Abstract
We examined the effect of ploidy on mitochondrial DNA (mtDNA) copy number in embryos and the amount of cell-free mitochondrial and nucleic DNA content (cf-mtDNA and cf-nDNA) in spent culture medium (SCM). Oocytes collected from the ovaries were matured, activated, incubated in medium containing cycloheximide (CHX) or CHX and cytochalasin B (CB) for 4.5 h to produce haploid or diploid embryos (H-group and D-group embryos). These embryos were cultured for 7 days, and the blastocysts and SCM were examined. The amount of mtDNA and nDNA was determined by real-time PCR. The rate of development to the blastocyst stage was higher for the D-group than for the H-group. Moreover, D-group blastocysts had less mtDNA compared to the H-group blastocysts. After activation, the mitochondrial content was constant before the blastocyst stage in D-group embryos, but increased earlier in H-group embryos. The amount of cf-mtDNA in the SCM of D-group blastocysts was greater than that of H-group blastocysts. However, when the cf-mtDNA in the SCM of 2 cell-stage embryos (day 2 post-activation) was examined, the amount of cf-mtDNA was greater in the H-group than in the D-group embryos. When D-group embryos were cultured for 7 days, a significant correlation was observed between the total cell number of blastocysts and cf-nDNA content in the SCM. Hence, although careful consideration is needed regarding the time point for evaluating mtDNA content in the embryos and SCM, this study demonstrates that mtDNA in the embryos and SCM was affected by the ploidy of the embryos.
Keywords: Blastocyst, Cell-free DNA, Mitochondrial number, Ploidy
Mitochondria are the main energy generators of cells and play a variety of roles in cellular homeostasis, including calcium regulation and apoptosis. The number of mitochondria increases during oocyte growth [1], and oocytes with a greater mitochondrial number have higher maturation, fertilization, and developmental competence [2,3,4]. Although the relationship between mitochondrial number and embryonic quality has been studied extensively in oocytes, studies in humans and mice have found that blastocysts with low mitochondrial content have high implantation capacity [5,6,7], while other studies have not detected this relationship [8,9,10]. In addition, blastocysts with chromosomal abnormalities have been reported to have greater mitochondrial numbers compared to euploid blastocysts [5, 11,12,13]; however, this relationship was not detected in other studies [7,8,9]. Therefore, more evidence is needed to confirm whether mitochondrial number can be used to evaluate embryonic quality and chromosomal abnormalities.
Cell-free DNA (cfDNA) is a potential noninvasive marker for predicting embryonic quality. cfDNA is derived from cellular secretions and apoptotic bodies and consists of nucleic and mitochondrial DNA (nDNA and mtDNA) [14]. Stifliani et al. [15] reported that a high mtDNA/nDNA ratio in spent culture medium (SCM) is associated with the implantation outcome and embryonic quality in humans. In contrast, they also reported that the amount of cell-free mtDNA (cf-mtDNA) in SCM was positively correlated to the extent of embryonic fragmentation [16]. Further, the amount of cfDNA content in SCM was successfully used to predict chromosomal abnormalities in embryos [17]. However, a low concordance was previously reported between the amount of cfDNA content in the medium and chromosomal abnormalities in embryos obtained from trophectoderm cell (TE) using biopsy analysis [18, 19]. The cf-mtDNA/cf-nDNA ratio in the medium was found to not differ between aneuploid and euploid embryos in humans [20]. However, further evidence is needed to evaluate the amount of cfDNA in the SCM of embryos.
Many factors have hampered the evaluation of mtDNA content in SCM. For example, IVF embryos have multiple sperms attached to the zona pellucida (ZP) and oolemma. Moreover, embryos that require careful handling often have cumulus cells attached to the ZP, which may affect the measurement of mtDNA copy number in the medium [21]. In addition, cryopreservation process has been previously found to affect the mtDNA content in SCM [22]. Other factors that may affect the secretion of mtDNA into the culture medium include the physical handling of oocytes, the presence of ZP, making a cut in ZP, and the invasive manipulation of embryos. In the present study, we created haploid and diploid parthenogenetically activated embryos and evaluated the amount of cf-nDNA and cf-mtDNA content in SCM and the mtDNA copy number in embryos. Furthermore, we investigated the factors that strongly affect the cfDNA content in SCM and evaluated whether embryo ploidy affects mtDNA content in embryos and cfDNA content in SCM.
Materials and Methods
Chemicals and media
All drugs used in this study were purchased from Nacalai Tesque (Kyoto, Japan), unless stated otherwise. The medium used for the in vitro maturation (IVM) of oocytes was TCM199 (Gibco, Paisley, UK), supplemented with 10% v/v porcine follicular fluid, 0.5 mM L-cysteine, 1 mM L-glutamine, 0.9 mM sodium pyruvate, 10 ng/ml epidermal growth factor (Sigma-Aldrich, St. Louis, MO, USA), 5% fetal calf serum, 10 IU/ml equine chorionic gonadotropin (ASKA Pharma, Tokyo, Japan), and 10 IU/ml human chorionic gonadotropin (Fuji Pharma, Tokyo, Japan). Porcine follicular fluid was aspirated from the antrum follicles (3–5 mm in diameter) of gilts, centrifuged (10,000 × g for 5 min), and stored at ‒20ºC until use. The medium used for the in vitro culture (IVC) of embryos was porcine zygote medium 3 (PZM3) [23], which contained polyvinyl alcohol (PVA). IVM was performed under atmospheric conditions of 5% CO2 and 95% air at 38.5ºC, and IVC was performed under atmospheric conditions of 5% O2, 5% CO2, and 90% N2 at 38.5ºC.
Oocyte collection and in vitro maturation
The ovaries were collected from gilts at a slaughterhouse, stored at 37ºC in phosphate-buffered saline (PBS) containing antibiotics, and transported to the laboratory within 1 h. Cumulus oocyte complexes (COCs) were aspirated from antral follicles (3–5 mm in diameter) using a syringe with a 21 G needle (Terumo, Tokyo, Japan). The COCs were cultured in IVM medium for 44 h (10 COCs/100 µl drops) and then subjected to activation.
Parthenogenetic activation of oocytes and in vitro culture of embryos
After IVM, the oocytes were denuded from the surrounding cumulus cells by vortexing in hyaluronidase (0.5% w/v) with IVC medium for 6 min. These oocytes were examined for the absence of cumulus cells under a stereo microscope (Olympus, Tokyo, Japan). The oocytes were then activated using a single electrical pulse of 100 V for 0.1 msec and CUY500P1 electrode (NEPA21; NepaGene, Chiba, Japan). The oocytes were divided into two groups: (i) oocytes incubated in IVC medium containing 10 µM cytochalasin B (CB) and 10 µM cycloheximide (CHX) for 4.5 h and (ii) oocytes incubated in IVC medium containing CHX for 4.5 h. The first group of oocytes was found to inhibit second polar body (PB) extrusion, resulting in diploid embryos (D-group embryos), while the second group of oocytes extruded second PB, resulting in haploid embryos (H-group embryos). Zygotes were individually cultured in 10 µl of IVC medium.
Validation of differential ploidy between oocytes treated with or without CB
After IVM, oocytes with first PB were selected under a stereo microscope and activated to create the D-group and H-group embryos described above. Eighteen hours after activation, the presumptive zygotes were stained with Hoechst 33342 and examined for the presence of a second PB under a florescence microscope (IX71; Olympus). Embryos with two PBs were defined as haploid zygotes, while oocytes with only one PB were defined as diploid zygotes. Oocytes with unclear images (polar bodies overlapped with another polar body or nucleus) were categorized as diploid zygotes (1PB). Three days after activation, cleaved embryos at the 8 cell-stage were morphologically selected using a stereomicroscope (Olympus), stained with Hoechst 33342, and observed under a fluorescence microscope (IX71; Olympus) to obtain fluorescence images of the embryos and under a Leica DMI 6000B microscope to determine the number of cells (nucleus number) using LAS AF software (Leica, Wetzlar, Germany). The fluorescence intensities were quantified using ImageJ software (NIH, Bethesda, MD, USA) and divided by the number of nuclei.
Removal of ZP from zygotes and incubation of ZP-free zygotes
The oocytes were activated and incubated with CB and CHX for 4.5 h to create D-group embryos. Eighteen hours after activation, half of the presumptive zygotes were treated with 0.1% proteinase for 1 min to remove ZP and washed five times in IVC medium. The ZP-free zygotes and non-treated zygotes were incubated individually in 10 µl of culture medium for 7 days. A total of 240 oocytes were activated, and the SCM of morphological clear blastocysts (clear cavity, inner cell mass) and dead embryos (cleaved but morphologically degraded before the blastocyst stage) were used.
Developmental ability of H-group and D-group embryos, mtDNA content in the blastocysts, and cf-mtDNA and cf-nDNA content in the corresponding SCM
Forty oocytes were activated and incubated with CHX and CB, or CHX for 4.5 h to create H-group and D-group embryos, followed by culturing for 7 days to examine the time taken to develop to the blastocyst stage (i.e., the developmental rate). At day 7, one or two blastocysts were examined for their total cell number, while other blastocysts and the corresponding medium was used for DNA extraction and real-time PCR. The experiment was repeated 20 times.
Correlation between total cell number and mitochondrial number in the embryos and cfDNA content in the SCM
Forty oocytes were activated and incubated with CHX and CB for 4.5 h to create D-group embryos, followed by culturing for 7 days. The experiment was repeated 12 times. In the first six trials, the total cell number of all of the developed blastocysts and the cf-mtDNA and cf-nDNA content in the corresponding SCM were measured. Then, the correlation among them was examined. In the second six trials, the mtDNA in blastocysts and the cf-mtDNA and cf-nDNA content in the corresponding SCM were measured; their correlations were also examined.
Kinetics of mtDNA content in embryos during development
Forty oocytes were activated and incubated with CHX or CB and CHX for 4.5 h to create H-group and D-group embryos, followed by culturing for 7 days. During the culture period, 2–4-cell stage embryos, 4–8-cell stage embryos, > 8-cell stage embryos, and blastocysts were randomly selected on days 2, 3, 4 and 7 after activation. The experiment was repeated six times, and the mtDNA copy number in these embryos was measured.
Amount of cf-mtDNA in the SCM of 2-cell stage embryos
In this experiment, 40 oocytes were activated and incubated with CHX or CB and CHX for 4.5 h to create H-group and D-group embryos, followed by culturing. Twenty-four hours after activation, the embryos were stained with Hoechst 33342 to evaluate the PB number. The medium of the 2-cell stage embryos with 2PB and 1PB was collected. The experiment was repeated six times, and pooled samples were used for measuring cf-mtDNA content in the SCM.
Collection of SCM and extraction of DNA from the medium and embryos
The embryos were washed in culture medium and individually transported in a tube containing DNA extraction buffer (final concentration: 20 mM Tris, 0.4 mg/ml proteinase K, 0.9% Nonidet P-40, and 0.9% Tween-20). The SCM of all the samples (10 µl) was transported in a tube and centrifuged to separate cellular debris for 30 sec (3000 rpm). The resulting supernatant (6 µl) was mixed with the same volume of (× 2) DNA extraction buffer. DNA extraction was conducted by heating the sample at 55ºC for 30 min, followed by incubation at 98ºC for 5 min.
Measurement of mtDNA and nucleic DNA content
The copy number of nucleic DNA and mtDNA was determined by polymerase chain reaction (PCR) using a real-time system (Bio-Rad Laboratories, Hercules, CA, USA) as described before [24,25,26,27]. Briefly, 6 µl of sample, 4 µl of primers (final concentration is 0.25 µM), and 10 µl of KAPA SYBER Fast (Kapa Biosystem, Cape Town, South Africa) were used for PCR. Real-time PCR was performed at 95ºC for 3 min, followed by 40 cycles at 98ºC for 5 sec and 60ºC for 11 sec. The primer set was designed using Primer-BLAST: forward 5′-ATCCAAGCACTATCCATCACCA-3′ and reverse 5′-CCGATGATTACGTGCAACCC-3′ (NC_000845.1, 10277-10431, 155 bp) were used for mtDNA; forward 5′-AGCAGAATCAACACCATCGGT-3′ and reverse 5′-TGGCTCCACCCATAGAATGC-3′ (NC_010457, Chr15, GCG Glucagon, 154 bp) were used for nucleic DNA. A standard curve was generated for each assay using 10-fold serial dilutions representing copies of the external standard (water, 2, 20, 200, 2000, 20,000, 200,000, and 2,000,000 copies), which was the PCR product of the corresponding gene cloned into a vector using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA). To obtain the copy number of the standard DNA, the concentration of the standard DNA, the molecular weight of the vector, and Avogadro’s number were used. Prior to its use, this product was sequenced for confirmation. In all trails, the CT value of the most diluted standard (2 copy) was greater than that of water. Although the efficiency of the amplification was over 1.98 in all trials, the measurement of the copy number of mtDNA or nucleic DNA was conducted once using a 96-well plate, taking into account intra-plate variation. Therefore, the total number of samples used for measurement was restricted to approximately 80 (standard, 12; water, 2–4; sample, 80).
Expression levels of TOMM20 in embryos
Forty oocytes were activated and incubated with CHX, CB, or CHX for 4.5 h to create H-group and D-group embryos, followed by culturing for 4 days. The embryos (> 8-cell stage) were randomly selected and fixed in 4% paraformaldehyde for one day. This embryo culture procedure was repeated eight times. The pooled embryos were immunoassayed against TOMM20. The incubation period was determined through the above experiment, where the mtDNA content in the embryos differed between the H-group and D-group. The primary antibody used was rabbit polyclonal anti-TOMM20 (ab31163; Abcam, Cambridge, UK), and the secondary antibody used was anti-rabbit IgG (H + L) F(ab')2 fragment (Alexa Fluor® 555 Conjugate; Cell Signaling Technology, Danvers, MA, USA). The embryos were mounted onto glass slides using an anti-fade reagent containing DAPI and were observed under a Leica DMI6000B microscope using LAS AF software (Leica). The fluorescence intensities were quantified using ImageJ software (NIH).
Measurement of mitochondrial reactive oxygen species
One hundred twenty oocytes were matured, activated, and incubated with CHX, CB, or CHX for 4.5 h to create the H-group and D-group embryos, followed by culturing for 4 days. Randomly selected embryos (> 8-cell stage) were examined for their reactive oxygen species (ROS) content using MitoSOX (mitochondrial superoxide indicator; Invitrogen). Fluorescent images of the embryos were captured using a Leica DMI6000B microscope using LAS AF software (Leica), and fluorescence intensity was calculated using ImageJ software (NIH).
Statistical analysis
The data were compared between D-groups and H-groups using Student’s t-test, and the rate of development (%) was arcsine transformed prior to the analysis. The ratios of PB extrusion were compared using the chi-square test. P value less than 0.05 was considered a significant difference.
Results
Effect of CB treatment on the ploidy of zygotes post-activation
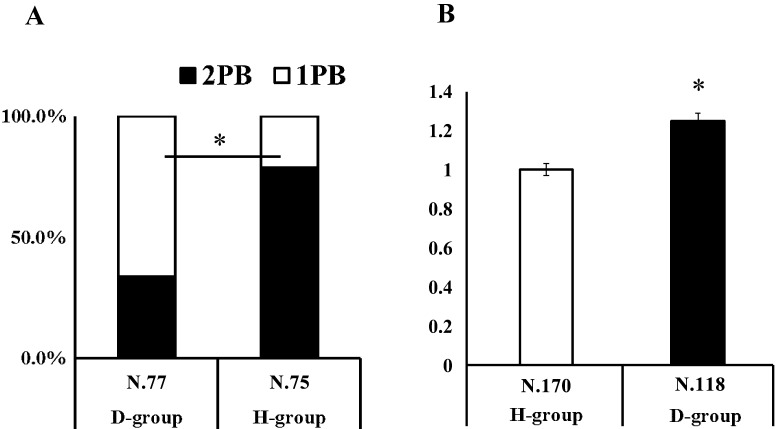
As shown in Fig. 1, the ratio of zygotes with 2PB was significantly higher for H-group zygotes than for D-group zygotes (78.7% vs. 33.8%; P < 0.05). In addition, the average fluorescence intensity of the nucleus was higher in the D-group than in the H-group (P < 0.05).
Fig. 1.
The oocytes were activated and incubated with cycloheximide (CHX) and cytochalasin B (CB) or with CHX for 4.5 h. The presence of polar body (PB) (A) and the intensity of nuclear staining by Hoechst 33342 (B) were compared between the D-group and H-group embryos. * indicates significant differences (P < 0.05). A: Ratio (%) of embryos with 2PB or 1PB after activation. B: Fluorescence intensity of embryos stained with Hoechst 33342. Average intensity of H-Group was defined as 1.0.
Effect of the presence of ZP on the cfDNA content of the SCM
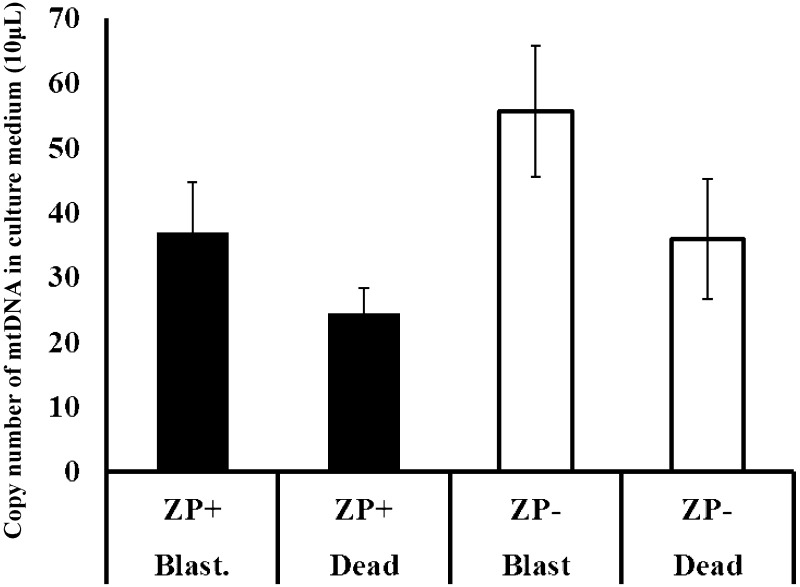
We evaluated whether ZP inhibited the dispersion of cf-mtDNA into the SCM from the embryos. As shown in Fig. 2, the presence of ZP tended to prevent the dispersion of cf-mtDNA into the medium, resulting in a higher amount of cf-mtDNA in the SCM of blastocysts without ZP than in blastocysts with ZP; however, the difference was not significant (P = 0.082). In addition, the amount of cf-mtDNA was lower in the SCM of dead embryos than that of blastocysts, but this difference was also not significant. Based on these results and taking into account the difficulties associated with handling embryos and the SCM of ZP-free embryos, non-treated (ZP-intact embryos) were used in subsequent experiments.
Fig. 2.
Effect of the presence of zona pellucida (ZP) on cell-free mitochondrial DNA (cf-mtDNA) content in the culture medium of D-group embryos with or without ZP after incubation for 7 days. The culture medium of blastocysts and dead embryos was examined.
Developmental ability, mtDNA copy number, and cf-mtDNA and cf-nDNA content in the SCM of the D-group and H-group embryos
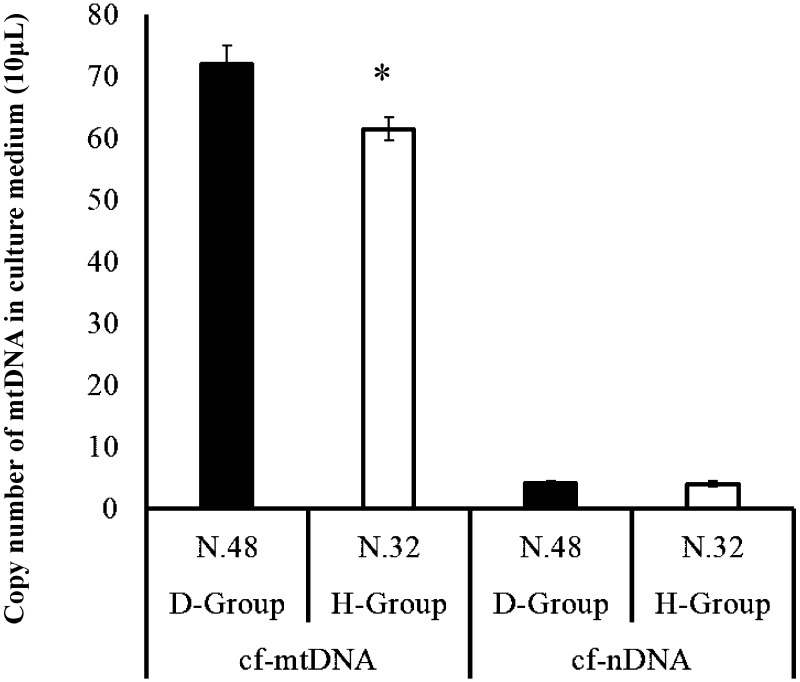
The developmental rate of oocytes to the blastocyst stage was significantly higher in the oocytes treated with CB (D-group) than in those not treated with CB (H-group). However, the total cell number of the blastocysts did not differ between the two groups (Table 1). The mtDNA copy number was greater for the H-group blastocysts than for the D-group blastocysts (Table 1). Furthermore, the amount of cf-mtDNA in the SCM was higher in the D-group than in the H-group (P < 0.05). However, the amount of cf-nDNA did not differ between the two groups (Fig. 3).
Table 1. Developmental ability and mitochondrial DNA (mtDNA) copy number of the D-group and H-group embryos.
| Group | No. of trials | No. of oocytes | No. of blastocyst | Rate of blastulation | No. of blastocyst | TCN | No. of blastocyst | mtDNA copy number |
|---|---|---|---|---|---|---|---|---|
| D | 20 | 800 | 94 | 11. 8 ± 1.3 a | 21 | 31.8 ± 1.9 | 48 | 219,494 ± 12,686 a |
| H | 20 | 800 | 42 | 5.1 ± 0.6 b | 10 | 33.9 ± 2.9 | 32 | 345,645 ± 27,700 b |
D-group and H-group embryos were cultured for 7 days when developmental rate to the blastocyst stage, and total cell number (TCN) and mitochondrial DNA copy number (mtDNA copy) in the blastocysts were examined. Data are shown as mean ± SEM. a–b P < 0.05.
Fig. 3.
Comparison of cell-free nuclear and mitochondrial DNA (cf-nDNA and cf-mtDNA) content in the culture medium of D-group and H-group blastocysts. After culturing H-group and D-group embryos individually for 7 days, the amount of cf-mtDNA and cf-nDNA was compared. * indicates significant differences between D-group and H group (P < 0.05).
Correlation between cf-DNA content in the medium and total cell number and mtDNA content in blastocysts
We also evaluated whether that the amount of cf-mtDNA and cf-nDNA in the SCM is affected by the cell number and mtDNA in the corresponding blastocysts. As shown in Table 2, the total cell number of blastocysts significantly correlated with the cf-nDNA content in the SCM but not with the cf-mtDNA content. However, no significant relationship between mtDNA in the blastocysts and cf-mtDNA and cf-nDNA in the SCM was observed.
Table 2. Correlation between cell free DNA (cf-DNA) content in the medium and total cell number and mitochondrial DNA (mtDNA) content in blastocysts.
| No. of samples (18) | Correlation coefficient |
|||
|---|---|---|---|---|
| TCN | cf-mtDNA | cf-nDNA | ||
| 31.7 ± 2.5 | TCN | – | 0.2 | 0.52 * |
| 107.8 ± 20.8 | cf-mtDNA | – | –0.22 | |
| 6.2 ± 1.2 | cf-nDNA | - | ||
| No. of samples (18) | Correlation coefficient |
|||
| Mt No. | cf-mtDNA | cf-nDNA | ||
| 217,319 ± 15,790 | Mt No. | – | 0.03 | 0.18 |
| 90.7 ± 9.1 | cf-mtDNA | - | 0.02 | |
| 9.6 ± 1.0 | cf-nDNA | - | ||
Data are presented as mean ± SEM. TCN, Total cell number of blastocysts; Mt No, Mitochondrial DNA copy number in blastocyst; cf-mtDNA, cell-free mitochondrial DNA copy number in culture medium; cf-ntDNA, cell-free nucleic DNA copy number in culture medium. * P < 0.05,
H-group embryos had a higher mtDNA copy number than D-group embryos
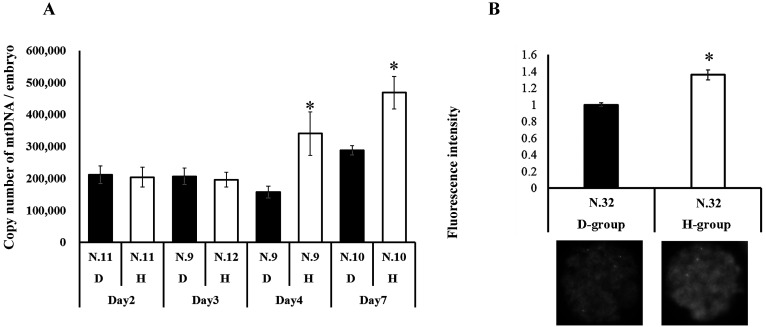
We addressed the kinetics of mtDNA in the embryos during embryo development from the zygote to blastocyst stages. As shown in Fig. 4A, the mtDNA content was constant at days 2–4 in the D-group embryos, whereas the mtDNA in the H-group embryos was significantly higher compared to that in the D-group embryos (day 4). Consistent with our results (Table 1), a high mtDNA content was observed in the H-group blastocysts. To validate the differential mitochondrial content in day 4 embryos between the H-group and D-group embryos, the expression levels of a mitochondrial membrane protein, TOMM20, in the embryos were compared using immunostaining. As a result, the expression levels were found to be significantly higher in the H-group embryos than in the D-group embryos (Fig. 4B).
Fig. 4.
Comparison of mitochondrial quantity between D-group and H-group embryos. A: Mitochondrial DNA copy number in embryos at 2, 3, 4, and 7 days after activation. * indicates significant differences between D-group and H-group within the same days after activation. B: Expression levels and representative pictures of TOMM20 in day 4 embryos. The average fluorescence intensity of D-group was defined as 1.0. * indicates significant differences between D-group and H-group (P < 0.05).
cf-mtDNA content differed in the SCM of 2-cell stage embryos
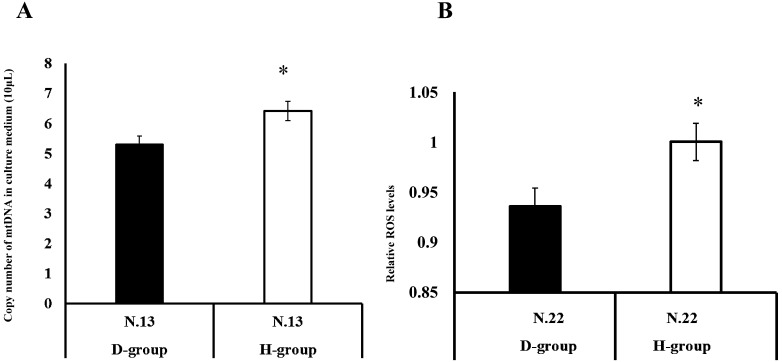
To determine whether the difference between cf-mtDNA content in the SCM between the H-group and D-group embryos was observed at an earlier time point, we determined the mtDNA content in the SCM of H-group and D-group 2-cell stage embryos. As a result, the levels of cf-mtDNA content in the SCM of 2-cell stage embryos were found to be much lower compared to those observed in the SCM of blastocysts (Fig. 3) but higher in the H-group than in the that in D-group embryos (Fig. 5A).
Fig. 5.
Comparison of cell-free mitochondrial DNA (cf-mtDNA) and reactive oxygen species (ROS) levels between the D-group and H-group embryos. A: Two-cell stage embryos (24 h) after activation were examined for ploidy (2PB or 1PB), and the cf-mtDNA content in the medium was compared between the D-group and H-group embryos. B: The levels of mitochondrial ROS in embryos (day 4 post-activation) were examined by MitoSOX staining. The expression levels of the H-group embryos were defined as 1.0. * P < 0.05. (The fluorescence intensity of the H-group embryos was defined as 1.0.)
Mitochondrial ROS content was higher in H-group embryos than in D-group embryos
Here, we examined the mitochondrial ROS content in day 4 embryos and found that the mitochondrial ROS levels were higher in the H-group embryos than in the D-group embryos (Fig. 5B).
Discussion
The present study findings showed that mtDNA copy number increased faster and was greater in the H-group embryos compared to that in the D-group embryos. Moreover, the amount of cf-mtDNA in the SCM differed between the H-group and D-group embryos. In addition, the amount of cf-nDNA in the medium appeared to be associated with the total cell number in the blastocysts.
Porcine oocytes can be activated by electric pulses followed by CHX and CB treatment [28]. CHX treatment inhibits the synthesis of proteins necessary for cyclin B, which reduces MPF activity, allowing the progression of cell cycle of zygotes following activation [29]. In contrast, CB treatment inhibits actin polymerization and suppresses polar body extrusion, resulting in diploid embryos. In this study, oocytes without CB treatment extruded the second polar body, giving rise to haploid embryos. A similar trial has been reported previously, in which porcine oocytes were activated with ethanol, followed by incubation in the presence or absence of CB for 5 h [30]. In the present study, differential ploidy was observed between the H-group and D-group embryos at 1 and 2 days post-activation. However, many in vitro-produced porcine blastocysts have previously been found to be polyploid, mixoploid, and haploid [31], indicating that ploidy may change during embryo development. Thus, the precise determination of all blastomeres in blastocysts is difficult. Therefore, careful consideration is required to evaluate the results.
Consistent with that in previous reports [30, 32], the developmental ability of the oocytes to the blastocyst stage was greater for diploid embryos in this study. The mtDNA copy number is becoming an increasingly attractive marker for oocyte quality due to accumulating evidence showing that oocytes with greater mtDNA copy numbers have a high capacity for maturation, fertilization, and development [2,3,4]. However, contrary to the finding for oocytes, there are many discrepancies in the literature regarding the relationship between mtDNA copy number and embryonic quality and ploidy [5, 7, 9, 10, 13]. In the present study, H-group blastocysts were found to have a greater mtDNA content than D-group blastocysts. The mtDNA copy number is constant during early porcine embryos [33] because mitochondrial biosynthesis is inactive in early cleavage-stage embryos [34]. Therefore, the mtDNA copy number in early cleavage-stage embryos reflects that in oocytes, and begins to increase before the blastocyst stage [4]. In line with this report, the mtDNA content in the D-group embryos was constant during the early cleavage-stage and increased at the blastocyst stage. However, in the H-group embryos, an increase in mtDNA occurred at an early time point (day 4 post-activation). Although the factors responsible for inducing early mitochondrial biosynthesis in the H-group embryos remain unclear, it is believed that embryonic stress (e.g., reactive oxygen stress) induces mitochondrial biosynthesis [35, 36], while differential ploidy induces unbalanced protein stress, resulting in ROS generation [37]. We found a greater mitochondrial ROS content in H-group embryos, indicating that the mitochondria in these embryos were affected by stress. However, in the present study, we did not evaluate the ROS content or other functions of individual mitochondrion. Therefore, further studies will be needed to elucidate the molecular mechanisms underlying our results.
Carbohydrate metabolism differs between inner cell mass (ICM) and TE [38]. For example, ICM has a low pyruvate consumption compared with TE. In addition, the mitochondrial content between TE and ICM has been suggested to differ [1]. These reports indicate that the differential mtDNA content between D-group and H-group blastocysts may be associated with different ICM and TE ratios. However, Ho et al. [11] reported that the mtDNA copy number per cell obtained from biopsied TE was comparable to that obtained from whole embryos. Furthermore, another study [39] found that the mtDNA copy number did not differ between ICM and TE. In addition, in present study, comparative total cell number was observed between the H-group and D-group blastocysts. Therefore, we speculated that differences in the mtDNA copy numbers arose due to other molecular mechanisms, which will need to be addressed in future studies.
The measurement of DNA in the SCM can be hampered by several factors. In the present study, we first addressed the effect of the presence of ZP on the amount of cfDNA content in the medium. The presence of ZP did not significantly affect the amount of cfDNA content in the medium, although a higher cfDNA content was detected in the medium of ZP-free embryos. Based on these results, we selected intact ZP embryos for use in subsequent experiments. However, the presence of ZP may have resulted in an underestimation of the cfDNA content in the medium. Next, we evaluated whether dead embryos had higher levels of cfDNA content. By comparing the cfDNA content in the medium between well-developed blastocysts and morphologically dead embryos, lower levels of cfDNA were observed in the medium of the dead embryos, although there was no significant difference. These results suggest that live developing embryos secrete cfDNA into the medium. Consistent with these findings, studies using cancer cell lines or granulosa cells have previously shown that live cells (not apoptotic and or necrotic cells) actively secrete cfDNA into media [40, 41]. It is generally assumed that the greater the mtDNA content and total cell number of the blastocysts, the greater the amount of cfDNA in the SCM. In the present study, a significant correlation was observed between the total cell number of the blastocysts and the cf-nDNA content in the medium. Kiersten et al. [42] reported that the amount of cfDNA content in blastocoel fluid positively correlated with the embryonic grade, determined by measuring dsDNA. However, the molecular mechanism underlying this relationship remains unclear.
cfDNA has been studied as a non-invasive marker for embryonic ploidy, and although the prediction of embryonic ploidy has been reported [43], there is insufficient evidence for the prediction of embryonic ploidy from the cfDNA content in the medium. In this context, TE biopsy analysis has also been reported [42]. Contrary to our expectations, we did not detect a significant difference in the cf-nDNA content in the medium between the H-group and D-group blastocysts. In this study, we used a primer set targeting a single-copy gene (located on chromosome 15). Further, the blastocyst developed in this study had a small total cell number (approximately 30). Therefore, the low frequency of nucleic DNA in embryos and the primer set examined may be insufficient to detect clear differences in cf-nDNA content. In addition to cf-nDNA, the cf-mtDNA content was higher and thus more easily detected by real-time PCR [24, 41]. It has been reported that the amount of cf-mtDNA content in the medium is associated with the ratio of embryonic fragmentation at days 2 and 3 [16]. In contrast, Stigliani et al. [15] showed that a high mtDNA/nuclear DNA ratio in the SCM of day 3 human embryos indicates a high potency of embryonic development. In our study, we found that the cf-mtDNA content in the medium was higher in the D-group blastocysts than in the H-group blastocysts, indicating that ploidy affects the amount of cf-mtDNA content in the medium. We further examined whether this difference occurred at an earlier developmental stage using the SCM of 2-cell stage embryos. As the results, the cf-mtDNA content was found to be lower than that in the SCM at the blastocyst stage. However, to our surprise, the cf-mtDNA content was higher in the H-group blastocysts than in the D-group blastocysts. These results suggest that there may be complex factors affecting cf-mtDNA secretion by embryos. When granulosa cells were treated with mitochondrial uncoupler inducing mitochondrial dysfunction, the amount of cf-mtDNA in the medium increased [44]. Vitrification tends to affect the levels of cf-mtDNA in the SCM of bovine embryos, as does the treatment of vitrified-warmed embryos with resveratrol, which was previously reported to improve embryonic development and increases cf-mtDNA content in the medium [22, 45]. In addition, it has been reported that the inhibition of autophagy increases the cf-mtDNA content and that the inhibition of either proteasome or intracellular vesicle formation decreases cf-tDNA content in the SCM of bovine granulosa cells [41]. Taken together, our results indicate that not only simple embryonic factors (cell number and mtDNA content) but also mitochondrial homeostasis should be considered when evaluating cf-DNA content in the medium.
Therefore, our findings suggest that embryonic ploidy affects the mtDNA content of blastocysts and the amount of cf-mtDNA content in the medium.
Conflict of interests
The authors declare no conflicts of interest.
Acknowledgments
We thank D Hamanaka, R Huruya, and M Takeoka for helping with the preparation of parthenogenetic embryos.
References
- 1.St John J. The control of mtDNA replication during differentiation and development. Biochim Biophys Acta 2014; 1840: 1345–1354. [DOI] [PubMed] [Google Scholar]
- 2.Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006; 85: 584–591. [DOI] [PubMed] [Google Scholar]
- 3.Zeng HT, Ren Z, Yeung WS, Shu YM, Xu YW, Zhuang GL, Liang XY. Low mitochondrial DNA and ATP contents contribute to the absence of birefringent spindle imaged with PolScope in in vitro matured human oocytes. Hum Reprod 2007; 22: 1681–1686. [DOI] [PubMed] [Google Scholar]
- 4.Cecchino GN, Garcia-Velasco JA. Mitochondrial DNA copy number as a predictor of embryo viability. Fertil Steril 2019; 111: 205–211. [DOI] [PubMed] [Google Scholar]
- 5.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet 2015; 11: e1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, Díaz-Gimeno P, Valbuena D, Simón C. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril 2015; 104: 534–41.e1. [DOI] [PubMed] [Google Scholar]
- 7.Jing Y, Li L, Li YY, Ouyang YC, Sun QY, Zhang CL, Li R. Embryo quality, and not chromosome nondiploidy, affects mitochondrial DNA content in mouse blastocysts. J Cell Physiol 2019; 234: 10481–10488. [DOI] [PubMed] [Google Scholar]
- 8.Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, Morrison L, Morin SJ, Scott RT., Jr Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod 2017; 32: 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, Viotti M. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril 2017; 107: 34–42.e3. [DOI] [PubMed] [Google Scholar]
- 10.Victor A, Griffin D, K Gardner D, Brake A, Zouves C, Barnes F, Viotti M. Births from embryos with highly elevated levels of mitochondrial DNA. Reprod Biomed Online 2019; 39: 403–412. [DOI] [PubMed] [Google Scholar]
- 11.Ho JR, Arrach N, Rhodes-Long K, Salem W, McGinnis LK, Chung K, Bendikson KA, Paulson RJ, Ahmady A. Blastulation timing is associated with differential mitochondrial content in euploid embryos. J Assist Reprod Genet 2018; 35: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Los Santos MJ, Diez Juan A, Mifsud A, Mercader A, Meseguer M, Rubio C, Pellicer A. Variables associated with mitochondrial copy number in human blastocysts: what can we learn from trophectoderm biopsies? Fertil Steril 2018; 109: 110–117. [DOI] [PubMed] [Google Scholar]
- 13.Lee YX, Chen CH, Lin SY, Lin YH, Tzeng CR. Adjusted mitochondrial DNA quantification in human embryos may not be applicable as a biomarker of implantation potential. J Assist Reprod Genet 2019; 36: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016; 35: 347–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stigliani S, Persico L, Lagazio C, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA in Day 3 embryo culture medium is a novel, non-invasive biomarker of blastocyst potential and implantation outcome. Mol Hum Reprod 2014; 20: 1238–1246. [DOI] [PubMed] [Google Scholar]
- 16.Stigliani S, Anserini P, Venturini PL, Scaruffi P. Mitochondrial DNA content in embryo culture medium is significantly associated with human embryo fragmentation. Hum Reprod 2013; 28: 2652–2660. [DOI] [PubMed] [Google Scholar]
- 17.Shamonki MI, Jin H, Haimowitz Z, Liu L. Proof of concept: preimplantation genetic screening without embryo biopsy through analysis of cell-free DNA in spent embryo culture media. Fertil Steril 2016; 106: 1312–1318. [DOI] [PubMed] [Google Scholar]
- 18.Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, Peinado V, Mercader A, Meseguer M, Blesa D, Moreno I, Valbuena D, Rubio C, Simon C. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Hum Reprod 2018; 33: 745–756. [DOI] [PubMed] [Google Scholar]
- 19.Yeung QSY, Zhang YX, Chung JPW, Lui WT, Kwok YKY, Gui B, Kong GWS, Cao Y, Li TC, Choy KW. A prospective study of non-invasive preimplantation genetic testing for aneuploidies (NiPGT-A) using next-generation sequencing (NGS) on spent culture media (SCM). J Assist Reprod Genet 2019; 36: 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Sun Y, Dong X, Zhou J, Sun F, Han T, Lei P, Mao R, Guo X, Wang Q, Li P, Qu T, Huang J, Li L, Huang T, Zhong Y, Gu J. Mitochondrial DNA and genomic DNA ratio in embryo culture medium is not a reliable predictor for in vitro fertilization outcome. Sci Rep 2019; 9: 5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Fang R, Chen L, Chen D, Xiao JP, Yang W, Wang H, Song X, Ma T, Bo S, Shi C, Ren J, Huang L, Cai LY, Yao B, Xie XS, Lu S. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci USA 2016; 113: 11907–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, Kin A, Aoki S, Nakamura S, Shirasuna K, Kuwayama T, Iwata H. Resveratrol enhances the clearance of mitochondrial damage by vitrification and improves the development of vitrified-warmed bovine embryos. PLoS One 2018; 13: e0204571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod 2002; 66: 112–119. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa K, Shibahara H, Shirasuna K, Kuwayama T, Iwata H. Cell-free DNA content in follicular fluid: A marker for the developmental ability of porcine oocytes. Reprod Med Biol 2019; 19: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itami N, Shirasuna K, Kuwayama T, Iwata H. Short-term heat stress induces mitochondrial degradation and biogenesis and enhances mitochondrial quality in porcine oocytes. J Therm Biol 2018; 74: 256–263. [DOI] [PubMed] [Google Scholar]
- 26.Itami N, Shirasuna K, Kuwayama T, Iwata H. Palmitic acid induces ceramide accumulation, mitochondrial protein hyperacetylation, and mitochondrial dysfunction in porcine oocytes. Biol Reprod 2018; 98: 644–653. [DOI] [PubMed] [Google Scholar]
- 27.Sato D, Itami N, Tasaki H, Takeo S, Kuwayama T, Iwata H. Relationship between mitochondrial DNA copy number and SIRT1 expression in porcine oocytes. PLoS One 2014; 9: e94488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Cui K, Li HL, Sun JM, Lu XR, Shen KY, Liu QY, Shi S. Comparison of chemical, electrical, and combined activation methods for in vitro matured porcine oocytes. In Vitro Cell Dev Biol Anim 2015; 51: 103–112. [DOI] [PubMed] [Google Scholar]
- 29.Hagemann LJ, Hillery-Weinhold FL, Leibfried Rutledge ML, First NL. Activation of murine oocytes with Ca2+ ionophore and cycloheximide. J Exp Zool 1995; 271: 57–61. [DOI] [PubMed] [Google Scholar]
- 30.Kim NH, Uhm SJ, Ju JY, Lee HT, Chung KS. Blastocoele formation and cell allocation to the inner cell mass and trophectoderm in haploid and diploid pig parthenotes developing in vitro. Zygote 1997; 5: 365–370. [DOI] [PubMed] [Google Scholar]
- 31.McCauley TC, Mazza MR, Didion BA, Mao J, Wu G, Coppola G, Coppola GF, Di Berardino D, Day BN. Chromosomal abnormalities in Day-6, in vitro-produced pig embryos. Theriogenology 2003; 60: 1569–1580. [DOI] [PubMed] [Google Scholar]
- 32.Jeong YJ, Cui XS, Kim BK, Kim IH, Kim T, Chung YB, Kim NH. Haploidy influences Bak and Bcl-xL mRNA expression and increases incidence of apoptosis in porcine embryos. Zygote 2005; 13: 17–21. [DOI] [PubMed] [Google Scholar]
- 33.Spikings EC, Alderson J, St John JC. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol Reprod 2007; 76: 327–335. [DOI] [PubMed] [Google Scholar]
- 34.Pikó L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 1987; 123: 364–374. [DOI] [PubMed] [Google Scholar]
- 35.Irrcher I, Ljubicic V, Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1alpha transcription in skeletal muscle cells. Am J Physiol Cell Physiol 2009; 296: C116–C123. [DOI] [PubMed] [Google Scholar]
- 36.Yoboue ED, Devin A. Reactive oxygen species-mediated control of mitochondrial biogenesis. Int J Cell Biol 2012; 2012: 403870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman DL, Thurgood LA, Gregory SL. The impact of aneuploidy on cellular homeostasis. Free Radic Res 2019; 53: 705–713. [DOI] [PubMed] [Google Scholar]
- 38.Gopichandran N, Leese HJ. Metabolic characterization of the bovine blastocyst, inner cell mass, trophectoderm and blastocoel fluid. Reproduction 2003; 126: 299–308. [DOI] [PubMed] [Google Scholar]
- 39.Noli L, Khorsandi SE, Pyle A, Giritharan G, Fogarty N, Capalbo A, Devito L, Jovanovic VM, Khurana P, Rosa H, Kolundzic N, Cvoro A, Niakan KK, Malik A, Foulk R, Heaton N, Ardawi MS, Chinnery PF, Ogilvie C, Khalaf Y, Ilic D. Effects of thyroid hormone on mitochondria and metabolism of human preimplantation embryos. Stem Cells 2020; 38: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Kong P, Ma G, Li L, Zhu J, Xia T, Xie H, Zhou W, Wang S. Characterization of the release and biological significance of cell-free DNA from breast cancer cell lines. Oncotarget 2017; 8: 43180–43191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kansaku K, Munakata Y, Shirasuna K, Kuwayama T, Iwata H. Mitochondrial cell-free DNA secreted from porcine granulosa cells. Zygote 2019; 27: 272–278. [DOI] [PubMed] [Google Scholar]
- 42.Rule K, Ferro J, Hoffman A, Williams J, Golshiri S, Padre R, Avila J, Coca C, Valdes K. Purdue manual dexterity testing: A cohort study of community-dwelling elderly. J Hand Ther 2020; S0894-1130(20)30005-3. [DOI] [PubMed] [Google Scholar]
- 43.Rubio C, Rienzi L, Navarro-Sánchez L, Cimadomo D, García-Pascual CM, Albricci L, Soscia D, Valbuena D, Capalbo A, Ubaldi F, Simón C. Embryonic cell-free DNA versus trophectoderm biopsy for aneuploidy testing: concordance rate and clinical implications. Fertil Steril 2019; 112: 510–519. [DOI] [PubMed] [Google Scholar]
- 44.Kansaku K, Munakata Y, Itami N, Shirasuna K, Kuwayama T, Iwata H. Mitochondrial dysfunction in cumulus-oocyte complexes increases cell-free mitochondrial DNA. J Reprod Dev 2018; 64: 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayashi T, Kansaku K, Abe T, Ueda S, Iwata H. Effects of resveratrol treatment on mitochondria and subsequent embryonic development of bovine blastocysts cryopreserved by slow freezing. Anim Sci J 2019; 90: 849–856. [DOI] [PubMed] [Google Scholar]