Abstract
MicroRNAs (miRNAs), as a series of important short-chain non-coding RNAs, play an important post-transcriptional role in many biological activities, including adipogenesis. miR-144 is significantly upregulated in type II diabetes (T2D), and is considered to be an important biomarker for T2D. However, although the occurrence of T2D is inextricably linked to adipogenesis, whether miR-144 directly regulates adipogenesis remains to be further explored. In this paper, we demonstrate that miR-144 has a higher expression level in a porcine high backfat group, and it has a significant positive effect on promoting the differentiation of pre-adipocytes. FoxO1 is a target gene of miR-144, and inhibits the differentiation of pre-adipocytes. On the other hand, we demonstrate that FoxO1 can bind to the AdipoQ gene promoter, then regulate the AdipoQ expression by binding to the FoxO1 binding site in the AdipoQ promoter -1,499 to -1,489 bp and -1,238 to -1,228 bp regions, especially the -1,499 to -1,489 bp region. Meanwhile, miR-144 and FoxO1 co-expressional research has also shown that both factors regulate adipogenesis. To sum up, our research indicates that miR-144 targets FoxO1, thus reducing its expression and inhibiting its promotional effect on adiponectin, thereby alleviating the inhibitory effect of adiponectin on adipogenesis.
Keywords: miRNAs, miR-144-3p, FoxO1, adiponectin, adipogenesis
Introduction
Obesity has gradually become one of the major threats to human health around the world. A batch of diseases such as type II diabetes (T2D), hypertension, and cardiovascular disease are closely related to being overweight or obesity (Kopelman, 2000; Goran et al., 2003). The adipose tissue developed can also trigger an immune mechanism which could create an inflammatory reaction (Hammarstedt et al., 2018). Adipocyte differentiation and proliferation lead to fat tissue expansion, and excessive fat tissue results in obesity-related metabolic syndromes. Hence, adipocytes are emerging as a significant target in the treatment of obesity-related metabolic syndromes in the clinical setting (Shen et al., 2018). In this context, it is very necessary to explore the molecular genetic mechanisms of adipocyte differentiation and proliferation.
Adipocyte differentiation and proliferation is controlled by an enormous and complicated interaction network, which involves hundreds of factors and genes. Among them, peroxisome proliferator activated receptor γ (PPARγ) has been confirmed to play a critical role in adipocyte development. As a downstream factor in the adipogenesis regulation network, PPARγ is involved in regulating insulin sensitivity (Barroso et al., 1999; Doney et al., 2004; Monajemi et al., 2007). Other important adipogenic factors include the CCAAT/enhancer binding proteins (C/EBP) family, such as C/EBPs α, β, γ, δ, ε, and ζ, which can form both homodimers and heterodimers to bind the promoter region of genes containing the CCAAT sequence to regulate the expression level (Hamm et al., 2001; Yu et al., 2015). Dexamethasone (DEX) and isobutyl-methylxanthine (IBMX) can promote C/EBPδ and C/EBPβ at the early stage of adipogenesis, respectively. Then, both of them trans-activate PPARγ and C/EBPα. PPARγ and C/EBPα regulate the expression of each other and both play central roles in adipogenesis (Cao et al., 1991).
Besides, there are other factors that play a regulatory role in adipogenesis, including microRNAs. Generally, microRNA (miRNA) is a non-coding RNA with ∼21 nucleotides. It differs in coding genes, for its length is too short to code, thus miRNAs play a post-transcriptional role by binding the 3′ untranslated region (3′UTR) of coding genes, or binding non-coding genes, for instance, long-coding RNAs (lncRNAS), circle RNAs (circRNAs), pseudogenes, and so on (Hamilton and Baulcombe, 1999; Grishok et al., 2001; Ketting et al., 2001). It regulates a series of biological activities, including adipogenesis. For instance, miR-27 inhibits lipoprotein lipase (LPL) which suppresses adipocyte differentiation (Zhang et al., 2014). miR-34 acts as an inhibitor of beige or brown fat formation (Fu et al., 2014). miR-130 suppresses adipogenesis by binding the 3′UTR of PPARγ to inhibit its expression (Pan et al., 2014). Nevertheless, miR-103 is one of the few miRNAs that has a positive effect on adipogenesis, it can promote 3T3-L1 cell differentiation by targeting MEF2D and activating the Akt/mTOR signal pathway (Li et al., 2015). Except for this miRNA, miR-17 (Han et al., 2017), miR-199 (Shi et al., 2014), miR-425 (Chen et al., 2017), and miR-7134 (Wang et al., 2017) all regulate adipogenesis.
miR-144 is highly upregulated in T2D and has the ability to impair insulin signaling and thus regulate adipogenesis, it has even been used as a biomarker of T2D (Karolina et al., 2011; Liang et al., 2018). However, whether miR-144 directly regulates adipogenesis is still controversial. There is a report that suggests that miR-144 targets C/EBPα and thus inhibits adipocyte differentiation (Itziar et al., 2017), but another report demonstrates that it can promote adipogenesis by targeting Klf3 and CtBP2 (Shen et al., 2018). Here, this paper tries to explore the molecular mechanism of miR-144 in adipogenesis regulation.
Materials and Methods
Experiment Animals
The animals used in this study were Erhualian piglets of 7 days old. All of them were from the Changzhou Erhualian Pig Production Cooperation (Changzhou, Jiangsu, China).
All animal experiments including the pre-adipocytes collected were approved by the Animal Ethics Committee of Nanjing Agricultural University.
Cell Culture
Subcutaneous adipose tissue was isolated from the piglets and soaked in phosphate-buffered saline (PBS). Adipose tissue was cut with scissor into 1 mm3 pieces, and then digested by 1 mg/mL collagenase type I (Invitrogen, Carlsbad, CA, United States) in a 37°C, 50 rpm/min shaking bath for over 2 h. Digestion was stopped by adding 1.5 times the volume of Dulbecco’s modified Eagle’s medium/Ham’s F-12 (DMEM-F12) growth medium (10% fetal bovine serum + 1% penicillin-streptomycin). The digested tissue was filtrated by 200 μm nylon mesh to collect the solution containing pre-adipocytes. The solution was centrifuged twice at 1,000 rpm/min for 10 min to collect the pre-adipocytes. The supernatant was removed and 4 ml of growth medium was added to resuspend the pre-adipocytes. The pre-adipocytes were cultured in the growth medium at 37°C with 5% CO2. The medium was replaced every 2 days.
Cell Transfection and Differentiation
Cells were cultured in 6- or 12-well plates until its density was of 85% confluence. Purpose plasmids or oligonucleotides were transfected to the cells using Lipofectamine 3000 (Invitrogen, Shanghai, China) following the protocol. All the purpose plasmids, siRNAs, and miRNA mimics and inhibitors are shown in Supplementary Table S1.
The adipogenic differentiation inducer medium (DIM) was used to stimulate the pre-adipocytes whose density was of 85% confluence for almost 4 days.
The pre-adipocytes were then induced for adipogenic differentiation using the DIM inducer, which comprised of 2.5 μM dexamethasone, 8.6 μM insulin, 0.1 mM 3-isobutyl-1 methylxanthine (IBMX), 1% penicillin-streptomycin, and 10% FBS in Dulbecco’s modified Eagle’s medium/Ham’s- High Glucose (DMEM-HG) (Sigma-Aldrich, Shanghai, China). After adipogenic differentiation induction for 4 days, the medium was then replaced with maintenance medium containing 8.9 μM insulin and 10% FBS-DMEM-HG until day 8. The above medium was replaced with fresh medium every 2 days.
RNA Isolation, Library Preparation, and RT-PCR
Total RNA was isolated using the Trizol reagent (TaKaRa, Dalian, China). The mRNAs and miRNAs cDNA libraries were reverse-transcribed by the PrimeScriptTM RT Master Mix (TaKaRa, Dalian, China) and the miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) (Vazyme, Nanjing, China), respectively. Quantitative real time PCR (q-PCR) was performed using the AceQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) and the miRNA Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), respectively. The relative level of RNA expression was normalized to GAPDH and U6 expression levels using the 2–ΔΔCt method. Every sample was performed in triplicate. All the primers used are shown in Supplementary Table S1.
Oil Red O Staining and Triglyceride Assay
Briefly, the differentiated porcine pre-adipocytes were gently washed three times with fresh 1×PBS and then fixed in 4% paraformaldehyde for 30 min. The fixed cells were washed three times with 1×PBS and stained with 60% saturated oil red O for 30 min (Sigma-Aldrich, Shanghai, China). Subsequently, the fixed cells were washed three times with 1×PBS. Images of the cells were captured using a Zeiss Axiovert 40 CFL inverted microscope (Thornwood, NY, United States).
Total triglyceride was quantified by the elution of oil red O with isopropanol and the absorbance was measured at the 510 nm wavelength.
Luciferase Reporter Assay
The porcine pre-adipocytes were used as luciferase reporter vectors. The cells were cultured in 12-well plates until they reached 85% confluence. pGL3 vector containing the AdipoQ promoter region and FoxO1-CDs, pmirGLO vector containing the miR-144 binding region of FoxO1 3′UTR, and miR-144 mimic/NC or miR-144 inhibitor/NC were, respectively, co-transfected into the pre-adipocytes using Lipofectamine 3000 (Invitrogen Shanghai, China). Firefly and Renilla luciferase activity were quantified using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP experiments used the ChIP Assay Kit (Boytime, Nanjing, China) following the manufacturer’s instruction. In brief, porcine pre-adipocytes were cross-linked with 1% formaldehyde at 37°C for 10 min. Cross-linking was quenched with 1× glycine for 5 min at RT. The ultrasonic cell smash machine VCX750 (Sonics, United States) was used to smash the cells and to obtain DNA fragments between 200 and 1,000 bp as verified by ethidium bromide electrophoresis. For immunoprecipitation, every 100 μl cell lysis buffer was incubated with 1 μg of antibody against FoxO1 (cat 383312 ZENBIO) at 4°C overnight. Immunoprecipitated complexes were isolated using 60 μl Protein A Agarose/SalmonSperm DNA at 4°C for 2 h, and were then washed with the following: low salt wash buffer, high salt wash buffer, LiCl wash buffer once, and TE buffer twice. Final ChIP DNA was subjected to PCR analysis using a AdipoQ promoter specific primer pair, as shown in Supplementary Table S1.
Western Blotting
In brief, porcine pre-adipocytes were lysed and the total protein was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Jiangsu, China), following the manufacturer’s instructions. A BCA Protein Assay kit (Beyotime, Jiangsu, China) was used to quantify the total extracted protein. Each protein sample (20 μg/well) was loaded into a 12% SDS-PAGE gel (Zoman, Beijing, China). Then the SDS-PAGE was transferred to a PVDF membrane (Millipore, Billerica, MA, United States) after 70 min of electrophoresis. Subsequently, the transferred membranes were blocked by TBST containing 5% non-fat dried milk for 2 h at 37°C, followed by overnight primary antibody incubation (ZenBioScience, Chengdu, China) at 4°C. Then, the immunoblot membranes were washed three times with 1×TBST for 15 min, and were incubated in horseradish peroxidase-conjugated secondary antibody for 2 h at 37°C. The blots were developed using the ECL Chemiluminescence Detection Kit (Vazyme, Nanjing, China), and were photographed using the VersaDoc 4000 MP system (Bio-Rad).
Bioinformatics Analysis
Four kinds of online software were used to predict the miR-144-3p target genes, including Targetscan1, PicTar2, miRmap3, and miRanda4. The precursor and mature sequences of miR-144 were obtained from miRBase5.
The GEO datasets series accession is GSE104441 (ID: 200104441). The GO map was analyzed by OmicShare Tools6.
Cis-element Cluster Finder7, Methprimer8, and JASPAR9 were used for promoter and transcription factors prediction.
Statistical Analysis
Statistical analysis was carried out with the SPSS software (21.0 version, IBM, United States). All data were presented as means ± standard error (SE). Two-tailed Student’s t-test was used to compare average difference between the groups. p < 0.05 was regarded as a statistically significant difference.
Results
MiR-144 Promotes Pre-adipocytes Differentiation
As T2D is closely related to adipogenesis, and miR-144 is significantly upregulated in T2D, the role it plays still needs to be further investigated. To macroscopically ascertain the role of miR-144 in regulating adipogenesis, we screened the published GEO data.
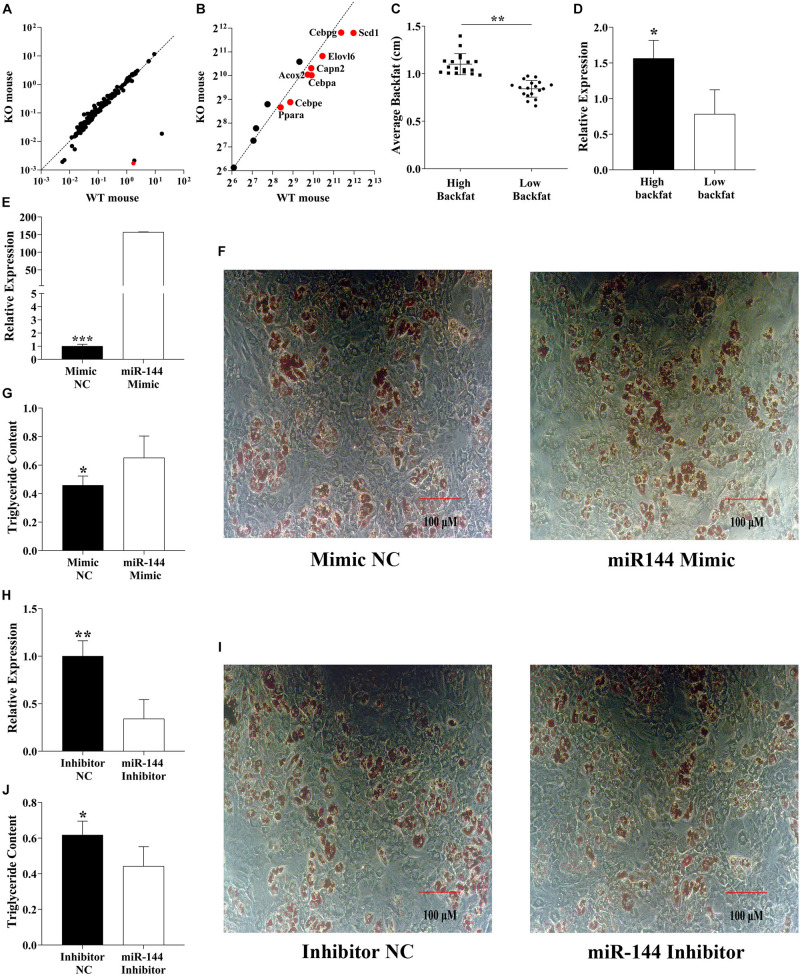
We found a miR-144 knock-out GEO dataset (GSE104441) (Figure 1A). By analysis, we found that some genes were involved in adipogenesis, mainly C/EBPα, C/EBPε, C/EBPγ, PPARα, CAPN2, and fatty acid synthesis-related genes, including ELOVL6 and SCD1 which were significantly downregulated (p < 0.05), as shown in Figure 1B.
FIGURE 1.
MiR-144 promotes pre-adipocyte differentiation. (A) Mouse knockout data from the GEO dataset (GSE104441) show the miR-144 specific knockout in mouse liver tissue. The red dot means miR-144. (B) Analysis of the adipogenic label genes in the GEO datasets (GSE104441). The above two dotted lines mean the standard line. (C,D) Detected miR-144 expressed in two backfat thickness groups. (E–G) Overexpressed miR-144-3p mimics in pre-adipocytes (E) then after 8-day differentiation inducer medium (DIM) stimulation, oil red O staining (F), and triglyceride assay (G), the results identified that miR-144 promoted pre-adipocyte differentiation. (H–J) Specific interference of miR-144 by its inhibitor confirms its regulatory function on adipogenesis. After statistical analysis, * in the figure indicates p < 0.05, **p < 0.01, and ***p < 0.001.
However, these data were sequenced from the liver rather than adipose tissue, so we detected the miR-144 expression level in porcine white adipose tissue (WAT). The samples were divided into two groups (every group had 18 individuals) by the thickness of backfat (0.84 and 1.10 cm, respectively, p < 0.01), miR-144 in the high backfat group had an abundant expression level (p < 0.05), as shown in Figures 1C,D.
To confirm the data we detected, we transfected miR-144 mimics and inhibitors to porcine pre-adipocytes, as shown in Figures 1E,H, respectively. After the transfection, the samples were stimulated with DIM for 8 days, then we analyzed the adipogenesis of the pre-adipocytes. The results show that miR-144 mimics promoted pre-adipocyte differentiation, and miR-144 inhibitors suppressed the differentiation, as shown in Figures 1F,G,I,J, respectively.
This result further proved that miR-144 promotes pre-adipocyte differentiation, which is consistent with another paper (Shen et al., 2018).
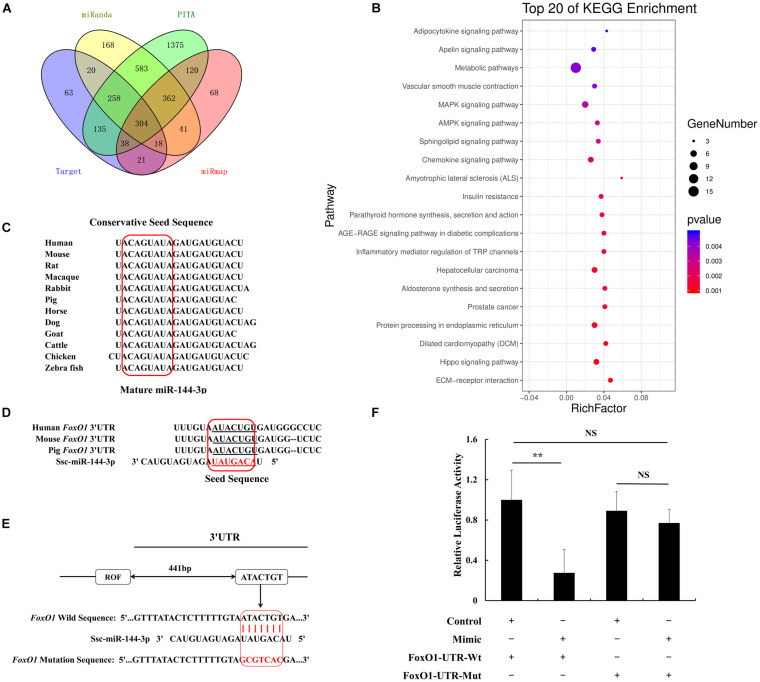
MiR-144 Targets 3′UTR of FoxO1
miRNA can bind the miRNA response elements (MREs) located at 3′UTR of target genes to play a post-transcriptional role. We chose four different frequently used prediction software, including TargetScan, PITA, miRanda, and miRmap to predict the target genes of miR-144. In total, 304 potential targets were retrieved (Figure 2A). We used the GO analysis to study whether the above genes were involved in the adipogenic signaling pathway, results show there was an insulin pathway among the top 20 pathways. Insulin resistance characterizes T2D which in turn identifies miR-144 as a biomarker of T2D (Lettieri-Barbato and Aquilano, 2020). Here, the insulin pathway by GO analysis, respectively, contained four genes, phosphatase and tensin homolog (PTEN), forkhead box O1 (FoxO1), protein kinase C epsilon (PRKCE), and protein phosphatase 1 catalytic subunit gamma (PPP1CC) (Figure 2B).
FIGURE 2.
MiR-144 targets FoxO1 by binding in its 3′UTR. (A) The Venn diagram of target genes prediction data. (B) Top 20 KEGG enrichment that predicted genes by Venn diagram. Dot means genes number, and the color means P-value. (C) Conservation estimation of mature miR-144 sequences within different species. (D) Conservation estimation of miRNA response elements (MREs) between human, mouse, and pig. (E) The location of miR-144 MREs in porcine FoxO1 gene 3′UTR. (F) The results of dual fluorescence activity detection of miR-144 targeting FoxO1. **p < 0.01.
Among them, FoxO1 has been demonstrated to be related to adipogenesis. Hence, FoxO1 was selected for this study. We found that mature miR-144 was conservative in the mass of species, as shown in Figure 2C. We estimated that the miR-144 MRE in FoxO1 would be located at 442–448 bp of its 3′UTR. The MRE in FoxO1 was found to be shared between humans, mice, and pigs, which was conservative as well (Figures 2D,E). We respectively, constructed a recombinant pmirGLO plasmid containing the wild and mutational MRE of FoxO1. According to the luciferase reporter assay data, we proved that miR-144 targeted FoxO1 (Figure 2F). The western blotting results by, respectively, transfecting miR-144 mimics and inhibitors also identified that miR-144 targeted FoxO1 (Figure 5H).
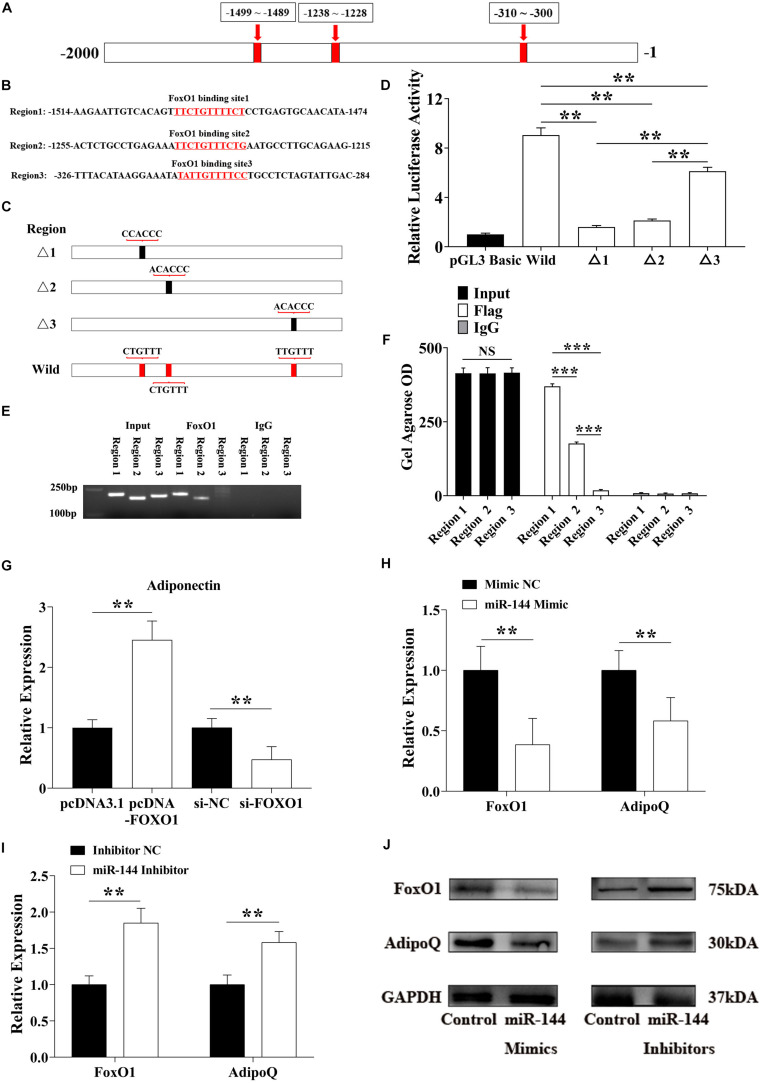
FIGURE 5.
miR-144 inhibits FoxO1 to reduce its regulation of adiponectin and promote adipogenesis. (A,B) The specific binding location of the FoxO1 binding site in the -2,000 bp region (A) and the response element at each site (B). (C) Constructed homologous recombinant pGL3 basic plasmid, including three mutational FoxO1 binding sites and a whole region contained three wild binding sites. (D) pGL3-regions directed luciferase gene reporter plasmid (pGL3-regions) was co-transfected with FoxO1 encoding pcDNA3.1+ plasmids in porcine pre-adipocytes. Luciferase activity was measured 48 h after transfection. (E,F) ChIP-PCR detected the FoxO1 binding sites in the AdipoQ promoter region (E), and the OD value was visualized by the ImageJ software (F). (G) Detected adiponectin expressed level after FoxO1 overexpression and interference. (H,I) Detected FoxO1 and adiponectin expressed level after miR-144 mimics (H) and inhibitors (I) overexpressed in pre-adipocytes. (J) Western blotting analysis of FoxO1 and adiponectin proteins expression level after miR-144 mimics and inhibitors overexpressed in pre-adipocytes. **p < 0.01 and ***p < 0.001, respectively.
FoxO1 Inhibits the Pre-adipocytes Differentiation
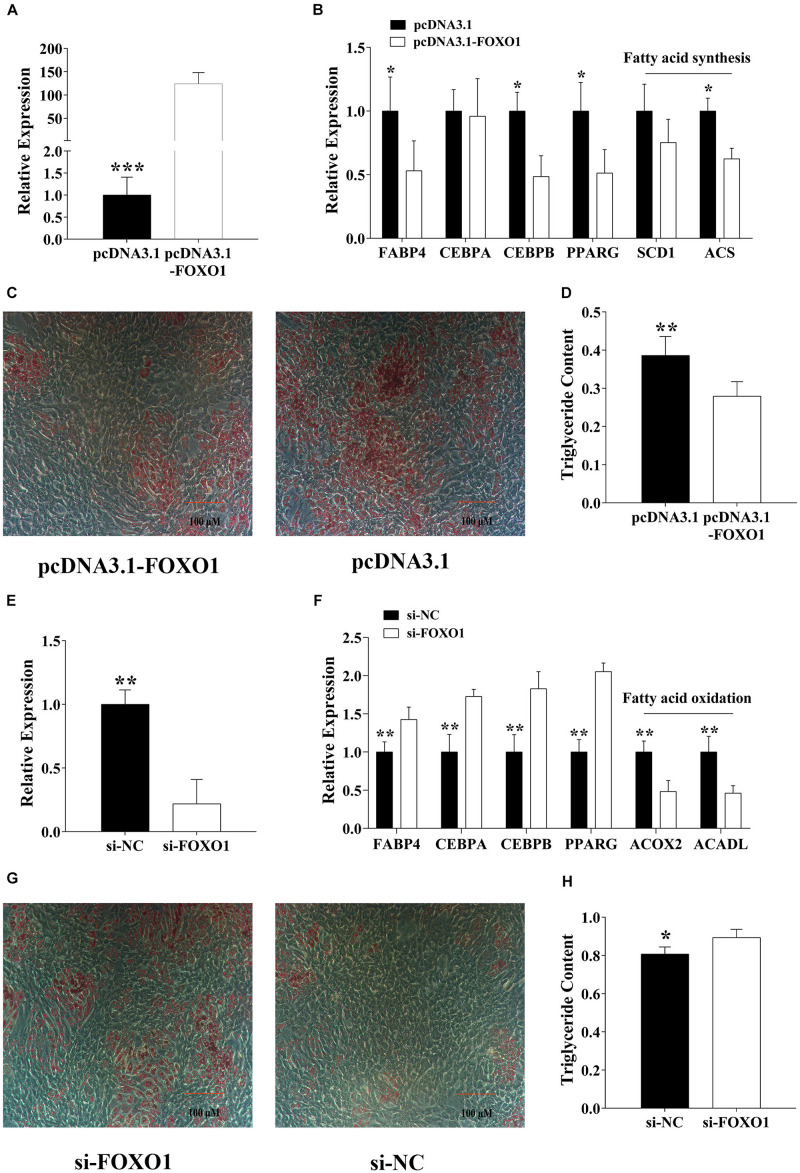
To identify the adipogenic role of FoxO1, we constructed a recombinant pcDNA3.1-FoxO1-CDs plasmid and transfected it to the pre-adipocytes by Lipofectamine 3000 for 48 h, then after 8-day DIM stimulation, we analyzed the adipogenesis (Figure 3A). Results show that, after FoxO1 transfection, the adipogenesis label genes, containing FABP4, C/EBPβ, and PPARγ were significantly downregulated, and fatty acid synthesis genes, for instance, SCD1 and ACS were also significantly downregulated (p < 0.05, Figure 3B). Oil red O staining and a triglyceride assay further identified that FoxO1 inhibited pre-adipocyte differentiation (Figures 3C,D). On the other hand, we also found that si-FoxO1 regulated pre-adipocytes (Figure 3E). In the detection of the expression level of the label genes, we found that all of them were upregulated (p < 0.05), and that FoxO1 was involved in fatty acid oxidation. Thus we detected that ACOX2 and ACADL, which were important label genes for fatty acid oxidation, were downregulated (p < 0.01).
FIGURE 3.
FoxO1 suppresses pre-adipocyte differentiation. (A) Overexpressed FoxO1 in pre-adipocytes. (B) Detected adipogenic label genes in pre-adipocytes after FoxO1 overexpression. (C,D) After 8-day differentiation inducer medium (DIM) stimulation, oil red O staining (C), and triglyceride assay (D). (E) Specific interference of FoxO1 by siRNA in pre-adipocytes. (F) After interference of FoxO1, the expression level of adipogenesis label genes in pre-adipocytes was detected. (G,H) After 8-day differentiation inducer medium (DIM) stimulation, oil red O staining (G), and triglyceride assay (H). *p < 0.05, **p < 0.01, and ***p < 0.001.
As mentioned above, we believe that FoxO1 suppressed pre-adipocyte differentiation by regulating fatty acid synthesis and oxidation.
MiR-144 and FoxO1 Co-regulate Adipogenesis
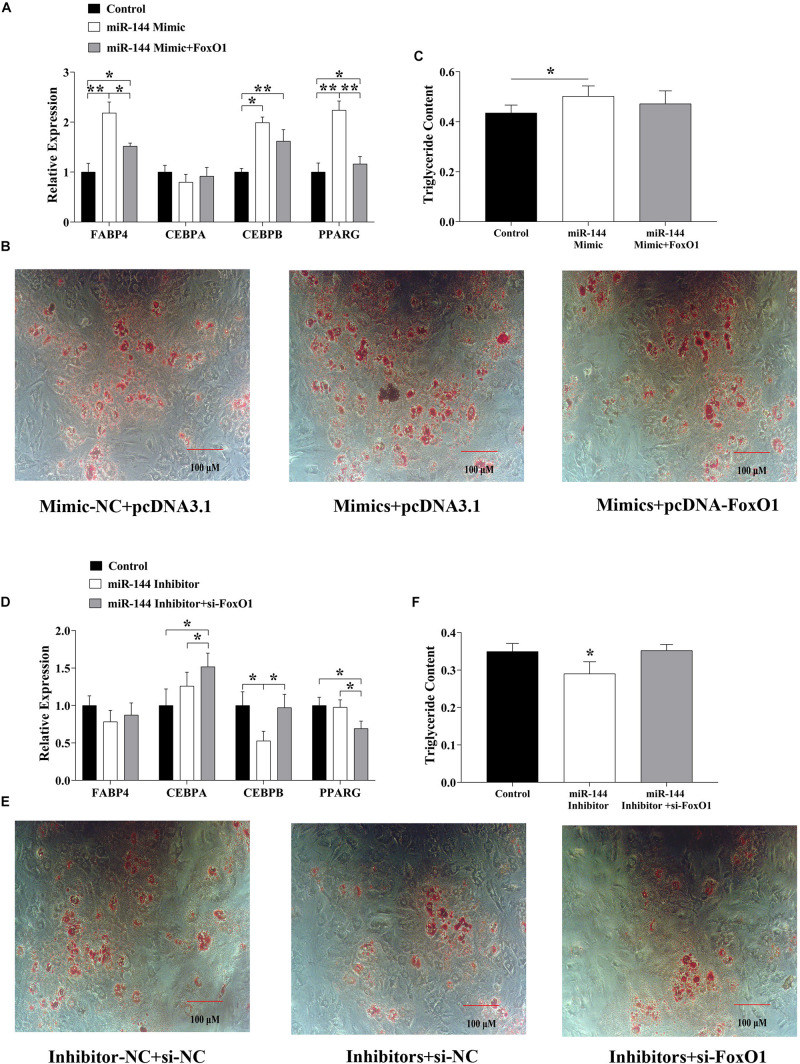
The above results have demonstrated that miR-144 promoted adipogenesis while FoxO1 inhibited it, meanwhile miR-144 targeted FoxO1. But how they both co-regulate adipogenesis still needs to be further explored. Hence, we grouped miR-144 mimics and FoxO1 inhibitors, and si-FoxO1 to identify their regulation by co-transfecting pre-adipocytes. As the results show, when we co-transfected miR-144 mimics and FoxO1, some adipogenic label genes, for instance, FABP4, C/EBPβ, and PPARγ were upregulated in the co-transfected group compared with the control group, while FABP4 and PPARγ were downregulated compared with the mimic group (p < 0.05), as shown in Figure 4A. Further adipogenic-induced results show that there was no significant difference between the control and the co-transfected group while it was significantly less than the mimic groups (Figures 4B,C). On the other side, when we co-transfected miR-144 inhibitors and si-FoxO1, the results identified that C/EBPα and C/EBPβ expressed in the co-transfected group was significantly more upregulated than the inhibitors group (p < 0.05), as shown in Figure 4D. Meanwhile, adipogenic-induced results also demonstrated that the co-transfected group recovered from the suppressed effect of adipogenesis found in the inhibitor group (Figures 4E,F).
FIGURE 4.
MiR-144 and FoxO1 co-regulated the adipogenesis. (A) Detected adipogenic label genes expressed in the miR-144 mimics and FoxO1 co-transfected group. (B,C) After 8-day differentiation inducer medium (DIM) stimulation, oil red O staining (B), and triglyceride assay (C). (D) Detected adipogenic label genes expressed in the miR-144 inhibitors and si-FoxO1 co-transfected group. (E,F) After 8-day differentiation inducer medium (DIM) stimulation, oil red O staining (E), and triglyceride assay (F). * p < 0.05, **p < 0.01.
MiR-144 Inhibits FoxO1 to Reduce Its Regulation of Adiponectin and Promote Adipogenesis
Adipose not only acts as an energy storage unit, but also as an important secretor. Adipose secrets a series of adipocytokines, among them, adiponectin is one of the most important. Adiponectin is the only adipocytokine to be found to play a negative regulatory role in adipogenesis so far.
In one report, FoxO1 bound the promoter of the AdipoQ gene in mice to activate the insulin signal pathway to promote fatty acid oxidation (Qiao and Shao, 2006). We chose the -2,000 bp region of the porcine AdipoQ gene transcriptional start site (TSS), and found three FoxO1 binding sites in this region (Figures 5A,B). To identify the most efficient promoter in the FoxO1 binding site, we constructed a homologous recombinant pGL3 basic plasmid, and found three mutational FoxO1 binding sites and a whole region containing three wild binding sites (Figure 5C). The co-transfection results revealed that binding site 1 and site 2 were, respectively, located at the −1,499 to −1,489 bp and −1,238 to −1,228 bp promoter regions. Both sites had high promoting efficiency, especial binding site 1 (Figure 5D). ChIP results further proved that only binding site 1 and site 2 had a PCR amplified band, the Gel Agarose OD showed that binding site 1 had significantly higher promoting efficiency than site 2 and site 3 (p < 0.01) (Figures 5E,F).
To confirm the results we detected, FoxO1 and si-FoxO1 were transfected to porcine pre-adipocytes, respectively. After 8-day DIM stimulation, endogenous adiponectin was detected, these results reconfirmed that FoxO1 promoted adiponectin expression by binding in its promoter region to regulate adipogenesis (Figure 5G). Moreover, as shown above, miR-144 inhibited FoxO1, here we, respectively, transfected miR-144 mimics and inhibitors to identify whether miR-144 affected adiponectin by inhibiting the FoxO1 expressed, and the results confirmed it (Figures 5H,I). Western blotting results further identified the above results (Figure 5J).
Discussion
Adipogenesis has attracted great attention in the field of human health. Extreme adipogenesis leads to an increase in weight and even obesity which seriously endangers the health of mankind. Obesity is the source of most metabolic diseases, including hypertension, cardiovascular disease, T2D, and so on (Kopelman, 2000; Goran et al., 2003). In one report, approximately 40% of the population of the world suffer the risk of obesity, among them, nearly 20% are teenagers (Reinehr, 2018). So further adipogenesis molecular mechanism exploration is essential to resolve various diseases that threaten human health caused by obesity. However, in livestock production, the focus of research on adipogenesis differs slightly from the medicine field. For a better taste, muscles with a certain amount of intramuscular fat (IMF) are often pursued, which differs completely from the field of human health, after all, IMF is one of the prevalent causes of T2D. Though the focus is different between the above fields, the inquiry into the mechanism is consistent between them.
With an adipogenesis consensus developed, it has been gradually verified that in addition to the classical molecular mechanism that regulates adipogenesis, the role that epigenetics play is also becoming increasingly recognized. The role of epigenetics includes but is not limited to DNA methylation, histone modification, chromatin remodeling, maternal effect, and non-coding RNA (ncRNA). Among them, the mechanisms of DNA methylation and ncRNAs have been more thoroughly studied. In general, DNA methylation inhibits the transcription of genes, leading to a reduction in expression level. While ncRNA involves long non-coding RNAs (lncRNAs), circle RNAs (circRNA), pseudogenes, and miRNA. Except for lncRNAs, miRNAs are generally known as the most high-profile ncRNAs.
It is well known that miRNAs are involved in a mass of biological activities including adipogenesis, among them, some famous miRNAs including miR-27 and miR-130, have been demonstrated to play an important role in adipogenesis (Eun Kyung et al., 2011; Pan et al., 2014; Zhang et al., 2014). In this study, we found that miR-144 was significantly upregulated in WAT (p < 0.01) and we identified that it promoted pre-adipocyte differentiation (Figure 1). This result is consistent with the conclusion in another study that miR-144 promotes adipogenesis (Shen et al., 2018). The function of miRNAs mainly depends on binding in the 3′UTR of its target genes in order to play a post-transcriptional role, we proved that FoxO1 was one of its targets (Figure 2).
The FoxOs family contains four members, FoxO1, FoxO3, FoxO4, and FoxO6, which are nearly expressed in all tissues. Furthermore, the FoxOs oversee a mass of cellular processes, for instance, differentiation, proliferation, metabolism, apoptosis, autophagy, and stress resistance in numerous tissues. FoxO1 was the first to be discovered in humans, meanwhile in homozygous knockout mice it led to embryonic lethality (Hosaka et al., 2004). FoxO1 plays a crucial role in ruling the cell cycle and cellular metabolism (Li et al., 2017). Above all, FoxO1 is massively expressed in adipose tissue, where it is involved in regulating adipocyte differentiation and trans-differentiation, oxidative stress defense, and lipid metabolism (Gómezlópez et al., 2007; Lettieri Barbato et al., 2014). FoxO1 regulates the process of adipogenesis, it achieves this by acting as a regulator in insulin signaling. In detail, fasting stimulates the activation of the lipolytic pathway and promotes the breakdown of triglycerides and the releases of free fatty acids from adipose tissue, FoxO1, in this context, transcribes genes involved in lipid catabolism by signaling pathway inhibition, whereas, in the feeding condition the reverse is seen (Assimacopoulos-Jeannet et al., 1995; Lettieri Barbato et al., 2014; Ioannilli et al., 2020).
We subsequently detected the regulatory effect of FoxO1 on adipogenesis. Consistent with our findings, FoxO1 has a significant inhibitory effect on adipogenesis (Figure 3). To confirm the co-regulation of miR-144 and FoxO1 on adipogenesis, we co-transfected miR-144 and FoxO1. The results further proved that miR-144 and FoxO1 adipogenesis regulation is unified (Figure 4). However, there was another paper that demonstrated that miR-144 targeted C/EBPα. As one of the most crucial regulators of adipogenesis, C/EBPα is recognized as a positive factor of adipogenesis, thus the function of miR-144 on adipogenesis seems to be debatable (Itziar et al., 2017). However, our results clearly show that miR-144 has a promoting effect on adipogenesis. Perhaps there are mechanisms that need to be further explored.
FoxO1 is involved in the insulin signaling pathway as a regulator. FoxO1 can bind to a series of targets, we can finally verify that adiponectin is its target as confirmed by our estimation. Adiponectin is widely recognized as an inhibitor of adipogenesis, therefore, we only needed to detect whether FoxO1 had the ability to bind to the adiponectin gene promoter region and facilitate its expression. Moreover, miR-144 inhibits FoxO1, hence miR-144 would regulate adiponectin by inhibiting FoxO1. The results demonstrated that these two have a unified regulatory effect on adipogenesis, which confirms the hypothesis. Meanwhile, we further determined the key binding site of FoxO1 in the adiponectin gene promoter region, the results identified that binding site 1 and site 2 have significant promoting efficiency, especially site 1, which provides a basis for subsequent research (Figure 5).
In addition, regarding the regulatory function of miRNA, one of the most interesting hypotheses at present is the competitive endogenous RNAs (ceRNAs) hypothesis (Cesana et al., 2011). The theoretical basis of this hypothesis is based on the shared MREs between different target genes. In this study, we did not consider the ceRNA effect, the main reason is we believe that the “competitiveness” caused by a single miRNA is very limited, and the regulatory role of the overall endogenous environment has yet to be considered (Denzler et al., 2014; Chiu et al., 2018). However, if there is a co-interaction net containing more than one miRNA, the ceRNA effect is a regulatory perspective that is worth exploring.
Conclusion
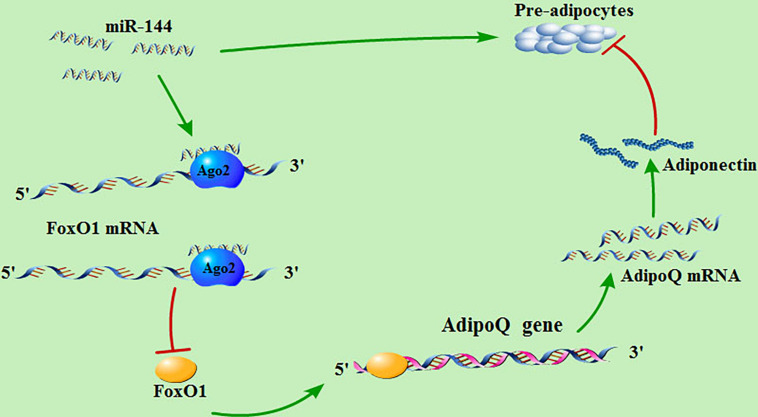
As mentioned above, our research indicates that miR-144 targets FoxO1, thus reducing its expression and inhibiting its promotional effect on adiponectin, thereby alleviating the inhibitory effect of adiponectin on adipogenesis (Figure 6).
FIGURE 6.
MiR-144 regulates the signaling pathway of adipogenesis in pre-adipocytes. miR-144 targets FoxO1, thus reducing its expression and inhibiting its promotional effect on adiponectin, thereby alleviating the inhibitory effect of adiponectin on adipogenesis.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of the Nanjing Agricultural University.
Author Contributions
WL conceived the idea, performed the analyses, and wrote the manuscript. WL and YT implemented the package. YZ and JZ contributed to the data analysis and checked the manuscript. WW and LZ checked the finished manuscript. JC conceived the idea, supervised the project analysis, and contributed to the manuscript preparation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31872334 and 31902132) and the Fundamental Research Funds for the Central Universities (Grant No. KJQN202040).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.603144/full#supplementary-material
References
- Assimacopoulos-Jeannet F. O., Brichard S., Rencurel F., Cusin I., Jeanrenaud B. (1995). In vivo effects of hyperinsulinemia on lipogenic enzymes and glucose transporter expression in rat liver and adipose tissues. Metab. Clin. Exp. 44 228–233. 10.1016/0026-0495(95)90270-8 [DOI] [PubMed] [Google Scholar]
- Barroso I., Gurnell M., Crowley V., Agostini M., Schwabe J., Soos M., et al. (1999). Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402 880–883. 10.1038/47254 [DOI] [PubMed] [Google Scholar]
- Cao Z., Umek R., McKnight S. (1991). Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5 1538–1552. 10.1101/gad.5.9.1538 [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. F., Yan X., Ying P., Yun G., Jin Q., Chu G. Y., et al. (2017). miR-425-5p inhibits differentiation and proliferation in porcine intramuscular preadipocytes. Int. J. Mol. Sci. 18:2101. 10.3390/ijms18102101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H., Martínez M., Komissarova E., Llobet-Navas D., Bansal M., Paull E., et al. (2018). The number of titrated microRNA species dictates ceRNA regulation. Nucleic Acids Res. 46 4354–4369. 10.1093/nar/gky286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R., Agarwal V., Stefano J., Bartel D. P., Stoffel M. (2014). Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54 766–776. 10.1016/j.molcel.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doney A., Fischer B., Leese G., Morris A., Palmer C. (2004). Cardiovascular risk in type 2 diabetes is associated with variation at the PPARG locus: a Go-DARTS study. Arterioscler. Thromb. Vasc. Biol. 24 2403–2407. 10.1161/01.ATV.0000147897.57527.e4 [DOI] [PubMed] [Google Scholar]
- Eun Kyung L., Jeong L. M., Kotb A., Wook K., Kim M. M., Subramanya S., et al. (2011). miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol. Cell. Biol. 31 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T., Seok S., Choi S., Huang Z., Suino-Powell K., Xu H. E., et al. (2014). MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell. Biol. 34 4130–4142. 10.1128/mcb.00596-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómezlópez G., Cañamero M., Gonzálezbarroso M. M., Rial E., Valverde ÁM., Serrano M. (2007). Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 15 382–394. 10.1016/j.cmet.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Goran M., Ball G., Cruz M. (2003). Obesity and risk of type 2 diabetes and cardiovascular disease in children and adolescents. J. Clin. Endocrinol. Metab. 88 1417–1427. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., Ha I., et al. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106 23–34. 10.1016/s0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Hamilton A. J., Baulcombe D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. 10.1126/science.286.5441.950 [DOI] [PubMed] [Google Scholar]
- Hamm J., Park B., Farmer S. (2001). A role for C/EBPbeta in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 276 18464–18471. 10.1074/jbc.M100797200 [DOI] [PubMed] [Google Scholar]
- Hammarstedt A., Gogg S., Hedjazifar S., Nerstedt A., Smith U. (2018). Impaired adipogenesis and dysfunctional adipose tissue in human hypertrophic obesity. Physiol. Rev. 98 1911–1941. 10.1152/physrev.00034.2017 [DOI] [PubMed] [Google Scholar]
- Han H., Gu S., Chu W., Sun W., Chen J. (2017). miR-17-5p regulates differential expression of NCOA3 in pig intramuscular and subcutaneous adipose tissue. Lipids 52 939–949. 10.1007/s11745-017-4288-4 [DOI] [PubMed] [Google Scholar]
- Hosaka T., Biggs W., Tieu D., Boyer A., Varki N., Cavenee W., et al. (2004). Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U.S.A. 101 2975–2980. 10.1073/pnas.0400093101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannilli L., Ciccarone F., Ciriolo M. (2020). Adipose tissue and FoxO1: bridging physiology and mechanisms. Cells 9:849 10.3390/cells9040849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itziar E., Arrate L., Jonatan M., Ana G., Portillo M. P., Cristina Ó. (2017). Potential miRNA involvement in the anti-adipogenic effect of resveratrol and its metabolites. PLoS One 12:e0184875. 10.1371/journal.pone.0184875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolina D., Armugam A., Tavintharan S., Wong M., Lim S., Sum C., et al. (2011). MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One 6:e22839. 10.1371/journal.pone.0022839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R. F., Fischer S. E., Bernstein E., Sijen T., Hannon G. J., Plasterk R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15 2654–2659. 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman P. (2000). Obesity as a medical problem. Nature 404 635–643. 10.1038/35007508 [DOI] [PubMed] [Google Scholar]
- Lettieri Barbato D., Aquilano K., Ciriolo M. R. (2014). FoxO1 at the nexus between fat catabolism and longevity pathways. Biochim. Biophys. Acta 1841 1555–1560. 10.1016/j.bbalip.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Lettieri-Barbato D., Aquilano K. (2020). Aging and immunometabolic adaptations to thermogenesis. Ageing Res. Rev. 63:101143. 10.1016/j.arr.2020.101143 [DOI] [PubMed] [Google Scholar]
- Li M., Liu Z., Zhang Z., Liu G., Sun S., Sun C. (2015). miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 396 235–244. 10.1515/hsz-2014-0241 [DOI] [PubMed] [Google Scholar]
- Li Y., Ma Z., Jiang S., Hu W., Li T., Di S., et al. (2017). A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 66 42–49. 10.1016/j.plipres.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Liang Y., Dong J., Zhang J., Wang S., He Y., Yan Y. (2018). Identification of neuroendocrine stress response-related circulating MicroRNAs as biomarkers for type 2 diabetes mellitus and insulin resistance. Front. Endocrinol. 9:132. 10.3389/fendo.2018.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monajemi H., Zhang L., Li G., Jeninga E., Cao H., Maas M., et al. (2007). Familial partial lipodystrophy phenotype resulting from a single-base mutation in deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-gamma. J. Clin. Endocrinol. Metab. 92 1606–1612. 10.1210/jc.2006-1807 [DOI] [PubMed] [Google Scholar]
- Pan S., Yang X., Jia Y., Li R., Zhao R. (2014). Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-γ expression. J. Cell. Physiol. 229 631–639. 10.1002/jcp.24486 [DOI] [PubMed] [Google Scholar]
- Qiao L., Shao J. (2006). SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein ? Transcriptional complex. J. Biol. Chem. 281 39915–39924. 10.1074/jbc.m607215200 [DOI] [PubMed] [Google Scholar]
- Reinehr T. (2018). Long-term effects of adolescent obesity: time to act. Nat. Rev. Endocrinol. 14 183–188. 10.1038/nrendo.2017.147 [DOI] [PubMed] [Google Scholar]
- Shen L., Li Q., Wang J., Zhao Y., Zhu L. (2018). miR-144-3p promotes adipogenesis through releasing C/EBPα from Klf3 and CtBP2. Front. Genet. 9:677. 10.3389/fgene.2018.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. E., Li Y. F., Jia L., Ji H. L., Song Z. Y., Cheng J., et al. (2014). MicroRNA-199a-5p affects porcine preadipocyte proliferation and differentiation. Int. J. Mol. Sci. 15 8526–8538. 10.3390/ijms15058526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li W., Bai Y., Yang W., Fang M. (2017). ssc-miR-7134-3p regulates fat accumulation in castrated male pigs by targeting MARK4 gene. Int. J. Biol. Sci. 13 189–197. 10.7150/ijbs.17386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., He K., Wang L., Hu J., Gu J., Zhou C., et al. (2015). Stk40 represses adipogenesis through translational control of CCAAT/enhancer-binding proteins. J. Cell Sci. 128 2881–2890. 10.1242/jcs.170282 [DOI] [PubMed] [Google Scholar]
- Zhang M., Wu J., Chen W., Tang S., Mo Z., Tang Y., et al. (2014). MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis 234 54–64. 10.1016/j.atherosclerosis.2014.02.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.