., Lin Z., Tsirigos A., Xuanmao J., Whitaker-Menezes D., Howell A., Lisanti M.P., Sotgia F. The milk protein α-casein functions as a tumor suppressor via activation of STAT1 signaling, effectively preventing breast cancer tumor growth and metastasis. Cell Cycle. Nov 1, 2012;11(21):3972–3982. doi: 10.4161/cc.22227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P.J.N. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. Jan-Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., Varambally S., Ualcan “. A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. Aug, 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S.J., Yoon B.H., Kim S.K., Kim S.Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genom. Jul 11, 2019;12(Suppl 5):101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z., Kang B., Li C., Chen T., Zhang Z.J. N.a. r. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Proteomics. Tissue-based map of the human proteome. Science. Jan 23, 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 23.Jézéquel P., Frénel J.S., Campion L., Guérin-Charbonnel C., Gouraud W., Ricolleau G., Campone M. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database. 2013:2013. doi: 10.1093/database/bas060. bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Díez-Villanueva A., Mallona I., Peinado M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chromatin. 2015;8:22. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman M., Craft B., Kamath A., Brooks A., Zhu J., Haussler D. ” bioRxiv; 2018. The UCSC Xena Platform for Cancer Genomics Data Visualization and Interpretation; p. 326470. [Google Scholar]
- 26.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. Apr 2, 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. Apr 15, 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno H., Kitada K., Nakai K., Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. Apr 24, 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisemann N., Waldmann A., Katalinic A. Epidemiology of breast cancer - current figures and trends. Geburtshilfe Frauenheilkd. Feb, 2013;73(2):130–135. doi: 10.1055/s-0032-1328075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardoso F., Kyriakides S., Ohno S., Penault-Llorca F., Poortmans P., Rubio I.T., Zackrisson S., Senkus E. “Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. Aug 1, 2019;30(8):1194–1220. doi: 10.1093/annonc/mdz173. [DOI] [PubMed] [Google Scholar]
- 31.Marmot M.G., Altman D.G., Cameron D.A., Dewar J.A., Thompson S.G., Wilcox M. The benefits and harms of breast cancer screening: an independent review. Br. J. Canc. Jun 11, 2013;108(11):2205–2240. doi: 10.1038/bjc.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazi U., Martinez-Pomares L. Influence of the mannose receptor in host immune responses. Immunobiology. 2009;214(7):554–561. doi: 10.1016/j.imbio.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Stahl P.D., Ezekowitz R.A. The mannose receptor is a pattern recognition receptor involved in host defense. Curr. Opin. Immunol. Feb, 1998;10(1):50–55. doi: 10.1016/s0952-7915(98)80031-9. [DOI] [PubMed] [Google Scholar]
- 34.Long J., Zhang C.J., Zhu N., Du K., Yin Y.F., Tan X., Liao D.F., Qin L. Lipid metabolism and carcinogenesis, cancer development. Am J Canc. Res. 2018;8(5):778–791. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y., Aupperlee M.D., Zhao Y., Tan Y.S., Kirk E.L., Sun X., Troester M.A., Schwartz R.C., Haslam S.Z. Pubertal and adult windows of susceptibility to a high animal fat diet in Trp53-null mammary tumorigenesis. Oncotarget. Dec 13, 2016;7(50):83409–83423. doi: 10.18632/oncotarget.13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byon C.H., Hardy R.W., Ren C., Ponnazhagan S., Welch D.R., McDonald J.M., Chen Y. Free fatty acids enhance breast cancer cell migration through plasminogen activator inhibitor-1 and SMAD4. Lab. Invest. Nov, 2009;89(11):1221–1228. doi: 10.1038/labinvest.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz P.M., Mo H., McConathy W.J., Sabnis N., Lacko A.G. The role of cholesterol metabolism and cholesterol transport in carcinogenesis: a review of scientific findings, relevant to future cancer therapeutics. Front. Pharmacol. Sep 25, 2013;4:119. doi: 10.3389/fphar.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]